Abstract
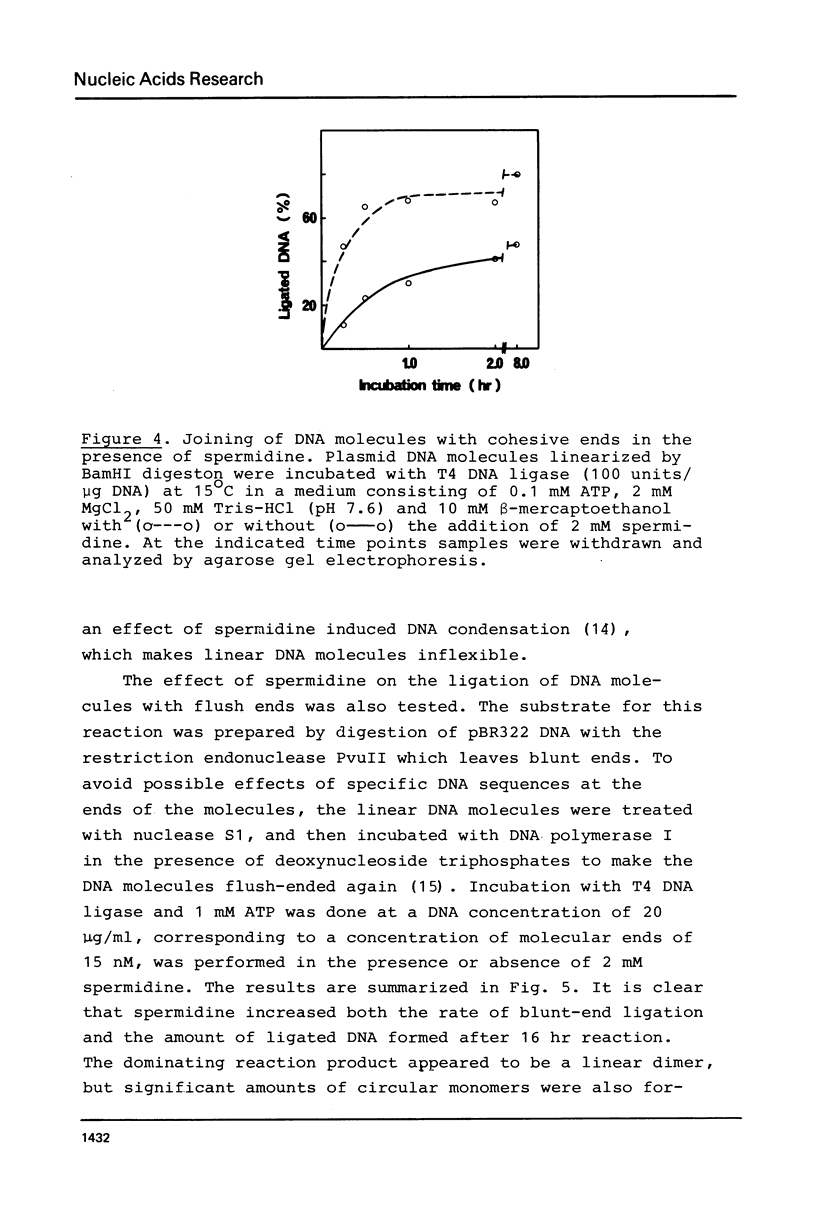
Single-strand gaps in DNA molecules were found to be a substrate for T4 DNA ligase. Sealing of the gaps was optimal at the same conditions as ligation of blunt-ended DNA molecules. Spermidine at a concentration of 2 mM stimulated the ligation of gaps, as well as the joining of DNA molecules with cohesive and blunt ends. In addition, spermidine reduced the optimal ATP concentration. The ligation of single-stranded gaps was a slow process, reaching a plateau after several hours at 25 degrees C. Approximately 10% of circular duplex plasmid pBR322 DNA molecules with a gap of 1-5 nucleotides could be converted to a covalently closed form. When such molecules were used for transformation of E. coli cells deletion mutants were obtained at a high frequency. The size and position of the gaps and the deletions were equivalent, confirming that T4 DNA ligase was sealing the gaps.
