Abstract
Signal transduction pathways are tightly controlled by positive and negative regulators. We have previously identified Odin (also known as ankyrin repeat and sterile alpha motif domain containing 1A; gene symbol AKNS1A) as a negative regulator of growth factor signaling; however, the mechanisms through which Odin regulates these pathways remain to be elucidated. To determine how Odin negatively regulates growth factor signaling, we undertook a proteomic approach to systematically identify proteins that interact with Odin using the SILAC strategy. In this study, we identified 18 molecules that were specifically associated in a protein complex with Odin. Our study established that the complete family of 14-3-3 proteins occur in a protein complex with Odin, which is also supported by earlier reports that identified a few members of the 14-3-3 family as Odin interactors. Among the novel protein interactors of Odin were CD2-associated protein, SH3 domain kinase binding protein 1 and DAB2 interacting protein. We confirmed 8 of the eighteen interactions identified in the Odin protein complex by co-immunoprecipitation experiments. Finally, a literature-based network analysis revealed that Odin interacting partners are involved in various cellular processes, some of which are key molecules in regulating receptor endocytosis.
Keywords: Odin, interacting proteins, SILAC, receptor tyrosine kinase
1. Introduction
Receptor tyrosine kinases (RTKs) are a large family of transmembrane receptors that contain a tyrosine kinase domain in their cytoplasmic domains. Oligomerization of RTKs induces transphosphorylation of their cytoplasmic domains, which leads to activation of their tyrosine kinase activity. Phosphorylated tyrosine residues on downstream molecules serve as docking sites for other cytoplasmic signaling molecules containing phosphotyrosine binding domains such as SH2 and PTB domains to form multimeric protein complexes.
The activation of RTKs induced by ligand binding is tightly controlled by ‘positive’ and ‘negative’ regulatory mechanisms. One such negative mechanism is removal of activated RTKs from the cell surface by the process of endocytosis in which the Cbl family of E3 ubiquitin ligases and other adapter proteins play a key role. Cbl contains an SH2 domain that binds to phosphotyrosine residues on activated RTKs and a RING finger domain that is responsible for ubiquitination of the RTKs. Cbl also serves as a scaffold protein to recruit other adapter proteins such as CD2-associated protein (CD2AP) and SH3 domain kinase binding protein 1 (SH3KBP1) in close proximity of RTKs. Endophilins, pre-associated with SH3KBP1, regulate clathrin-coated vesicles that can induce negative curvature and invagination of the plasma membrane during the early step of RTK internalization. This mechanism has been observed during internalization of several RTKs, including EGFR, PDGFR, c-Met and c-Kit [1–3]. Internalized RTKs are sorted into multivesicular bodies and targeted for endosomal degradation. Overall, the negative regulatory pathways of RTKs that attenuate and terminate signals have not been studied well.
Using a mass spectrometry-based proteomic approach, we previously identified a number of signaling molecules undergoing tyrosine phosphorylation in the EGFR signaling pathway, including Odin [4–6]. Odin is a ubiquitously expressed 130 kDa cytosolic protein that contains six ankyrin repeat domains at its N-terminus, two sterile alpha motifs (SAM) and a phosphotyrosine binding domain (PTB) at its C-terminus. Our group has previously shown that overexpression of Odin causes inhibition of c-Fos promoter activity and that microinjection of Odin cDNA into NIH3T3 cells inhibits PDGF-induced mitogenesis [5]. We have also examined the effects of loss of expression of Odin by generating Odin-deficient mice. Although, Odin-deficient mice do not display any obvious phenotype, mouse embryonic fibroblasts (MEFs) generated from these mice exhibit a hyperproliferative phenotype compared to wild-type-derived MEFs [7] implicating Odin as a negative regulator in growth factor signaling. More recently, Odin was identified as a downstream scaffold protein in EphA receptor signaling and suggested to play a pivotal role in EphA receptor signaling [8, 9]. However, the molecular mechanisms by which Odin affects cellular proliferation/growth mediated by RTK signaling are still poorly characterized.
