Abstract
This paper describes software for neuroanatomical knowledge synthesis based on neural connectivity data. This software supports a mature methodology developed since the early 1990s. Over this time, the Swanson laboratory at USC has generated an account of the neural connectivity of the sub-structures of the hypothalamus, amygdala, septum, hippocampus, and bed nucleus of the stria terminalis. This is based on neuroanatomical data maps drawn into a standard brain atlas by experts. In earlier work, we presented an application for visualizing and comparing anatomical macro connections using the Swanson third edition atlas as a framework for accurate registration. Here we describe major improvements to the NeuARt application based on the incorporation of a knowledge representation of experimental design. We also present improvements in the interface and features of the data mapping components within a unified web-application. As a step toward developing an accurate sub-regional account of neural connectivity, we provide navigational access between the data maps and a semantic representation of area-to-area connections that they support. We do so based on an approach called “Knowledge Engineering from Experimental Design” (KEfED) model that is based on experimental variables. We have extended the underlying KEfED representation of tract-tracing experiments by incorporating the definition of a neuronanatomical data map as a measurement variable in the study design. This paper describes the software design of a web-application that allows anatomical data sets to be described within a standard experimental context and thus indexed by non-spatial experimental design features.
Keywords: knowledge engineering, neural connectivity, tract-tracing, neuroanatomical mapping
1. Introduction
Neural connectivity may be described at three different scales: macro connections (representations of large-scale projections between gray matter regions); mesoconnections (descriptions of connections between defined neuron types); and micro-connections (accounts of connections between individual neurons; Swanson and Bota, 2010). The process of evolving the underlying knowledge representation of neural connectivity from the “macro” to the “meso” level is the high-level goal that motivates this current paper. Currently, neural connectivity in humans must be inferred from structural, diffusion, and functional magnetic resonance imaging data (Hagmann et al., 2007; Feldt et al., 2011). Tract-tracing techniques (Zaborszky et al., 2006; Lanciego and Wouterlood, 2011) permit direct observation of the structure of neuronal connections in the histologically prepared tissue of experimental animals. In general, each type of experiment generates data that needs to be interpreted in different ways. In particular, macro connectivity data tends to be described either (a) as spatial maps (often in the context of standardized atlases) or (b) as a symbolic representation of connections between named areas.
A framework for the logical representation of neural connectivity data has been succinctly proposed by Swanson and Bota (2010), in which they provide a terminology for standard neuroanatomical entities (such as “gray matter region,” “fiber-tract,” “amacrine cell,” etc.). They also include terms pertaining to a modeling framework for neural connectivity (such as “wiring diagram,” “node,” and “connection”). The definitions of a “wiring diagram” and its constituents (nodes and connections) are defined to be general-purpose so that they may be re-used for the representation of macro-, meso-, or micro-connections.
The central contribution of this paper is based on tract-tracing studies that describe the topographic organization of macro connections by manually mapping the distribution of axonal fibers generated in tract-tracing experiments directly onto the plates of a neuroanatomical atlas, see (Dong et al., 2001b; Dong and Swanson, 2003, 2004a, 2006a) for examples that will be referred to later in this paper. This process of hand-drawing maps into a standard brain atlas provides an accurate registration of anatomical data into a standard atlas-based framework when performed carefully and correctly by experts (Swanson, 2004; Burns et al., 2006).This representation is only the first step of the larger goal of building systems to understand the substructure of a connection’s fields of origin and termination in a knowledge engineering interface.
It is also important to note that a logical representation in a wiring diagram is an interpretation and that a geometrical representation in a data map is an observation1. In this paper, we exploit this distinction by using a knowledge engineering approach that incorporates the data maps into a knowledge representation for tract-tracing experimental observations. This permits us to use the reasoning framework of the knowledge engineering system to generate an interpretive “connection matrix” (that is topologically equivalent to the concept of a “wiring diagram” defined in Swanson and Bota, 2010) which has been implemented in the “BioScholar” system described in (Russ et al., 2011)2.
We have implemented the interaction between spatial and semantic components directly as an executable neuroinformatics software application where an end-user may navigate between geometric data maps and connection matrix entries that pertain to them. The system is fully functional and open source (but the atlas delineations are copyright-protected and are therefore access-restricted). This software is described here as a practical methodology for understanding the spatial structure of macro connections within a framework for reasoning that allows users to describe experimental design. This is only an initial step toward constructing neuroinformatics infrastructure capable of representing the types of experiments to examine and model meso connections (see Discussion).
1.1. Biomedical knowledge engineering tools: NeuARt, BioScholar, and BAMS
We built this application by combining three preexisting independent systems which had their own particular technical focus: NeuARt and BioScholar were developed within the Biomedical Knowledge Engineering Group at ISI and the Brain Architecture Management System (BAMS) was developed by Mihail Bota and Larry Swanson at the University of Southern California (USC).
NeuArt is a system for viewing hand-drawn, atlas-based maps derived from histological labeling in brain tissue (Dashti et al., 2001). It was implemented as a downloadable Java application (Burns et al., 2006)3 and has now been re-engineered as a Flex-based web-application. It provides a flexible windowing framework that permits the inclusion and manipulation of different types of User Interface (UI) component frames. This is similar in concept to the Eclipse integrated development environment4.
The field of online neuroanatomical mapping systems includes several notable development efforts from large, well-supported teams such as Elsevier’s Brain Navigator5, the International Neuroinformatics Coordinating Facility’s (INCF’s) “Scalable Brain Atlas” system6, the Allen Institute’s Mouse, and Human Brain Atlases7, The University of California, Los Angeles (UCLA) Laboratory of NeuroImaging’s (LONI) Mouse Brain Atlasing Toolkit8 (MBAT) and other systems from academic developers. The combined NeuARt/BioScholar application is original for two reasons. Firstly, it is based on a practical methodology that has been developed “in-house” for working neuroanatomists. Secondly, the tools that we are developing have a knowledge engineering methodology that goes beyond simply dealing with the geometrical relationships of brain structures and is concerned with the argumentation structure of scientific observations and interpretations.
“BioScholar” is a knowledge engineering and management system for biomedical experimental observations derived either directly from data or curated from the literature (Russ et al., 2011). The key distinction of this system is that the underlying knowledge representation (KR) is based on a model called “Knowledge Engineering from Experimental Design” (KEfED). In this model, we focus directly on the independent and dependent variables of a given study, so that we may make a clear distinction between statements being made that are interpretive and those that are observational. We have built and deployed web-based software for generating KEfED models and then instantiated them with data. BioScholar is hosted as a BIRN capability project (see text footnote 2); it is open source and is available as a multi-platform download for collaborators, developers, and end users from http://code.google.com/p/bioscholar.
