Abstract
Background
Atopic dermatitis is a chronic inflammatory skin disease associated with increased susceptibility to recurrent skin infections.
Objective
To determine why a subset of atopic dermatitis patients have an increased risk of developing disseminated viral skin infections.
Methods
Human subjects with atopic dermatitis with a history of eczema herpeticum and various control groups were enrolled. Vaccinia virus expression was measured by PCR and immunofluorescent staining in skin biopsies from each study group after incubation with vaccinia virus. Transgenic mice with a constitutively active STAT6 gene were characterized for response to vaccinia virus skin inoculation. Genotyping for ten STAT6 single nucleotide polymorphisms (SNP) was performed in a European American sample (n=444).
Results
Vaccinia virus gene and protein expression were significantly increased in the skin of eczema herpeticum subjects, as compared to other subject groups, following incubation with vaccinia virus in vitro. Antibody neutralization of IL-4 and IL-13 resulted in lower vaccinia virus replication in subjects with a history of eczema herpeticum. Mice that expressed a constitutively active STAT6, compared to wild type mice, had increased mortality and satellite lesion formation following vaccinia virus skin inoculation. Significant associations were observed between STAT6 SNPs and eczema herpeticum (rs3024975, rs841718, rs167769, and rs703817) and IFNγ production. The strongest association was observed for a 2-SNP (CT) haplotype (ADEH+ vs ADEH−, 24.9% vs 9.2% P = 5.17×10−6).
Conclusion
The STAT6 gene increases viral replication in the skin of atopic dermatitis patients with a history of eczema herpeticum. Further genetic association studies and functional investigations are warranted.
Keywords: Atopic dermatitis, STAT6, eczema, virus, infection
INTRODUCTION
Atopic dermatitis (AD) is a genetically complex, chronic inflammatory skin disease that affects approximately 20% of all children and often persists in adults.1,2 The skin of AD patients is characterized by a defective skin barrier and increased T helper 2 (Th2) responses.3 Additionally, some patients with AD suffer from recurrent bacterial and disseminated viral skin infections such as eczema herpeticum (EH).4
Vaccinia virus (VV) was routinely used for vaccination against smallpox until the worldwide vaccination program ceased in the late 1970’s following the global eradication of smallpox.5 While the vaccine was effective at conferring protective immunity in the general public, vaccination was associated with increased numbers of severe adverse effects in a subset of patients with AD. The re-emergence of orthopoxviruses as a human health concern prompted health officials to reinstitute voluntary smallpox vaccination in first line responders;6 however, the Centers for Disease Control and Prevention recommended that individuals with allergic skin disease refrain due to the potential for severe adverse events.7 Based on this recommendation, greater than 30% of our military refrained from vaccination posing a significant health concern.8
The National Institute of Allergy and Infectious Diseases (NIAID) established the Atopic Dermatitis Vaccinia Network (ADVN) to further characterize the immune response in the skin of AD patients following VV inoculation into the skin and to identify genetic biomarkers for those AD patients with an increased propensity towards disseminated viral skin infections such as EH. In a previous study, we reported that skin from patients with severe AD showed increased VV replication.9 More recently, we found that AD subjects with a history of EH (ADEH+) had significantly higher serum IgE levels than AD subjects without a history of EH (ADEH−).10 Because IL-4 and IL-13, which play a key role in isotype switching to IgE synthesis, act via STAT6, we speculated that abnormalities in STAT6 function due to genetic variation could be associated with ADEH+ and increased VV replication in the skin of such patients.
METHODS
Study Subjects
For initial studies of viral replication in the skin of ADEH+ vs. control group (see Table I), skin biopsies were obtained from 55 subjects with active AD (ADEH−), 20 subjects with AD and a history of eczema herpeticum (ADEH+), 38 asthmatics (AS), 35 healthy individuals with no history of skin disease (N), 36 subjects with psoriasis (PS), and 36 subjects with a history of AD (QAD). The mean ± standard error (SEM) for age, total serum IgE, and number of positive RAST tests are summarized for each subject group in Table I. Table II describes the racial distribution among skin biopsy donors. Additional ADEH− (n=17), ADEH+ (n=6), and healthy subjects (n=19) were recruited to test the effect of Th2 cytokines or neutralization of Th2 cytokines on viral replication in the skin.
Table I.
Characteristics of Skin Biopsy Donors
| Subject Group | Age Years (mean ± SEM) |
Serum IgE (KU/L) (Geometric mean ± SEM1) |
# of IgE allergen positive ImmunoCAP tests (mean ± SEM) |
|---|---|---|---|
| ADEH− (N = 44) | 32.6 ± 1.98 | 137.4± 51.97 | 5.2 ± 0.59 |
| ADEH+ (N = 18) | 21.6 ± 4.14 | 536.4 ± 288.97 | 6.7 ± 1.06 |
| Asthmatic (N = 30) | 31.4 ± 1.99 | 77.05 ± 29.40 | 3.8 ± 0.60 |
| Normal (N = 42) | 29.2 ± 1.18 | 24.62 ± 7.19 | 2.0 ± 0.40 |
| Psoriasis (N = 29) | 40.4 ± 2.48 | 28.21 ± 10.89 | 1.6 ± 0.45 |
| Quiescent AD (N = 28) | 34.3 ± 2.15 | 54.50 ± 20.09 | 2.8 ± 0.53 |
Back-transformed standard errors computed using the Delta method
Table II.
Racial Distribution Among Skin Biopsy Donors
| Race | N |
|---|---|
| Indian | 5 |
| Asian | 5 |
| Caucasian | 166 |
| Other | 12 |
| Multi-Racial | 3 |
Genotyping was conducted on 278 unrelated European American AD patients (of whom 112 had ADEH+) and 166 healthy controls participating in the ADVN. Clinical characteristics for this sample has been described previously.11
Patients had not received topical corticosteroids for a period of one week prior to enrollment nor systemic immunosuppressives for one month before enrollment into this study. This study was approved by the institutional review boards at National Jewish Health, Johns Hopkins Medical Institute, Oregon Health & Sciences University, University of Rochester Medical Center, University of California San Diego, and Children’s Hospital Boston. All subjects gave written informed consent prior to participation in these studies.
Virus Source and Culture
The Wyeth/ACAM 2000 strain of VV was obtained from the Centers for Disease Control and Prevention (Atlanta, GA). The Western Reserve VV strain 1354 was obtained from the ATCC (Manassas, VA) for use in murine studies.
Human Skin Explant Cultures
Two-mm punch biopsies were collected from the non-lesional skin of each enrolled subject. Skin biopsies were cultured in the presence of media alone (RPMI supplemented with 10% FCS) or 2.5 × 105 pfu VV for 24 hours. Following the exposure period, media was removed and biopsies were submerged in 10% buffered formalin for immunofluorescent staining or Tri-Reagent (Molecular Research Center Inc., Cincinnati, OH) for DNA analysis.
In some experiments, monoclonal anti-human IL-4 (1 mcg/ml; R&D Systems, Minneapolis, MN) and monoclonal anti-human IL-13 (1 mcg/ml; R&D Systems) were added to the skin biopsies from ADEH− (n=20) and ADEH+ (n=10) for 24 hours to neutralize Th2 cytokines prior to infection with VV. Following the viral exposure, biopsies were cultured for an additional 24 hours and prepared for DNA isolation to measure viral copies.
VV DNA Analysis
DNA was prepared from human skin explants according to the manufacturer’s guidelines (Molecular Research Center Inc.). Viral copies were quantified by real-time PCR using primers specific for vaccinia ribonucleotide reductase (Vvl4L) as previously described.13
VV Immunofluorescent Staining
Paraffin-embedded tissues were cut at 5 mm on frosted microscope slides. Using toluene and a series of ethanol washes, slides were deparaffinized and then rehydrated. Skin sections were then blocked with 5% BSA in Super Block (ScyTek Laboratories, Logan, UT) containing 10% non-immune donkey serum (Jackson Laboratories, West Grove, PA) for 60 min. Slides were then stained with a rabbit anti-A27 antibody (Abcam, Cambridge, MA) directed against an early viral protein located in the nucleus and cytoplasm of infected cells or control rabbit IgG (Santa Cruz Biotechnology, Inc., Santa Cruz, CA) at 4°C overnight. Slides were washed with PBS/Tween 0.05%, followed by incubation with a Cy3-conjugated donkey anti-rabbit IgG (Jackson Laboratories).
Immunohistochemical staining was visualized with confocal microscopy (Leica, Wetzlar Germany). Slides were coded to ensure patient anonymity. Images were collected at 40×, and levels of mean fluorescence intensity (MFI) were measured with Slidebook 4.1 (Intelligent Imaging innovations, Denver, CO). MFI was determined for each exposure group and was reported as mean MFI ± SE.
Mice
STAT-6VT transgenic mice with constitutively active STAT6 were generated at Indiana University as previously described.12 Briefly, the V547 and T548 of the human STAT6 gene were mutated to alanines under the transcriptional control of the CD2 locus control region. C57Bl/6 mice were purchased from The Jackson Laboratories (Bar Harbor, ME) to generate hemizygous STAT-6VT and wild type littermate mice. All mice were maintained in specific pathogen-free conditions and experiments were approved by the Indiana University and National Jewish Health Institutional Animal Care and Use Committees.
