Abstract
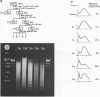
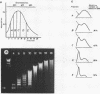
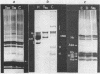
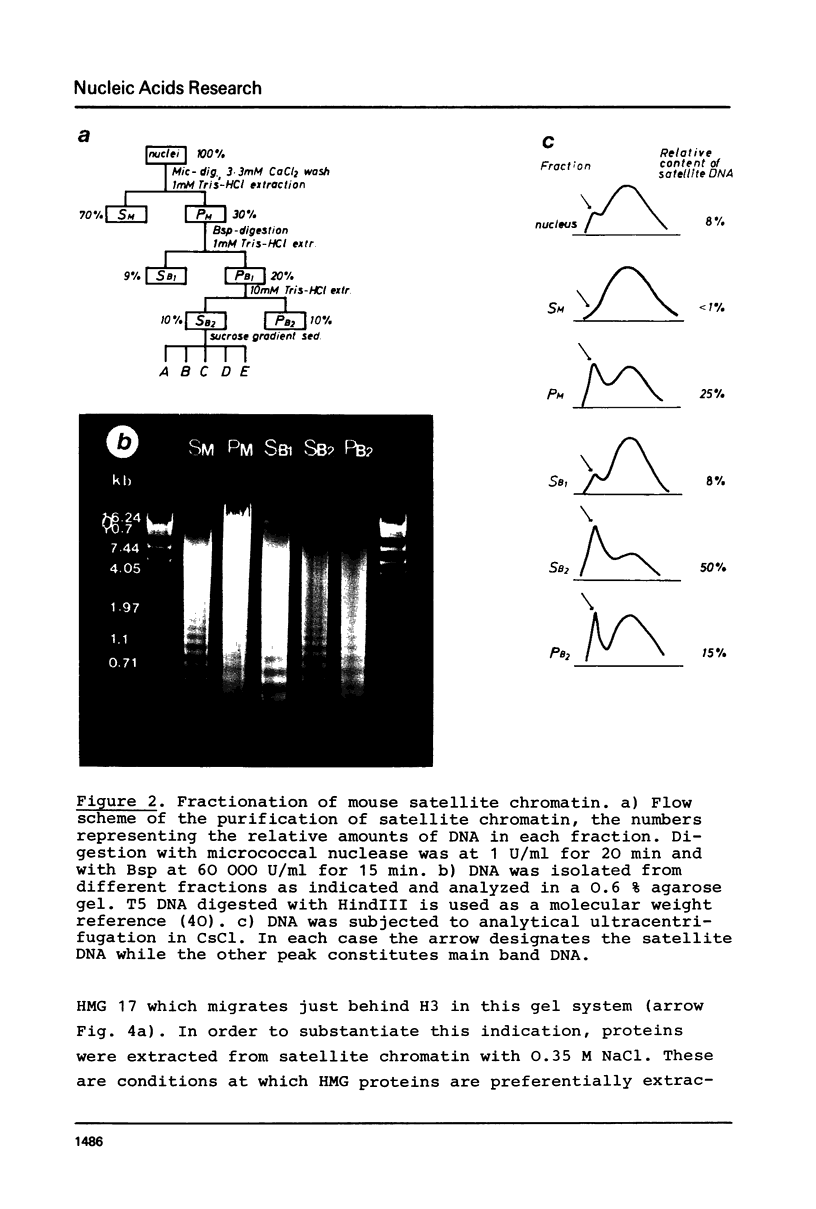
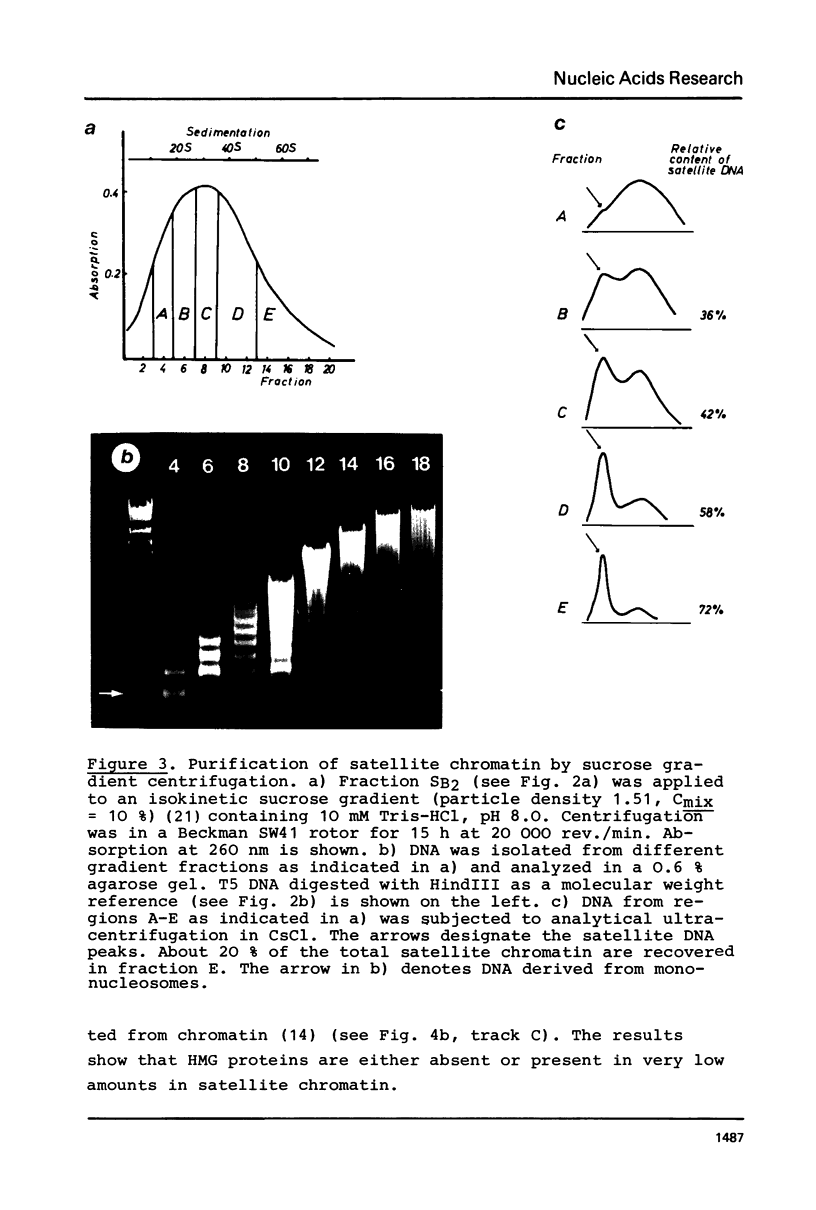
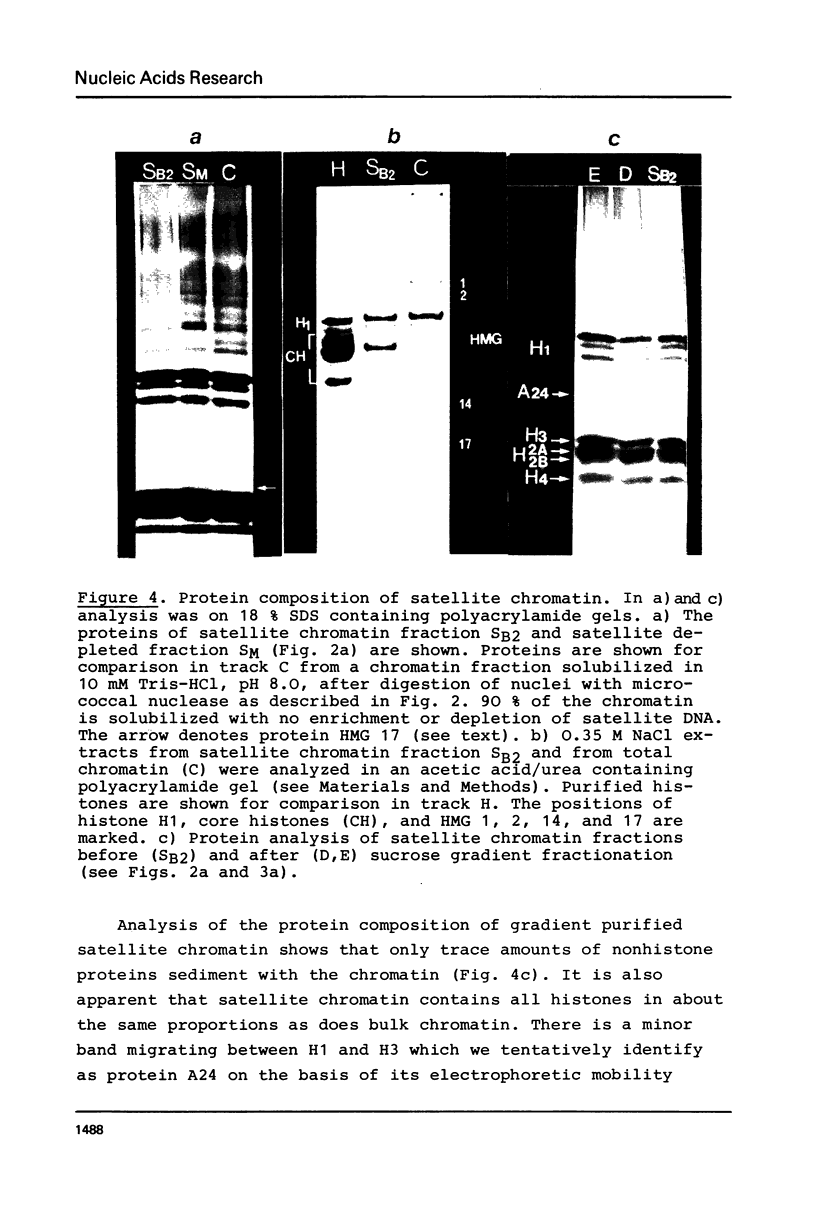
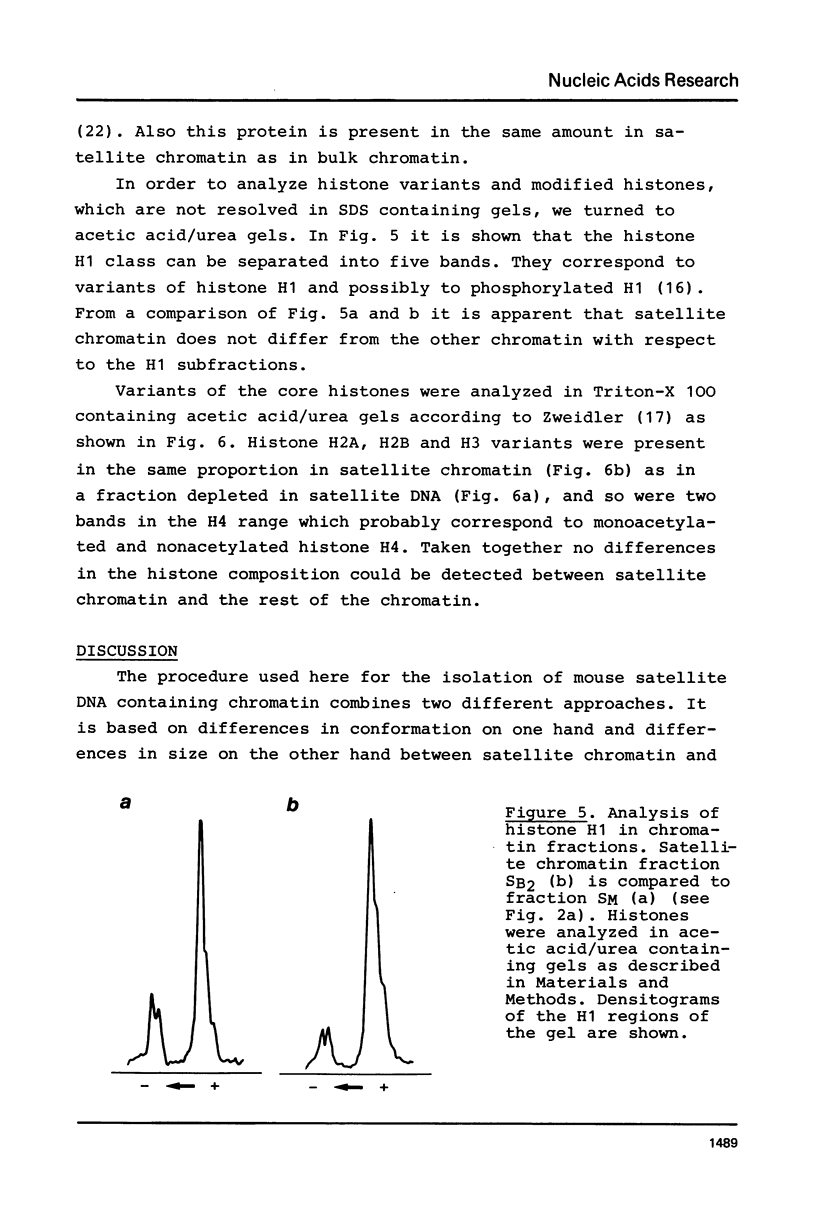
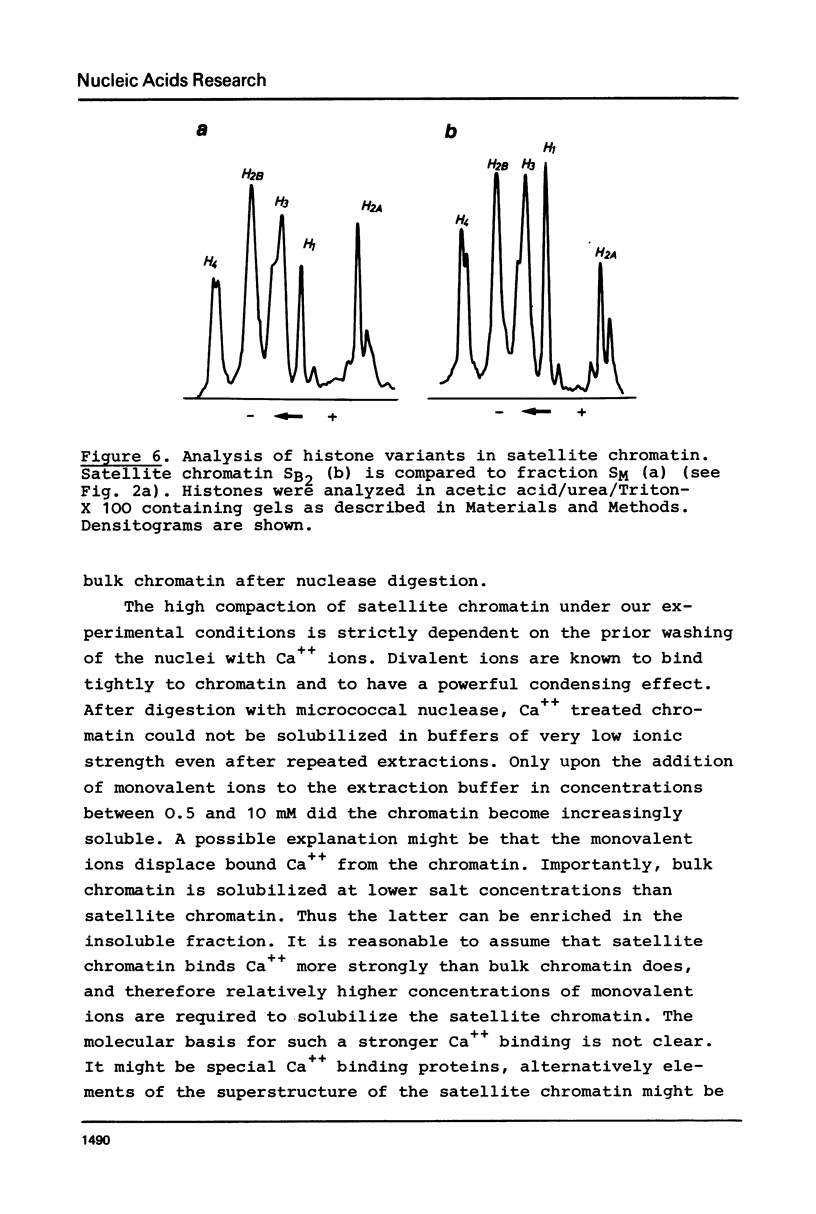
A purification scheme for satellite DNA containing chromatin from mouse liver has been developed. It is based on the highly condensed state of the satellite chromatin and also takes advantage of its resistance to digestion by certain restriction nucleases. Nuclei are first treated with micrococcal nuclease and the satellite chromatin enriched 3-5 fold by extraction of the digested nuclei under appropriate conditions. Further purification is achieved by digestion of the chromatin with a restriction nuclease that leaves satellite DNA largely intact but degrades non-satellite DNA extensively. In subsequent sucrose gradient centrifugation the rapidly sedimenting chromatin contains more than 70% satellite DNA. This material has the same histone composition as bulk chromatin. No significant differences were detected in an analysis of minor histone variants. Nonhistone proteins are present only in very low amounts in the satellite chromatin fraction, notably the HMG proteins are strongly depleted.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Baker W. K. Position-effect variegation. Adv Genet. 1968;14:133–169. [PubMed] [Google Scholar]