BAMS is a mature online data repository of neuroanatomical information including brain parts, molecules, cell types, and projections9. It is built on a MySQL database with a PHP web-interface and contains manually curated information corresponding to all the data maps currently described in NeuARt. The system’s originator (Dr. Mihail Bota) provided direct access to data so that we could then import it into our KEfED model representation. Thus, this work is concerned with the construction of a unified spatio-semantic neuroanatomical mapping system.
1.2. Overview of the KEfED formalism
The KEfED model is specifically designed to be a domain-independent representation of experimental “observational assertions” where the main challenge is providing a mechanism for representing the semantics of different variables (constants, parameters, measurements, and calculations). The relationship between “tract-tracing” experiments and neural macro connectivity provides an example of this distinction (Burns, 2001; Russ et al., 2011).
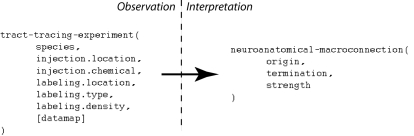
An experimental design may contain many variables that are not directly needed to interpret the experimental observations given a specific interpretive model (Burns et al., 2008). The “primary observations” of a tract-tracing experiment may be summarized as follows: the experimental design involves performing an injection of a tracer chemical into a specific injection-region. The experimental animal is then euthanized and its brain processed histologically to reveal the presence of labeling which itself would include an account of the labeling’s location, type, and density. Data captured in this form can then be interpreted to provide a semantic representation of a macro connection, defined here as consisting of the region where the projection has its origin, its termination, and its strength (see Figure 1). If the tracer chemical is an “anterograde” tracer then active axonal transport is assumed to have carried the tracer from the injection site to the regions containing transported labeling. If the tracer chemical is “retrograde” then the tracer will be carried from the terminals back to the neurons’ somata. “Strength” refers to a qualitative measure of the relative magnitude of the projections (inputs and/or outputs) labeled by a tracer injection site. This is different from the notion of “connection strength” which implies a measure that addresses the number of synapses involved in a specific macro-connection and so we adopt a more conservative definition.
Figure 1.
A simplified view illustrating how a small number of variables contributing to the primary observations of a tract-tracing experiment map to the variables supporting an interpretive assertion describing a macro connection. Note the presence of an additional variable (“data map”) assigned to the histological maps that may be viewed within the NeuARt system.
2. Materials and Methods
We built this application by combining two preexisting systems: BioScholar and NeuARt. The Bioscholar system is a web-based domain-independent biocuration system based on KEfED models. The NeuARt system is a web-based neuroinformatics system for accessing and comparing neuroanatomical data registered onto brain atlases. The integration process, described below, was accomplished through our group’s design and reuse of our software development framework.
We produced a dataset that included the whole set of tract-tracing experiments currently included in the NeuARt application (which effectively summarizes the entire connectivity output of the Swanson laboratory over a period of 15 years). This dataset describes 78 experiments that reported 3000 axonal projections from 58 gray matter source regions into 383 target regions. The dataset also included 1059 images depicting detailed labeling data produced by these experiments and registered against a brain atlas. This dataset was composed from preexisting databases and images collections.
Both the BioScholar and NeuARt systems are built on a Spring10 server with a Flex 4.0 client11, using the BlazeDS remoting architecture, SOAP web services and bare HTTP (for image files) for client-server communication. This remoting approach allows efficient server-client communication since the Java class schema operating on the server exactly mirrors the Action script class schema operating on the client. Data is stored as SVG files on disk or within a Persevere NoSQL database. The system uses the svgweb12 library to view atlas images and maps.
2.1. Bioscholar/NeuARt integration
The final combined application was composed of the Bioscholar and NeuARt client components since each application was web-based with their GUI behavior completely contained in the client (and implemented in Flex). Flex is a versatile, cross-platform “Rich Internet Application” (RIA) framework with a sophisticated event-handler model for user interactions. Contrary to most conventional web-based frameworks where there is a tight integration between client and server, here the presentation of data and user interaction can be contained and managed by the client programs, so that the overall integration was achieved entirely within the client.
The base client component for the combined application was based on a spatial view from the NeuARt client with added semantic views from Bioscholar. These semantic elements were a representation of a summarized connection matrix and associated experimental evidence formatted as KEfED data. We added navigational tools between the underlying knowledge and relevant maps and removed the ability for users to edit the KEfED tables. We packaged the Bioscholar client application as a library to be used in the context of the NeuARt application.
2.2. Data maps
The data maps themselves were derived from a collection of Adobe Illustrator (AI) files provided directly by members of the Swanson laboratory. These files are stacks of drawings that show the observed injection sites and labeling locations hand-drawn onto atlas plates. The methodology to generate these maps was initially developed in Dr. Swanson’s laboratory and is described in (Swanson, 2004). This methodology has lead to a large number of neuroanatomy publications in leading journals (Watts and Swanson, 1987; Watts et al., 1987; Canteras and Swanson, 1992a,b; Canteras et al., 1992a,b, 1994, 1995, 2010; Risold et al., 1994, 1997; Risold and Swanson, 1995, 1997a,b; Petrovich et al., 1996, 2001; Thompson et al., 1996; Petrovich and Swanson, 1997; Thompson and Swanson, 1998, 2003; Dong et al., 2001b; Dong and Swanson, 2003, 2004a,b, 2006b,c; Goto and Swanson, 2004; Goto et al., 2005; Swanson, 2005; Cenquizca and Swanson, 2007; Hahn and Swanson, 2010) system. The maps are semi-qualitative drawings of the appearance of experiments within the atlas structure. Estimates of the positional accuracy of these expert-generated maps varies by structure and number of local landmarks (or fiducial markers) but is generally on the order of 100 μm or less (R. Thompson, unpublished observation).
This data set consisted of 73 vector images (one for each coronal level in the atlas), with a large number of individual layers that each correspond to a named experiment (e.g., “ADP_PHAL” denotes a case where the tracer Phaseolus vulgaris Leucoagglutinin or “PHAL” was injected into the anterodorsal preoptic nucleus). The labels for these experiment layers were consistent across all images and could be used to aggregate all data corresponding to a single experiment. Given the large number of maps available, we automatically extracted all the atlas and data map images from these documents by converting each level into Scalable Vector Graphics (SVG) documents using scripts written in the AI application. We then extracted each graphic layers corresponding to an atlas level or an experimental map into separated SVG files, which could be then loaded into the NeuARt database.