Stable isotope labeling by amino acids in cell culture (SILAC) has been previously used to identify specific protein-protein interactions [10–12]. The advantages of the SILAC method are that it not only uses mass spectrometry for comprehensive identification of protein complexes but can also allows differentiation of true interacting proteins from proteins that bind non-specifically. To better understand the function of Odin, we employed a SILAC-based proteomic approach to characterize the interactome of Odin in activated EGFR signaling. Our quantitative mass spectrometric analysis identified 18 out of 193 proteins as true interacting partners of Odin. Further, Western blotting results confirmed eight protein-protein interactions. By searching Human Protein Reference Database (HPRD) [13, 14] for known interactions among 18 novel protein interactors of Odin, we constructed an interaction network of the Odin protein complex, in which several Odin interacting partners also bind to each other. This literature-based functional analysis also revealed that proteins involved in endocytosis are enriched in the Odin protein complex suggesting that Odin may play a role in endocytosis of EGFR as a negative regulator.
2. Materials and Methods
2.1. Reagents
Stable isotope containing amino acids, 13 C6-arginine and 13C6-lysine, were purchased from Cambridge Isotope Laboratories (Andover, MA, USA). Arginine and lysine-free Dulbecco’s modified Eagle’s medium (DMEM), fetal bovine serum (FBS) and antibiotics (Pen Strep) were purchased from Invitrogen (Carlsbad, CA, USA). Protein G agarose beads, anti-FLAG M2 affinity gel, mouse monoclonal anti-FLAG M2-Peroxidase (HRP) antibody and FLAG peptides were purchased from Sigma-Aldrich (St. Louis, MO, USA). Sequencing grade modified porcine trypsin was purchased from Promega (Madison, WI, USA). Antibodies against 14-3-3γ (sc-731), 14-3-3ζ (sc-1019), 14-3-3ε (sc-1020), CD2AP (sc-9137), talin (sc-7534), heat shock 70kDa protein 9 (GRP75, sc-13967) and EGFR (sc-03) were purchased from Santa Cruz Biotechnology (Santa Cruz, CA, USA). Antibodies against SH3KBP1 (CIN85, Cat#231006) were purchased from CALBIOCHEM. Antibodies against ARHGAP10 (PSGAP) were gifts from Dr. Wen-Cheng Xiong [15]. Wild type (WT) and kinase dead mutant (KD, K745M) EGFR were gifts from Dr. Philip Cole.
2.2. Stable isotope labeling by amino acids in cell culture
Two sets of human embryonic kidney 293T (HEK 293T) cells were cultured in Dulbecco’s modified Eagle’s medium containing light (12C6-arginine and 12C6-lysine) or heavy (13C6-arginine and 13C6-lysine) amino acid supplemented with 10% FBS plus antibiotics, separately. For each set, 20 of 10-cm dishes of HEK 293T cells were used. The set grown in light medium was transfected with human EGF receptor and empty vector (pCMV-Tag4A) while the set grown in heavy medium was transfected with human EGF receptor and FLAG tagged Odin (in pCMV-Tag4A) using a Lipofectamine 2000 transient transfection kit (Invitrogen). Twenty four hours after transfection, both sets of HEK 293T cells were starved for 18 hours in serum-free DMEM and lysed in lysis buffer (50 mM Tris pH 7.4, 150 mM NaCl, 1% N-octylglucoside, 1 mM sodium orthovanadate) in the presence of cocktail protease inhibitors (Roche).
2.3. Immunoprecipitation for mass spectrometry analysis
The protein concentration of the lysates from both sets of cells was measured by Lowry method. Equal amounts of light and heavy lysates were precleared using Protein G-agarose beads (Sigma), filtered through a 0.45 μm sterile filter and incubated with anti-FLAG affinity gel for 6 hours, separately. After washing, the immunoprecipitates from both sets were mixed and the bound proteins were eluted using 1 mg/ml FLAG peptides (Sigma). The eluates were dialyzed, concentrated and resolved by 10% SDS-PAGE. The gel was stained using a colloidal blue staining kit (Invitrogen).
