Abstract
Estrogen receptor α (ERα) acts as a transcription factor by virtue of the ability of its DNA-binding (DB) domain, comprised of a tandem pair of zinc fingers, to recognize the estrogen response element (ERE) within the promoters of target genes. Herein, using an array of biophysical methods, we probe structural consequences of the replacement of zinc within the DB domain of ERα with various environmental metals and their effects on the thermodynamics of binding to DNA. Our data reveal that while the DB domain reconstituted with divalent ions of zinc, cadmium, mercury and cobalt binds to DNA with affinities in the nanomolar range, divalent ions of barium, copper, iron, lead, manganese, nickel and tin are unable to regenerate DB domain with DNA-binding potential though they can compete with zinc for coordinating the cysteine ligands within the zinc fingers. We also show that the metal-free DB domain is a homodimer in solution and that the binding of various metals only results in subtle secondary and tertiary structural changes, implying that metal-coordination may only be essential for DNA-binding. Collectively, our findings provide mechanistic insights into how environmental metals may modulate the physiological function of a key nuclear receptor involved in mediating a plethora of cellular functions central to human health and disease.
Keywords: Estrogen receptor α, Zinc fingers, Environmental metals, Isothermal titration calorimetry, Analytical light scattering, Circular dichroism, Steady-state fluorescence, Steady-state absorbance
INTRODUCTION
Estrogen receptor α (ERα) is a member of a family of ligand-modulated transcription factors that have come to be known as nuclear receptors (NRs) (Evans, 1988; Thornton, 2001; Escriva et al., 2004; McKenna et al., 2009). ERα mediates the action of estrogens such as estradiol in a diverse array of cellular processes and its hyperactivation leads to the genesis of large fractions of breast cancer (Heldring et al., 2007). ERα is constructed on a modular architecture, also shared by other members of the NR family, comprised of a central DNA-binding (DB) domain flanked between an N-terminal trans-activation (TA) domain and a C-terminal ligand-binding (LB) domain. Upon the binding of estrogens to the LB domain, ERα translocates to the nucleus and binds as a homodimer with a two-fold axis of symmetry to the estrogen response element (ERE), containing the AGGTCAnnnTGACCT consensus sequence, located within the promoters of target genes (Klinge, 2001). DNA-binding is accomplished through a pair of tandem C4-type Zinc fingers located within the DB domain, with each finger containing a Zn2+ ion coordinated in a tetrahedral arrangement by four highly conserved cysteine residues to generate the Zn2+[Cys]4 metal-protein complex (Schwabe et al., 1990; Schwabe et al., 1993) (Figure 1). Importantly, while the first Zinc finger (ZF-I) within each monomer of DB domain recognizes the hexanucleotide sequence 5′-AGGTCA-3′ within the major groove at each end of the ERE duplex, the second Zinc finger (ZF-II) is responsible for the homodimerization of DB domain.
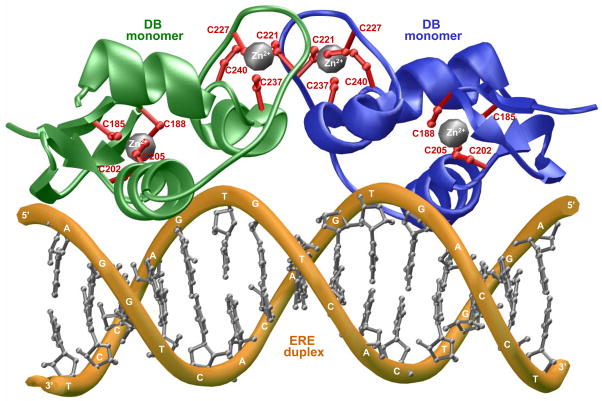
Figure 1.
3D structural model of the DB domain of human ERα in complex with ERE duplex containing the AGGTCAcagTGACCT consensus sequence based on the crystal structure (PDB# 1HCQ) determined by Rhodes and co-workers (Schwabe et al., 1993). The structural model was built as described earlier (Deegan et al., 2010) and rendered using RIBBONS (Carson, 1991). Note that the DB domain binds to DNA as a homodimer with a two-fold axis of symmetry. One monomer of DB domain is shown in green and the other in blue. The Zn2+ divalent ions are depicted as gray spheres and the sidechain moieties of cysteine residues being coordinated in red. The DNA backbone is shown in yellow and the bases are colored gray for clarity.
Upon binding to the promoters of target genes through its DB domain, the LB domain of ERα recruits a multitude of cellular proteins in an estrogen-dependent manner, such as transcription factors, co-activators and co-repressors, to the site of DNA transcription and thereby allowing it to exert its action at the genomic level in a concerted fashion (Ham and Parker, 1989; Darimont et al., 1998). The TA domain is believed to be responsive to growth factors acting through MAPK signaling and thus it further synergizes the action of various co-activators and co-repressors recruited by the LB domain at the site of DNA transcription (Kato et al., 1995; Warnmark et al., 2003). In this manner, ERα and other nuclear receptors mediate a diverse array of cellular functions from embryonic development to metabolic homeostasis and their aberrant function has been widely implicated in disease (Brzozowski et al., 1997; Gottlieb et al., 2004; Gurnell and Chatterjee, 2004; Noy, 2007; Sonoda et al., 2008; McEwan, 2009).
Discovered more than a quarter of century ago (Miller et al., 1985), the zinc finger is one of the most common motifs found in transcription factors (Green et al., 1998; Wolfe et al., 2000). Several lines of evidence suggest that metals other than zinc can serve as coordination sites for cysteine ligands within zinc fingers with important consequences on cellular processes involved in gene expression, DNA repair and genomic stability (Freedman et al., 1988; Thiesen and Bach, 1991; Hartwig, 2001; Bal et al., 2003; Blessing et al., 2004; Kopera et al., 2004; Hartwig et al., 2010). In an effort to further our understanding of the interaction of metals with zinc fingers, we analyze here structural consequences of the replacement of zinc within the DB domain of ERα with various environmental metals and their effects on the thermodynamics of binding to DNA using an array of biophysical methods. Our data reveal that while the DB domain reconstituted with divalent ions of zinc, cadmium, mercury and cobalt binds to DNA with affinities in the nanomolar range, divalent ions of barium, copper, iron, lead, manganese, nickel and tin are unable to regenerate DB domain with DNA-binding potential though they can compete with zinc for coordinating the cysteine ligands within the zinc fingers. We also show that the metal-free DB domain is a homodimer in solution and that the binding of various metals only results in subtle secondary and tertiary structural changes, implying that metal-coordination may only be essential for DNA-binding. Collectively, our findings provide mechanistic insights into how environmental metals may modulate the physiological function of a key nuclear receptor involved in mediating a plethora of cellular functions central to human health and disease.
