Abstract
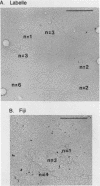
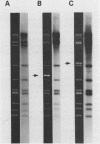
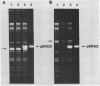
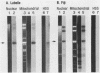
Mitochondria from two Neurospora intermedia strains (P4O5-Labelle and Fiji N6-6) were found to contain plasmid DNAs in addition to the standard mitochondrial DNA species. The plasmid DNAs consist of monomeric circles (4.1-4.3 kbp and 5.2-5.3 kbp for Labelle and Fiji, respectively) and oligomers in which monomers are organized as head-to-tail repeats. DNA-DNA hybridization experiments showed that the plasmids have no substantial sequence homology to mtDNA, to each other, or to a previously characterized mitochondrial plasmid from N. crassa strain Mauriceville-lc (Collins et al. Cell 24, 443-452, 1981). The intramitochondrial location of the plasmids was established by cell fractionation and nuclease protection experiments. In sexual crosses, the plasmids showed strict maternal inheritance, the same as Neurospora mitochondrial DNA. The plasmids may represent a novel class of mitochondrial genetic elements.
Full text
PDF



















Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bernard U., Goldthwaite C., Küntzel H. Physical map of Neurospora crassa mitochondrial DNA and its transcription unit for ribosomal RNA. Nucleic Acids Res. 1976 Nov;3(11):3101–3108. doi: 10.1093/nar/3.11.3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard U., Pühler A., Mayer F., Küntzel H. Denaturation map of the circular mitochondrial genome of Neurospora crassa. Biochim Biophys Acta. 1975 Aug 21;402(2):270–278. doi: 10.1016/0005-2787(75)90047-7. [DOI] [PubMed] [Google Scholar]
- Bertrand H., Collins R. A., Stohl L. L., Goewert R. R., Lambowitz A. M. Deletion mutants of Neurospora crassa mitochondrial DNA and their relationship to the "stop-start" growth phenotype. Proc Natl Acad Sci U S A. 1980 Oct;77(10):6032–6036. doi: 10.1073/pnas.77.10.6032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins R. A., Stohl L. L., Cole M. D., Lambowitz A. M. Characterization of a novel plasmid DNA found in mitochondria of N. crassa. Cell. 1981 May;24(2):443–452. doi: 10.1016/0092-8674(81)90335-4. [DOI] [PubMed] [Google Scholar]
- Cummings D. J., Belcour L., Grandchamp C. Mitochondrial DNA from Podospora anserina. II. Properties of mutant DNA and multimeric circular DNA from senescent cultures. Mol Gen Genet. 1979 Mar 27;171(3):239–250. doi: 10.1007/BF00267578. [DOI] [PubMed] [Google Scholar]
- Dale R. M. Sequence homology among different size classes of plant mtDNAs. Proc Natl Acad Sci U S A. 1981 Jul;78(7):4453–4457. doi: 10.1073/pnas.78.7.4453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambowitz A. M., LaPolla R. J., Collins R. A. Mitochondrial ribosome assembly in Neurospora. Two-dimensional gel electrophoretic analysis of mitochondrial ribosomal proteins. J Cell Biol. 1979 Jul;82(1):17–31. doi: 10.1083/jcb.82.1.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambowitz A. M. Preparation and analysis of mitochondrial ribosomes. Methods Enzymol. 1979;59:421–433. doi: 10.1016/0076-6879(79)59103-4. [DOI] [PubMed] [Google Scholar]
- Lazarus C. M., Earl A. J., Turner G., Küntzel H. Amplification of a mitochondrial DNA sequence in the cytoplasmically inherited 'ragged' mutant of Aspergillus amstelodami. Eur J Biochem. 1980 May;106(2):633–641. doi: 10.1111/j.1432-1033.1980.tb04611.x. [DOI] [PubMed] [Google Scholar]
- Levings C. S., 3rd, Kim B. D., Pring D. R., Conde M. F., Mans R. J., Laughnan J. R., Gabay-Laughnan S. J. Cytoplasmic Reversion of cms-S in Maize: Association with a Transpositional Event. Science. 1980 Aug 29;209(4460):1021–1023. doi: 10.1126/science.209.4460.1021. [DOI] [PubMed] [Google Scholar]
- Lizardi P. M., Luck D. J. Absence of a 5S RNA complnent in the mitochondrial ribosomes of Neurospora crassa. Nat New Biol. 1971 Feb 3;229(5):140–142. doi: 10.1038/newbio229140a0. [DOI] [PubMed] [Google Scholar]
- Mannella C. A., Goewert R. R., Lambowitz A. M. Characterization of variant Neurospora crassa mitochondrial DNAs which contain tandem reiterations. Cell. 1979 Dec;18(4):1197–1207. doi: 10.1016/0092-8674(79)90232-0. [DOI] [PubMed] [Google Scholar]
- Mannella C. A., Lambowitz A. Interaction of wild-type and poky mitochondrial DNA in heterokaryons of Neurospora. Biochem Biophys Res Commun. 1978 Feb 14;80(3):673–679. doi: 10.1016/0006-291x(78)91621-2. [DOI] [PubMed] [Google Scholar]
- Mannella C. A., Pittenger T. H., Lambowitz A. M. Transmission of mitochondrial deoxyribonucleic acid in Neurospora crassa sexual crosses. J Bacteriol. 1979 Mar;137(3):1449–1451. doi: 10.1128/jb.137.3.1449-1451.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMaster G. K., Carmichael G. G. Analysis of single- and double-stranded nucleic acids on polyacrylamide and agarose gels by using glyoxal and acridine orange. Proc Natl Acad Sci U S A. 1977 Nov;74(11):4835–4838. doi: 10.1073/pnas.74.11.4835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pring D. R., Levings C. S., Hu W. W., Timothy D. H. Unique DNA associated with mitochondria in the "S"-type cytoplasm of male-sterile maize. Proc Natl Acad Sci U S A. 1977 Jul;74(7):2904–2908. doi: 10.1073/pnas.74.7.2904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reich E., Luck D. J. Replication and inheritance of mitochondrial DNA. Proc Natl Acad Sci U S A. 1966 Jun;55(6):1600–1608. doi: 10.1073/pnas.55.6.1600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigby P. W., Dieckmann M., Rhodes C., Berg P. Labeling deoxyribonucleic acid to high specific activity in vitro by nick translation with DNA polymerase I. J Mol Biol. 1977 Jun 15;113(1):237–251. doi: 10.1016/0022-2836(77)90052-3. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Sutcliffe J. G. pBR322 restriction map derived from the DNA sequence: accurate DNA size markers up to 4361 nucleotide pairs long. Nucleic Acids Res. 1978 Aug;5(8):2721–2728. doi: 10.1093/nar/5.8.2721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson R. D., Kemble R. J., Flavell R. B. Variations in mitochondrial DNA organisation between normal and male-sterile cytoplasms of maize. Nucleic Acids Res. 1980 May 10;8(9):1999–2008. doi: 10.1093/nar/8.9.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward B. L., Anderson R. S., Bendich A. J. The mitochondrial genome is large and variable in a family of plants (cucurbitaceae). Cell. 1981 Sep;25(3):793–803. doi: 10.1016/0092-8674(81)90187-2. [DOI] [PubMed] [Google Scholar]