Abstract
Quantitative mass spectrometry-based proteomics is highly versatile, but not easily multiplexed. Isobaric labeling strategies allow mass spectrometry-based multiplexed proteome quantification; however, ratio distortion due to protein quantification interference is a common effect. We present a multi-proteome model (mixture of human and yeast proteins) in a 6-plex isobaric labeling system to fully document the interference effect, and we report that a multistage MS3-based approach almost completely eliminates interference.
Mass spectrometry-based proteomics is commonly used in a non-array based format.1 While this approach provides versatility, it also limits the technology’s throughput by hindering parallel multiplexing of quantitative experiments. This shortcoming was addressed by the development of the isobaric labeling strategy for quantitative proteomics, including isobaric tag for relative and absolute quantitation (iTRAQ),2 and tandem mass tags (TMT).3,4 Relative protein concentrations in up to eight samples can be determined in a single experiment through tagging peptides with stable isotope labeled reagents.5 Peptide ions are indistinguishable in full-MS spectra but are quantified based on small m/z fragment reporter ions after the peptide ion is isolated and analyzed in a tandem mass spectrometry (MS2) experiment (Fig. 1a). However, it was demonstrated recently that the accuracy and precision of quantitative data from such experiments suffer because contaminating near isobaric ions are co-isolated and co-fragmented together with the target ions (Fig. 1b); thus skewing reporter ion intensities.6–9 When analyzing a complex protein mixture, in which only a fraction of the proteins show expression changes, the interference effect causes an underestimation of these differences in expression. Intensity differences between the reporter ions when expressed as ratios tend towards unity due to interfering peptide ions derived from proteins with unchanging expression. We believe that fully addressing the issues caused by the interference effect will broaden acceptance of multiplexed MS-based proteomics. Here we describe the use of a multi-proteome model sample to accurately measure the extent of the interference effect on quantitative data from a large-scale proteomics experiment. We used this model to evaluate several analytical strategies as potential solutions to the outlined difficulties and we show that collecting an additional isolation and fragmentation event (MS3 scan) overcomes these problems by eliminating the interference effect.
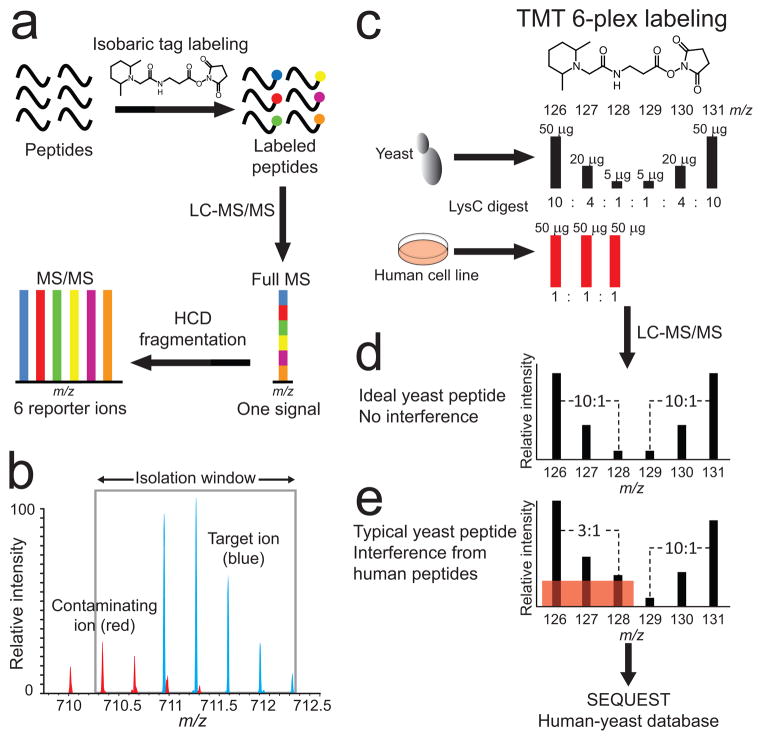
Figure 1.
Isobaric labeling, interference and interference modeling. (a) Quantitative mass spectrometry-based proteomics experiments with stable isotope containing isobaric tags (TMT or iTRAQ) produce differentially labeled peptide ions that are indistinguishable in full-MS spectra. Quantification is based on low m/z reporter fragment ion intensities after a target ion is isolated and fragmented in an MS2 experiment. (b) Both accuracy and precision of peptide quantification are affected when contaminating peptide ions (red) are co-isolated with the target peptide ion (blue), a phenomenon that we term the interference effect. (c) Accurate measurement of the interference effect by a multi-proteome model using TMT six-plex labeled human cell line and S. cerevisiae Lys-C digests. The first three channels model interference, where yeast peptides were combined to create three different ratios (10:1, 4:1, 2.5:1) amongst unchanging interfering human peptides (1:1:1). In the last three channels, yeast peptides were combined in the same ratios (mirrored relative to the first three channels) without any human peptide interference. (d) An ideal yeast peptide MS2 spectrum without human peptide interference in the first three channels would have identical and mirrored TMT reporter ion intensities in the last three channels. (e) A typical yeast peptide has some degree of interference in the first three reporter ion channels (red shaded box), which results in ratio distortion towards 1:1:1.