- Bernstine E. G. Satellite DNA content of chromatin fractions isolated from Eco R1-digested mouse liver nuclei. Exp Cell Res. 1978 Apr;113(1):205–208. doi: 10.1016/0014-4827(78)90101-5. [DOI] [PubMed] [Google Scholar]
- Billings P. C., Orf J. W., Palmer D. K., Talmage D. A., Pan C. G., Blumenfeld M. Anomalous electrophoretic mobility of Drosophila phosphorylated H1 histone: is it related to the compaction of satellite DNA into heterochromatin? Nucleic Acids Res. 1979;6(6):2151–2164. doi: 10.1093/nar/6.6.2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brutlag D. L. Molecular arrangement and evolution of heterochromatic DNA. Annu Rev Genet. 1980;14:121–144. doi: 10.1146/annurev.ge.14.120180.001005. [DOI] [PubMed] [Google Scholar]
- Goodwin G. H., Nicolas R. H., Johns E. W. An improved large scale fractionation of high mobility group non-histone chromatin proteins. Biochim Biophys Acta. 1975 Oct 20;405(2):280–291. doi: 10.1016/0005-2795(75)90094-x. [DOI] [PubMed] [Google Scholar]
- Greene P. J., Heyneker H. L., Bolivar F., Rodriguez R. L., Betlach M. C., Covarrubias A. A., Backman K., Russel D. J., Tait R., Boyer H. W. A general method for the purification of restriction enzymes. Nucleic