IFNγ ELISPOT
The differential immune responses (ex vivo) to HSV have been investigated by measuring IFNγ production in isolated peripheral blood mononuclear cells (PBMCs) from a subset of the ADVN sample (64 subjects), and clinical characteristics of participants have been previously described.41 IFNγ production was examined by using enzyme-linked immunosorbent spot (ELISPOT) adapted from the protocol as previously described.40
Genotyping and Quality Control
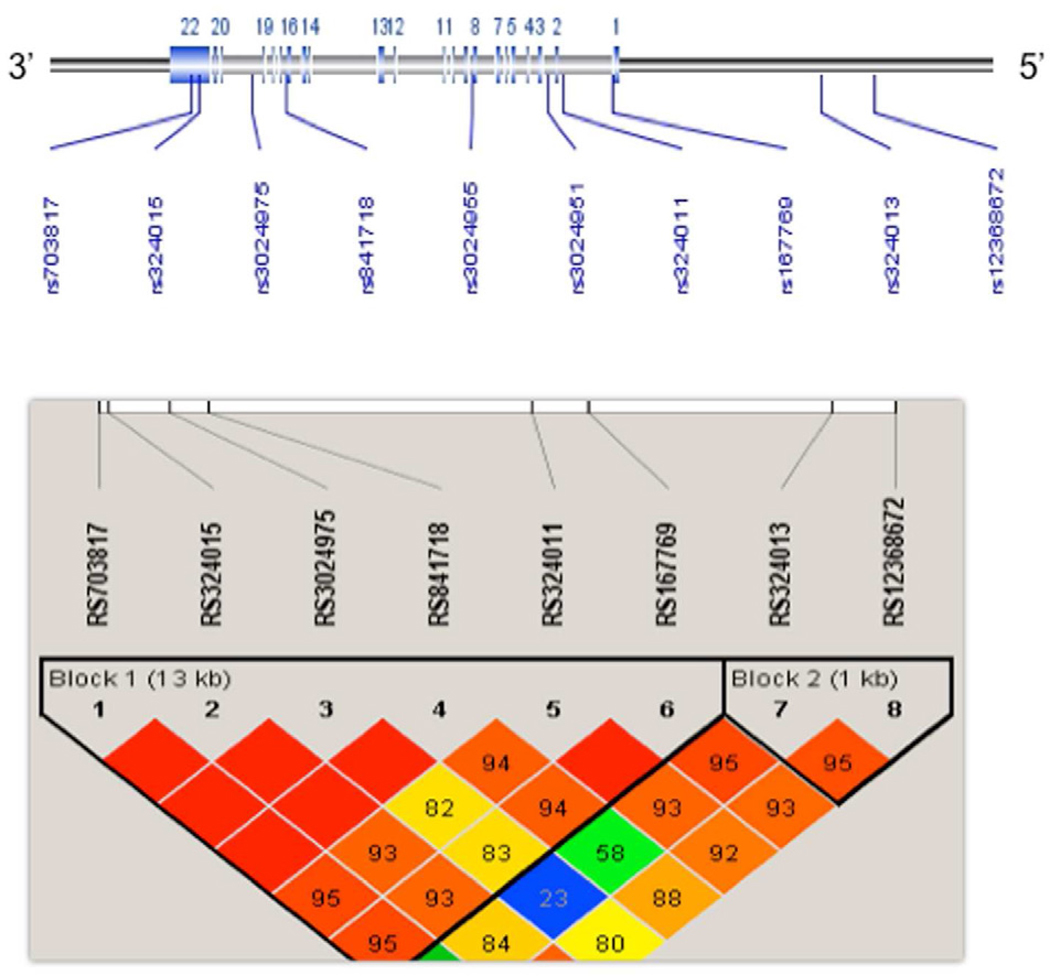
Because no common variants were detected in the coding region of STAT6, we selected six tagging single nucleotide polymorphisms (SNPs) and four SNPs consisting of promoter SNP rs12368672, 3'-untranslated region (UTR) SNP rs703817, and two intronic SNPs close to the initially indentified region. These SNPs were distributed in a 22.6-kb region on chromosome 12q13 covering the STAT6 gene (Figure 4A). The strategy for the tagging SNP selection has been previously described.38 SNPs were genotyped using TaqMan Allelic discrimination Assays on the 7900HT Sequence Detection System (Applied Biosystems, Foster City, CA). Potential confounding due to population sub-structure was assessed using genotype data from the 74 Ancestry Informative Markers (AIMs) as described previously.11
Figure 4.
Analysis of STAT6 polymorphisms in European American samples. A) STAT6 gene structure and distribution of the genotyped SNPs across the gene (22.6 kb) on chromosome 12q13 Haplotype block structure of STAT6 SNPs in European American healthy controls (n = 166) was presented. The intensity of shading represents D' (a measure of LD) generated using HAPLOVIEW software, with red (100) to green reflecting higher to lower D’ values. Both rs3024951 and rs3024955 were excluded in LD plot because of their low allele frequencies, B) Haplotype results showing Omnibus P-values constructed across sliding windows of sizes 2–5 for eight common SNPs and ADEH+. Black vertical lines represent all individual SNP tests, and colored horizontal lines represent 2-, 3, 4, 5, haplotype tests. *see detailed data in Table IV.
Statistical Analyses
Statistical analysis of viral data was conducted using Graph Pad Prism, version 5.03 (San Diego, CA) and SAS version 9.1. Comparisons of quantitative variables (e.g., viral replication) among diagnostic groups were made using one-way analysis of variance (ANOVA), followed by t-tests for pairwise comparisons. The Tukey-Kramer method was used to control the family-wise Type I error rate at level 0.05. Survival analysis comparisons across groups of mice were computed with log rank tests. Significant differences were conferred at P<0.05. Genetic association analysis was performed using the Cochran–Armitage trend test under an additive model using PLINK software.14 Associations with the log-transformed total serum IgE (tIgE), Eczema Area and Severity Index (EASI) score, and log-transformed spot forming units (SFU)/106 PBMCs, were performed using a linear regression analysis adjusted for confounding variables (age and gender). To report tIgE statistics, means of log-transformed values were back-transformed (geometric means), and back-transformed standard errors were estimated using the Delta method. Haplotype analyses were performed using sliding windows of 2–5 SNPs where empiric P-values for haplotype frequency differences were generated over 10,000 permutations. Analyses of the effect of anti-Th2 cytokines were conducted using paired t tests (e.g., effect of anti-Th2 antibodies versus media control on viral replication). Our power calculation using QUANTO version 1.1 program39 demonstrated that the study population provided sufficient power (80%) to detect a genotypic odds ratio (OR) of 2.06, even if the allele frequency with disease is only 10%.