2.4. Liquid chromatography tandem mass spectrometry and data analysis
The colloidal blue stained gel was excised into 36 bands and each band was processed using an in-gel trypsin digestion protocol as described previously [16]. The resulting peptides were extracted and analyzed by reversed phase liquid chromatography tandem mass spectrometry (LC-MS/MS). Briefly, the peptides were injected into an in-house prepared trap column (2 cm long, 75 μm inner diameter) packed with C18 particles (ODS-A YMC, 5–10 μm, 120 Å), using an Agilent 1100 autosampler (Agilent Technologies, Palo Alto, CA). The peptides were eluted from the trap column onto an analytical column (2 cm long, 75 μm inner diameter) packed with C18 particles (ODS-A YMC, 5 μm, 120 Å), using a 5–60% acetonitrile gradient and analyzed on a QSTAR Pulsar mass spectrometer (Applied Biosystems/MDS Sciex, Foster City, CA). The mass spectrometric data were searched by the Mascot search engine (Matrix Science, Boston, MA) against the RefSeq database (version 22) downloaded from National Center for Biotechnology Information. The search parameters included a maximum of one missed cleavage; carbamidomethylation of cysteine as a fixed modification; N-terminal acetylation; oxidation of methionine, phosphorylation of serine, threonine and tyrosine as variable modifications; monoisotopic peptide tolerance of ±0.5 Da; MS/MS tolerance of ±0.3 Da. For all protein identifications, an FDR cutoff of 1% was used. Fold changes of peptides were quantitated by using MSQuant (version 1.4.3a39) [17] and further verified by manual interpretation of MS spectra.
2.4. Immunoprecipitation and Western blot analysis
HEK 293T cells were transfected with empty vector or Odin expression vector using a Lipofectamine 2000 transient transfection kit (Invitrogen). Twenty four hours after transfection, cells were starved for 18 hours in serum-free DMEM and lysed in lysis buffer. Cell lysates were subjected to immunoprecipitation by incubation with anti-FLAG affinity gel for 6 hours. The immunoprecipitates were washed three times and boiled in SDS-PAGE sample buffer with β-mercaptoethanol for 5 min. The eluted immunoprecipitates were resolved by NuPAGE 4–12% Bis-Tris gel (Invitrogen). Proteins in the gel were transferred onto nitrocellulose membrane. After blocking with 5% nonfat dry milk in PBS-T buffer, the membranes were incubated with the relevant primary antibodies overnight at 4°C and probed using the corresponding HRP-conjugated secondary antibodies. All films were developed with ECL reagents (Amersham Bioscience).
3. Results and Discussion
3.1 Odin undergoes tyrosine phosphorylation in receptor tyrosine kinase signaling
Odin has previously been shown to undergo tyrosine phosphorylation upon EGF or PDGF stimulation and to play an inhibitory role in PDGF-mediated cellular proliferation [5, 7]. We first examined the tyrosine phosphorylation of Odin in HEK 293T cells in response to RTK signaling. Odin was coexpressed with wild type (WT) or kinase dead (KD, K745M) EGFR by transient transfection. As shown in Figure 1A, overexpression of WT, but not KD, EGFR induced changes in the tyrosine phosphorylation profile of the whole cell lysates. Odin was subjected to immunoprecipitation and the tyrosine phosphorylation status of Odin was examined by Western blotting with anti-phosphotyrosine antibodies. As shown in Figure 1B, and consistent with our previous data [5], WT but not KD EGFR induced tyrosine phosphorylation of Odin. These results confirmed that Odin was phosphorylated on tyrosine residues in activated RTK signaling. We used this system to study the interactome of tyrosine phosphorylated Odin in EGFR signaling.