Acids Res. 1978 Jul;5(7):2373–2380. doi: 10.1093/nar/5.7.2373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath P., Hörz W. The compaction of mouse heterochromatin as studied by nuclease digestion. FEBS Lett. 1981 Nov 2;134(1):25–28. doi: 10.1016/0014-5793(81)80542-x. [DOI] [PubMed] [Google Scholar]
- Hsieh T., Brutlag D. L. A protein that preferentially binds Drosophila satellite DNA. Proc Natl Acad Sci U S A. 1979 Feb;76(2):726–730. doi: 10.1073/pnas.76.2.726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyde J. E., Igo-Kemenes T., Zachau H. G. The non-histone proteins of the rat liver nucleus and their distribution amongst chromatin fractions as produced by nuclease digestion. Nucleic Acids Res. 1979 Sep 11;7(1):31–48. doi: 10.1093/nar/7.1.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hörz W., Zachau H. G. Characterization of distinct segments in mouse satellite DNA by restriction nucleases. Eur J Biochem. 1977 Mar 1;73(2):383–392. doi: 10.1111/j.1432-1033.1977.tb11329.x. [DOI] [PubMed] [Google Scholar]
- Hörz W., Zachau H. G. Deoxyribonuclease II as a probe for chromatin structure. I. Location of cleavage sites. J Mol Biol. 1980 Dec 15;144(3):305–327. doi: 10.1016/0022-2836(80)90093-5. [DOI] [PubMed] [Google Scholar]