MATERIALS and METHODS
Protein preparation
The DB domain (residues 176–250) of human ERα was cloned into pET101 bacterial expression vector with a C-terminal polyhistidine (His)-tag using Invitrogen TOPO technology. The recombinant protein was expressed in bacteria supplemented with 50μM ZnCl2 and purified to apparent homogeneity on a Ni-NTA affinity column followed by treatment on a Hiload Superdex 200 size-exclusion chromatography (SEC) column coupled in-line with GE Akta FPLC system as described previously (Deegan et al., 2010; Deegan et al., 2011). Zinc divalent ions were stripped by the treatment of purified protein in Tris buffer (50mM Tris, 200mM NaCl and 10mM β-mercaptoethanol at pH 8.0) containing 8M urea and 10mM EDTA. After denaturation of protein overnight, EDTA was removed under denatured conditions by dialysis in acetate buffer (50mM Sodium acetate, 200mM NaCl and 10mM β-mercaptoethanol at pH 6.0) containing 8M urea. Further dialysis of protein in acetate buffer containing one of the metal chlorides at a final protein-to-metal molar ratio of 1:10 led to simultaneous removal of urea and reconstitution of the DB domain with the corresponding metal divalent ions. It is important to note that the reconstitution of DB domain at pH 6.0 was necessary to prevent the formation of insoluble salts such as lead chloride. Additionally, acetate buffer was preferred over phosphate buffer due to insolubility of various metal phosphates. Metal-reconstituted DB domain was extensively dialyzed in an appropriate buffer to remove excess metal ions. Protein concentration was determined by the fluorescence-based Quant-It assay (Invitrogen) and spectrophotometrically using an extinction coefficient of 14,940 M−1cm−1. Results from both assays were in an excellent agreement.
DNA synthesis
21-mer DNA oligos containing the ERE consensus site AGGTCAnnnTGACCT were commercially obtained from Sigma Genosys. The complete nucleotide sequences of the sense and antisense oligos constituting the ERE duplex is presented below:
5′-cccAGGTCAcagTGACCTgcg-3′
3′-gggTCCAGTgtcACTGGAcgc-5′
Oligo concentrations were determined spectrophotometrically on the basis of their extinction coefficients derived from their nucleotide sequences. Sense and antisense oligos were annealed together to generate the ERE duplex as described earlier (Deegan et al., 2010; Deegan et al., 2011).
ITC measurements
Isothermal titration calorimetry (ITC) measurements were performed on a Microcal VP-ITC instrument and data were acquired and processed using fully automized features in Microcal ORIGIN. All measurements were repeated at least three times. Briefly, protein and DNA samples were prepared in 50mM Sodium phosphate buffer containing 5mM β-mercaptoethanol at pH 7.0 and de-gassed using the ThermoVac accessory for 5min. The experiments were initiated by injecting 25 × 10μl aliquots of 50–100μM of ERE duplex from the syringe into the calorimetric cell containing 1.8ml of 2–5μM of DB domain of ERα reconstituted with various metals at 25°C. The change in thermal power as a function of each injection was automatically recorded using the ORIGIN software and the raw data were further processed to yield binding isotherms of heat release per injection as a function of molar ratio of ERE duplex to dimer-equivalent DB domain. The heats of mixing and dilution were subtracted from the heat of binding per injection by carrying out a control experiment in which the same buffer in the calorimetric cell was titrated against the ERE duplex in an identical manner. Control experiments with scrambled dsDNA oligos generated similar thermal power to that obtained for the buffer alone, implying that there was no non-specific binding of DB domains to non-cognate DNA. To extract various thermodynamic parameters, the binding isotherms were iteratively fit to a built-in one-site model by non-linear least squares regression analysis using the ORIGIN software as described previously (Wiseman et al., 1989; Deegan et al., 2010).
ALS experiments
Analytical light scattering (ALS) experiments were conducted on a Wyatt miniDAWN TREOS triple-angle static light scattering detector and Wyatt QELS dynamic light scattering detector coupled in-line with a Wyatt Optilab rEX differential refractive index detector and interfaced to a Hiload Superdex 200 size-exclusion chromatography column under the control of a GE Akta FPLC system within a chromatography refrigerator at 10°C. The DB domain of ERα pre-treated with EDTA to strip divalent zinc ions and upon reconstitution with divalent ions of various metals was loaded onto the column at a flow rate of 1ml/min and the data were automatically acquired using the ASTRA software. All protein samples were prepared in 50mM Sodium phosphate buffer containing 5mM β-mercaptoethanol at pH 7.0 and the starting concentrations injected onto the column were between 20–50μM. The angular- and concentration-dependence of static light scattering (SLS) intensity of each protein species resolved in the flow mode was measured by the Wyatt miniDAWN TREOS detector. The SLS data were analyzed according to the following built-in Zimm equation in ASTRA software (Zimm, 1948; Wyatt, 1993):
| [1] |
where Rθ is the excess Raleigh ratio due to protein in the solution as a function of protein concentration c (mg/ml) and the scattering angle θ (42°, 90° and 138°), Mobs is the observed molecular mass of each protein species, A2 is the second virial coefficient, λ is the wavelength of laser light in solution (658nm), Rg is the radius of gyration of protein, and K is given by the following relationship:
| [2] |
where n is the refractive index of the solvent, dn/dc is the refractive index increment of the protein in solution and NA is the Avogadro’s number (6.02×1023mol−1). If we assume that c → 0 and θ → 0, then Eq [1] reduces to:
| [3] |
Thus, under dilute protein concentrations (c → 0) and at low scattering angles (θ → 0), the y-intercept of Eq [1] equates to 1/Mobs. Accordingly, the weighted average value for Mobs was obtained from the y-intercept of linear fits of a range of (Kc/Rθ)−sin2(θ/2) plots as a function of protein concentration along the elution profile of each protein species using SLS measurements at three scattering angles. The time- and concentration-dependence of dynamic light scattering (DLS) intensity fluctuation of each protein species resolved in the flow mode was measured by the Wyatt QELS detector positioned at 90° with respect to the incident laser beam. The DLS data were iteratively fit using non-linear least squares regression analysis to the following built-in equation in ASTRA software: software (Koppel, 1972; Berne and Pecora, 1976; Chu, 1991):
| [4] |
where G(τ) is the autocorrelation function of dynamic light scattering intensity fluctuation I, τ is the delay time of autocorrelation function, Γ is the decay rate constant of autocorrelation function, α is the initial amplitude of autocorrelation function at zero delay time, and β is the baseline offset (the value of autocorrelation function at infinite delay time). Thus, fitting the above equation to a range of G(τ)-τ plots as a function of protein concentration along the elution profile of each protein species computes the average value of Γ using DLS measurements at a scattering angle of 90°. Accordingly, the translational diffusion coefficient (Dt) of each protein species was calculated from the following relationship:
| [5] |
where λ is the wavelength of laser light in solution (658nm), n is the refractive index of the solvent and θ is the scattering angle (90°). Additionally, the hydrodynamic radius (Rh) of each protein species was calculated from the Stokes-Einstein relationship:
| [6] |
where kB is Boltzman’s constant (1.38×10−23JK−1), T is the absolute temperature and η is the solvent viscosity. It should be noted that, in both the SLS and DLS measurements, protein concentration (c) along the elution profile of each protein species was automatically quantified in the ASTRA software from the change in refractive index (Δn) with respect to the solvent as measured by the Wyatt Optilab rEX detector using the following relationship:
| [7] |
where dn/dc is the refractive index increment of the protein in solution.
CD analysis
Circular dichroism (CD) analysis was conducted on a Bio-Logic MOS450/SFM400 spectropolarimeter thermostatically controlled with a water bath at 25°C and data were acquired using the BIOKINE software. Briefly, experiments were conducted on 20–50μM of DB domain of ERα pre-treated with EDTA to strip divalent zinc ions and upon reconstitution with divalent ions of various metals in 50mM Sodium phosphate buffer at pH 7.0. For far-UV measurements in the 190–250nm wavelength range, experiments were conducted in a quartz cuvette with a 2-mm pathlength. For near-UV measurements in the 250–350nm wavelength range, experiments were conducted in a quartz cuvette with a 10-mm pathlength. All data were recorded with a slit bandwidth of 2nm at a scan rate of 3nm/min. Data were normalized against reference spectra to remove the contribution of buffer. The reference spectra were obtained in a similar manner on a 50mM Sodium phosphate buffer at pH 7.0. Each data set represents an average of at least four scans acquired at 1nm intervals. Data were converted to molar ellipticity, [θ], as a function of wavelength (λ) of electromagnetic radiation using the following equation:
| [8] |
where Δε is the observed ellipticity in mdeg, c is the protein concentration in μM and l is the cuvette pathlength in cm. All data were processed and analyzed using the Microcal ORIGIN software.