2.3. Generating the tract-tracing experiments dataset
We edited the KEfED model design for tract-tracing experiments developed previously in (Russ et al., 2011) by adding a new measurement variable to the final processing step, denoting the act of drawing these maps. We then extracted all available data from the BAMS database pertaining to the publications for which data maps were available. Some data cleaning and transformation was required to port this data into a modified KEfED model, matching terminology between BAMS and our models. This process generated the rules for relations that were used in our reasoning framework and in our base data store. The connection matrix itself (all available interpretive connection reports) was created by running a brute-force search that iterated over the brain regions of interest to find all evidence in the data for a connection. This process was time-consuming, taking about 24 h to run.
3. Results
3.1. Organization of the interface and basic NeuARt functionality
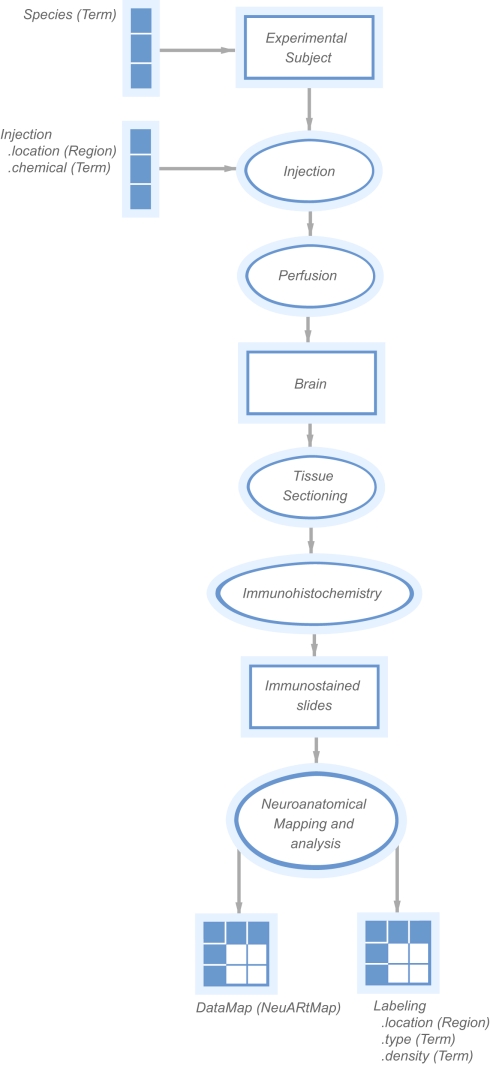
This latest version of the NeuARt application is based on a combined semantic representation of experimental design and a spatial representation of neuroanatomical maps. Under the KEfED formalism, the mechanism for this interaction is relatively straightforward: extend the existing KEfED model for tract-tracing experiments by adding a “Data Map” measurement as part of the experimental design (see Figure 2). The underlying logic of the experiments’ relationships to their interpretations remain unchanged, but the NeuARt system now provides a specialized viewing tool for connectivity data in a neuroanatomical context. This also serves as a framework for presenting connectivity data.
Figure 2.
A “KEfED” model for tract-tracing studies as it appears within the KEfED editor system. This forms the basis of the interaction between the spatial and semantic representations of these experiments.
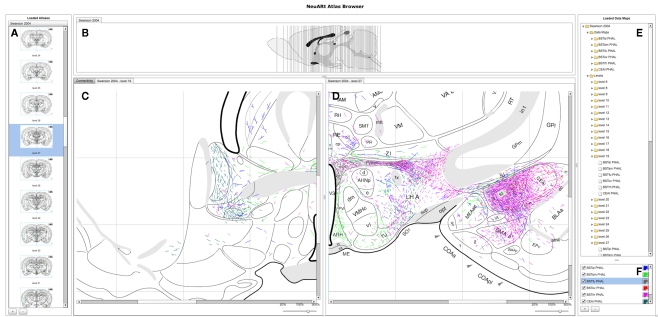
The basic user interface of the NeuARt system has four panes arranged in a west-north-central-east configuration (see Figure 3): (A) the west pane shows coronal sections of open atlases (the system can contain several atlases); (B) the north pane shows a sagittal view of the selected atlas showing each coronal section’s rostro-caudal position; (C/D) the central pane serves as the main viewing area for the various components generated within the application; the east pane has two sub sections (E) lists all maps and coronal levels, providing interactive links to the data and (F) controls for displaying particular data maps and showing their color mapping. This basic configuration mirrors that of the original NeuARt II application (Burns et al., 2006).
Figure 3.
A screenshot of the “standard” configuration of the NeuARt system specifically tailored to view neuroanatomical maps drawn in register with a brain atlas. This view shows detail of level 27 of (Swanson, 1998) with data maps pertaining to axonal labeling originating from the following subnuclei of the BST: oval (BSTov), fusiform (BSTfu; Dong et al., 2001b), rhomboid (BSTrh; Dong and Swanson, 2003), anterolateral (BSTal; Dong and Swanson, 2004a), and anteromedial (BSTam; Dong and Swanson, 2006a). We also include maps from PHAL injections into the lateral part of the Central nucleus of the Amygdala (CEAl) on the opposite side (Petrovich and Swanson, 1997). (A) Coronal sections for each open atlas in an “accordion” control. (B) A sagittal view of the open atlas (vertical lines are navigable). (C) Level 19 of (Swanson, 1998) in the main, vertically split central panel. (D) Level 27 of (Swanson, 1998) in the main, vertically split central panel. (E) List of the loaded data maps. (F) Control for selecting the maps to show and their color mapping.
The system permits any of the maps in the system to be loaded directly by clicking the button marked with a [ + ] symbol to the bottom right. Each map denotes a single experimental case mapped over several atlas plates. Two basic components display these maps (a) views of an individual level with all data maps superimposed and (b) a view of all available levels within a single experimental case. These components appear as tabbed panes in the central panel which may itself be divided either vertically or horizontally to allow simultaneous viewing of multiple atlas plates. The viewer may also toggle the visibility of the maps to provide an uncluttered view. This configuration provides a functional architecture that allows other components to be added as needed to the central panel. The general-purpose nature of this central panel allowed us to implement the additional semantic functions from BioScholar.