RESULTS
Increased Viral Replication in ADEH
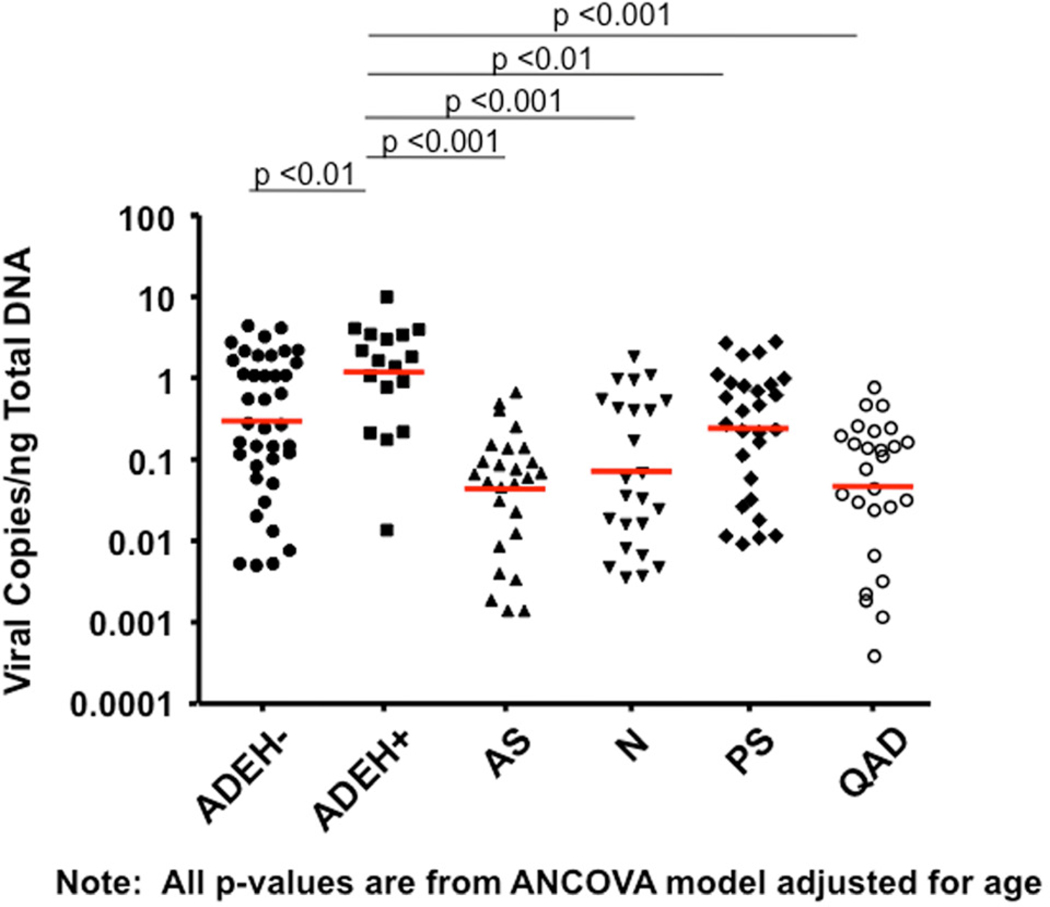
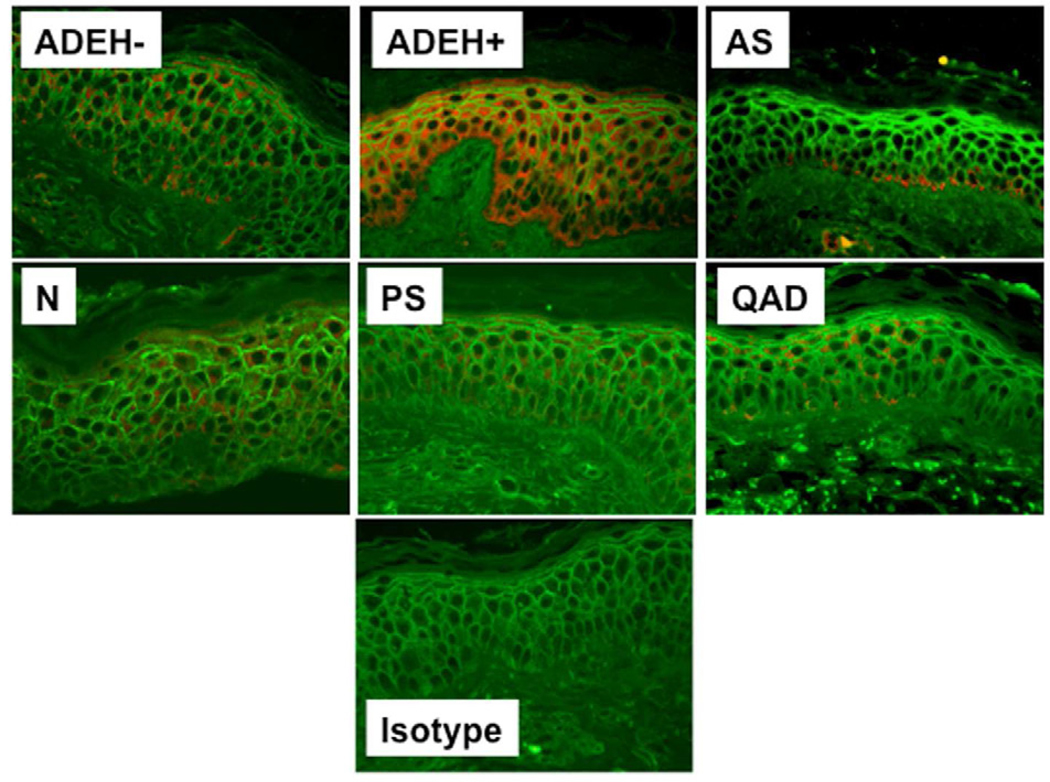
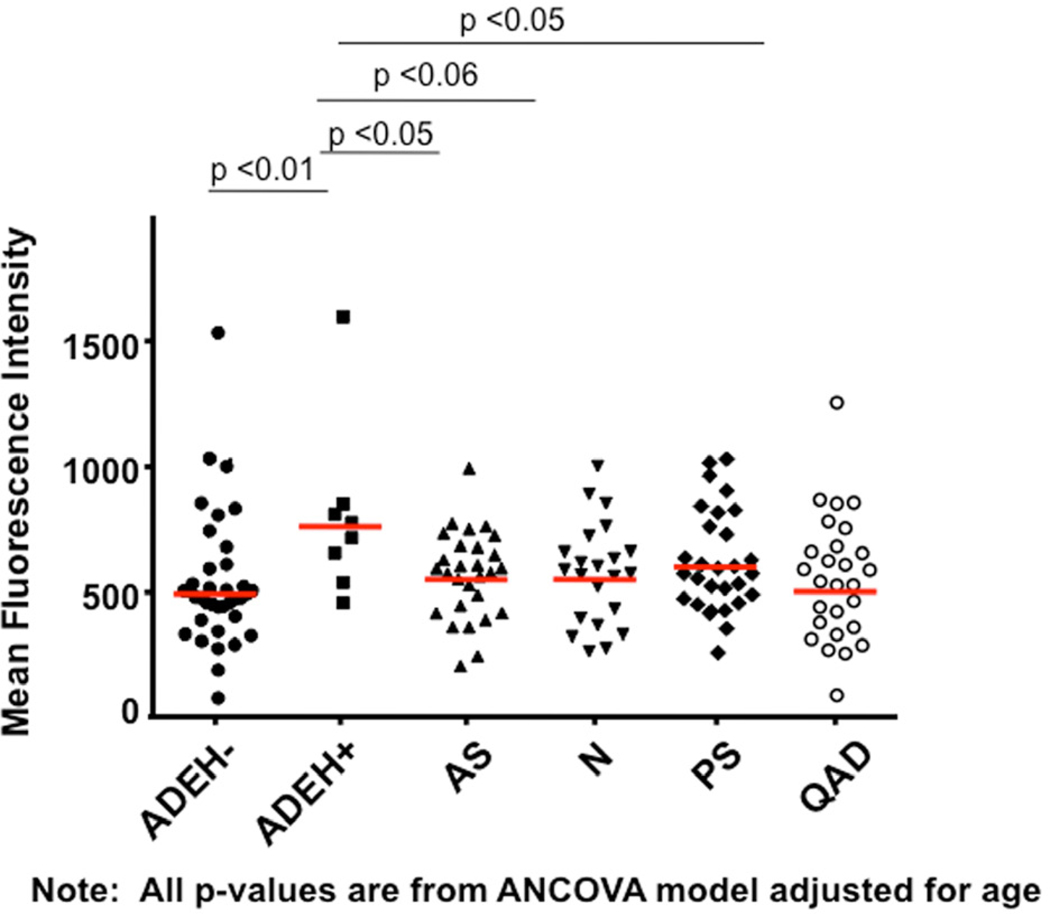
Skin biopsies were collected from all subject groups in Table I to compare the ability of VV to replicate ex vivo in ADEH+ skin vs. various control groups. Biopsies were treated with the standard human dose of VV used in smallpox vaccinations (2×105 pfu). Figure 1A demonstrates that skin from ADEH+ subjects supported significantly greater VV replication (mean: 2.42 ± 0.63 VV viral copies/ng total DNA) than ADEH− (1.03 ± 0.20; P = 0.01), asthmatic (0.12 ± 0.03; P <0.001), normal (0.34 ± 0.10; P <0.001), psoriasis (0.70 ± 0.16; P <0.01), and quiescent AD (0.15 ± 0.04; p<0.001). Additionally, VV replication was significantly greater in ADEH− skin compared to skin from asthmatics (P <0.001), normal (P <0.01), and quiescent AD (P <0.01). Immunofluorescent staining for an early viral protein confirmed higher levels of VV replication in the skin of ADEH+ as compared to other subject groups (Figure 1, B&C).
Figure 1.
ADEH+ skin supports significantly greater VV replication. A) DNA was isolated from media or VV stimulated non-lesional skin and analyzed for VV gene expression by real-time RT-PCR. B) Immunofluorescent staining for A27L. C) The MFI for VV expression in the basal keratinocytes of each biopsy. *, ** and *** indicate significant differences of P <0.05, P <0.01 and P <0.001, respectively.