Figure 1.
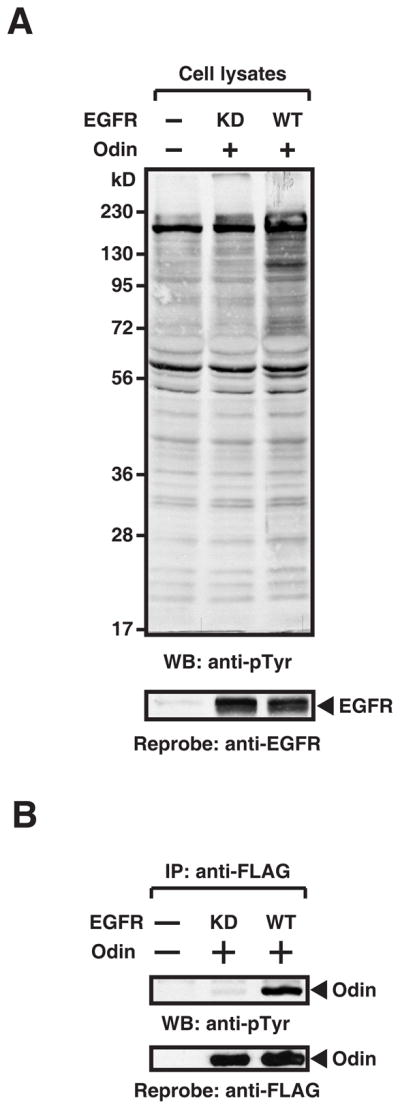
Tyrosine phosphorylation of Odin is induced by EGFR signaling. (A) Wild type (WT) but not kinase dead (KD) EGFR, induces tyrosine phosphorylation. Tyrosine phosphorylation status of HEK 293T cells expressing proteins the indicated constructs was assessed by Western blotting with anti-phosphotyrosine antibodies (4G10). The level of expression of WT and KD EGFR was determined by using anti-EGFR antibodies (bottom panel). (B) Odin is tyrosine phosphorylated in WT but not KD EGFR-expressing cells. Odin was immunoprecipitated from HEK 293T cells expressing the EGFR constructs as indicated in the figure. Tyrosine phosphorylation of Odin was assessed by Western blotting with anti-phosphotyrosine antibodies (4G10) and the total amount of Odin was detected by reprobing the membrane with anti-FLAG antibodies.
3.2. A proteomic screen of Odin interacting partners using the SILAC strategy
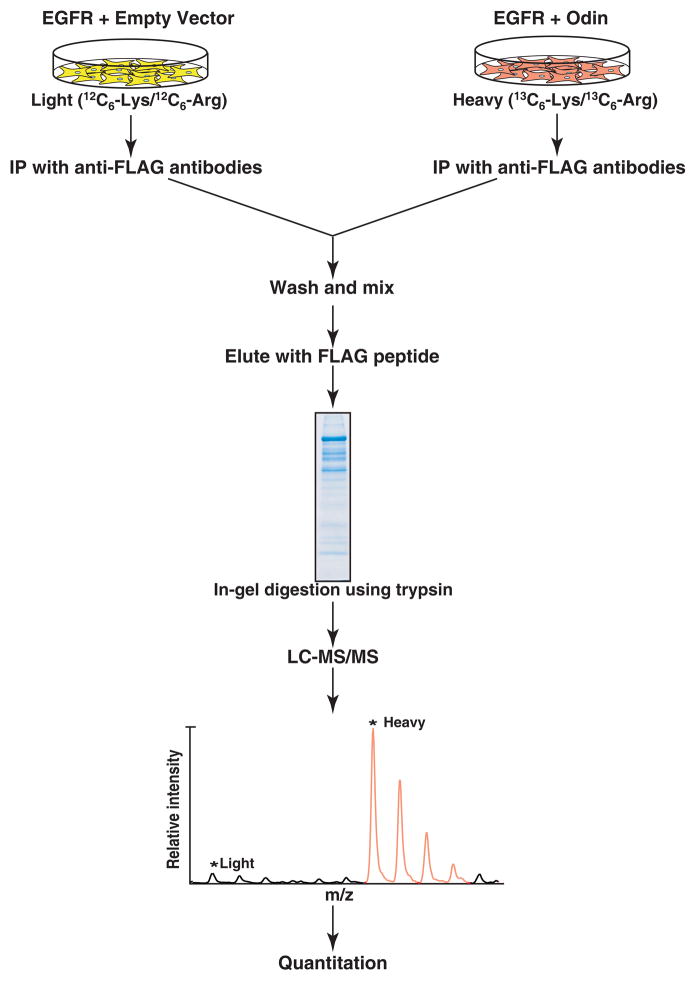
MS-based identification of protein complexes, although a powerful approach to analyze protein-protein interactions, can generate false positive results because proteins non-specifically bound to antibodies or agarose matrix usually co-purify with the ‘bait’ protein. To selectively identify proteins that interact with Odin, we employed a SILAC-based quantitative proteomic approach to carry out one affinity purification experiment for the Odin protein complex. The basic principle of this approach is illustrated in Figure 2. Two populations of HEK 293T cells were grown in either light or heavy medium. Cells grown in the heavy medium were transfected with Odin while those grown