We used six-plexed TMT reagents to build a model multi-proteome peptide mixture sample from Lys-C protein digests of Saccharomyces cerevisiae (yeast) and the human HeLa cell line (Fig. 1c and Supplementary Fig. 1). Peptides tagged with the six different TMT reagents generate reporter ions (quantification channels) at m/z 126, 127, 128, 129, 130, and 131 when subjected to MS2. We labeled yeast peptides with all six reagents and mixed the differentially labeled peptides so that the ratios of reporter ions 126/127 and 131/130 were 2.5:1; the ratio of 127/128 and 130/129 were 4:1; and the ratio of 126/128 and 131/129 were 10:1. We then labeled human peptides with TMT reagents 126, 127, and 128. We combined equal amounts (1:1:1) of the differentially tagged human peptides and added this peptide mixture to the yeast peptides so that the total amount of human peptides in channels 126, 127, and 128 was twice the amount of yeast peptides in the same channels. We used this sample to accurately measure the interference effect. With no interference, ratios of channels 126/127, 127/128, and 126/128 are expected to be equal to the ratios of channels 131/130, 130/129, and 131/129, respectively, for each yeast peptide ion selected for MS2 (Fig. 1d). Interference from human peptide ions on yeast ions in channels 126, 127, and 128 was responsible for a leveling out of yeast reporter ion intensities so that ratios measured for channels 126, 127, and 128 were less than those measured for channels 129, 130, and 131 where no interference from human peptides can occur (Fig. 1e). Comparable model samples to study the interference effect have been reported previously;6–9 however, our approach of mixing whole proteomes, instead of spiking protein standard digests into background peptide samples, maximizes the number of observable individual interference events.
We fractionated the described sample using strong cation exchange (SCX) chromatography10 and analyzed fractions by liquid chromatography (LC)-MS2 on an LTQ Orbitrap Velos mass spectrometer, where MS2 experiments were performed using higher-energy collision dissociation (HCD)11 to generate high resolution and high mass accuracy MS2 spectra. This resulted in the identification of 2,089 yeast proteins from 8,771 non-redundant peptide sequences and 6,127 human proteins from 34,885 peptides at a protein false discovery rate of 1.5% (Table 1 and Supplementary Data).12,13 Yeast peptide ions were detected with varying degrees of human peptide interference (Fig. 2a and Supplementary Fig. 2) and we also observed yeast peptide interference in human peptide ion MS2 spectra (Fig. 2a). For yeast peptide ions the median compression of expected 10:1 ratios was 3.2-fold, 2.2-fold for 4:1 ratios, and 1.3-fold for 2.5:1 ratios (Fig. 2b–c). The loss of quantification accuracy was accompanied by a loss in precision, where the 10:1 log2 ratio distribution standard deviation was 0.5 for channels without interference and 0.8 for channels with human peptide interference, an increase of 60%.
Table 1. MS2 versus MS3 identifications and quantifications.
A comparison of the number of peptide and protein identifications and quantifications from 20 SCX fractions of the human yeast multi-proteome model using the MS2 and the MS3 methods.
| Total peptides | Unique peptides | Total proteins | % MS2 methodc | ||
|---|---|---|---|---|---|
| Identificationsa | MS2 method | 117,849 | 43,656 | 8,216 | |
| MS3 method | 96,202 | 38,122 | 7,578 | 92% | |
|
| |||||
| Quantificationsb | MS2 method | 109,967 | 41,804 | 8,803 | |
| MS3 method | 75,330 | 33,849 | 7,089 | 88% | |
All human and yeast identifications.
All human and yeast quantifications. See Supplementary Table 1 for a more detailed breakdown of the data. TMT reporter ions had to be present (signal > 0) in all six channels for yeast peptides and for channels 126, 127 and 128 for human peptides.