SSA experiments
Steady-state absorbance (SSA) spectra were collected on a Jasco V-630 spectrophotometer using a quartz cuvette with a 10-mm pathlength at 25 °C. Briefly, experiments were conducted on 5μM of DB domain of ERα pre-treated with EDTA to strip divalent zinc ions and upon reconstitution with divalentions of various metals in 50mM Sodium phosphate buffer at pH 7.0. All data were recorded in the 200–350nm wavelength range using a 1.5-nm slit bandwidth. Data were normalized against reference spectra to remove the contribution of buffer. The reference spectra were obtained in a similar manner on a 50mM Sodium phosphate buffer at pH 7.0.
SSF measurements
Steady-state fluorescence (SSF) spectra were collected on a Jasco FP-6300 spectrofluorometer using a quartz cuvette with a 10-mm pathlength at 25 °C. Briefly, experiments were conducted on 5μM of DB domain of ERα pre-treated with EDTA to strip divalent zinc ions and upon reconstitution with divalent ions of various metals in 50mM Sodium phosphate buffer at pH 7.0. Excitation wavelength was 295nm and emission was acquired from 310nm to 500nm. All data were recorded using a 2.5-nm bandwidth for both excitation and emission. Data were normalized against reference spectra to remove the contribution of buffer. The reference spectra were obtained in a similar manner on a 50mM Sodium phosphate buffer at pH 7.0.
RESULTS and DISCUSSION
Binding of the DB domain of ERα to DNA is restored upon substitution of zinc with only specific divalent metal ions
To determine the extent to which environmental metals may be able to replace zinc within the zinc fingers of ERα, we measured the binding of ERE duplex to the DB domain pre-treated with EDTA to remove divalent zinc ions (as a control) and upon reconstitution with divalent ions of various metals using ITC (Figure 2). Our data reveal that while the DB domain reconstituted with divalent ions of zinc, cadmium, mercury and cobalt binds to DNA with affinities in the nanomolar range (Table 1), divalent ions of barium, copper, iron, lead, manganese, nickel and tin are unable to regenerate DB domain with DNA-binding potential. These data are consistent with previous studies demonstrating that DB domain of ERα regenerated with divalent ions of zinc, cadmium and cobalt binds DNA but not that regenerated with divalent ions of copper and nickel (Predki and Sarkar, 1992; Predki and Sarkar, 1994).
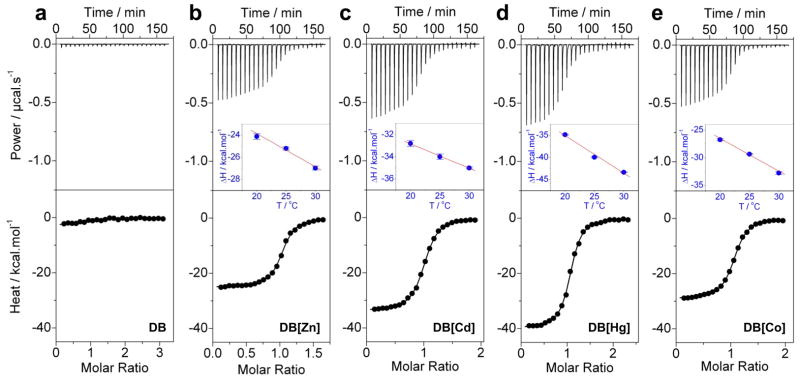
Figure 2.
Representative ITC isotherms for the binding of ERE duplex to the DB domain of ERα pre-treated with EDTA to strip divalent zinc ions (a) and upon reconstitution with divalent ions of zinc (b), cadmium (c), mercury (d) and cobalt (e). The upper panels show the raw ITC data expressed as change in thermal power with respect to time over the period of titration. In the lower panels, change in molar heat is expressed as a function of molar ratio of ERE duplex to dimer-equivalent DB domain. The solid lines in the lower panels represent the fit of data to a one-site model, based on the binding of a ligand to a macromolecule assuming the law of mass action, using the ORIGIN software (Wiseman et al., 1989; Deegan et al., 2010). The insets in top panels represent ΔH-T plots for the corresponding protein-DNA complexes. The solid line in red represents linear fit to the corresponding data.
Table 1.
Thermodynamic parameters obtained from ITC measurements at 25°C for the binding of ERE duplex to the DB domain of ERα pre-treated with EDTA to strip divalent zinc ions (DB) and upon reconstitution with divalent ions of zinc (DB[Zn]), cadmium (DB[Cd]), mercury (DB[Hg]) and cobalt (DB[Co])
| Kd/nM | ΔH/kcal.mol−1 | TΔS/kcal.mol−1 | ΔG/kcal.mol−1 | ΔCp/kcal.mol−1.K−1 | |
|---|---|---|---|---|---|
| DB | NB | NB | NB | NB | NB |
| DB[Zn] | 68 ± 8 | −25.20 ± 0.15 | −15.40 ± 0.21 | −9.80 ± 0.07 | −0.28 ± 0.04 |
| DB[Cd] | 69 ± 5 | −33.99 ± 0.25 | −24.20 ± 0.20 | −9.78 ± 0.04 | −0.22 ± 0.04 |
| DB[Hg] | 59 ± 4 | −39.95 ± 0.13 | −30.07 ± 0.16 | −9.88 ± 0.04 | −0.85 ± 0.01 |
| DB[Co] | 81 ± 2 | −29.35 ± 0.10 | −19.66 ± 0.10 | −9.69 ± 0.01 | −0.60 ± 0.01 |
The values for the affinity (Kd) and enthalpy change (ΔH) accompanying the binding of ERE duplex to the DB domain reconstituted with various metals were obtained from the fit of a one-site model, based on the binding of a ligand to a macromolecule using the law of mass action, to the corresponding ITC isotherms as described earlier (Wiseman et al., 1989; Deegan et al., 2010). Free energy of binding (ΔG) was calculated from the relationship ΔG=RTlnKd, where R is the universal molar gas constant (1.99 cal/mol/K) and T is the absolute temperature (K). Entropic contribution (TΔS) to binding was calculated from the relationship TΔS=ΔH−ΔG. Heat capacity change (ΔCp) was calculated from the slope of ΔH-T plot for the corresponding protein-DNA complex (Figure 2). Binding stoichiometries generally agreed to within ±10%. Errors were calculated from at least three independent measurements. All errors are given to one standard deviation. Note that the DB domain pre-treated with EDTA to strip divalent zinc ions and upon reconstitution with divalent metal ions of barium, copper, iron, lead, manganese, nickel and tin showed no binding (NB) to the ERE duplex.