3.2. Spatio-semantic interactions
We present our connectivity data summary as a “matrix” representing a directed graph of projections between the brain regions under study. As described in (Russ et al., 2011), each cell in this matrix represents an interpretive assertion about a macro connection. Within our demonstration system, we focus on representing the same connectivity data reported within the Swanson Laboratory’s complete set of maps. The number of individual regions of origin and termination are quite large in this set and would generate a very large, sparse connection matrix that would be unreadable. We instead show the connections as a filtered, sortable list that the user may manipulate to show a specific subset of connections (see Figure 4A and Figure 5A; Dong et al., 2001b). This table contains three columns: Source, Target, and Strength.
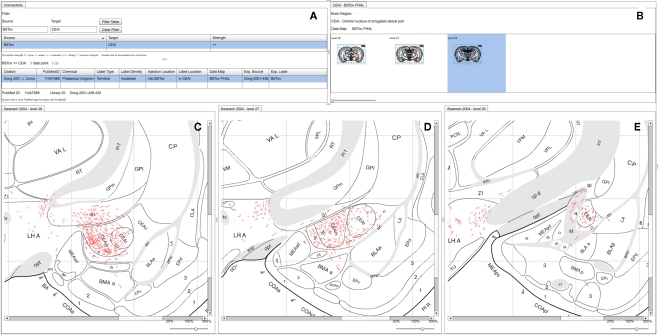
Figure 4.
A screenshot of the combined BioScholar/NeuARt application’s central panel showing all maps pertaining to a single connection from a single piece of evidence (in this case from BSTov to the CEAl) within the central panel of the application. (A) The “connectivity matrix” panel with both interpretive summary and evidence table. (B) Data Map for the BSTov PHAL case limited to levels where CEAl can be found. (C) Level 26 of (Swanson, 1998) with BSTov PHAL data overlaid. (D) Level 27 of (Swanson, 1998) with BSTov PHAL data overlaid. (E) Level 29 of (Swanson, 1998) with BSTov PHAL data overlaid. Note, due to its time-consuming nature, data mapping generally cannot be performed on every available level.
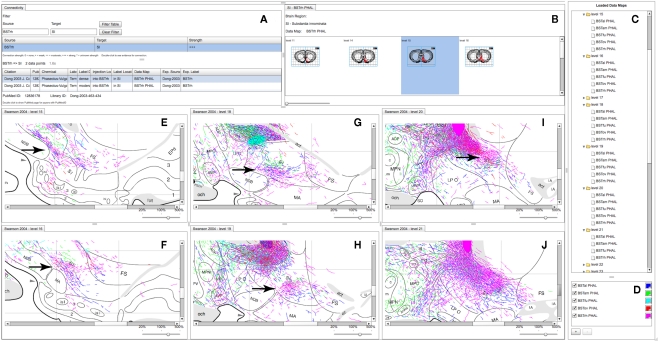
Figure 5.
A partial screenshot of the combined BioScholar/NeuARt application showing multiple maps possibly pertaining to the topography of the projection from multiple subnuclei of the BST to the SI. (A) The connection matrix panel showing interpretive and observational data pertaining to connections from BSTrh to SI. (B) One row of levels from the Data Map for the BSTrh PHAL case limited to levels where SI can be found. (C) List of Data Maps and Levels in the system (currently with all levels in the main panel expanded to show the maps displayed there. (D) Main controller for adding new Data Maps within NeuARt. (E–J) A selection of levels where fibers originating from BSTrh are shown in SI. These panels also include fibers from the other subnuclei shown in (D).
Given a list of specific connections of interest, the user double-clicks on a specific interpretive assertion about a specific connection. The system then retrieves all the supporting evidence concerning that connection by executing a number of queries to an underlying knowledge base, implemented within the “PowerLoom®” architecture13. We are able to provide support for geometric reasoning to retrieve connection reports where either the injection site or the labeled region have relationships to the regions implicated in the connection of interest. An example of these relationships could be that the region where labeling was found is a subregion of the termination region of the connection, or that the injection site only impinged upon the connection’s region of origin.
The supporting evidence is presented to the end-user as a table. Citation and PubMed ID refer to the article reporting the experiment. Chemical denotes the tracer used (e.g., Phaseolus vulgaris Leucoagglutinin). Label Type describes the type of cellular entity being labeled, typically cells for retrograde studies and axonal fibers for anterograde studies. Label Density is a seven-point ordinal scale to mimic the way that scientists typically describe labeling in publications (e.g., “sparse,” “moderate,” “dense,” etc.). Injection location describes the injection site as a region defined by relations to the atlas-based regions. Label Location describes the brain regions where the labeling was observed. Data map connects to the NeuARt collection of images that document the experiment’s results. Finally, Exp. Source and Exp. Label provide metadata to denote the paper and the experiment that our representation is based on. In our interface, the spatio-semantic interaction is instantiated by providing navigational links from the connection reports to their corresponding maps.
3.3. Example usage scenarios
We present three scenarios to illustrate possible usage of the system as a tool for studying neural connectivity. The underlying cyto- and chemo-architecture of the bed nucleus of the stria terminalis (BST, “a rather obscure non-cortical component of the cerebral hemisphere" Dong and Swanson, 2003) was elucidated in 1989 leading to the delineation of a large number of individual nuclei within this complex cell group (Ju and Swanson, 1989; Ju et al., 1989). From 1997 to 2006, Dr. Hong Wei Dong performed an exhaustive series of tract-tracing studies describing the neural efferent connections of the constituent cell-groups of the BST generating 15 complete histological data maps, published in eight large-scale, comprehensive anatomical research articles (Dong et al., 2000, 2001a,b; Dong and Swanson, 2003, 2004a, 2006a,b,c). We use a subset of these maps to illustrate some features of the combined NeuARt/BioScholar interface.
3.3.1. Tracing circuits using maps
Figure 3 shows a high-level view of the basic configuration of the web-application with level 19 and 27 shown with data from a number of experiments. These maps are illustrative of specific features published as figures within individual studies and could not be otherwise synthesized into a single view. Figure 3 shows the utility of the basic functionality of the NeuARt system by illustrating the connectional topography between five BST subnuclei (BSTov, BSTfu, BSTrh, BSTal, and BSTam) and the lateral part of the central nucleus of the amygdala across two atlas plates. As an illustrative example, this showcases both the process of synthesizing maps to reveal connectivity systems and also the complexity of the spatial data. Within the figure, the output connectivity of BSTrh to the CEA obscures the other labeling. The end-user may currently toggle the visibility of maps to examine patterns by interacting with the live system.