The proportion of identified or quantified proteins using the MS3 method compared to the MS2 method
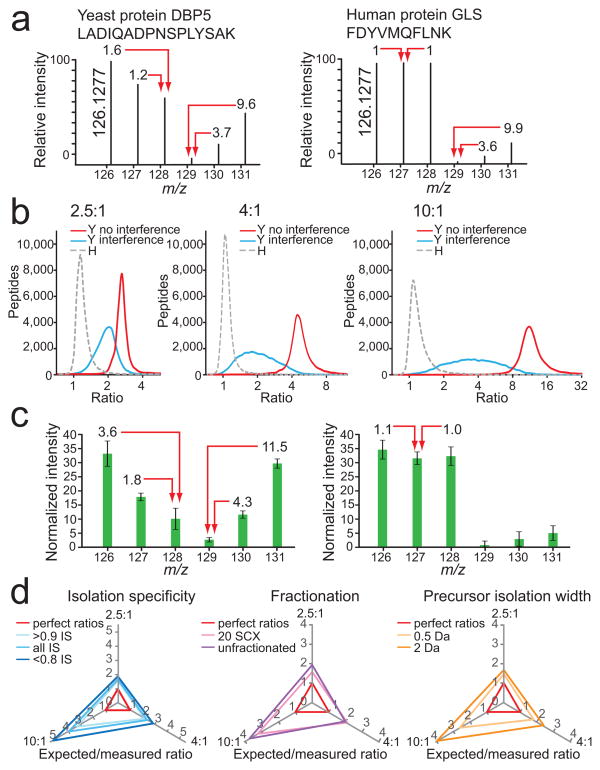
Figure 2.
Evaluation and attempted removal of the interference effect. (a) An example of strong distortion of reporter ion intensities for a yeast peptide ion by interference of human peptide ions (left) and the effect of interference by yeast peptides on the reporter ion intensities of a human peptide (right; signals in channels 129, 130, and 131). (b) Ratiodistributions (log2) of yeast (Y) peptides in channels with human (H) peptide interference (blue line), without human peptide interference (red line), and human peptides only (gray dashed line) for the predicted ratios 2.5:1 (left), 4:1 (middle), and 10:1 (right). LC-MS2 data were collected after separating the yeast and human whole cell lysate digest into 20 fractions using SCX. (c) Averaged normalized relative intensities for each TMT reporter ion channel for yeast peptides (left), and human peptides (right) from the dataset described in (b). Error bars represent one standard deviation for 20,272 and 81,180 yeast and human peptides, respectively. (d) There was minimal improvement of yeast peptide ratios in channels with human peptide interference (126, 127, and 128) with post-acquisition data filtering of isolation specificity (IS; left), fractionation (middle) and precursor isolation width (right). Expected ratios were divided by measured ratios, where perfect ratios would have a value of 1 for all three axes (red triangle). The greater the distance of each tested condition from the red perfect ratio triangle, the greater the influence of human peptide interference on yeast peptide ratio distortion. Increasing dynamic range results in greater ratio distortion.
With the ability to accurately measure the extent of the interference effect in a high-complexity multi-proteome model, we evaluated four analytical strategies that could potentially eliminate or reduce the interference effect: (i) estimation of the extent of interference based on full-MS data, (ii) sample fractionation, (iii) reduction of the precursor ion isolation width, and (iv) MS3 data acquisition.
We measured interference by examining for visible interfering peaks in the isolation window for each full-MS. We estimated the number and intensity of contaminating peptide ions by a measure we termed isolation specificity (IS), which represents the relative fraction (0–1) of target ion intensity compared to the complete ion intensity within the precursor ion isolation window (see Online Methods for a detailed description). We found that 94% of yeast peptide ions showed an IS ≥ 0.9 with more than or equal to 90% of the ion intensity assigned to the target ion (Supplementary Fig. 3), but that the measure was poor in predicting interference. Comparing median reporter ion ratios for peptide ions with an IS of < 0.8 with those for peptide ions with an IS of ≥ 0.9, we observed an increase from 1.9 to 3.0 for an expected ratio of 10, from 1.4 to 1.8 for an expected ratio of 4, and from 1.3 to 1.6 for an expected ratio of 2.5 (Fig. 2d). The modest reduction of interference for peptide ions with high IS indicates that an estimation of the effect based on full-MS data is insufficient in predicting or eliminating the interference problem.
Using a widely applied fractionation strategy, we then separated a whole cell lysate digest sample into 20 SCX fractions, which, intriguingly, resulted in an even lower reduction of the interference effect (Fig. 2d). We measured median ratio changes from 2.7 to 3.1 (expected ratio of 10), from 2.0 to 1.9 (expected ratio of 4), and from 1.3 to 1.6 (expected ratio of 2.5). We note that more extensive approaches for fractionation may yield different results with respect to ratio interference.