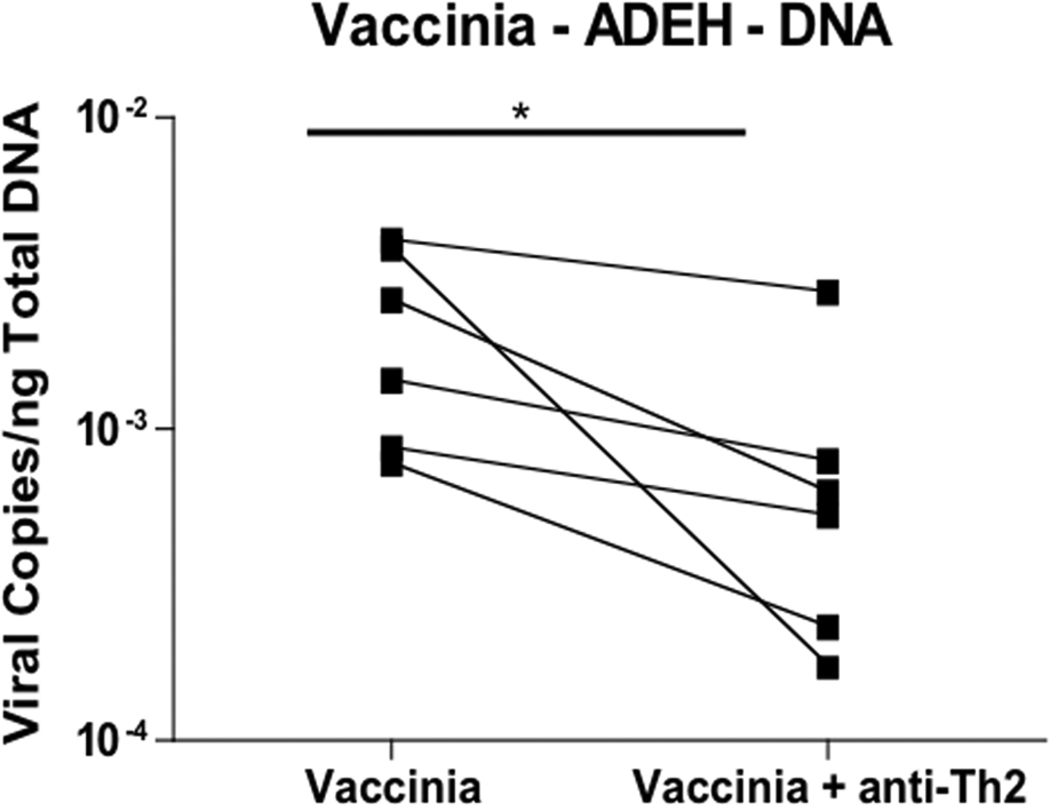
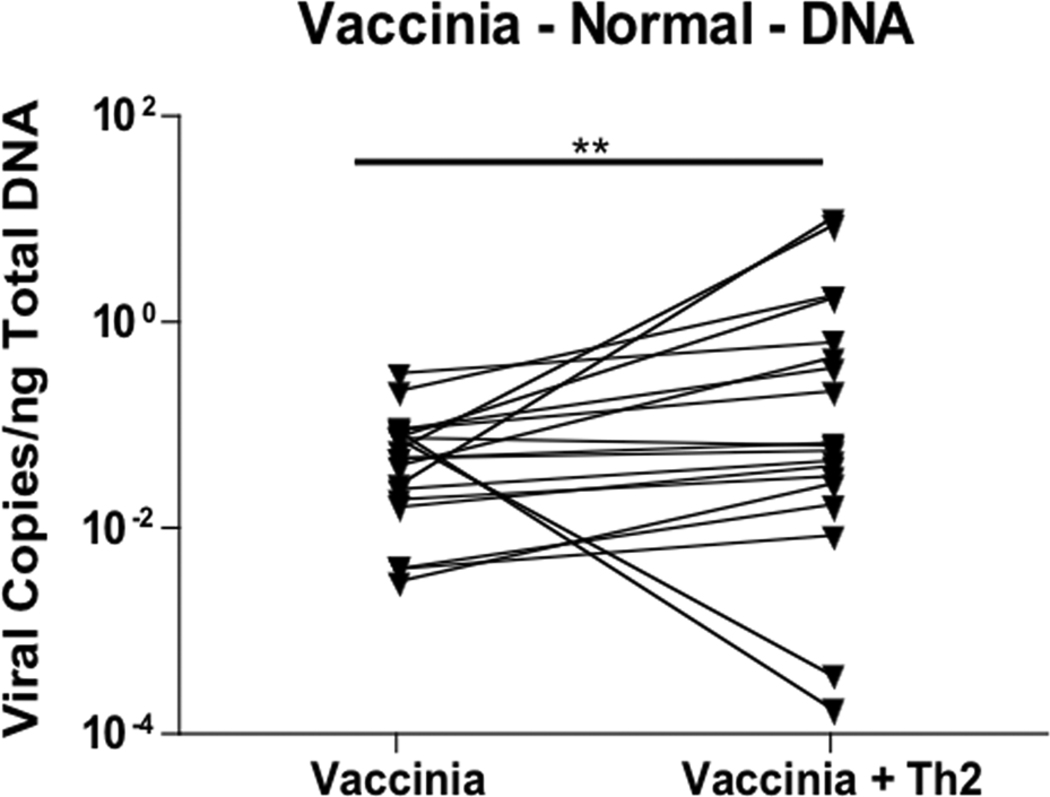
ADEH+ patients are characterized by a significantly greater Th2 response (i.e. elevated serum IgE levels and increased number of positive RAST tests) than ADEH− patients.10 Because the Th2 response is characterized by increased IL-4 and IL-13, we pre-incubated skin biopsies from ADEH− and ADEH+ patients with antibodies against IL-4 and IL-13 to determine whether neutralization of Th2 cytokines would inhibit VV replication. Pre-treating ADEH− skin biopsies with neutralizing antibodies did not significantly inhibit VV replication (Figure 2A). In contrast, VV replication was significantly lower in ADEH+ skin biopsies pre-treated with neutralizing antibodies (0.0009 ± 0.0005 VV copies/ng DNA; P = 0.053) compared to control treated skin biopsies (0.0024 ± 0.0007 VV copies/ng DNA; Figure 2B).
Figure 2.
IL-4 and IL-13 modulate VV replication. VV expression in non-lesional skin from ADEH− (panel A; n=17) and ADEH+ (panel B; n=6) subjects infected with VV following pre-treatment with/without neutralizing antibodies to IL-4 and IL-13. * indicates significant difference of P <0.05.
Viral Responses in STAT-6VT Mice
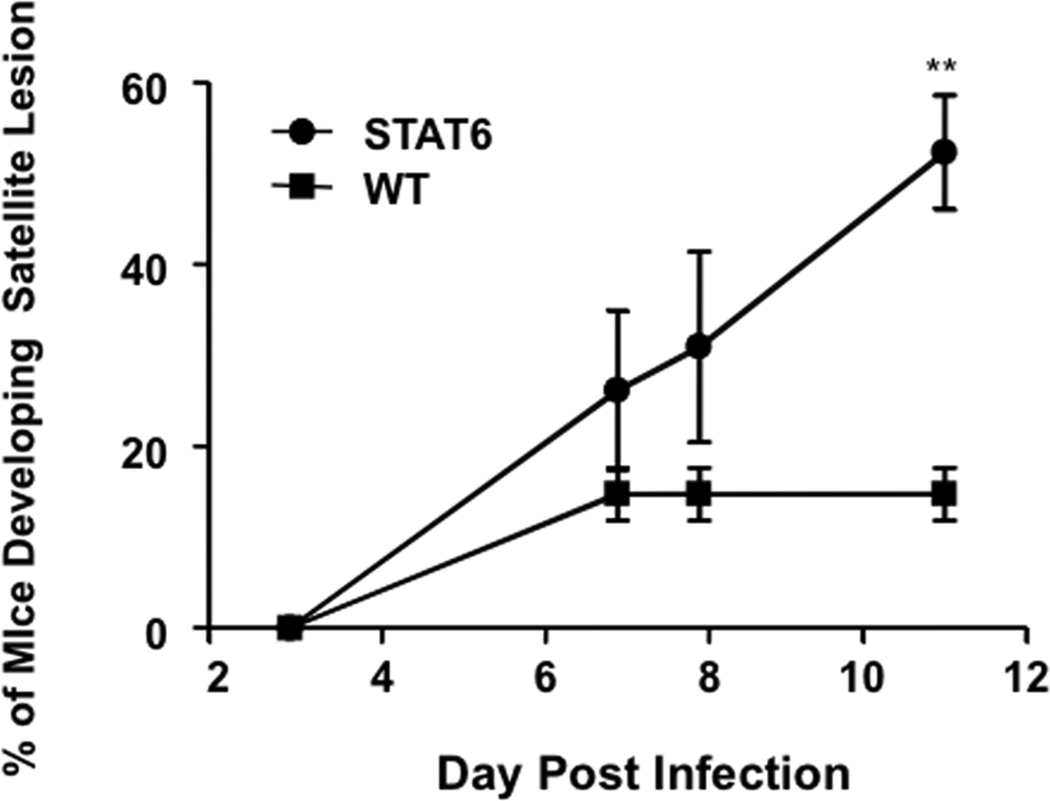
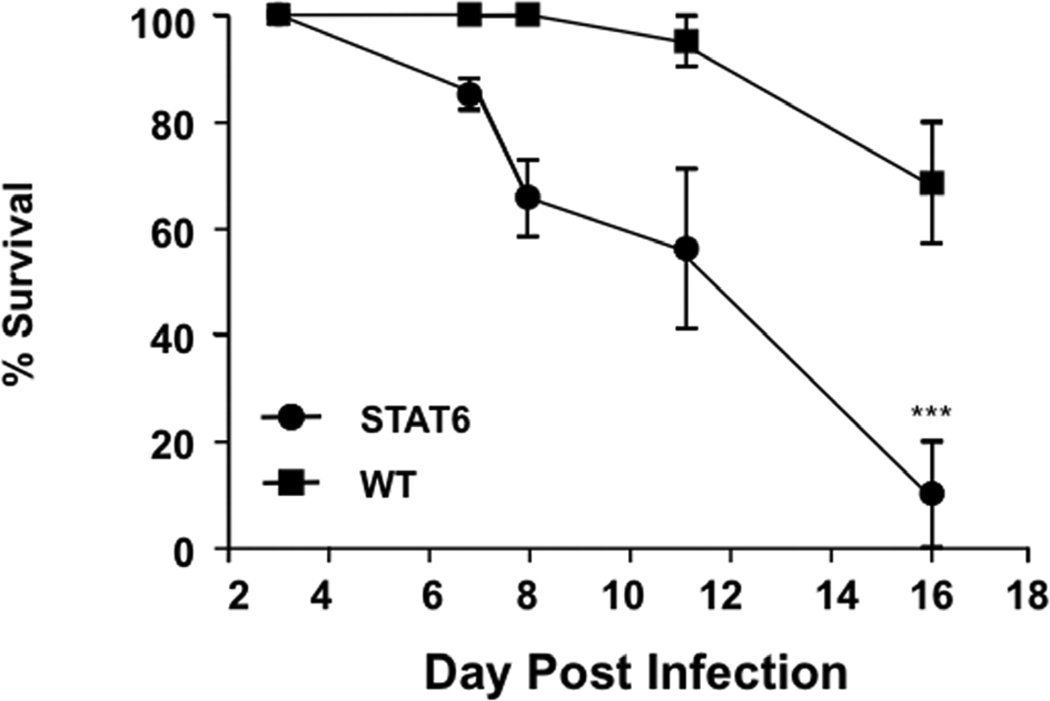
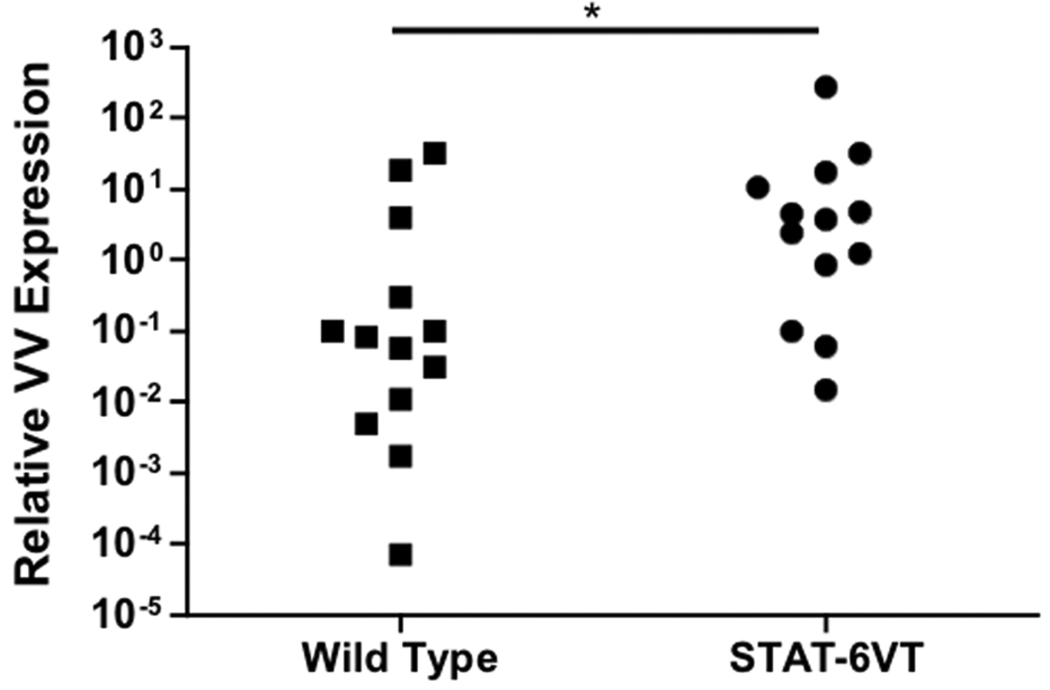
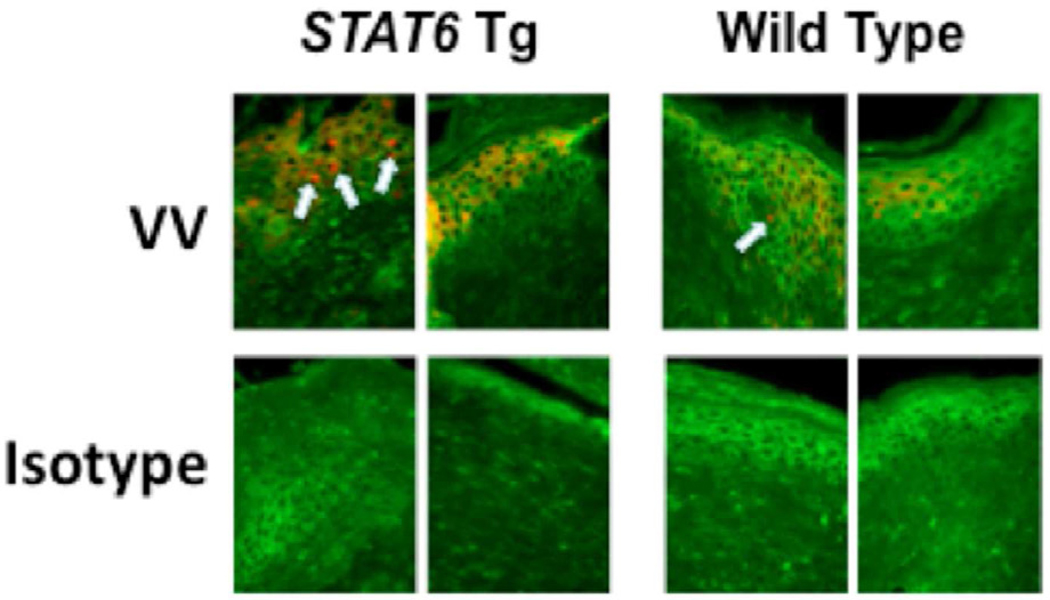
To determine whether constitutive activation of STAT-6 modulates the response to VV, we inoculated four week old STAT-6VT and wild type mice with 5 × 106 pfu of VV (WR1354). Following infection, mice were monitored for death and the development of satellite lesions. At the time of inoculation, none of the mice had any areas of skin irritation or rash. Interestingly, increased morbidity and mortality was observed in VV infected STAT-6VT mice. This was evidenced by satellite lesions in significantly more STAT-6VT mice on day 11 (50.0% of mice) as compared to wild type mice (14.3 %; P = 0.022 by Fisher’s Exact Test) (Figure 3A). Satellite lesions are an indicator of systemic infection that can be fatal. Supporting this notion, we found higher rates of survival in wild type mice compared to STAT-6VT mice, reaching significance on day 16 (wild type: 68.5% survival; STAT-6VT: 10.0% survival; P <0.001 by log rank test) (Figure 3B). In a separate set of experiments, we further evaluated the primary lesions of mice on day seven to determine whether STAT-6 over-expression potentiated VV pathogenesis. VV