in the light medium were transfected with empty vector. Both populations of cells were transfected with EGFR to activate the EGF signaling pathway. Twenty four hours after transfection, proteins occurring in a complex with Odin were isolated by immunoprecipitation and resolved by SDS-PAGE as described in the Materials and Methods section. The proteins were subjected to in-gel digestion followed by LC-MS/MS analysis as described in the Materials and Methods section.
Figure 2.
A schematic illustration of the SILAC methodology to identify proteins in a protein complex with Odin. Cells grown in light medium were transfected with human EGFR and empty vector as control while cells grown in heavy medium were transfected with human EGFR and FLAG-tagged Odin. Cell lysates were subjected to immunoprecipitation using anti-FLAG antibodies. After washing, the immunoprecipitates were mixed, and the bound proteins were eluted by FLAG peptides. The proteins were resolved by SDS-PAGE. The gel was stained and the protein bands excised, digested with trypsin, and analyzed by LC-MS/MS. The absence of light MS ion peak indicates a true protein interactor of Odin.
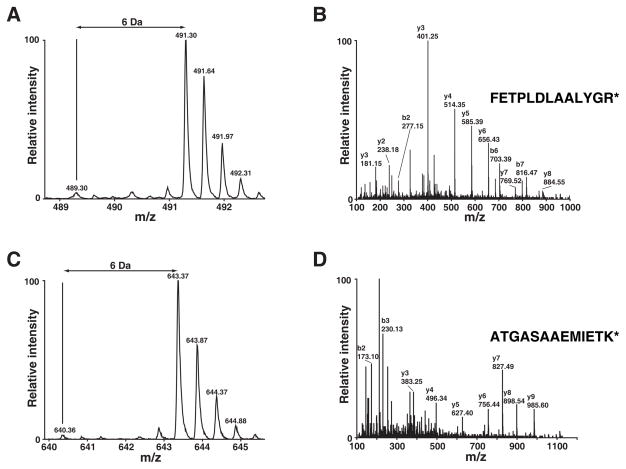
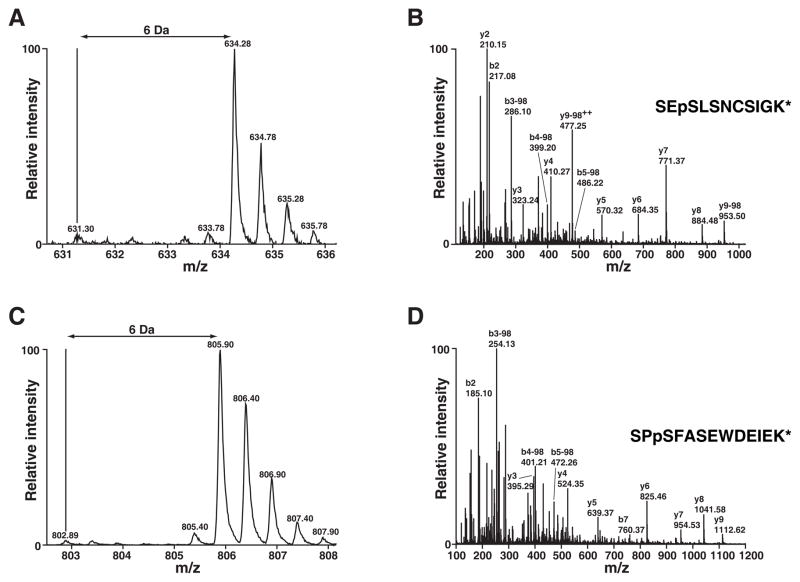
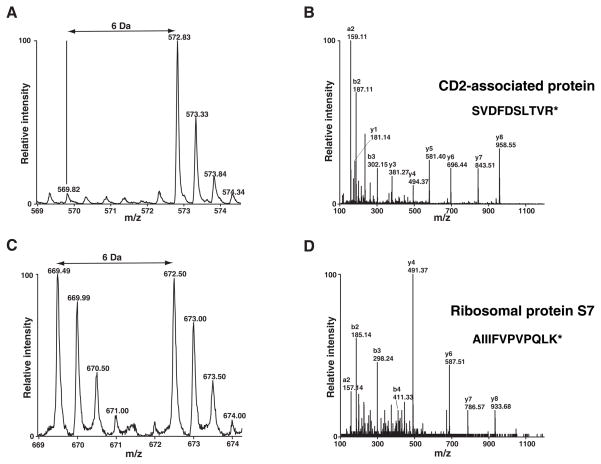
The acquired MS spectra were searched against NCBI RefSeq databases using the Mascot search engine. By using 1% FDR as a cutoff, we identified 927 unique peptides that led to the identification 194 proteins including Odin in the immunoprecipitates after affinity purification of Odin (Supplementary Table 1). As expected and shown in Figure 3, Odin was completely labeled with heavy