Narrowing of the precursor ion isolation width from 2 m/z to 0.5 m/z had a higher impact in reducing the interference effect, although it was not completely eliminated (Fig. 2d). Median yeast peptide ratios from one SCX fraction increased from 2.5 to 3.8 (expected ratio of 10), from 1.7 to 2.2 (expected ratio of 4), and from 1.5 to 1.7 (expected ratio of 2.5). The reduction of precursor ion width from 2 to 0.5 m/z resulted in a 22% reduction of non-redundant peptide identifications (Supplementary Fig. 4).
When applying MS3 in mass spectrometry-based proteomics, first a peptide ion is isolated and fragmented, followed by the selection of a fragment ion that is again isolated and fragmented to produce an MS3 spectrum.14 When applying such a strategy to quantitative studies using isobaric labeling, fragment ions from contaminating co-isolated and co-fragmented ions span a large MS2 m/z space before a relatively small m/z window containing a high intensity target fragment ion is selected for MS3. This strategy offers a potentially very effective way for reducing the interference effect. We therefore designed an MS3 method where a target ion identified in a full-MS Orbitrap scan was first selected for collision induced dissociation (CID)-MS2 experiments using the linear ion trap (Fig. 3a), which was used for peptide ion identifications. The most intense MS2 fragment ion was then selected for HCD-MS3, yielding quantitative data. We selected fragment ions for MS3 experiments from a limited m/z range (110–160% relative to the precursor ion m/z) to avoid the selection of neutral loss products as well as other fragment ions, which were common to target and contaminant ion MS2 spectra (Supplementary Fig. 5).
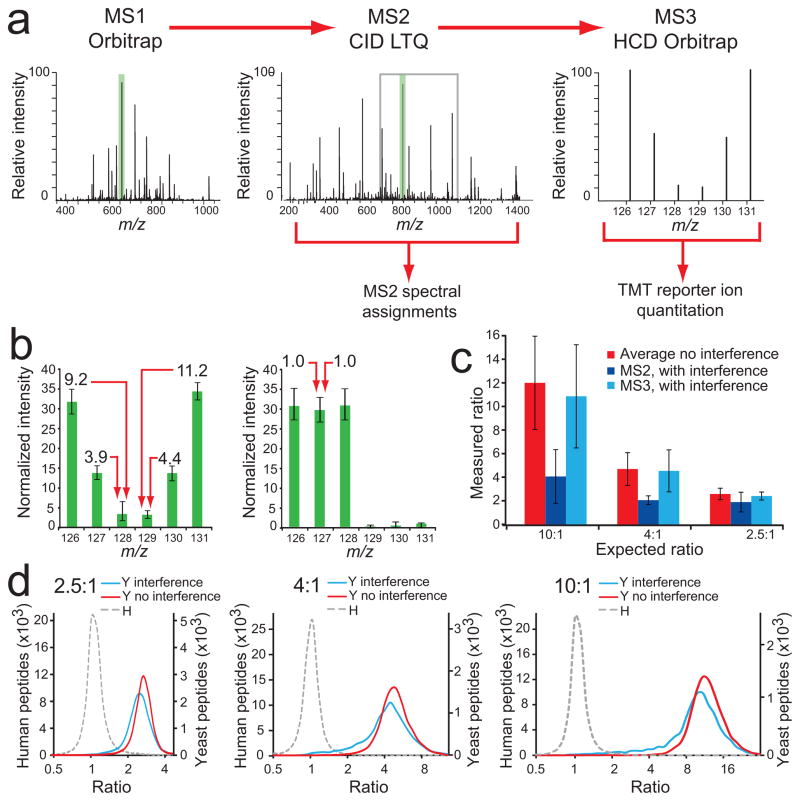
Figure 3.
An MS3-based method eliminates the interference effect. (a) Our MS3 method separates quantification from peptide identification. Fast LTQ-CID-MS2 experiments are used for peptide sequence assignments, followed by the selection of the most intense fragment ion from MS2 (green shading) for HCD-MS3, within a 110–160% m/z range relative to the precursor ion m/z value (gray box), where TMT reporter ion intensities are measured in the Orbitrap. (b) Normalized intensities for each TMT reporter ion channel for yeast peptides (left), and human peptides (right). Almost no interference effect was measured, except for forward false positives (1.5%). Error bars represent one standard deviation for 8,919 and 65,595 yeast and human peptides, respectively (c) Yeast peptide ratios from dataset combining 20 SCX fractions. Ratios were substantially compressed in channels containing interference when quantification was based on MS2 spectra. Using MS3 for quantification, measured yeast peptide ratios from channels also containing human peptides (126, 127, 128) were very close to ratios measured for channels that contained only yeast peptide ions (129, 130, 131). Error bars represent one standard deviation for 29,813, 22,640 and 7,173 peptides for Average, MS2 and MS3 analyses, respectively. (d) Ratio distribution (log2 scale) of yeast (Y) peptides in channels with human (H) peptide interference (blue line), without human peptide interference (red line), and for human peptides (gray dashed line) for 2.5:1 (left), 4:1 (middle), and 10:1 (right) combinations.