In this illustrative case, although not explicitly shown in the figure, we had used our system’s query capability to discover the experiments supporting the connectivity between BST and CEA. In the following two examples we show how our toolset exploits the semantics of the KEfED data to support this type of queries.
3.3.2. Viewing all the data for an individual experiment pertaining to a specific connection
An organizing feature of the organization of the BSTov is its connections with the central nucleus of the amygdala (see Figure 10 of Dong et al., 2001a). Although a block-and-line wiring diagram could provide a summary of this projection, it cannot express any details about the spatial organization of the projection. The view shown in Figure 4 provides a view of data pertaining to this single macro connection across all the levels for which data is available. Panel (A) of Figure 4 shows the “connectivity matrix” summary table with its filter set to search for the required connection. The summary table shows a single report denoting the interpretative assertion concerning the presence of a connection from BSTov to CEAl. The “Evidence Table” below lists one single experimental report from (Dong et al., 2001a) describing a PHAL injection into BSTov with anterograde labeling found in CEAl. The system, provides navigational tools from this summary to either the injection site or the labeling involved in the connection. It is possible to show a panel which lists each level where label is found in this case (with expandable thumbnails). Clicking on each thumbnail will generate panels with more detailed data (see Figures 4C–E).Note that the system does not yet lay these panels out automatically, and users must manually compose the view themselves.
This example shows that the connection to CEAm is denser than CEAl and illustrate the route that fibers take to arrive in the nucleus. It is possible to ascertain the rostro-caudal organization of the projection and provide the context of surrounding nuclei to the overall connection.
3.3.3. Examining the substructure of labeling within a larger structure
Neuroanatomical atlases allow expert users to examine the substructure of named regions. This is specifically useful when data is specifically mapped to atlas plates so that patterns between cases become evident. Figure 5 shows maps across the substantia innominata (SI) arising from injections in multiple sub-regions of the BST where it is possible to identify a small area of consistent labeling on level 19 (see the arrow in Figure 5H). The description of the results of this experiment describes this labeling as part of a “rostral pathway” from BSTrh and is described as entering SI in the “rostral half and continuing to branch and generate boutons” (excerpted from page 441 of Dong and Swanson, 2003). The spatial view provides additional detail to this description. The maps in the NeuARt system can provide a view of fibers originating from other BST subnuclei within the same subregion of SI (BSTal, BSTam, and BSTov). For this to be a practical working solution, neuroanatomists would undoubtedly have to examine the experimental tissue in depth across multiple cases to be sure that any significant topography or pattern could be found. The purpose of NeuARt would be to identify possible candidates for further examination.
4. Discussion and Conclusion
The goal of the work underlying this paper is to describe a practical software tool to permit an end-user to access and use maps drawn onto atlas plates and then to use these maps in a semantic framework pertaining to specific experimental designs and interpretive models. We focus on tract-tracing experiments since the model is well-developed and there are available data for both maps and tract-tracing data. Our methodology is generalizable to other experimental types if they use the same hand-drawn maps and can be formulated in terms of the KEfED model. We are working to create a suite of components to serve as the basis for a general-purpose development platform for knowledge engineering tools. Flex code may also be used to generate elements that can be used by other sites and we have deployed the KEfED editor component as an element within a Ruby-on-Rails data management application (Jacobs et al., 2009).
The neuroinformatics atlasing community has produced several systems in recent years. Notable examples include: MBAT (Lee et al., 2010), the Allen Brain Atlas (ABA)14 (Ng et al., 2009), and the Scalable Brain Atlas15. Since NeuARt and KEfED were originally conceived as in-house tools for neuroanatomists directly targeting tract-tracing data, there are some differences to these other efforts. MBAT is an application toolkit that permits an investigator to query external resources (both data- and literature-centric), to register images against atlases and to compare and analyze images. It is an extensible framework where either in-house or third party tools can be included as plugins and it is a java application that must be installed locally in the user’s system. Since NeuARt uses a client/server architecture to store data in a central repository to be served up to a rich internet application, it is conceivable that both the data and tooling of NeuARt could be made available to the MBAT toolkit as a plugin in the future.
There are many more similarities between the Allen Brain Atlas (ABA) and the NeuArt systems. Both ABA and NeuARt use atlases to index data and in both cases, the data was produced and registered to the atlas under a well-defined, “in-house” mapping process. The important distinction is that ABA is automated and therefore scales to process orders-of-magnitude more data. For this same reason, it lacks the detailed involvement of a human expert to place each feature from the data carefully into the atlas space. Our manual approach may be considered a more accurate method of data registration but carries a significant cost in processing speed. In the current paper, we also show the use of a representation of the logic of the underlying experiment. In neuroanatomical databases in general, this information would typically be included in the metadata associated with each mapped case. We have here taken the additional step of incorporating this knowledge into a reasoning framework that provides summarized evidence for each interpretative assertion of the system (i.e., the output of our semantic system is not an account of localized labeling density but an assertion about macro connections between brain structures).
The INCF’s Scalable Brain Atlas is used to browse volume delineations of reference atlases and is notable due to it’s incorporation of the Waxholm reference atlas which serves as an elegant computational vehicle for standardization based on three-dimensional coordinates taken from a standard brain. The INCF is also developing a service-oriented infrastructure to support the publication, discovery, and invocation of atlas-based resources (Hawrylycz et al., 2011). Translation of our data into this framework remains an important step to making the Swanson atlas maps useful within this broader community. An important barrier to the development of all of these tools is the closed-source nature of the most widely used atlases. There is a need for licenses that allow open development and dissemination of tools to be made available to neuroinformatics researchers.
Neuronal data is usually inherently indexed by the neuroanatomical location of phenomena or entities under study. Invariably, these location details are not easily machine-accessible, being expressed either as non-standardized terms embedded in the natural-language-based prose of a research article or as camera-lucida drawings, maps, and photographs within a paper’s figures. This information, embedded in neuroscience papers, represents a massive outlay of funding, time, and expertise that is currently inaccessible to searching, comparing, and analyzing with current tools. This is an untapped resource that we hope to harness by using the system described in this paper as a possible biocuration vehicle. All of the data in this study was obtained from previously existing sources. It could be possible to use the interface itself to encourage end users to draw maps, to link photographs or other binary data types to specific brain locations.