replication was significantly higher in the primary lesions of STAT-6VT mice (26.84 ± 20.67 ng VV/ng GAPDH; P <0.05) compared to wild type mice (4.19 ± 2.69; Figure 3C). Increased VV replication was confirmed using immunofluorescent staining for A27L (Figure 3D). The mean fluorescence intensity for A27L was significantly greater in primary lesions of six STAT-6VT mice (277.0 ± 30.0 MFI; P =<0.01) compared to primary lesions from six wild type mice (183.3 ± 13.4).
Figure 3.
Constitutive STAT6 expression predisposes mice to disseminated VV infection. A) Satellite lesions, B) Mortality rate, C) VV gene expression, and D) VV protein staining in wild type and STAT-6VT mice following infection with 5×106 pfu of VV. VV replication was visualized by staining for the surface protein A27L (red) and wheat germ agglutinin (green) to visualize the epidermis. *, ** and *** indicate significant differences of P <0.05, P <0.01 and P <0.001, respectively.
Association of STAT6 Variants with risk of ADEH
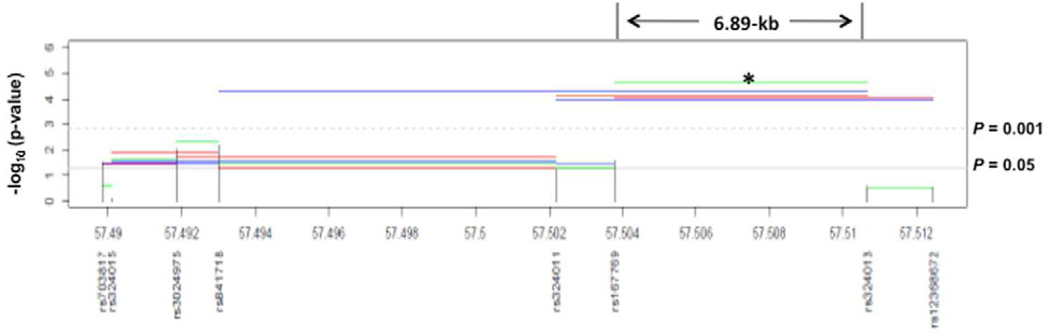
IL-4 and IL-13 potentiate Th2 responses by signaling through STAT6.15,16 To test for association between STAT6 SNPs and ADEH+, we genotyped a total of ten SNPs in a European American sample. Two linkage disequilibrium (LD) blocks were identified, consisting of one ~13 kb block covering the STAT6 gene and one ~1 kb block covering part of the promoter region (Figure 4A). As seen in Table III, significant associations with ADEH+ were observed for three intronic SNPs (rs3024975, odds ratio (OR), 2.14, 95% confidential interval (CI), 1.15–4.01, P = 0.009; rs841718, OR, 1.66, 95%CI, 1.14–2.41, P = 0.006; rs167769, OR, 0.65, 95%CI, 0.43–0.98, P = 0.027), and one 3'-UTR SNP rs703817 (OR, 1.40, 95%CI, 0.92–2.15, P = 0.029). The associations were further enhanced by 2–5 marker haplotype analysis with Omnibus P values from 2.47×10−5 to 0.037, Figure 4B). Among these haplotype analyses, the strongest signal was observed for a 2-SNP haplotype (CT) comprised of SNP rs167769 and rs324013 spanning a region <6.89-kb in the promoter region of STAT6 (ADEH+ vs ADEH−, 24.9 vs 9.2%, P = 5.17×10−6, Table IV). A suggestive association was also observed for SNP rs324011 (OR, 0.70, 95%CI, 0.47–1.01, P = 0.06). In addition, we tested for association between STAT6 SNPs and tIgE and EASI, but no association was found for any of those tested SNPs. Similarly, no associations were observed between the STAT6 variants and risk of AD.