Although the replacement of zinc with cadmium, mercury and cobalt restores DNA-binding with very similar affinities, the underlying thermodynamic forces display remarkable contrast (Table 1). Thus, while the binding of DB domain reconstituted with various metals is universally driven by favorable enthalpic changes accompanied by entropic penalty, the binding of DB domain reconstituted with cadmium and mercury results in the release of 5–15 kcal/mol of additional enthalpic contribution to the overall free energy relative to reconstitution with zinc and cobalt. We believe that such enthalpic advantage is due to the fact that the DB domain reconstituted with cadmium and mercury is only partially structured and that it only becomes fully structured upon binding to DNA in a binding-coupled-folding manner. To shed further light on this phenomenon, we also measured heat capacity changes associated with the binding of various metal-coordinated DB domains to DNA (Figure 2 and Table 1). It has been previously reported that a large negative heat capacity change (ΔCp) associated with protein-DNA interactions is indicative of protein folding coupled to DNA-binding (Spolar and M.T. Record, 1994). Remarkably, our analysis reveals that while a large negative ΔCp indeed accompanies the binding of mercury-coordinated DB domain to DNA, ΔCp associated with the binding of cadmium-coordinated DB domain is smaller than that observed for DB domain reconstituted with zinc and cobalt. In fact, ΔCp accompanying the binding of cobalt-coordinated DB domain to DNA is similar to that observed for mercury-coordinated DB domain. However, it is important to note that such discrepancies in the values of ΔCp do not necessarily contradict the corresponding enthalpic contributions to the overall free energy of binding of various metal-coordinated DB domains to DNA. On the contrary, it is believed that factors other than protein folding can also contribute toΔCp. In particular, factors such as entrapment of water molecules and counterions within interfacial cavities as well as proton-linked equilibria during the formation of macromolecular complexes may also contribute to heat capacity changes (Cooper et al., 2001; Cooper, 2005). Thus, the large negative values of ΔCp reported here may not solely reflect the folding of DB domain upon binding to DNA but they could also arise from other factors. We have previously shown that the binding of DB domain of ERα to DNA is coupled to proton uptake (Deegan et al., 2010). Interestingly, this phenomenon is not affected by the replacement of zinc with divalent ions of cadmium, mercury and cobalt. As shown in Figure 3, the observed enthalpies for the binding of various metal-reconstituted DB domains to DNA are strongly dependent on buffer, implying that DNA-binding is coupled to proton uptake irrespective of the nature of metal coordination. Our analysis also suggests that the binding of metal-reconstituted DB domains to DNA involves a net uptake of two protons in agreement with our previous study (Deegan et al., 2010).
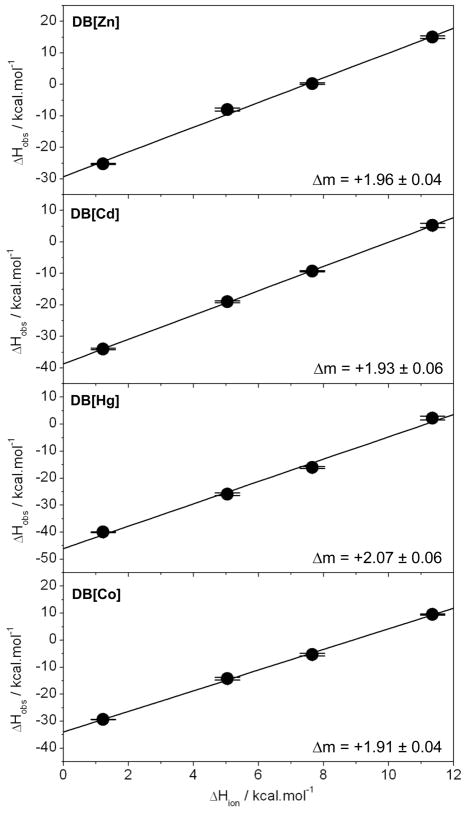
Figure 3.
Dependence of observed enthalpy (ΔHobs) as a function of ionization enthalpy (ΔHion) of various buffers upon the binding of ERE duplex to the DB domain of ERα reconstituted with divalent ions of zinc (DB[Zn]), cadmium (DB[Cd]), mercury (DB[Hg]) and cobalt (DB[Co]). The ΔHion of various buffers used were +1.22 kcal/mol (Phosphate), +5.02 kcal/mol (Hepes), +7.64 kcal/mol (Ticine) and +11.35 kcal/mol (Tris) (Fukada and Takahashi, 1998; Kozlov and Lohman, 2000; Ortiz-Salmeron et al., 2001). The solid lines within each panel represent linear fits to data points and the net change in the number of protons (Δm) uptaken per DB monomer upon binding to DNA was calculated from the corresponding slopes as described earlier (Deegan et al., 2010). Error bars were calculated from at least three independent measurements. All errors are given to one standard deviation.
Importantly, the metal-coordination of cysteine ligands in a tetrahedral arrangement to generate the M2+[Cys]4 metal-protein complex may be important for the proper folding of the DB domain such that it can recognize the ERE duplex in a specific manner. This is further corroborated by the knowledge that the divalent ions of zinc, cadmium, mercury and cobalt are all capable of coordinating their ligands with tetrahedral geometries (Rulisek and Vondrasek, 1998). However, the fact that divalent ions of metals such as nickel and manganese can also adopt tetrahedral geometry implies that the factors other than coordination geometry may also hold key to determining whether a particular divalent metal ion can replace zinc within the zinc fingers of the DB domain. Of particular importance are the ionic radii and internuclear coordinaton distances of various divalent metal ions in complex with their ligands. Interestingly, the ionic radii of hydrated divalent ions of all divalent metal ions analyzed here fall in the 100–150pm range (Marcus, 1988), and there seems to be no correlation between their ionic radii and their ability to replace zinc within the zinc fingers of the DB domain. To what extent the hydration shell, or the extent to which a divalent metal ion becomes hydrated in solution, may be an important determinant of its ability to coordinate a given ligand also remains debatable. In short, it is not clear from our studies as to why some divalent ions can replace zinc within the zinc fingers of the DB domain while others cannot on basis of their physicochemical properties such as coordination geometry, ionic radius and internuclear coordination distance. Our study thus clearly warrants further investigation of the precise mechanisms driving protein-metal interactions.
Substitution of zinc with other divalent metal ions does not affect hydrodynamic properties of the DB domain of ERα
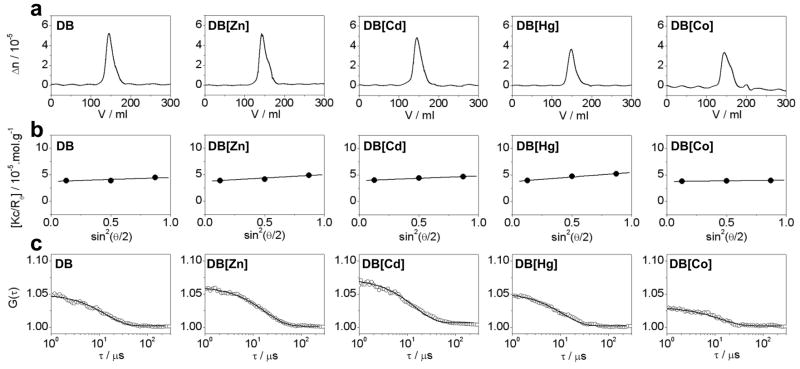
To further investigate how the substitution of zinc with cadmium, mercury and cobalt affects the hydrodynamic properties of the DB domain of ERα, we conducted ALS analysis based on first principles of hydrodynamics with no assumptions (Figure 4). It is generally believed that the DB domain is monomeric in solution and that it only homodimerizes upon binding to DNA (Schwabe et al., 1990; Schwabe et al., 1993). Contrary to this school of thought, our analysis reveals that the DB domain predominantly exists as a homodimer in solution even in the absence of DNA (Table 2). Analysis on a non-reducing SDS-PAGE further confirmed that the ability of DB domain to homodimerize in solution was not an artifact of intermolecular disulfide bridges.
Figure 4.