Although our system deals only with knowledge at the level of macro connections (connections between brain regions), it provides a workable initial neuroinformatics infrastructure capable of being extended to describe meso connections (connections between cell types). In general, the logical basis for inferring knowledge about the connectivity of specific cell types is to combine tract-tracing experiments with another experiment type. This could involve, for example, concurrent immunohistochemical labeling of the tract tracers and one or more relevant neurotransmitters (or other neurochemicals) in the same tissue (see, e.g., Brinschwitz et al., 2010; Schofield, 2010) for examples of how this logic is applied to establish the projections of neuronal cell types). Another example can be found in functional neuroanatomical studies that seek to identify the behavioral or physiological relevance of given projection (see, e.g., Petrovich et al., 2005; Perez-Manso et al., 2006). These studies often use in situ hybridization histochemistry for the immediate-early gene c-fos as a marker of neuronal activation and record the number of retrogradely labeled neurons that express c-fos in response to the experimental manipulation. Finally, the NeuARt system provides a neuroinformatics platform that supports a third type of study for investigating networks of neural connections. This double-coinjection (DCI) tracing strategy is a recent development that utilizes combinations of tract tracers to examine meso connections systematically (Thompson and Swanson, 2010). In this approach two conventional tract tracers, one anterograde and one retrograde, are combined in solution and injected from the same pipette. When two coinjections are used in concert (four different tracers in total), all possible interactions between the two sites are labeled; that is, any given brain region sends and/or receives from one, both, or neither of the brain sites injected. The DCI approach exploits the ability to label multiple interactions directly in each experiment as a means of establishing registration across experiments. Together these features provide a framework that allows a comprehensive description of brain structure to be generated in an iterative manner from the synthesis of multiple incomplete descriptions.
Double- (triple- and quadruple-) labeling studies such as these are technically challenging and increasingly common in an effort to better understand the structural organization of brain circuits at the mesoscopic scale (Smith, 2007; Lanciego and Wouterlood, 2011). NeuARt’s existing data maps provide a valuable collection to help scientists plan these types of experiments (by providing widespread access to the spatial organization of macro connections in systems of interest). Additionally, neuroinformatics databases for knowledge from such studies would be challenging to encode with standard methods, but the flexibility of the KEfED approach could provide a vehicle to capture these data (and any maps generated from them). This, coupled with clear criteria for how specific cell types are defined based on experimental criteria (which variables need to be measured with which values), could support the definition of a system for representing meso connections.
We have presented a practical software system to show spatial and semantic views of tract-tracing experiments in a preliminary way as a demonstration of feasibility. This platform now permits us to (a) tailor the more detailed functionality of the tool to improve the end-user anatomists experience of using the system, (b) add new analysis functionality to discover patterns within the maps and interpret these patterns within the logical representation of neural connectivity and (c) broaden the scope of this methodology to new experimental types other than tract-tracing studies. Our software is open source and accessible via http://www.neuart.org/.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This research is funded by the U.S. National Institutes of Health under the “NeuArt” project (MH079068-01A2), “BioScholar” project grant (GM083871). This work was also supported in part by the NIH through the following NCRR grant: the Biomedical Informatics Research Network (1 U24 RR025736-01). Special thanks to Mihail Bota for providing access to the underlying BAMS database. Thanks to Larry Swanson, Joel Hahn, and other members of the Swanson laboratory for their support.
Footnotes
1The argument could be made that the Swanson-laboratory maps are also interpretations since a single case was selected to be representative of a class of experiments and then hand drawn over atlas plates by experts in an interpretative process. For the purposes of the work we describe here, we consider these maps to be direct observations.
3Available for download from http://sourceforge.net/projects/neuroscholar/
References
- Brinschwitz K., Dittgen A., Madai V., Lommel R., Geisler S., Veh R. (2010). Glutamatergic axons from the lateral habenula mainly terminate on GABAergic neurons of the ventral midbrain. Neuroscience 168, 463–476 10.1016/j.neuroscience.2010.03.050 [DOI] [PubMed] [Google Scholar]
- Burns G., Cheng W.-C., Thompson R. F., Swanson L. (2006). The NeiuARt II system: a viewing tool for neuroanatomical data based on published neuroanatomical atlases. BMC Bioinformatics 7, 531. 10.1186/1471-2105-7-531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns G. A. (2001). Knowledge management of the neuroscientific literature: the data model and underlying strategy of the neuroscholar system. Philos. Trans. R. Soc. Lond. B Biol. Sci. 356, 1187–1208 10.1098/rstb.2001.0909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns G. A., Feng D., Hovy E. H. (2008). “Intelligent approaches to mining the primary research literature: techniques, systems, and examples,” in Computational Intelligence in Medical Informatics, eds Kelemen A., Abraham A., Chen Y., Liang Y. (Berlin: Springer; ), 17–50 [Google Scholar]
- Canteras N. S., Ribeiro-Barbosa E. R., Goto M., Cipolla-Neto J., Swanson L. W. (2010). The retinohypothalamic tract: comparison of axonal projection patterns from four major targets. Brain Res. Rev. 65, 0165–0173 [DOI] [PubMed] [Google Scholar]
- Canteras N. S., Simerly R. B., Swanson L. W. (1992a). Connections of the posterior nucleus of the amygdala. J. Comp. Neurol. 324, 143–179 10.1002/cne.903240205 [DOI] [PubMed] [Google Scholar]