We used Lys-C to digest proteins in this study because of its C-terminal lysine residue cleavage action. TMT reagents are amine reactive and will label a typical Lys-C peptide on both the C and the N termini. Consequently, both b- and y-type fragment ions from an MS2 experiment will carry a TMT tag, and MS3 experiments on almost any fragment ion will produce TMT reporter ions. This is not always the case when tryptic peptides are analyzed and therefore, we recommend the use of Lys-C for digesting protein samples when using the described MS3 method. In principle, the MS3 experiment could be performed on any ion trap instrument, though we have not tested this.
The MS3 analysis of the multi-proteome sample separated by SCX in 20 fractions resulted in ratios that were very similar for yeast peptide ions with human peptide interference and yeast peptide ions with no interference (Fig. 3b). Median ratios for yeast peptides with and without human peptide interference, respectively, were 10.5 and 11.7 (expected ratio of 10), 4.4 and 4.7 (Fig. 3c–d). The data indicate that the MS3 method almost completely eliminated the interference effect and improved accuracy and precision (Supplementary Fig. 6). As the method required the acquisition of two spectra per peptide ion, we expected fewer peptide and protein identifications in comparison to the HCD-MS2 method. Indeed, there was a 13% reduction of identified non-redundant peptide sequences, and 8% fewer proteins were assigned (Table 1 and Supplementary Data). Considering both human and yeast identifications, there was a 12% reduction in the number of proteins quantified using the MS3 method. The results can be further separated by species and by the number of channels containing signal (Supplementary Table 1). The 12% decrease in quantified proteins reflects both an expected sensitivity loss through quantification using MS3 and a reduction in acquisition speed. However, we believe that the quality of the quantitative data provided by the MS3 method justifies the loss in analytical depth, and we expect further method and instrument developments to narrow the sensitivity gap between the MS2 and the MS3 methods.
In summary, we present a multi-proteome sample strategy to accurately evaluate the extent of interference when using isobaric labeling in quantitative mass spectrometry-based proteomics and with the MS3 method, we provide an experimental solution to remove interference, thus eliminating the ratio distortion problem. We believe that the method will clear the way for wider usage of multiplexed quantitative proteomics.
ONLINE METHODS
Cell culture, harvest, and lysis
Wild type yeast (Saccharomyces cerevisiae) strain SUB62 (MATa his3-Δ200 lys2–801 leu2–3, 112 trp1–1 ura3–52) was cultured in synthetic complete medium. At OD600 1, cells were harvested by addition of sodium azide to a final concentration of 10 mM, followed by centrifugation at 1,700 × g (Eppendorf), washed once with cold deionized water, pelleted and snap frozen at −80 °C. Pellets were thawed and resuspended in modified RIPA buffer containing 150 mM NaCl, 1% Nonidet P-40, 0.5% sodium deoxycholate, 0.1% SDS, 50 mM Tris-HCl pH 8.0, 5 mM DTT, and yeast specific protease inhibitor (Sigma-Aldrich). Cells were mechanically lysed using a minibead beater (BioSpec Products) as previously described,10 where cells were beaten with 1 mL of chilled zirconia beads (Biospec Products) in four 30 s cycles, with 30 s rest on ice between cycles.
HeLa cells were maintained in Dulbecco’s modified Eagle’s medium (Invitrogen) supplemented with 10% dialyzed fetal bovine serum (HyClone, Thermo Scientific). For cell lysis, the media was removed, the cells harvested by scraping, the cell pellet was washed with ice-cold PBS and then lysed with ice-cold lysis buffer composed of 10 mM K2HPO4 pH 7.5, 1 mM EDTA, 10 mM MgCl2, 50 mM β-glycerophosphate, 5 mM EGTA, 0.5% Nonidet P-40, 0.1% Brij 35, 0.1% sodium deoxycholate, 1 mM sodium orthovanadate, 1 mM PMSF, 5 μg/ml leupeptin and 5 μg/ml pepstatin A. The lysate was centrifuged at 20,000 × g for 10 min to remove cell debris.
Sample preparation, protein digestion, and peptide TMT-labeling
Protein content was measured using a BCA assay (Thermo Scientific); disulfide bonds were reduced with DTT and cysteine residues alkylated with iodoacetamide as previously described.10 Protein lysates were cleaned up by methanol-chloroform precipitation and digested overnight with Lys-C (Wako) in a 1/100 enzyme/protein ratio in 4 M urea and 50 mM Tris-HCl, pH 8.8. The digest was acidified with formic acid (FA) to a pH of ~ 2–3 and subjected to C18 solid-phase extraction (SPE) (Sep-Pak, Waters).