Full text
PDF









Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolivar F., Rodriguez R. L., Greene P. J., Betlach M. C., Heyneker H. L., Boyer H. W., Crosa J. H., Falkow S. Construction and characterization of new cloning vehicles. II. A multipurpose cloning system. Gene. 1977;2(2):95–113. [PubMed] [Google Scholar]
- Carbon J., Shenk T. E., Berg P. Biochemical procedure for production of small deletions in simian virus 40 DNA. Proc Natl Acad Sci U S A. 1975 Apr;72(4):1392–1396. doi: 10.1073/pnas.72.4.1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferretti L., Sgaramella V. Specific and reversible inhibition of the blunt end joining activity of the T4 DNA ligase. Nucleic Acids Res. 1981 Aug 11;9(15):3695–3705. doi: 10.1093/nar/9.15.3695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gosule L. C., Schellman J. A. DNA condensation with polyamines I. Spectroscopic studies. J Mol Biol. 1978 May 25;121(3):311–326. doi: 10.1016/0022-2836(78)90366-2. [DOI] [PubMed] [Google Scholar]
- Green C., Tibbetts C. Targeted deletions of sequences from closed circular DNA. Proc Natl Acad Sci U S A. 1980 May;77(5):2455–2459. doi: 10.1073/pnas.77.5.2455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenfield L., Simpson L., Kaplan D. Conversion of closed circular DNA molecules to single-nicked molecules by digestion with DNAase I in the presence of ethidium bromide. Biochim Biophys Acta. 1975 Oct 15;407(3):365–375. doi: 10.1016/0005-2787(75)90104-5. [DOI] [PubMed] [Google Scholar]
- Higgins N. P., Cozzarelli N. R. DNA-joining enzymes: a review. Methods Enzymol. 1979;68:50–71. doi: 10.1016/0076-6879(79)68006-0. [DOI] [PubMed] [Google Scholar]
- Johnson P. H., Grossman L. I. Electrophoresis of DNA in agarose gels. Optimizing separations of conformational isomers of double- and single-stranded DNAs. Biochemistry. 1977 Sep 20;16(19):4217–4225. doi: 10.1021/bi00638a014. [DOI] [PubMed] [Google Scholar]
- Jovin T. M., Englund P. T., Bertsch L. L. Enzymatic synthesis of deoxyribonucleic acid. XXVI. Physical and chemical studies of a homogeneous deoxyribonucleic acid polymerase. J Biol Chem. 1969 Jun 10;244(11):2996–3008. [PubMed] [Google Scholar]
- Lehman I. R. DNA ligase: structure, mechanism, and function. Science. 1974 Nov 29;186(4166):790–797. doi: 10.1126/science.186.4166.790. [DOI] [PubMed] [Google Scholar]
- Loening U. E. The fractionation of high-molecular-weight ribonucleic acid by polyacrylamide-gel electrophoresis. Biochem J. 1967 Jan;102(1):251–257. doi: 10.1042/bj1020251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osland A., Kleppe K. Polyamine induced aggregation of DNA. Nucleic Acids Res. 1977 Mar;4(3):685–695. doi: 10.1093/nar/4.3.685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panet A., van de Sande J. H., Loewen P. C., Khorana H. G., Raae A. J., Lillehaug J. R., Kleppe K. Physical characterization and simultaneous purification of bacteriophage T4 induced polynucleotide kinase, polynucleotide ligase, and deoxyribonucleic acid polymerase. Biochemistry. 1973 Dec 4;12(25):5045–5050. doi: 10.1021/bi00749a003. [DOI] [PubMed] [Google Scholar]
- Peden K. W., Pipas J. M., Pearson-White S., Nathans D. Isolation of mutants of an animal virus in bacteria. Science. 1980 Sep 19;209(4463):1392–1396. doi: 10.1126/science.6251547. [DOI] [PubMed] [Google Scholar]
- Raae A. J., Kleppe K. T4 polynucleotide ligase catalyzed joining on triple-stranded nucleic acids. Biochemistry. 1978 Jul 11;17(14):2939–2942. doi: 10.1021/bi00607a037. [DOI] [PubMed] [Google Scholar]
- Raae A. J., Kleppe R. K., Kleppe K. Kinetics and effect of salts and polyamines on T4 polynucleotide ligase. Eur J Biochem. 1975 Dec 15;60(2):437–443. doi: 10.1111/j.1432-1033.1975.tb21021.x. [DOI] [PubMed] [Google Scholar]
- Seeburg P. H., Shine J., Martial J. A., Baxter J. D., Goodman H. M. Nucleotide sequence and amplification in bacteria of structural gene for rat growth hormone. Nature. 1977 Dec 8;270(5637):486–494. doi: 10.1038/270486a0. [DOI] [PubMed] [Google Scholar]
- Sgaramella V., Ehrlich S. D. Use of the T4 polynucleotide ligase in the joining of flush-ended DNA segments generated by restriction endonucleases. Eur J Biochem. 1978 May 16;86(2):531–537. doi: 10.1111/j.1432-1033.1978.tb12336.x. [DOI] [PubMed] [Google Scholar]
- Shenk T. E., Rhodes C., Rigby P. W., Berg P. Biochemical method for mapping mutational alterations in DNA with S1 nuclease: the location of deletions and temperature-sensitive mutations in simian virus 40. Proc Natl Acad Sci U S A. 1975 Mar;72(3):989–993. doi: 10.1073/pnas.72.3.989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shortle D., Nathans D. Local mutagenesis: a method for generating viral mutants with base substitutions in preselected regions of the viral genome. Proc Natl Acad Sci U S A. 1978 May;75(5):2170–2174. doi: 10.1073/pnas.75.5.2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugino A., Goodman H. M., Heyneker H. L., Shine J., Boyer H. W., Cozzarelli N. R. Interaction of bacteriophage T4 RNA and DNA ligases in joining of duplex DNA at base-paired ends. J Biol Chem. 1977 Jun 10;252(11):3987–3994. [PubMed] [Google Scholar]
- Sutcliffe J. G. pBR322 restriction map derived from the DNA sequence: accurate DNA size markers up to 4361 nucleotide pairs long. Nucleic Acids Res. 1978 Aug;5(8):2721–2728. doi: 10.1093/nar/5.8.2721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace R. B., Johnson P. F., Tanaka S., Schöld M., Itakura K., Abelson J. Directed deletion of a yeast transfer RNA intervening sequence. Science. 1980 Sep 19;209(4463):1396–1400. doi: 10.1126/science.6997991. [DOI] [PubMed] [Google Scholar]
- Westergaard O., Brutlag D., Kornberg A. Initiation of deoxyribonucleic acid synthesis. IV. Incorporation of the ribonucleic acid primer into the phage replicative form. J Biol Chem. 1973 Feb 25;248(4):1361–1364. [PubMed] [Google Scholar]