TABLE III.
Associations between STAT6 SNPs and ADEH
| dbSNP | Position | Role | Risk allele |
MAF | OR (95% CI) |
P- value |
|
|---|---|---|---|---|---|---|---|
| ADEH+ | ADEH− | ||||||
| rs12368672 | 57512470 | Promoter | G | 0.321 | 0.376 | 0.78 (0.49–1.24) | 0.276 |
| rs324013 | 55796928 | Promoter | T | 0.453 | 0.503 | 0.82 (0.56–1.19) | 0.285 |
| rs167769 | 55790042 | Intron | C | 0.302 | 0.400 | 0.65 (0.43–0.98) | 0.027 |
| rs324011 | 55788449 | Intron | C | 0.323 | 0.406 | 0.70 (0.47–1.01) | 0.060 |
| rs841718 | 55779263 | Intron (boundary) | A | 0.505 | 0.381 | 1.66 (1.14–2.41) | 0.006 |
| rs3024975 | 55778130 | Intron | G | 0.141 | 0.071 | 2.14 (1.15–4.02) | 0.009 |
| rs324015 | 55776367 | Downstream | C | 0.261 | 0.252 | 1.05 (0.68–1.62) | 0.818 |
| rs703817 | 57489828 | Downstream | C | 0.552 | 0.467 | 1.40 (0.92–2.15) | 0.029 |
MAF: minor allele frequency. OR (95%CI): Odds ratio (95% confidence interval)
TABLE IV.
Association of the 2-SNP haplotype (rs167769 and rs324013) in STAT6 and ADEH. MAF: minor allele frequency.
| Haplotype | rs167769 | rs324013 | MAF | OR (95% CI) | P-value | |
|---|---|---|---|---|---|---|
| ADEH+ | ADEH− | |||||
| 1 | C | C | 0.45 | 0.51 | 0.77 (0.43–1.40) | 0.210 |
| 2 | T | T | 0.31 | 0.40 | 0.66 (0.31–1.24) | 0.038 |
| 3 | C | T | 0.25 | 0.09 | 3.33 (1.39–8.55) | 5.17×10−6 |
Omnibus P-value = 2.47 ×10−5
STAT6 variants are associated with IFNγ ELISPOT values
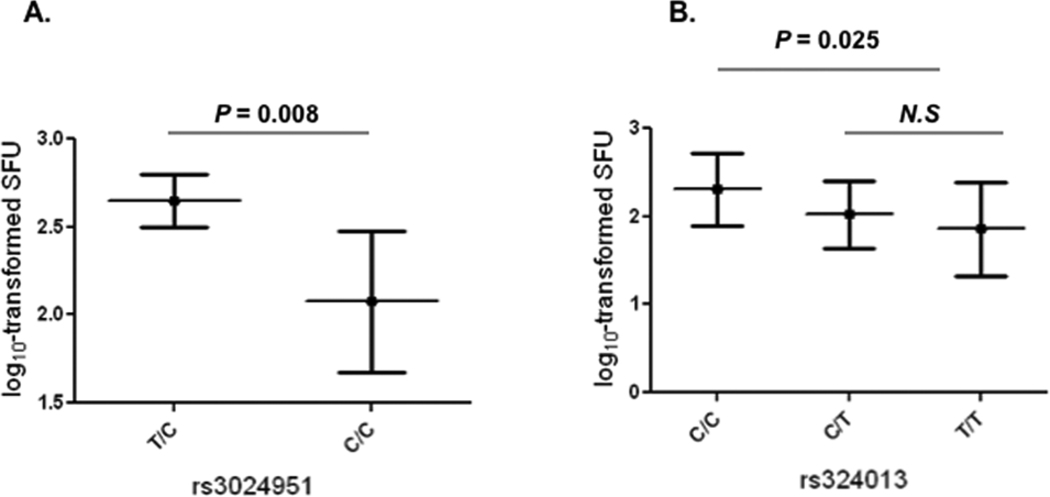
In previous studies, we have observed a reduced IFNγ production in patients with ADEH+ when compared with AD patients without EH (ADEH−) and healthy controls.41 To explore the functional link of STAT6 variants to disease, we tested if patients carrying these SNPs have differential levels of IFNγ. We found a significant association for SNP rs3024951 and IFNγ levels (P = 0.031) when analyses were made for all these SNPs and IFNγ levels in HSV-stimulated PBMCs from all subjects (n=64). Moreover, when analyses were restricted to AD patients (n =44), the significance was further enhanced (TC vs TT, P = 0.008, Figure 5A). In addition, we observed a significant association for a promoter SNP rs324013. Interestingly, SNP rs324913 T allele, which has been shown to be the strongest association with an increased risk of ADEH+ as part of 2-marker haplotype, showed lower levels of IFNγ as compared to others (log10-tranformed SFU, CC vs CT vs TT: 2.31 vs 2.02 vs 1.86, P =0.025, Figure 5B).
Figure 5.
Association of STAT6 SNPs with IFNγ production in HSV-stimulated PBMCs from AD patients (n=44) as determined by the log10-transformed mean SFC/106 cells. A) The association was observed for SNP rs3024951 (TC [n=7] vs CC [n=32)] P = 0.008), B) the association was observed for SNP rs324013 (CC [n=13] vs CT [n=18] +TT [n = 8], P = 0.025). N.S.: not significant.
DISCUSSION
Eczema vaccinatum is a severe, life-threatening adverse event that develops in a subset of AD patients following smallpox vaccination. Our current study is the first to demonstrate that the skin of ADEH+ patients supports VV replication to a greater extent than skin from ADEH− and QAD as well as non-AD patient groups including asthma and psoriasis. Increased VV replication was confirmed by analyzing the number of VV DNA copies and immunofluorescent staining for A27L, a protein essential for transport of intracellular virus particles.18 We hypothesize that increased VV replication in skin explants can predict those patients who are at risk of developing a disseminated viral skin infection following smallpox vaccination.