Representative ALS chromatograms for the DB domain of ERα pre-treated with EDTA to strip divalent zinc ions (DB) and upon reconstitution with divalent ions of zinc (DB[Zn]), cadmium (DB[Cd]), mercury (DB[Hg]) and cobalt (DB[Co]). (a) Elution profiles as monitored by the differential refractive index (Δn) plotted as a function of elution volume (V) for the indicated species. (b) Partial Zimm plots obtained from analytical SLS measurements at a specific protein concentration for indicated species. The solid lines through the data points represent linear fits. (c) Autocorrelation function plots obtained from analytical DLS measurements at a specific protein concentration for the indicated species. The solid lines through the data points represent non-linear least squares fits to Eq [4].
Table 2.
Hydrodynamic parameters obtained from ALS measurements for the DB domain of ERα pre-treated with EDTA to strip divalent zinc ions (DB) and upon reconstitution with divalent ions of zinc (DB[Zn]), cadmium (DB[Cd]), mercury (DB[Hg]) and cobalt (DB[Co])
| Mobs/kD | Dt/μm2.s−1 | Rh/Å | Associativity | |
|---|---|---|---|---|
| DB | 26 ± 1 | 102 ± 3 | 24 ± 1 | Dimer |
| DB[Zn] | 27 ± 1 | 104 ± 9 | 23 ± 2 | Dimer |
| DB[Cd] | 26 ± 1 | 102 ± 5 | 25 ± 1 | Dimer |
| DB[Hg] | 27 ± 1 | 103 ± 4 | 23 ± 1 | Dimer |
| DB[Co] | 26 ± 1 | 108 ± 5 | 23 ± 1 | Dimer |
Mobs, Dt and Rh are respectively the observed molecular mass, translational diffusion coefficient and hydrodynamic radius for each indicated species. Note that the calculated molecular mass of the recombinant DB domain from amino acid sequence alone is 13 kD. Errors were calculated from at least three independent measurements. All errors are given to one standard deviation.
Strikingly, the DB domain not only homodimerizes in the absence of DNA but even the metal-coordination does not appear to be obligatory. Thus, the metal-free DB domain not only appears to behave as a homodimer in solution in a manner akin to when reconstituted with divalent metal ions but its hydrodynamic radius also does not seem to change, implying that the protein possesses a globular fold even in the absence of metal-coordination. It is however important to note that our ALS measurements were conducted on DB domains in the 20–50μM range. Although all DB domains behaved as homodimers within this concentration range, it is conceivable that the DB domain in the absence or presence of metal-coordination may behave as a monomer at protein concentrations in the submicromolar range. It should however be noted that ALS measurements for the DB domain outside the 20–50μM concentration range were not feasible. Thus, while the ALS signal-to-noise ratio starts to get poorer at protein concentrations below 20μM making hydrodynamic analysis less reliable, the DB domain appears to precipitate on the SEC column at concentrations above 50μM. Although it has not been possible at this stage, our future efforts will further explore the ability of DB domain to homodimerize in solution using alternative methodologies such as analytical ultracentrifugation and native mass spectrometry. Nonetheless, these observations suggest strongly that the DB domain may be able to attain a globular fold alone and that metal-coordination may only be essential for DNA-binding. Importantly, these findings are consistent with the knowledge that the LB domain of ERα also behaves as a homodimer in solution (Brandt and Vickery, 1997), suggesting that both the LB and DB domains contribute to the dimerization of ERα in solution (Notides and Nielsen, 1974; Notides et al., 1981).
Substitution of zinc with other divalent metal ions results in subtle secondary and tertiary structural changes within the DB domain of ERα
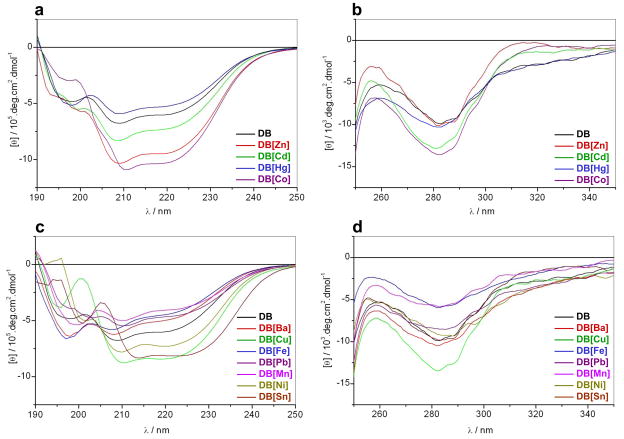
To further understand how the binding of various metals changes secondary and tertiary structural features of the DB domain of ERα, we conducted CD analysis of DB domain in complex with various divalent metal ions (Figure 5). Consistent with our ALS analysis above, the metal-free DB domain indeed displays spectral features in the far-UV region characteristic of a mixed αβ-fold, with bands centered around 210nm and 220nm (Figure 5a). The metal-free DB domain also displays near-UV spectral features characteristic of a globular fold with bands centered around 260nm and 280nm (Figure 5b), due respectively to Phe and Trp/Tyr residues. Upon the addition of divalent ions of zinc and cobalt, the far-UV and near-UV spectra of the DB domain undergo sharp enhancement in intensity, implying that the binding of zinc and cobalt induces substantial folding of the protein. In contrast, addition of cadmium results in only slight changes in the spectral intensities of the DB domain in the far-UV and near UV-regions, suggesting that cadmium-coordination also results in some degree of folding but is not sufficient to lead to fully structured protein. Strikingly, in the presence of mercury, the far-UV and near-UV spectral intensities of the DB domain undergo slight reduction. This salient observation implies that the coordination of mercury has little or no effect on the secondary or tertiary structure of the DB domain in agreement with our thermodynamic data (Table 1). We believe that this is most likely due to the preference of mercury to coordinate its ligands with linear geometry (Rulisek and Vondrasek, 1998). Accordingly, the cysteine ligands within the DB domain may be coordinated by mercury with a linear geometry as opposed to tetrahedral arrangement necessary for its proper folding as observed in the case of coordination with zinc, cobalt and cadmium divalent ions. In light of our thermodynamic data (Table 1), it is conceivable that the binding of DNA causes mercury to switch its coordination from linear to tetrahedral geometry so as to allow the DB domain to undergo proper folding necessary for DNA recognition.
Figure 5.
Representative far-UV (a and c) and near-UV (b and d) CD spectra for the DB domain of ERα pre-treated with EDTA to strip divalent zinc ions (DB) and upon reconstitution with divalent ions of zinc (DB[Zn]), cadmium (DB[Cd]), mercury (DB[Hg]), cobalt (DB[Co]), barium (DB[Ba]), copper (DB[Cu]), iron (DB[Fe]), lead (DB[Pb]), manganese (DB[Mn]), nickel (DB[Ni]) and tin (DB[Sn]). Note that the spectra with divalent metal ions that can regenerate the DB domain with DNA-binding potential are presented in (a–b), while spectra for divalent metal ions that fail to do so in (c–d).