- Canteras N. S., Simerly R. B., Swanson L. W. (1992b). Projections of the ventral premammillary nucleus. J. Comp. Neurol. 324, 195–212 10.1002/cne.903240205 [DOI] [PubMed] [Google Scholar]
- Canteras N. S., Simerly R. B., Swanson L. W. (1994). Organization of projections from the ventromedial nucleus of the hypothalamus: a Phaseolus vulgaris-leucoagglutinin study in the rat. J. Comp. Neurol. 348, 41–79 10.1002/cne.903480103 [DOI] [PubMed] [Google Scholar]
- Canteras N. S., Simerly R. B., Swanson L. W. (1995). Organization of projections from the medial nucleus of the amygdala: a phal study in the rat. J. Comp. Neurol. 360, 213–245 10.1002/cne.903600203 [DOI] [PubMed] [Google Scholar]
- Canteras N. S., Swanson L. W. (1992a). Projections of the ventral subiculum to the amygdala, septum, and hypothalamus: a phal anterograde tract-tracing study in the rat. J. Comp. Neurol. 324, 180–194 10.1002/cne.903240205 [DOI] [PubMed] [Google Scholar]
- Canteras N. S., Swanson L. W. (1992b). The dorsal premammillary nucleus: an unusual component of the mammillary body. Proc. Natl. Acad. Sci. U.S.A. 89, 10089–10093 10.1073/pnas.89.21.10089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cenquizca L. A., Swanson L. W. (2007). Spatial organization of direct hippocampal field CA1 axonal projections to the rest of the cerebral cortex. Brain Res. Rev. 56, 1–26 10.1016/j.brainresrev.2007.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dashti A. E., Burns G. A. P. C., Simmons D. M., Swanson L., Ghandeharizadeh S., Shahabi C., Stone J., Jia S. (2001). “The neuroanatomical rat brain viewer (NeuART),” in Computing the Brain: A Guide to Neuroinformatics, eds Grethe J. S., Arbib M. A. (San Diego: Academic Press; ), 189–202 [Google Scholar]
- Dong H., Petrovich G. D., Swanson L. W. (2000). Organization of projections from the juxtacapsular nucleus of the bst: a phal study in the rat. Brain Res. 859, 1–14 10.1016/S0006-8993(99)02246-5 [DOI] [PubMed] [Google Scholar]
- Dong H. W., Petrovich G. D., Swanson L. W. (2001a). Topography of projections from amygdala to bed nuclei of the stria terminalis. Brain Res. Brain Res. Rev. 38, 192–246 10.1016/S0165-0173(01)00079-0 [DOI] [PubMed] [Google Scholar]
- Dong H. W., Petrovich G. D., Watts A. G., Swanson L. W. (2001b). Basic organization of projections from the oval and fusiform nuclei of the bed nuclei of the stria terminalis in adult rat brain. J. Comp. Neurol. 436, 430–455 10.1002/cne.1079 [DOI] [PubMed] [Google Scholar]
- Dong H. W., Swanson L. W. (2003). Projections from the rhomboid nucleus of the bed nuclei of the stria terminalis: implications for cerebral hemisphere regulation of ingestive behaviors. J. Comp. Neurol. 463, 434–472 10.1002/cne.10758 [DOI] [PubMed] [Google Scholar]
- Dong H. W., Swanson L. W. (2004a). Organization of axonal projections from the anterolateral area of the bed nuclei of the stria terminalis. J. Comp. Neurol. 468, 277–298 10.1002/cne.10949 [DOI] [PubMed] [Google Scholar]
- Dong H.-W., Swanson L. W. (2004b). Projections from bed nuclei of the stria terminalis, posterior division: implications for cerebral hemisphere regulation of defensive and reproductive behaviors. J. Comp. Neurol. 471, 396–433 10.1002/cne.20002 [DOI] [PubMed] [Google Scholar]
- Dong H. W., Swanson L. W. (2006a). Projections from bed nuclei of the stria terminalis, anteromedial area: cerebral hemisphere integration of neuroendocrine, autonomic, and behavioral aspects of energy balance. J. Comp. Neurol. 494, 142–178 10.1002/cne.20788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong H. W., Swanson L. W. (2006b). Projections from bed nuclei of the stria terminalis, dorsomedial nucleus: implications for cerebral hemisphere integration of neuroendocrine, autonomic, and drinking responses. J. Comp. Neurol. 494, 75–107 10.1002/cne.20788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong H. W., Swanson L. W. (2006c). Projections from bed nuclei of the stria terminalis, magnocellular nucleus: implications for cerebral hemisphere regulation of micturition, defecation, and penile erection. J. Comp. Neurol. 494, 108–141 10.1002/cne.20788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldt S., Bonifazi P., Cossart R. (2011). Dissecting functional connectivity of neuronal microcircuits: experimental and theoretical insights. Trends Neurosci. 34, 225–236 10.1016/j.tins.2011.02.007 [DOI] [PubMed] [Google Scholar]
- Goto M., Canteras N., Burns G., Swanson L. (2005). Projections from the subfornical region of the lateral hypothalamic area. J. Comp. Neurol. 493, 412–438 10.1002/cne.20764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto M., Swanson L. W. (2004). Axonal projections from the parasubthalamic nucleus. J. Comp. Neurol. 469, 581–607 10.1002/cne.11036 [DOI] [PubMed] [Google Scholar]
- Hagmann P., Kurant M., Gigandet X., Thiran P., Wedeen V. J., Meuli R., Thiran J.-P. (2007). Mapping human whole-brain structural networks with diffusion mri. PLoS ONE 2, e597. 10.1371/journal.pone.0000597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn J. D., Swanson L. W. (2010). Distinct patterns of neuronal inputs and outputs of the juxtaparaventricular and suprafornical regions of the lateral hypothalamic area in the male rat. Brain Res. Rev. 64, 14–103 10.1016/j.brainresrev.2010.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawrylycz M., Baldock R. A., Burger A., Hashikawa T., Johnson G. A., Martone M., Ng L., Lau C., Larsen S. D., Nissanov J., Puelles L., Ruffins S., Verbeek F., Zaslavsky I., Boline J. (2011). Digital atlasing and standardization in the mouse brain. PLoS Comput. Biol. 7, e1001065. 10.1371/journal.pcbi.1001065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs G. A., Llovet P., Judson I., White L., Heimbuch R., Jeager C., Burns G. (2009). “MILO: a general purpose data repository for disease foundations and individual laboratories,” in Society for Neuroscience Annual Meeting, Chicago, IL [Google Scholar]
- Ju G., Swanson L. W. (1989). Studies on the cellular architecture of the bed nuclei of the stria terminalis in the rat: I. Cytoarchitecture. J. Comp. Neurol. 280, 587–602 10.1002/cne.902800409 [DOI] [PubMed] [Google Scholar]
- Ju G., Swanson L. W., Simerly R. B. (1989). Studies on the cellular architecture of the bed nuclei of the stria terminalis in the rat: II. Chemoarchitecture. J. Comp. Neurol. 280, 603–621 10.1002/cne.902800409 [DOI] [PubMed] [Google Scholar]