Isobaric labeling of the peptides was accomplished with 6-plex TMT reagents (Thermo Scientific). Reagents, 0.8 mg, were dissolved in 40 μl acetonitrile (ACN), and 10 μl of the solution was added to 100 μg of peptides dissolved in 100 μl of 50 mM HEPES, pH 8.5. We found that the generation of unidentified and unwanted side reaction products – singly charged ions with m/z of 303.26, 317.26, and 331.29 – was prevented by using a 200 mM HEPES pH 8.5 buffer instead of the triethylammonium bicarbonate (TEAB) buffer recommended by the manufacturer. After 1 h at room temperature, the reaction was quenched by adding 8 μl of 5% hydroxylamine. Yeast peptides were labeled with all six reagents (126–131), human peptides were labeled with reagents 126, 127, and 128. Labeled peptides from yeast and human were separately mixed in the ratios described in the main manuscript and subjected to C18 SPE on Sep-Pak cartridges. After individual LC-MS2 analysis of both samples (Supplementary Fig. 5), they were mixed to generate the final multi-proteome digest mixture. Two samples were prepared using the ratios presented in Fig. 1: one was used to study the influence of isolation specificity and precursor ion isolation width on the interference effect (see below and main manuscript), while all other experiments used the second sample. One sample was prepared using the ratios given in Supplementary Fig. 1.
Strong cation exchange (SCX) chromatography sample fractionation
Sample fractionation was performed by strong cation exchange (SCX) chromatography as previously described.10 Briefly, the sample was resuspended in SCX buffer A (7 mM KH2PO4, pH 2.6, 30% ACN) and separated over a 4.6 × 200 mm polySULFOETHYL A HPLC column (5 μm, 200 Å, PolyLC). The separation was performed by applying a two-buffer (SCX A and B) gradient from 0 to 50% SCX buffer B (7 mM KH2PO4, 350 mM KCl, pH 2.6, 30% ACN) in 47 min at a flow rate of 0.9 ml/min, followed by 50 to 100% SCX buffer A to buffer B in 4.5 min using an Agilent 1100 quaternary pump outfitted with a degasser and a photodiode array detector (PDA) (Thermo Scientific). Samples were collected in 30 s increments into a 96-well plate, and dried under vacuum. Fractions were then redissolved with 1% FA and – based on the intensity from the SCX chromatographic UV trace – combined into a total of 20 samples, which were desalted by C18 SPE, and dried under vacuum.
Liquid chromatography electrospray ionization tandem mass spectrometry (LC-MS2)
All LC-MS2 experiments were performed on an LTQ Orbitrap Velos (Thermo Fischer Scientific) equipped with a Famos autosampler (LC Packings) and an Agilent 1100 binary HPLC pump (Agilent Technologies). Peptides were separated on a 100 μm I.D. microcapillary column packed first with approximately 0.5 cm of Magic C4 resin (5 μm, 100 Å, Michrom Bioresources) followed by 20 cm of Maccel C18AQ resin (3 μm, 200 Å, Nest Group). Separation was achieved by applying a gradient from 9 to 32% ACN in 0.125% FA over 150 min at approximately 300 nl/min. Electrospray ionization was enabled through applying a voltage of 1.8 kV through a PEEK junction at the inlet of the microcapillary column.
The LTQ Orbitrap Velos was operated in data-dependent mode for both MS2 and MS3 methods. For the MS2 method, the survey scan was performed in the Orbitrap in the range of 300–1500 m/z at a resolution of 3 × 104, followed by the selection of the ten most intense ions (TOP10) for HCD-MS2 fragmentation using a precursor isolation width window of 2 m/z. The AGC settings were 3 × 106 and 2.5 × 105 ions for survey and MS2 scans, respectively. Ions were selected for MS2 when their intensity reached a threshold of 500 counts and an isotopic envelope was assigned. Maximum ion accumulation times were set to 1000 ms for survey MS scans and to 250 ms for MS2 scans. The normalized collision energy for HCD-MS2 experiments was set to 45% at a 30 ms activation time. Singly charged ion species and ions for which a charge state could not be determined were not subjected to MS2. Ions within a 10 ppm m/z window around ions selected for MS2 were excluded from further selection for fragmentation for 120 s.