Taken together, our CD data are consistent with the notion that the DB domain may be able to attain a globular fold alone and that metal-coordination may only be essential for DNA-binding. It should also be noted here that the failure of DB domain regenerated with divalent ions of barium, copper, iron, lead, manganese, nickel and tin to bind DNA does not necessarily imply failure to replace zinc within the zinc fingers of the DB domain. Indeed, it has been previously reported that the divalent ions of copper can replace zinc within the zinc fingers of DB and that such substitution is met with profound effects on the physiological action of ERα (Young et al., 1977; Fishman and Fishman, 1988; Hutchens et al., 1992; Predki and Sarkar, 1992). It is thus conceivable that divalent ions of barium, copper, iron, lead, manganese, nickel and tin can replace zinc within the zinc fingers of DB domain but their inability to coordinate cysteine ligands in a tetrahedral arrangement results in improper folding such that the DB domain can no longer recognize the target DNA. This is indeed further supported by our CD measurements of DB domain in complex with divalent metal ions of barium, copper, iron, lead, manganese, nickel and tin (Figures 5c and 5d). Evidently, the addition of these metal divalent ions results in marked changes in both the secondary and tertiary structural features of the DB domain, implying that these metals can also coordinate the protein but in a non-productive manner. In light of these observations, we believe that metals such as barium, copper, iron, lead, manganese, nickel and tin may also be able to compete with zinc for coordinating to the DB domain within living cells with important consequences on the physiological action of ERα.
DB domain of ERα reconstituted with various metals displays distinct spectroscopic properties
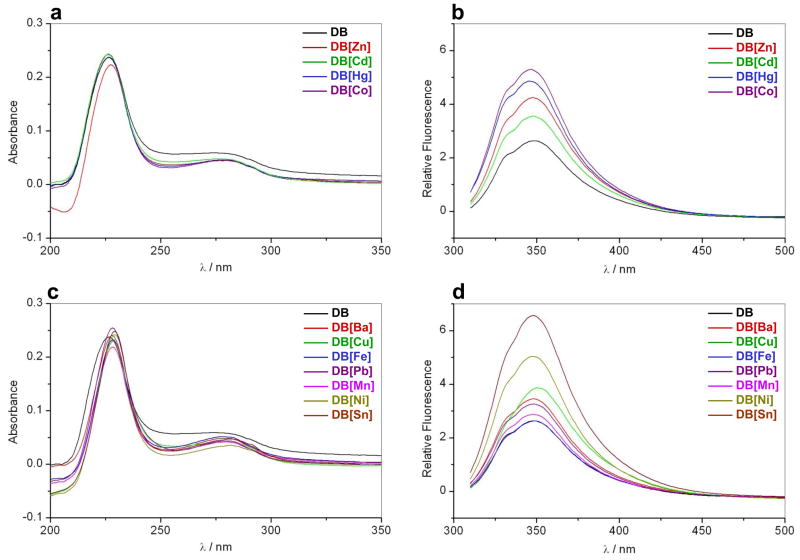
In an attempt to further analyze how reconstitution of DB domain of ERα with various divalent metal ions results in structural changes, we also measured SSA and SSF spectra (Figures 6a and 6b). As expected, the metal-free DB domain displays characteristic absorbance spectral features in the UV region with maxima centered around 225nm and 280nm (Figure 6a), due respectively to peptide bonds and Trp/Tyr/Phe residues. Additionally, the DB domain reconstituted with various metals also displays similar spectroscopic features but the spectral intensity of the 280-nm band appears to undergo reduction, implying that metal-binding most likely induces structural changes within the DB domain, due for example to burial of Trp/Tyr/Phe residues. Such metal-mediated modulation of spectral features is further indicative of specific metal-protein interactions as noted previously (Bal et al., 2003; Kopera et al., 2004). Consistent with our absorbance measurements, the DB domain reconstituted with various metals also displays fluorescence properties distinct from those observed for the metal-free DB domain (Figure 6b). Thus, the increase in fluorescence intensity and the emission wavelength maximum (λmax) undergoing a slight blue-shift in the DB domain reconstituted with various metals relative to metal-free DB domain is indicative of the transfer of Trp to a more hydrophobic environment and thereby suggesting that the protein undergoes conformational changes upon metal-coordination.
Figure 6.
Representative SSA (a and c) and SSF (b and d) spectra for the DB domain of ERα pre-treated with EDTA to strip divalent zinc ions (DB) and upon reconstitution with divalent ions of zinc (DB[Zn]), cadmium (DB[Cd]), mercury (DB[Hg]), cobalt (DB[Co]), barium (DB[Ba]), copper (DB[Cu]), iron (DB[Fe]), lead (DB[Pb]), manganese (DB[Mn]), nickel (DB[Ni]) and tin (DB[Sn]). Note that the spectra with divalent metal ions that can regenerate the DB domain with DNA-binding potential are presented in (a–b), while spectra for divalent metal ions that fail to do so in (c–d).
Interestingly, mercury-coordinated DB domain also undergoes substantial enhancement in fluorescence, implying that coordination of mercury also results in structural changes within the DB domain. However, this observation is at odds with our CD data above, wherein the CD spectral features of mercury-coordinated DB domain did not differ much from those observed for the metal-free DB domain. It is however conceivable that mercury-coordination of cysteine ligands within the DB domain with Cys-Hg2+-Cys linear geometry primarily results in perturbation of environment around Trp, which could also account for the enhancement of fluorescence observed here. Notably, our SSA and SSF measurements also suggest that the addition of divalent ions of barium, copper, iron, lead, manganese, nickel and tin also results in changes in spectroscopic properties of DB domain (Figures 6c and 6d), implying that although these metals cannot regenerate DB domain with DNA-binding potential, they are nonetheless capable of competing with zinc for the zinc fingers in agreement with our CD data.
CONCLUSIONS
In the industrialized world, the environment has come to play an increasing role on human health due to the presence of obnoxious chemicals in a wide variety of sources. In particular, over the past decade or so, it has become clear that metal ions absorbed from various environmental sources can potentiate the transcriptional activity of ERα within the body leading to the development and progression of breast cancer (Garcia-Morales et al., 1994; Martin et al., 2003; Darbre, 2006). Although it is generally believed that such metals up-regulate ERα by virtue of their ability to mimic the action of endogenous estrogens, our data presented here suggest that some of these metals may also be able to replace zinc within the zinc fingers of the DB domain and thereby modulate its binding to the promoters of target genes (Klinge, 2001). In particular, our data suggest that metals such as barium, copper, iron, lead, manganese, nickel and tin may coordinate to cysteine ligands within the DB domain in a manner that it can longer bind to DNA. It is thus quite possible that these metals may influence the physiological action of ERα by virtue of their ability to compete with zinc for coordinating to DB domain though their concentration is likely to be much lower than zinc within living cells. More importantly, given that the hyperactivation of ERα is linked to the genesis of large fractions of breast cancer (Deroo and Korach, 2006; Heldring et al., 2007), deciphering the molecular basis of how environmental metals modulate the transcriptional activity of ERα bears the potential to not only expand our biomedical knowledge but may also lead to the development of novel anti-cancer therapies harboring greater efficacy coupled with low toxicity. Toward this goal, our present study provides mechanistic insights into how environmental metals may replace zinc within the zinc fingers of ERα and thus bears important consequences on understanding its physiological function in human health and disease.
Acknowledgments
This work was supported by funds from the National Institutes of Health (Grant# R01-GM083897) and the USylvester Braman Family Breast Cancer Institute to AF. CBM is a recipient of a postdoctoral fellowship from the National Institutes of Health (Award# T32-CA119929). BJD and AF are members of the Sheila and David Fuente Graduate Program in Cancer Biology at the Sylvester Comprehensive Cancer Center of the University of Miami.