- Igo-Kemenes T., Greil W., Zachau H. G. Prepartation of soluble chromatin and specific chromatin fractions with restriction nucleases. Nucleic Acids Res. 1977 Oct;4(10):3387–3400. doi: 10.1093/nar/4.10.3387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johns E. W. The isolation and purification of histones. Methods Cell Biol. 1977;16:183–203. doi: 10.1016/s0091-679x(08)60100-4. [DOI] [PubMed] [Google Scholar]
- Kiss A., Sain B., Csordás-Tòth E., Venetianer P. A new sequence-specific endonuclease (Bsp) from Bacillus sphaericus. Gene. 1977 Jul;1(5-6):323–329. doi: 10.1016/0378-1119(77)90037-3. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Levinger L., Barsoum J., Varshavsky A. Two-dimensional hybridization mapping of nucleosomes. comparison of DNA and protein patterns. J Mol Biol. 1981 Mar 5;146(3):287–304. doi: 10.1016/0022-2836(81)90389-2. [DOI] [PubMed] [Google Scholar]
- Lipchitz L., Axel R. Restriction endonuclease cleavage of satellite DNA in intact bovine nuclei. Cell. 1976 Oct;9(2):355–364. doi: 10.1016/0092-8674(76)90125-2. [DOI] [PubMed] [Google Scholar]
- Manuelidis L. A simplified method for preparation of mouse satellite DNA. Anal Biochem. 1977 Apr;78(2):561–568. doi: 10.1016/0003-2697(77)90118-x. [DOI] [PubMed] [Google Scholar]
- Mathew C. G., Goodwin G. H., Igo-Kemenes T., Johns E. W. The protein composition of rat satellite chromatin. FEBS Lett. 1981 Mar 9;125(1):25–29. doi: 10.1016/0014-5793(81)80988-x. [DOI] [PubMed] [Google Scholar]