amino acids as evidenced by the lack of light version of peptides. Additionally, two known phosphorylation sites on Odin (Ser 647/Ser 663) [18, 19] were also detected (Figure 4). By using MSQuant, we carried out relative quantitation analysis of each peptide (Supplementary Table 1) and obtained the quantitation of the corresponding proteins (Supplementary Table 2). We only considered proteins where only the heavy amino acid labeled peptides were detected as components of the Odin protein complex and ignored cases where an observable signal existed for the light amino acid labeled peptides. The proteins that satisfied our criteria are listed in Table 1. The MS spectrum of one representative peptide from one protein in this category, CD2 associated protein, is shown in Figure 5A and its MS/MS spectrum in Figure 5B. Proteins where both heavy and light amino acid labeled peptides (Supplementary Table 2) were detected are likely to be contaminating proteins which bind to antibodies and/or the agarose matrix non-specifically. The MS spectrum of one representative peptide from one protein in this category, ribosomal protein S7, is shown in Figure 5C and its MS/MS spectrum in Figure 5D. We also identified proteins where only light amino acid labeled peptides were detected (Supplementary Table 1 and 2). These proteins are exogenous contaminating proteins and are mostly keratins.
Figure 3.
Odin expression is confirmed by heavy SILAC label. MS spectra of two representative peptides from Odin are shown in panels A and C. Panels B and D show the corresponding MS/MS spectra along with the peptide sequence. * indicates 13C6-Lys or 13C6-Arg.
Figure 4.