ABBREVIATIONS
- ALS
Analytical light scattering
- CD
Circular dichroism
- DB
DNA-binding
- DLS
Dynamic light scattering
- ERα
Estrogen receptor α
- ERE
Estrogen response element
- ITC
Isothermal titration calorimetry
- LB
Ligand-binding
- MAPK
Mitogen-activated protein kinase
- NR
Nuclear receptor
- PAGE
polyacrylamide gel electrophoresis
- SDS
Sodium dodecyl sulfate
- SEC
Size-exclusion chromatography
- SLS
Static light scattering
- SSA
Steady-state absorbance
- SSF
Steady-state fluorescence
- TA
Trans-activation
- ZF
Zinc finger
References
- Bal W, Schwerdtle T, Hartwig A. Mechanism of nickel assault on the zinc finger of DNA repair protein XPA. Chem Res Toxicol. 2003;16:242–248. doi: 10.1021/tx025639q. [DOI] [PubMed] [Google Scholar]
- Berne BJ, Pecora R. Dynamic Light Scattering. New York: Wiley; 1976. [Google Scholar]
- Blessing H, Kraus S, Heindl P, Bal W, Hartwig A. Interaction of selenium compounds with zinc finger proteins involved in DNA repair. Eur J Biochem. 2004;271:3190–3199. doi: 10.1111/j.1432-1033.2004.04251.x. [DOI] [PubMed] [Google Scholar]
- Brandt ME, Vickery LE. Cooperativity and dimerization of recombinant human estrogen receptor hormone-binding domain. J Biol Chem. 1997;272:4843–4849. doi: 10.1074/jbc.272.8.4843. [DOI] [PubMed] [Google Scholar]
- Brzozowski AM, Pike AC, Dauter Z, Hubbard RE, Bonn T, Engstrom O, Ohman L, Greene GL, Gustafsson JA, Carlquist M. Molecular basis of agonism and antagonism in the oestrogen receptor. Nature. 1997;389:753–758. doi: 10.1038/39645. [DOI] [PubMed] [Google Scholar]
- Carson M. Ribbons 2.0. J Appl Crystallogr. 1991;24:958–961. [Google Scholar]
- Chu B. Laser Light Scattering: Basic Principles and Practice. Boston: Academic; 1991. [Google Scholar]
- Cooper A. Heat capacity effects in protein folding and ligand binding: a re-evaluation of the role of water in biomolecular thermodynamics. Biophys Chem. 2005;115:89–97. doi: 10.1016/j.bpc.2004.12.011. [DOI] [PubMed] [Google Scholar]
- Cooper A, Johnson CM, Lakey JH, Nollmann M. Heat does not come in different colours: entropy-enthalpy compensation, free energy windows, quantum confinement, pressure perturbation calorimetry, solvation and the multiple causes of heat capacity effects in biomolecular interactions. Biophys Chem. 2001;93:215–230. doi: 10.1016/s0301-4622(01)00222-8. [DOI] [PubMed] [Google Scholar]
- Darbre PD. Metalloestrogens: an emerging class of inorganic xenoestrogens with potential to add to the oestrogenic burden of the human breast. J Appl Toxicol. 2006;26:191–197. doi: 10.1002/jat.1135. [DOI] [PubMed] [Google Scholar]
- Darimont BD, Wagner RL, Apriletti JW, Stallcup MR, Kushner PJ, Baxter JD, Fletterick RJ, Yamamoto KR. Structure and specificity of nuclear receptor-coactivator interactions. Genes Dev. 1998;12:3343–3356. doi: 10.1101/gad.12.21.3343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deegan BJ, Bhat V, Seldeen KL, McDonald CB, Farooq A. Genetic variations within the ERE motif modulate plasticity and energetics of binding of DNA to the ERalpha nuclear receptor. Arch Biochem Biophys. 2011 doi: 10.1016/j.abb.2011.01.002. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deegan BJ, Seldeen KL, McDonald CB, Bhat V, Farooq A. Binding of the ERalpha nuclear receptor to DNA is coupled to proton uptake. Biochemistry. 2010;49:5978–5988. doi: 10.1021/bi1004359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deroo BJ, Korach KS. Estrogen receptors and human disease. J Clin Invest. 2006;116:561–570. doi: 10.1172/JCI27987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escriva H, Bertrand S, Laudet V. The evolution of the nuclear receptor superfamily. Essays Biochem. 2004;40:11–26. doi: 10.1042/bse0400011. [DOI] [PubMed] [Google Scholar]
- Evans RM. The steroid and thyroid hormone receptor superfamily. Science. 1988;240:889–895. doi: 10.1126/science.3283939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fishman JH, Fishman J. Copper and endogenous mediators of estradiol action. Biochem Biophys Res Commun. 1988;152:783–788. doi: 10.1016/s0006-291x(88)80106-2. [DOI] [PubMed] [Google Scholar]
- Freedman LP, Luisi BF, Korszun ZR, Basavappa R, Sigler PB, Yamamoto KR. The function and structure of the metal coordination sites within the glucocorticoid receptor DNA binding domain. Nature. 1988;334:543–546. doi: 10.1038/334543a0. [DOI] [PubMed] [Google Scholar]
- Fukada H, Takahashi K. Enthalpy and heat capacity changes for the proton dissociation of various buffer components in 0.1 M potassium chloride. Proteins. 1998;33:159–166. [PubMed] [Google Scholar]
- Garcia-Morales P, Saceda M, Kenney N, Kim N, Salomon DS, Gottardis MM, Solomon HB, Sholler PF, Jordan VC, Martin MB. Effect of cadmium on estrogen receptor levels and estrogen-induced responses in human breast cancer cells. J Biol Chem. 1994;269:16896–16901. [PubMed] [Google Scholar]
- Gottlieb B, Beitel LK, Wu J, Elhaji YA, Trifiro M. Nuclear receptors and disease: androgen receptor. Essays Biochem. 2004;40:121–136. doi: 10.1042/bse0400121. [DOI] [PubMed] [Google Scholar]
- Green A, Parker M, Conte D, Sarkar B. Zinc Finger Proteins: A Bridge Between Transition Metals and Gene Regulation. J Trace Element Exp Med. 1998;11:103–118. [Google Scholar]
- Gurnell M, Chatterjee VK. Nuclear receptors in disease: thyroid receptor beta, peroxisome-proliferator-activated receptor gamma and orphan receptors. Essays Biochem. 2004;40:169–189. doi: 10.1042/bse0400169. [DOI] [PubMed] [Google Scholar]
- Ham J, Parker MG. Regulation of gene expression by nuclear hormone receptors. Curr Opin Cell Biol. 1989;1:503–511. doi: 10.1016/0955-0674(89)90012-4. [DOI] [PubMed] [Google Scholar]
- Hartwig A. Zinc finger proteins as potential targets for toxic metal ions: differential effects on structure and function. Antioxid Redox Signal. 2001;3:625–634. doi: 10.1089/15230860152542970. [DOI] [PubMed] [Google Scholar]
- Hartwig A, Schwerdtle T, Bal W. Biophysical analysis of the interaction of toxic metal ions and oxidants with the zinc finger domain of XPA. Methods Mol Biol. 2010;649:399–410. doi: 10.1007/978-1-60761-753-2_25. [DOI] [PubMed] [Google Scholar]
- Heldring N, Pike A, Andersson S, Matthews J, Cheng G, Hartman J, Tujague M, Strom A, Treuter E, Warner M, Gustafsson JA. Estrogen receptors: how do they signal and what are their targets. Physiol Rev. 2007;87:905–931. doi: 10.1152/physrev.00026.2006. [DOI] [PubMed] [Google Scholar]