The survey MS scan settings were identical for the MS3 method, where the ten most intense ions were first isolated for ion trap CID-MS2 at a precursor ion isolation width of 2 m/z, using an AGC setting of 2 × 103, a maximum ion accumulation time of 150 ms, and wide band activation. Directly following each MS2 experiment, the most intense fragment ion in an m/z range between 110–160% of the precursor m/z was selected for HCD-MS3. The fragment ion isolation width was set to 4 m/z, the MS3 AGC was 20 × 104 and the maximum ion time 250 ms. We note that we chose an isolation width of 4 m/z to avoid space charging because our preliminary experiments had very high AGC settings for the MS3 scan; however, at the AGC setting we have used here, there is no difference between a 2 or 4 m/z window (data not shown). Normalized collision energy was set to 35% and 60% at an activation time of 20 ms and 50 ms for MS2 and MS3 scans, respectively. It is important to note that charge state screening has to be disabled to allow fragment ions to be selected for MS3; this setting nevertheless allows for charge state-based exclusion of singly charged ions and ions for which no charge state was determined from MS2.
Data processing, MS2 spectra assignment, and data filtering
A suite of in-house developed software tools was used to convert mass spectrometric data from the RAW file to the mzxml format, as well as to correct erroneous assignments of peptide ion charge state and monoisotopic m/z.13 We modified the ReAdW.exe to include ion accumulation time in the output during conversion to the mzXML file format (http://sashimi.svn.sourceforge.net/viewvc/sashimi/). Assignment of MS2 spectra was performed using the Sequest algorithm15 by searching the data against a protein sequence database including all entries from the human ipi database (version 3.6) followed by sequences of proteins encoded by all known S. cerevisiae ORFs, and known contaminants such as porcine trypsin. This forward (target) database component was followed by a decoy component including all listed protein sequences in reversed order. The total number of entries in the database was 173,754. Protein sequences from the human database were listed before those from yeast so that a peptide included in both databases was always assigned to a human protein and did not intervene with measuring the interference effect. Searches were performed using a 50 ppm precursor ion tolerance, where both peptide termini were required to be consistent with Lys-C specificity, while allowing up to two missed cleavages. Six-plex TMT tags on lysine residues and peptide N termini (+ 229.162932 Da) and carbamidomethylation of cysteine residues (+57.02146 Da) were set as static modifications, oxidation of methionine residues (+ 15.99492 Da) as a variable modification. An MS2 spectral assignment false discovery rate (FDR) of less than 1% was achieved by applying the target-decoy database search strategy.12 Filtering was performed using a linear discrimination analysis method to create one combined filter parameter from the following peptide ion and MS2 spectra properties: Sequest parameters XCorr and ΔCn, peptide ion mass accuracy and charge state, predicted low pH (2.7) in-solution charge of peptide, and peptide length. Linear discrimination scores were used to assign probabilities to each MS2 spectrum for being assigned correctly and these probabilities were further used to filter the dataset with an MS2 spectra assignment FDR of less than 1% to obtain a protein identification FDR of less than 1.5%.13
Quantitative data analysis and data presentation
For quantification, a 0.06 m/z window centered on the theoretical m/z value of each reporter ion was monitored for ions, and the intensity of the signal closest to the theoretical m/z value was recorded. Reporter ion intensities were denormalized by multiplication with the ion accumulation time for each MS2 or MS3 spectrum and adjusted based on the overlap of isotopic envelopes of all reporter ions. Intensity distributions of isotopic envelopes were as provided by the manufacturer, and validated from MS2 data of yeast whole cell lysate digest individually labeled with each of the six TMT reagents.
Logarithmic (log2) reporter ion ratio distribution graphs were created by using the histogram function in Excel. Bin sizes for data collected by MS2 and MS3 methods were 0.1 log2, and 0.15 log2, respectively. For the depiction of reporter ion intensities in column graphs, absolute values were converted to relative intensities. In a first step this was done separately for channels 126, 127, and 128, and channels 129, 130, and 131. The calculated values were then corrected considering the difference of the summed intensities found in channels 126/127/128 and 129/130/131. Data were excluded from calculating the plotted average values and standard deviation, when measured ratios were lower than 25% and 10% for expected ratios of 10:1 and 4:1, respectively. Values shown in the radar plots presented in Fig. 3 were calculated by dividing expected ratios by the median of ratios measured in all assigned MS2 spectra. Values were plotted in Excel using the radar plot function. Perfect ratios are represented as an equilateral triangle, where a value of 1 (expected/measured) is expected in all three ratio combinations. For the measured versus expected ratio column graph in Fig. 3c, only yeast peptides were considered, and measurements are given as median values with standard deviations. For this figure, MS3 spectra with denormalized reporter ion intensities (see above) ≤ 5 × 104 in any channel were not considered in the data analysis.