Two serine phosphorylation sites identified in Odin. MS spectra of two phosphopeptides from Odin are shown in panels A and C. Panels B and D show the corresponding MS/MS spectra along with the peptide sequence. pS refers to phosphoserine. * indicates 13C6-Lys or 13C6-Arg.
Table 1.
A list of proteins identified as components of the Odin protein complex
| Gene Symbol | Accession Number | Protein | |
|---|---|---|---|
| 1 | YWHAG | gi|21464101 | Tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, gamma polypeptide (14-3-3 gamma) |
| 2 | YWHAZ | gi|4507953 | Tyrosine 3/tryptophan 5-monooxygenase activation protein, zeta polypeptide (14-3-3 zeta) |
| 3 | YWHAB | gi|4507949 | Tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, beta polypeptide (14-3-3 beta) |
| 4 | YWHAH | gi|4507951 | Tyrosine 3/tryptophan 5-monooxygenase activation protein, eta polypeptide (14-3-3 eta) |
| 5 | YWHAE | gi|5803225 | Tyrosine 3/tryptophan 5-monooxygenase activation protein, epsilon polypeptide (14-3-3 epsilon) |
| 6 | SFN | gi|5454052 | Stratifin (14-3-3 sigma) |
| 7 | SH3KBP1 | gi|13994242 | SH3-domain kinase binding protein 1 (CIN85) |
| 8 | CD2AP | gi|11321634 | CD2-associated protein |
| 9 | RASAL2 | gi|25121936 | RAS protein activator like 2 |
| 10 | YWHAQ | gi|5803227 | Tyrosine 3/tryptophan 5-monooxygenase activation protein, theta polypeptide (14-3-3 theta) |
| 11 | CAPZB | gi|4826659 | F-actin capping protein beta subunit |
| 12 | DAB2IP | gi|94967023 | DAB2 interacting protein |
| 13 | TLN2 | gi|22035665 | Talin 2 |
| 14 | GART | gi|4503915 | Phosphoribosylglycinamide formyltransferase, phosphoribosylglycinamide synthetase, phosphoribosylaminoimidazole synthetase |
| 15 | VAPA | gi|94721250 | Vesicle-associated membrane protein (VAMP)-associated protein A |
| 16 | ARHGAP10 | gi|50843948 | Rho GTPase activating protein 10 (ARHGAP10) |
| 17 | HSPA9 | gi|24234688 | Mortalin (heat shock 70 kDa protein 9) |
| 18 | UACA | gi|56550047 | Uveal autoantigen |
Figure 5.
Proteins identified by the SILAC strategy. MS (panel A) and MS/MS (panel B) spectra of a representative peptide, SVDFDSLTVR, from CD2 associated protein are shown. MS (panel C) and MS/MS (panel D) spectra of a representative peptide, AIIIFVPVPQLK, from ribosomal protein S7 are shown. * indicates 13C6-Lys or 13C6-Arg.
3.3. Constituents of the Odin protein complex identified in this study
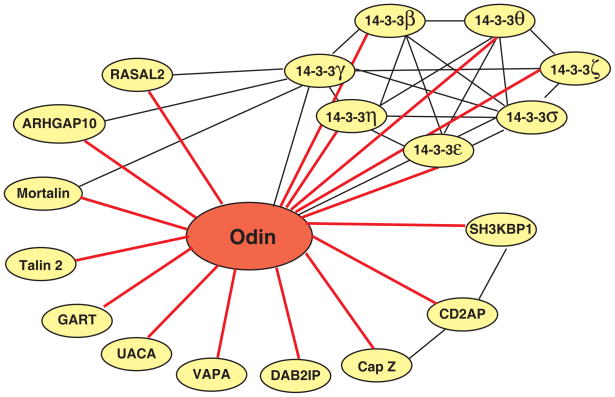
In all, we identified 18 proteins that specifically associated with Odin (Table 1). First we searched HPRD for known interactions among these Odin interactors [13, 14] and constructed an interaction network in the Odin protein complex. As shown in Figure 6, the known interactions among the Odin interacting molecules are shown by black lines whereas 16 novel interactions identified in this study are indicated by red lines. Further, for each one of these proteins, we carried out an extensive literature curation to determine whether they have previously been demonstrated to play a role in RTK signaling. Below, we will briefly describe the known functions of some of these proteins.