- Mathew C. G., Goodwin G. H., Johns E. W. Quantitative analysis of non-histone chromosomal proteins HMG 14 and HMG 17 by polyacrylamide gel electrophoresis. J Chromatogr. 1980 Sep 26;198(1):80–83. doi: 10.1016/s0021-9673(00)81200-5. [DOI] [PubMed] [Google Scholar]
- Mazrimas J. A., Balhorn R., Hatch F. T. Separation of satellite DNA chromatin and main band DNA chromatin from mouse brain. Nucleic Acids Res. 1979 Oct 25;7(4):935–946. doi: 10.1093/nar/7.4.935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarty K. S., Jr, Vollmer R. T., McCarty K. S. Improved computer program data for the resolution and fractionation of macromolecules by isokinetic sucrose density gradient sedimentation. Anal Biochem. 1974 Sep;61(1):165–183. doi: 10.1016/0003-2697(74)90343-1. [DOI] [PubMed] [Google Scholar]
- Merril C. R., Dunau M. L., Goldman D. A rapid sensitive silver stain for polypeptides in polyacrylamide gels. Anal Biochem. 1981 Jan 1;110(1):201–207. doi: 10.1016/0003-2697(81)90136-6. [DOI] [PubMed] [Google Scholar]
- Musich P. R., Brown F. L., Maio J. J. Subunit structure of chromatin and the organization of eukaryotic highly repetitive DNA: nucleosomal proteins associated with a highly repetitive mammalian DNA. Proc Natl Acad Sci U S A. 1977 Aug;74(8):3297–3301. doi: 