Determination of isolation specificity
In an attempt to estimate the intensity of contaminating near isobaric ions for each target peptide ion selected for MS2 we calculated an isolation specificity value. This was done by first recording all ion intensities in survey MS data within the isolation width m/z range (± 1 m/z) around target ion m/z values. Using a 10 ppm tolerance, the summed intensity of ions assigned to the target ion isotopic envelope (including 13C ions) was then divided by the total ion intensity. The calculated isolation specificity was graded on a scale from 0 to 1, where 0 represented 100% interference (none of the signal belonged to the precursor); and 1 represented 0% interference (the entire signal belonged to the precursor).
Supplementary Material
Supplementary Figure 1. Validation of the human-yeast interference model.
Supplementary Figure 2. Yeast peptide interference examples and the inverse human-yeast interference model.
Supplementary Figure 3. The effect of isolation specificity thresholds on post-experimental data filtering.
Supplementary Figure 4. The effect of decreasing precursor isolation specificity on peptide and protein identifications using an HCD-MS2 method.
Supplementary Figure 5. Fragment ion selection for MS3.
Supplementary Figure 6. The precision of quantitative data from the MS3 method is superior to that from the MS2 method over a broad range of protein abundances.
Supplementary Table 1. A comparison of the number of peptide and protein identifications and quantifications from 20 SCX fractions of the human yeast multi-proteome model using the MS2 and MS3 methods.
Supplementary Data. All yeast and human identification and quantification data.
Acknowledgments
This work was supported in part by US National Institutes of Health grants (HG3456 and GM67945) to S.P.G.
Footnotes
Author contributions
L.T., S.P.G., and W.H. designed the experiments, analyzed the data, and wrote the paper. L.T., and W.H. performed the experiments. R.R. developed software for data analysis.
References
- 1.Aebersold R, Mann M. Nature. 2003;422:198–207. doi: 10.1038/nature01511. [DOI] [PubMed] [Google Scholar]
- 2.Ross PL, et al. Mol Cell Prot. 2004;3:1154–1169. doi: 10.1074/mcp.M400129-MCP200. [DOI] [PubMed] [Google Scholar]
- 3.Dayon L, et al. Anal Chem. 2008;80:2921–2931. doi: 10.1021/ac702422x. [DOI] [PubMed] [Google Scholar]
- 4.Thompson A, et al. Anal Chem. 2003;75:1895–1904. doi: 10.1021/ac0262560. [DOI] [PubMed] [Google Scholar]
- 5.Choe L, et al. Proteomics. 2007;7:3651–3660. doi: 10.1002/pmic.200700316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bantscheff M, et al. Mol Cell Prot. 2008;7:1702–1713. doi: 10.1074/mcp.M800029-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Karp NA, et al. Mol Cell Prot. 2010;9:1885–1897. doi: 10.1074/mcp.M900628-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ow SY, et al. J Prot Res. 2009;8:5347–5355. doi: 10.1021/pr900634c. [DOI] [PubMed] [Google Scholar]
- 9.Shirran SL, Botting CH. J Proteomics. 2010;73:1391–1403. doi: 10.1016/j.jprot.2010.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Villén J, Gygi SP. Nat Protoc. 2008;3:1630–1638. doi: 10.1038/nprot.2008.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Olsen JV, et al. Nat Methods. 2007;4:709–712. doi: 10.1038/nmeth1060. [DOI] [PubMed] [Google Scholar]
- 12.Elias JE, Gygi SP. Nat Methods. 2007;4:207–214. doi: 10.1038/nmeth1019. [DOI] [PubMed] [Google Scholar]
- 13.Huttlin EL, et al. Cell. 2010;143:1174–1189. doi: 10.1016/j.cell.2010.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Beausoleil SA, et al. Proc Natl Acad Sci USA. 2004;101:12130–12135. doi: 10.1073/pnas.0404720101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eng JK, McCormack AL, Yates JR., III J Am Soc Mass Spectrom. 1994;5:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. Validation of the human-yeast interference model.
Supplementary Figure 2. Yeast peptide interference examples and the inverse human-yeast interference model.
Supplementary Figure 3. The effect of isolation specificity thresholds on post-experimental data filtering.
Supplementary Figure 4. The effect of decreasing precursor isolation specificity on peptide and protein identifications using an HCD-MS2 method.
Supplementary Figure 5. Fragment ion selection for MS3.
Supplementary Figure 6. The precision of quantitative data from the MS3 method is superior to that from the MS2 method over a broad range of protein abundances.
Supplementary Table 1. A comparison of the number of peptide and protein identifications and quantifications from 20 SCX fractions of the human yeast multi-proteome model using the MS2 and MS3 methods.
Supplementary Data. All yeast and human identification and quantification data.