Previously, we have shown that over-expression of IL-4 and IL-13 in AD skin supports increased VV replication in human keratinocytes.22,23 IL-4 and IL-13 have previously been shown to signal through STAT615,16 which binds to consensus target sequences and inhibits transcription of innate immune response genes.24,25 Specifically, we have demonstrated that IL-4 and IL-13 activate STAT-6 signaling thereby inhibiting the induction of AMPs that have activity against VV.22,25 In the current study, we demonstrate that neutralization of IL-4 and IL-13 in ADEH+ skin limits VV replication. It is unethical to test vaccination in AD patients; therefore, we used transgenic mice as a surrogate to evaluate whether constitutive STAT-6 activation predisposes mice to disseminated VV skin infection. Transgenic mice over-expressing constitutively active STAT-6 in T cells are predisposed towards Th2 responses and allergic inflammation, as previously described.12,34 These mice spontaneously develop eczematoid skin rashes, over-express IL-4 and IL-13 in the skin, and exhibit increased serum IgE levels due to increased Th2 development in vivo.35 It has previously been shown, using knockout mice, that VV replication is impaired in IL-4 deficient mice36 consistent with increased mortality in C57Bl/6 mice infected with an IL-4 expressing Ectromelia virus (mousepox).37 In our experiments, constitutive activation of STAT-6 increased the morbidity and mortality in mice following VV infection. This was evidenced by increased satellite lesion development and decreased survival in STAT-6VT mice. Given the importance of STAT-6 in IL-4 and IL-13 mediated VV replication in AD skin,9 we sought further support for this molecule in susceptibility of disseminated viral skin infections among atopic dermatitic patients by genotyping genetic polymorphisms encompassing the STAT6 gene and testing for association with ADEH+, and associated phenotypes in European American patients participating in the multicenter ADVN.10 Indeed, genetic linkage studies have previously identified chromosomal region 12q13-24, where STAT6 is located, to be linked with atopy-related diseases.26 Genetic variants in STAT6 have been associated with atopy,27 tIgE,12,29,30 and asthma12,31 in diverse populations. In particular, a polymorphism (rs324011) in intron 2 of STAT6, that was associated with elevated serum IgE, was suggested to alter NF-κB binding, STAT6 promoter activity, and mRNA expression.29,32 In this study, in addition to those significant associations observed for four STAT6 SNPs (rs3024975, rs841718, rs167769, rs703817) and ADEH+, we provided suggestive evidence of an association between ADEH and SNP rs324011. This marker was in strong LD with the strongest ADEH− associated SNP (rs841718) within the same LD block (D'=0.94, Figure 3B). The association with ADEH was further enhanced by haplotype analysis, where we observed that haplotype CT (a 2-marker haplotype, rs167769 C-rs324013T) across a region of 6.87-kb was significantly more common among in ADEH patients compared to AD patients without EH (24.9% vs and 9.2%; P = 5.17×10−6). Interestingly, this haplotype encompasses the major functional region in the promoter of STAT6. We contend that either this haplotype or an untyped genetic variant(s) within this locus may confer susceptibility to an increased risk of ADEH and have a functional impact on disease. Although STAT6 SNPs have been repeatedly associated with tIgE,29,30,33 none of those identified SNPs for ADEH+ in this study were associated with this trait (data not shown), suggesting that the genetic associations are specific to disease risk. To test for the possibility that the associations observed between STAT6 SNPs and ADEH were for manifestation of an HSV exacerbation in an AD population rather than the disease markers, we investigated the association between HSV infection and the associated SNPs among healthy controls, none of which showed association with HSV infection (data not shown), suggesting that the associations with ADEH were not confounded by HSV infection. Although the genetic association studies herein were intended to support the vaccinia virus expression studies and the transgenic murine studies, a shortcoming of these analyses is a lack of replication in an independent sample. In addition, to explore whether abnormalities in STAT6 function coexist with an FLG-dependent barrier defect, we performed a gene-gene interaction analysis (GxG) and found that there was no increased association for the combined STAT6 SNPs and FLG mutations as compared to SNP analyzed individually. The results suggested that the associations observed for STAT6 SNPs might be independent of FLG mutations, but this needs to be replicated in a relatively large population with a comprehensive coverage of STAT6. One of the major strengths in this study is that we observed significant associations between variants in STAT6 and IFNγ production in HSV-stimulated PBMCs from AD patients. Among all subjects, both SNP rs3024951 and rs324013 showed differential IFNγ production in response to HSV exposure. Although the precise mechanism is not clear, our findings suggest that these STAT6 SNPs may be involved in the regulation of IFNγ production. Indeed, SNP rs324013 T allele, one part of 2-marker haplotype CT (rs167769 C-rs324013T), showed the strongest association with an increased risk of ADEH+ but reduced levels of IFNγ production, suggesting that this SNP, or SNPs within the region covering the haplotype, may play a role in mediating HSV induced IFNγ production, and subsequently lead to the development of ADEH+. As a result, it will be essential to investigate additional SNPs in and around the identified region (~6.87-kb) in the future.
We recognized that this study has several limitations. In addition to the absence of a replication population, the sample available for these studies is relatively small. However, ADEH is a rare disease (~3%), and the sample used for this study reflects intense recruitment efforts after nearly five years from multiple medical centers. As demonstrated under “Methods”, we were in fact sufficiently powered to detect a true association given the frequency of the SNPs associated with ADEH+ (i.e., >10–20%); second, our observations in these initial studies relying heavily upon a tagging SNP approach are the basis for further studies. As a next step, we aim to identify the causal STAT6 variant(s) by an independent replication with sufficient sample size, deep resequencing the candidate region elucidated herein, and ultimately determining the functional relevance to risk of ADEH+.
In this current study, we demonstrate that skin from ADEH+ patients supports VV replication. Using STAT-6VT transgenic mice, we demonstrate a functional correlation between STAT6 activity in T cells and disseminated VV infection. Furthermore, we provide evidence of an association between STAT6 SNPs and the risk of ADEH and IFNγ production in a multicenter case-control study. Although the mechanisms remain unclear, our findings suggest that STAT6 SNPs may be involved in the regulation of IFNγ production and also serve as important genetic markers in determining those AD patients with greater susceptibility to develop disseminated VV infection following smallpox vaccination. This is the first study to implicate STAT6 as a potential candidate gene for ADEH. However, replication in an independent population with sufficient sample size, evidence of association between STAT6 SNPs and skin IL-4 and IL-13 expression, and evidence of functional relevance to disease, are clearly needed in the future studies. Additionally, given that FLG in our previous studies have been shown to have a significant impact on ADEH, further analysis on an interaction between FLG and STAT6 is essential. Taken together, our data suggest that the STAT6 gene increases viral replication in the skin of atopic dermatitis patients with a history of eczema herpeticum. Further genetic association studies and functional investigations are warranted.
Key Messages.
Increased IL-4 and IL-13 expression enhances viral replication in atopic skin
Activation of the STAT6 gene enhances viral replication in atopic dermatitis
Polymorphisms in the STAT6 gene are associated with propensity to eczema herpeticum
ACKNOWLEDGEMENTS
The authors thank Maureen Sandoval for her assistance in the preparation of this manuscript and the nursing staff in the CTRC for their help in patient recruitment and sample collection.
This work was supported by NIH/NIAID contracts N01 AI40029 and N01 AI40030; MHK supported by NIH/NIAID grants U19 AI070448 and PO1 AI056097
Supported in part by the Colorado Clinical Translational Science Award grant 1 UL1 RR025780 from NCRR/NIH
NON STANDARD ABBREVIATIONS
- AD
Atopic dermatitis
- ADVN
Atopic Dermatitis Vaccinia Network
- ADEH+
Atopic dermatitis with a history of eczema herpeticum
- ADEH−
Atopic dermatitis without a history of eczema herpeticum
- AIMs
Ancestry Informative Markers
- AMP
Anti-microbial peptide
- AS
Asthma
- CT
2-SNP haplotype
- EASI
Eczema Area and Severity Index
- EH
Eczema herpeticum
- EV
Eczema vaccinatum
- tIgE
total serum IgE
- LD
Linkage disequilibrium
- N
No history of skin disease
- NIAID
National Institute of Allergy and Infectious Diseases
- PS
Psoriasis
- QAD
Quiescent atopic dermatitis
- SNPs
single nucleotide polymorphisms
- STAT
Signal transducer and activator of transcription
- Th2
T helper 2
- VV
Vaccinia virus
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Leung DY, Boguniewicz M, Howell MD, Nomura I, Hamid QA. New insights into atopic dermatitis. J Clin Invest. 2004;113:651–657. doi: 10.1172/JCI21060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barnes KC. An update on the genetics of atopic dermatitis: scratching the surface in 2009. J Allergy Clin Immunol. 2010;125:16–29. doi: 10.1016/j.jaci.2009.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McGirt LY, Beck LA. Innate immune defects in atopic dermatitis. J Allergy Clin Immunol. 2006;118:202–208. doi: 10.1016/j.jaci.2006.04.033. [DOI] [PubMed] [Google Scholar]
- 4.Boguniewicz M, Leung DY. Recent insights into atopic dermatitis and implications for management of infectious complications. J Allergy Clin Immunol. 2010;125:4–13. doi: 10.1016/j.jaci.2009.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Declaration of Global Eradication of Smallpox. World Health Organization:Weekly Epidemiologic Records. 1980;55:148.