10.1073/pnas.74.8.3297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omori A., Igo-Kemenes T., Zachau H. G. Different repeat lengths in rat satellite I DNA containing chromatin and bulk chromatin. Nucleic Acids Res. 1980 Nov 25;8(22):5363–5375. doi: 10.1093/nar/8.22.5363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seale R. L. Rapid turnover of the histone-ubiquitin conjugate, protein A24. Nucleic Acids Res. 1981 Jul 10;9(13):3151–3158. doi: 10.1093/nar/9.13.3151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takatsuka Y., Kohno M., Higashi K., Hirano H., Sakamoto Y. Redistribution of chromatin containing ribosomal cistrons during liver regeneration. Exp Cell Res. 1976 Nov;103(1):191–199. doi: 10.1016/0014-4827(76)90255-x. [DOI] [PubMed] [Google Scholar]
- Zachau H. G., Igo-Kemenes T. Face to phase with nucleosomes. Cell. 1981 Jun;24(3):597–598. doi: 10.1016/0092-8674(81)90084-2. [DOI] [PubMed] [Google Scholar]
- Zweidler A. Resolution of histones by polyacrylamide gel electrophoresis in presence of nonionic detergents. Methods Cell Biol. 1978;17:223–233. [PubMed] [Google Scholar]
- von Gabain A., Hayward G. S., Bujard H. Physical mapping of the HindIII, EcoRI, Sal and Sma restriction endonuclease cleavage fragments from bacteriophage T5 DNA. Mol Gen Genet. 1976 Feb 2;143(3):279–290. doi: 10.1007/BF00269404. [DOI] [PubMed] [Google Scholar]