- Lanciego J. L., Wouterlood F. G. (2011). A half century of experimental neuroanatomical tracing. J. Chem. Neuroanat. 42, 157–183 10.1016/j.jchemneu.2011.07.001 [DOI] [PubMed] [Google Scholar]
- Lee D., Ruffins S., Ng Q., Sane N., Anderson S., Toga A. (2010). MBAT: a scalable informatics system for unifying digital atlasing workflows. BMC Bioinformatics 11, 608. 10.1186/1471-2105-11-608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng L., Bernard A., Lau C., Overly C. C., Dong H.-W., Kuan C., Pathak S., Sunkin S. M., Dang C., Bohland J. W., Bokil H., Mitra P. P., Puelles L., Hohmann J., Anderson D. J., Lein E. S., Jones A. R., Hawrylycz M. (2009). An anatomic gene expression atlas of the adult mouse brain. Nat. Neurosci. 12, 356–362 10.1038/nn.2281 [DOI] [PubMed] [Google Scholar]
- Perez-Manso M., Barroso-Chinea P., Aymerich M., Lanciego J. (2006). “Functional” neuroanatomical tract tracing: analysis of changes in gene expression of brain circuits of interest. Brain Res. 1072, 91–98 10.1016/j.brainres.2005.12.018 [DOI] [PubMed] [Google Scholar]
- Petrovich G. D., Canteras N. S., Swanson L. W. (2001). Combinatorial amygdalar inputs to hippocampal domains and hypothalamic behavior systems. Brain Res. Brain Res. Rev. 38, 247–289 10.1016/S0165-0173(01)00080-7 [DOI] [PubMed] [Google Scholar]
- Petrovich G. D., Holland P. C., Gallagher M. (2005). Amygdalar and prefrontal pathways to the lateral hypothalamus are activated by a learned cue that stimulates eating. J. Neurosci. 25, 8295–8302 10.1523/JNEUROSCI.2480-05.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrovich G. D., Risold P. Y., Swanson L. W. (1996). Organization of projections from the basomedial nucleus of the amygdala: a phal study in the rat. J. Comp. Neurol. 374, 387–420 [DOI] [PubMed] [Google Scholar]
- Petrovich G. D., Swanson L. W. (1997). Projections from the lateral part of the central amygdalar nucleus to the postulated fear conditioning circuit. Brain Res. 763, 247–254 10.1016/S0006-8993(96)01361-3 [DOI] [PubMed] [Google Scholar]
- Risold P. Y., Canteras N. S., Swanson L. W. (1994). Organization of projections from the anterior hypothalamic nucleus: a Phaseolus vulgaris-leucoagglutinin study in the rat. J. Comp. Neurol. 348, 1–40 10.1002/cne.903480102 [DOI] [PubMed] [Google Scholar]
- Risold P. Y., Swanson L. W. (1995). Evidence for a hypothalamothalamocortical circuit mediating pheromonal influences on eye and head movements. Proc. Natl. Acad. Sci. U.S.A. 92, 3898–3902 10.1073/pnas.92.9.3898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risold P. Y., Swanson L. W. (1997a). Chemoarchitecture of the rat lateral septal nucleus. Brain Res. Brain Res. Rev. 24, 91–113 10.1016/S0165-0173(97)00007-6 [DOI] [PubMed] [Google Scholar]
- Risold P. Y., Swanson L. W. (1997b). Connections of the rat lateral septal complex. Brain Res. Brain Res. Rev. 24, 115–195 10.1016/S0165-0173(97)00007-6 [DOI] [PubMed] [Google Scholar]
- Risold P. Y., Thompson R. H., Swanson L. W. (1997). The structural organization of connections between hypothalamus and cerebral cortex. Brain Res. Brain Res. Rev. 24, 197–254 10.1016/S0165-0173(97)00007-6 [DOI] [PubMed] [Google Scholar]
- Russ T., Ramakrishnan C., Hovy E. H., Bota M., Burns G. A. P. C. (2011). Knowledge engineering tools for reasoning with scientific observations and interpretations: a neural connectivity use case. BMC Bioinformatics 12, 351. 10.1186/1471-2105-12-351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schofield B. R. (2010). Projections from auditory cortex to midbrain cholinergic neurons that project to the inferior colliculus. Neuroscience 166, 231–240 10.1016/j.neuroscience.2009.12.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith S. (2007). Circuit reconstruction tools today. Curr. Opin. Neurobiol. 17, 601–608 10.1016/j.conb.2007.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson L. (2004). Brain Maps: Structure of the Rat Brain, 3 Edn San Diego: Elsevier Academic Press [Google Scholar]
- Swanson L. W. (1998). Brain Maps: Structure of the Rat Brain, 2 Edn San Diego: Elsevier Academic Press [Google Scholar]
- Swanson L. W. (2005). Anatomy of the soul as reflected in the cerebral hemispheres: neural circuits underlying voluntary control of basic motivated behaviors. J. Comp. Neurol. 493, 122–131 10.1002/cne.20733 [DOI] [PubMed] [Google Scholar]
- Swanson L. W., Bota M. (2010). Foundational model of structural connectivity in the nervous system with a schema for wiring diagrams, connectome, and basic plan architecture. Proc. Natl. Acad. Sci. U.S.A. 107, 20610–20617 10.1073/pnas.1015128107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson R. H., Canteras N. S., Swanson L. W. (1996). Organization of projections from the dorsomedial nucleus of the hypothalamus: a pha-l study in the rat. J. Comp. Neurol. 376, 143–173 [DOI] [PubMed] [Google Scholar]
- Thompson R. H., Swanson L. W. (1998). Organization of inputs to the dorsomedial nucleus of the hypothalamus: a reexamination with fluorogold and phal in the rat. Brain Res. Brain Res. Rev. 27, 89–118 10.1016/S0165-0173(98)00010-1 [DOI] [PubMed] [Google Scholar]
- Thompson R. H., Swanson L. W. (2003). Structural characterization of a hypothalamic visceromotor pattern generator network. Brain Res. Brain Res. Rev. 41, 153–202 10.1016/S0165-0173(02)00232-1 [DOI] [PubMed] [Google Scholar]
- Thompson R. H., Swanson L. W. (2010). Hypothesis-driven structural connectivity analysis supports network over hierarchical model of brain architecture. Proc. Natl. Acad. Sci. U.S.A. 107, 15235–15239 10.1073/pnas.1001999107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watts A. G., Swanson L. W. (1987). Efferent projections of the suprachiasmatic nucleus: II. Studies using retrograde transport of fluorescent dyes and simultaneous peptide immunohistochemistry in the rat. J. Comp. Neurol. 258, 230–252 10.1002/cne.902580205 [DOI] [PubMed] [Google Scholar]
- Watts A. G., Swanson L. W., Sanchez-Watts G. (1987). Efferent projections of the suprachiasmatic nucleus: I. Studies using anterograde transport of Phaseolus vulgaris leucoagglutinin in the rat. J. Comp. Neurol. 258, 204–229 10.1002/cne.902580205 [DOI] [PubMed] [Google Scholar]
- Zaborszky L., Wouterlood F. G., Lanciego J. L. (2006). Neuroanatomical Tract-Tracing 3: Molecules, Neurons, and Systems. New York: Springer [Google Scholar]