- 6.Reynolds MG, Davidson WB, Curns AT, Conover CS, Huhn G, Davis JP, et al. Spectrum of infection and risk factors for human monkeypox, United States, 2003. Emerg Infect Dis. 2007;13:1332–1339. doi: 10.3201/eid1309.070175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rotz LD, Dotson DA, Damon IK, Becher JA. Vaccinia (smallpox) vaccine: recommendations of the Advisory Committee on Immunization Practices (ACIP), 2001. MMWR Recomm Rep. 2001;50:1–25. [PubMed] [Google Scholar]
- 8.Grabenstein JD, Winkenwerder W., Jr US military smallpox vaccination program experience. JAMA. 2003;289:3278–3282. doi: 10.1001/jama.289.24.3278. [DOI] [PubMed] [Google Scholar]
- 9.Howell MD, Gallo RL, Boguniewicz M, Jones JF, Wong C, Streib JE, et al. Cytokine milieu of atopic dermatitis skin subverts the innate immune response to vaccinia virus. Immunity. 2006;24:341–348. doi: 10.1016/j.immuni.2006.02.006. [DOI] [PubMed] [Google Scholar]
- 10.Beck LA, Boguniewicz M, Hata T, Schneider LC, Hanifin J, Gallo R, et al. Phenotype of atopic dermatitis subjects with a history of eczema herpeticum. J Allergy Clin Immunol. 2009;124:260–269. doi: 10.1016/j.jaci.2009.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gao PS, Rafaels NM, Hand T, Murray T, Boguniewicz M, Hata T, et al. Filaggrin mutations that confer risk of atopic dermatitis confer greater risk for eczema herpeticum. J Allergy Clin Immunol. 2009;124:507–513. doi: 10.1016/j.jaci.2009.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bruns HA, Schindler U, Kaplan MH. Expression of a constitutively active STAT6 in vivo alters lymphocyte homeostasis with distinct effects in T and B cells. J Immunol. 2003;170:3478–3487. doi: 10.4049/jimmunol.170.7.3478. [DOI] [PubMed] [Google Scholar]
- 13.Freyschmidt EJ, Mathias CB, MacArthur DH, Laouar A, Narasimhaswamy M, Weih F, et al. Skin inflammation in RelB(−/−) mice leads to defective immunity and impaired clearance of vaccinia virus. J Allergy Clin Immunol. 2007;119:671–679. doi: 10.1016/j.jaci.2006.12.645. [DOI] [PubMed] [Google Scholar]
- 14.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Takeda K, Kamanaka M, Tanaka T, Kishimoto T, Akira S. Impaired IL-13-mediated functions of macrophages in STAT6-deficient mice. J Immunol. 1996;157:3220–3222. [PubMed] [Google Scholar]
- 16.Takeda K, Tanaka T, Shi W, Matsumoto M, Minami M, Kashiwamura S, et al. Essential role of STAT6 in IL-4 signalling. Nature. 1996;380:627–630. doi: 10.1038/380627a0. [DOI] [PubMed] [Google Scholar]
- 17.Duetsch G, Illig T, Loesgen S, Rohde K, Klopp N, Herbon N, et al. STAT6 as an asthma candidate gene: polymorphism-screening, association and haplotype analysis in a Caucasian sib-pair study. Hum Mol Genet. 2002;11:613–621. doi: 10.1093/hmg/11.6.613. [DOI] [PubMed] [Google Scholar]
- 18.Sanderson CM, Hollinshead M, Smith GL. The vaccinia virus A27L protein is needed for the microtubule-dependent transport of intracellular mature virus particles. J Gen Virol. 2000;81:47–58. doi: 10.1099/0022-1317-81-1-47. [DOI] [PubMed] [Google Scholar]
- 19.Hamid Q, Boguniewicz M, Leung DY. Differential in situ cytokine gene expression in acute versus chronic atopic dermatitis. J Clin Invest. 1994;94:870–876. doi: 10.1172/JCI117408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nomura I, Goleva E, Howell MD, Hamid QA, Ong PY, Hall CF, et al. Cytokine milieu of atopic dermatitis, as compared to psoriasis, skin prevents induction of innate immune response genes. J Immunol. 2003;171:3262–3269. doi: 10.4049/jimmunol.171.6.3262. [DOI] [PubMed] [Google Scholar]
- 21.Howell MD, Novak N, Bieber T, Pastore S, Girolomoni G, Boguniewicz M, et al. Interleukin-10 downregulates anti-microbial peptide expression in atopic dermatitis. J Invest Dermatol. 2005;125:738–745. doi: 10.1111/j.0022-202X.2005.23776.x. [DOI] [PubMed] [Google Scholar]
- 22.Howell MD, Boguniewicz M, Pastore S, Novak N, Bieber T, Girolomoni G, et al. Mechanism of HBD-3 deficiency in atopic dermatitis. Clin Immunol. 2006;121:332–338. doi: 10.1016/j.clim.2006.08.008. [DOI] [PubMed] [Google Scholar]
- 23.Howell MD. The role of human beta defensins and cathelicidins in atopic dermatitis. Curr Opin Allergy Clin Immunol. 2007;7:413–417. doi: 10.1097/ACI.0b013e3282a64343. [DOI] [PubMed] [Google Scholar]
- 24.Ohmori Y, Hamilton TA. Interleukin-4/STAT6 represses STAT1 and NF-kappa B-dependent transcription through distinct mechanisms. J Biol Chem. 2000;275:38095–38103. doi: 10.1074/jbc.M006227200. [DOI] [PubMed] [Google Scholar]
- 25.Albanesi C, Fairchild HR, Madonna S, Scarponi C, De Pita O, Leung DY, et al. IL-4 and IL-13 negatively regulate TNF-alpha- and IFN-gamma-induced beta-defensin expression through STAT-6, suppressor of cytokine signaling (SOCS)-1, and SOCS-3. J Immunol. 2007;179:984–992. doi: 10.4049/jimmunol.179.2.984. [DOI] [PubMed] [Google Scholar]
- 26.Ober C, Cox NJ, Abney M, Di Rienzo A, Lander ES, Changyaleket B, et al. Genome-wide search for asthma susceptibility loci in a founder population. The Collaborative Study on the Genetics of Asthma. Hum Mol Genet. 1998;7:1393–1398. doi: 10.1093/hmg/7.9.1393. [DOI] [PubMed] [Google Scholar]
- 27.Amoli MM, Hand S, Hajeer AH, Jones KP, Rolf S, Sting C, et al. Polymorphism in the STAT6 gene encodes risk for nut allergy. Genes Immun. 2002;3:220–224. doi: 10.1038/sj.gene.6363872. [DOI] [PubMed] [Google Scholar]
- 28.Gao PS, Mao XQ, Roberts MH, Arinobu Y, Akaiwa M, Enomoto T, et al. Variants of STAT6 (signal transducer and activator of transcription 6) in atopic asthma. J Med Genet. 2000;37:380–382. doi: 10.1136/jmg.37.5.380a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Weidinger S, Gieger C, Rodriguez E, Baurecht H, Mempel M, Klopp N, et al. Genome-wide scan on total serum IgE levels identifies FCER1A as novel susceptibility locus. PLoS Genet. 2008;4 doi: 10.1371/journal.pgen.1000166. e1000166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Weidinger S, Klopp N, Wagenpfeil S, Rummler L, Schedel M, Kabesch M, et al. Association of a STAT 6 haplotype with elevated serum IgE levels in a population based cohort of white adults. J Med Genet. 2004;41:658–663. doi: 10.1136/jmg.2004.020263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gao PS, Heller NM, Walker W, Chen CH, Moller M, Plunkett B, Roberts MH, Schleimer RP, Hopkin JM, Huang SK. Variation in dinucleotide (GT) repeat sequence in the first exon of the STAT6 gene is associated with atopic asthma and differentially regulates the promoter activity in vitro. J Med Genet. 2004;41:535–539. doi: 10.1136/jmg.2003.015842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schedel M, Frei R, Bieli C, Cameron L, Adamski J, Lauener R, et al. An IgE-associated polymorphism in STAT6 alters NF-kappaB binding, STAT6 promoter activity, and mRNA expression. J Allergy Clin Immunol. 2009;124:583–589. doi: 10.1016/j.jaci.2009.06.024. [DOI] [PubMed] [Google Scholar]
- 33.Vercelli D. Advances in asthma and allergy genetics in 2007. J Allergy Clin Immunol. 2008;122:267–271. doi: 10.1016/j.jaci.2008.06.008. [DOI] [PubMed] [Google Scholar]
- 34.Kaplan MH, Sehra S, Chang HC, O'Malley JT, Mathur AN, Bruns HA. Constitutively active STAT6 predisposes toward a lymphoproliferative disorder. Blood. 2007;110:4367–4369. doi: 10.1182/blood-2007-06-098244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sehra S, Yao Y, Howell MD, Nguyen ET, Kansas GS, Leung DY, et al. IL-4 regulates skin homeostasis and the predisposition toward allergic skin inflammation. J Immunol. 2010;184:3186–3190. doi: 10.4049/jimmunol.0901860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.van Den Broek M, Bachmann MF, Kohler G, Barner M, Escher R, Zinkernagel R, et al. IL-4 and IL-10 antagonize IL-12-mediated protection against acute vaccinia virus infection with a limited role of IFN-gamma and nitric oxide synthetase 2. J Immunol. 2000;164:371–378. doi: 10.4049/jimmunol.164.1.371. [DOI] [PubMed] [Google Scholar]
- 37.Jackson RJ, Ramsay AJ, Christensen CD, Beaton S, Hall DF, Ramshaw IA. Expression of mouse interleukin-4 by a recombinant ectromelia virus suppresses cytolytic lymphocyte responses and overcomes genetic resistance to mousepox. J Virol. 2001;75:1205–1210. doi: 10.1128/JVI.75.3.1205-1210.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gao PS, Rafaels NM, Mu D, Hand T, Murray T, Boguniewicz M, et al. Genetic variants in thymic stromal lymphopoietin are associated with atopic dermatitis and eczema herpeticum. J Allergy Clin Immunol. 2010;125:1403–1407. doi: 10.1016/j.jaci.2010.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gauderman WJ. Sample size requirements for association studies of gene-gene interaction. Am J Epidemiol. 2002;155:478–484. doi: 10.1093/aje/155.5.478. [DOI] [PubMed] [Google Scholar]
- 40.Janetzki S, Cox JH, Oden N, Ferrari G. Standardization and validation issues of the ELISPOT assay. Methods Mol Biol. 2005;302:51–86. doi: 10.1385/1-59259-903-6:051. [DOI] [PubMed] [Google Scholar]
- 41.Leung DYM, Gao PS, Grigoryev DN, Rafaels NM, Streib JE, Howell MD, et al. Human atopic dermatitis complicated by eczema herpeticum is associated with abnormalities in gamma interferon response. J Allergy Clin Immunol. 2011;127:957–963. doi: 10.1016/j.jaci.2011.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]