- Hutchens TW, Allen MH, Li CM, Yip TT. Occupancy of a C2–C2 type ‘zinc-finger’ protein domain by copper. Direct observation by electrospray ionization mass spectrometry. FEBS Lett. 1992;309:170–174. doi: 10.1016/0014-5793(92)81088-4. [DOI] [PubMed] [Google Scholar]
- Kato S, Endoh H, Masuhiro Y, Kitamoto T, Uchiyama S, Sasaki H, Masushige S, Gotoh Y, Nishida E, Kawashima H, et al. Activation of the estrogen receptor through phosphorylation by mitogen-activated protein kinase. Science. 1995;270:1491–1494. doi: 10.1126/science.270.5241.1491. [DOI] [PubMed] [Google Scholar]
- Klinge CM. Estrogen receptor interaction with estrogen response elements. Nucleic Acids Res. 2001;29:2905–2919. doi: 10.1093/nar/29.14.2905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopera E, Schwerdtle T, Hartwig A, Bal W. Co(II) and Cd(II) substitute for Zn(II) in the zinc finger derived from the DNA repair protein XPA, demonstrating a variety of potential mechanisms of toxicity. Chem Res Toxicol. 2004;17:1452–1458. doi: 10.1021/tx049842s. [DOI] [PubMed] [Google Scholar]
- Koppel DE. Analysis of Macromolecular Polydispersity in Intensity Correlation Spectroscopy. J Chem Phys. 1972;57:4814–4820. [Google Scholar]
- Kozlov AG, Lohman TM. Large contributions of coupled protonation equilibria to the observed enthalpy and heat capacity changes for ssDNA binding to Escherichia coli SSB protein. Proteins Suppl. 2000;4:8–22. doi: 10.1002/1097-0134(2000)41:4+<8::aid-prot20>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- Marcus Y. Ionic Radii in Aqueous Solutions. Chem Rev. 1988;88:1475–1498. [Google Scholar]
- Martin MB, Reiter R, Pham T, Avellanet YR, Camara J, Lahm M, Pentecost E, Pratap K, Gilmore BA, Divekar S, et al. Estrogen-like activity of metals in MCF-7 breast cancer cells. Endocrinology. 2003;144:2425–2436. doi: 10.1210/en.2002-221054. [DOI] [PubMed] [Google Scholar]
- McEwan IJ. Nuclear receptors: one big family. Methods Mol Biol. 2009;505:3–18. doi: 10.1007/978-1-60327-575-0_1. [DOI] [PubMed] [Google Scholar]
- McKenna NJ, Cooney AJ, DeMayo FJ, Downes M, Glass CK, Lanz RB, Lazar MA, Mangelsdorf DJ, Moore DD, Qin J, et al. Minireview: Evolution of NURSA, the Nuclear Receptor Signaling Atlas. Mol Endocrinol. 2009;23:740–746. doi: 10.1210/me.2009-0135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller J, McLachlan AD, Klug A. Repetitive zinc-binding domains in the protein transcription factor IIIA from Xenopus oocytes. Embo J. 1985;4:1609–1614. doi: 10.1002/j.1460-2075.1985.tb03825.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Notides AC, Lerner N, Hamilton DE. Positive cooperativity of the estrogen receptor. Proc Natl Acad Sci U S A. 1981;78:4926–4930. doi: 10.1073/pnas.78.8.4926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Notides AC, Nielsen S. The molecular mechanism of the in vitro 4 S to 5 S transformation of the uterine estrogen receptor. J Biol Chem. 1974;249:1866–1873. [PubMed] [Google Scholar]
- Noy N. Ligand specificity of nuclear hormone receptors: sifting through promiscuity. Biochemistry. 2007;46:13461–13467. doi: 10.1021/bi7018699. [DOI] [PubMed] [Google Scholar]
- Ortiz-Salmeron E, Yassin Z, Clemente-Jimenez MJ, Las Heras-Vazquez FJ, Rodriguez-Vico F, Baron C, Garcia-Fuentes L. Thermodynamic analysis of the binding of glutathione to glutathione S-transferase over a range of temperatures. Eur J Biochem. 2001;268:4307–4314. doi: 10.1046/j.1432-1327.2001.02349.x. [DOI] [PubMed] [Google Scholar]
- Predki PF, Sarkar B. Effect of replacement of “zinc finger” zinc on estrogen receptor DNA interactions. J Biol Chem. 1992;267:5842–5846. [PubMed] [Google Scholar]
- Predki PF, Sarkar B. Metal replacement in “zinc finger” and its effect on DNA binding. Environ Health Perspect. 1994;102(Suppl 3):195–198. doi: 10.1289/ehp.94102s3195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rulisek L, Vondrasek J. Coordination geometries of selected transition metal ions (Co2+, Ni2+, Cu2+, Zn2+, Cd2+, and Hg2+) in metalloproteins. J Inorg Biochem. 1998;71:115–127. doi: 10.1016/s0162-0134(98)10042-9. [DOI] [PubMed] [Google Scholar]
- Schwabe JW, Chapman L, Finch JT, Rhodes D. The crystal structure of the estrogen receptor DNA-binding domain bound to DNA: how receptors discriminate between their response elements. Cell. 1993;75:567–578. doi: 10.1016/0092-8674(93)90390-c. [DOI] [PubMed] [Google Scholar]
- Schwabe JW, Neuhaus D, Rhodes D. Solution structure of the DNA-binding domain of the oestrogen receptor. Nature. 1990;348:458–461. doi: 10.1038/348458a0. [DOI] [PubMed] [Google Scholar]
- Sonoda J, Pei L, Evans RM. Nuclear receptors: decoding metabolic disease. FEBS Lett. 2008;582:2–9. doi: 10.1016/j.febslet.2007.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spolar RS, Record MTJ. Coupling of local folding to site-specific binding of proteins to DNA. Science. 1994;263:777–784. doi: 10.1126/science.8303294. [DOI] [PubMed] [Google Scholar]
- Thiesen HJ, Bach C. Transition metals modulate DNA-protein interactions of SP1 zinc finger domains with its cognate target site. Biochem Biophys Res Commun. 1991;176:551–557. doi: 10.1016/s0006-291x(05)80219-0. [DOI] [PubMed] [Google Scholar]
- Thornton JW. Evolution of vertebrate steroid receptors from an ancestral estrogen receptor by ligand exploitation and serial genome expansions. Proc Natl Acad Sci U S A. 2001;98:5671–5676. doi: 10.1073/pnas.091553298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warnmark A, Treuter E, Wright AP, Gustafsson JA. Activation functions 1 and 2 of nuclear receptors: molecular strategies for transcriptional activation. Mol Endocrinol. 2003;17:1901–1909. doi: 10.1210/me.2002-0384. [DOI] [PubMed] [Google Scholar]
- Wiseman T, Williston S, Brandts JF, Lin LN. Rapid measurement of binding constants and heats of binding using a new titration calorimeter. Anal Biochem. 1989;179:131–137. doi: 10.1016/0003-2697(89)90213-3. [DOI] [PubMed] [Google Scholar]
- Wolfe SA, Nekludova L, Pabo CO. DNA recognition by Cys2His2 zinc finger proteins. Annu Rev Biophys Biomol Struct. 2000;29:183–212. doi: 10.1146/annurev.biophys.29.1.183. [DOI] [PubMed] [Google Scholar]
- Wyatt PJ. Light Scattering and the Absolute Characterization of Macromolecules. Anal Chim Acta. 1993;272:1–40. [Google Scholar]
- Young PC, Cleary RE, Ragan WD. Effect of metal ions on the binding of 17beta-estradiol to human endometrial cytosol. Fertil Steril. 1977;28:459–463. doi: 10.1016/s0015-0282(16)42498-2. [DOI] [PubMed] [Google Scholar]
- Zimm BH. The Scattering of Light and the Radial Distribution Function of High Polymer Solutions. J Chem Phys. 1948;16:1093–1099. [Google Scholar]