Figure 6.
A schematic illustration of interactome network of Odin. Protein-protein interactions indicated by black lines were those reported in the literature. Novel protein-protein interactions indicated by red lines indicate the proteins identified as components of the Odin protein complex in this study.
CD2-associated protein and SH3 domain kinase binding protein 1 both belong to a family of adapter proteins, which contain three SH3 domains, a proline-rich region and a coiled-coil domain. These molecules bind to numerous proteins and serve to organize protein interaction networks in a signaling-specific manner in diverse biological processes, such as T-cell activation [20] and apoptosis [21]. As shown in Figure 6, CD2AP and SH3KBP1 have been shown to bind to each other. Both CD2AP and SH3KBP1 are also important in regulation of RTK signaling by modulating receptor endocytosis. Specifically, in EGFR signaling, the Cbl family of ubiquitin ligases are recruited to activated EGFR, leading to translocation of CD2AP and SH3KBP1 close to EGFR thereby promoting its internalization [1]. This internalization mechanism has also been observed in other RTK signaling pathways, including PDGFR, c-Met and c-Kit [2, 3]. Endocytosis of RTK also involves regulation of cytoskeleton. F-actin capping protein regulates growth of the actin filament by capping the barbed end of growing actin filaments. We identified the beta subunit of F-actin capping protein (CAPZB) as an interacting protein of Odin. Interestingly, it has previously been shown that CD2AP and SH3KBP1 also interact with various subunits of F-actin capping protein including CAPZB [22]. VAMP (vesicle-associated membrane protein)-associated protein A (VAPA) is a type IV membrane protein present in the plasma membrane and in intracellular vesicles. We identified VAPA as an interacting protein of Odin. It has previously been proposed to be involved in vesicle fusion during cargo transport in cells [23–25]. Rho GTPase activating protein 10 (ARHGAP10) is a RhoGAP domain-containing protein, which is implicated in Arf1-mediated and dynamin- and clathrin-independent endocytosis [26, 27]. Previous proteomic analysis identified ARHGAP10 as a component of the 14-3-3 protein complexes (Figure 6) [28]. Taken together, identification of several of these endocytosis-related molecules as interacting proteins of Odin in EGF receptor signaling strongly suggests that Odin may play a role in endocytosis of activated RTKs.
14-3-3 adapter proteins are highly conserved proteins expressed in a wide range of organisms and tissues. Seven members have been described for this protein family – β, γ, ε, ζ, σ, θ and η. These proteins can assemble into homo- or hetero-dimers, as demonstrated in Figure 6. A survey of proteins that interact with 14-3-3 proteins [13, 29] reveals that they interact with a wide range of protein kinases, phosphatases and other signaling proteins. In fact, recent proteomic studies have led to the discovery of >200 proteins that interact with the 14-3-3 proteins [28, 30–32]. Notably, Odin has previously been identified as an interacting protein of 14-3-3 γ and σ [30, 31]. In our studies, all seven members of the 14-3-3 adapter protein family were found to be specifically associated with the Odin protein complex. Phosphorylation plays a regulatory role in the assembly of the 14-3-3 proteins complexes. Muslin et al. first demonstrated that phosphorylation of target proteins is critical for 14-3-3 binding [33]. Yaffe and colleagues further identified two 14-3-3 protein binding motifs: RSXpSXP (mode I) and RXY/FXpSXP (mode II) (pS refers to phosphoserine) [34]. Recent studies have expanded this list of 14-3-3 protein binding motifs [35–37]. Interestingly, using Phosphomotif Finder [38], we discovered that one known phosphorylation site on Odin (Ser 647), also identified in our study, was localized to a 14-3-3 binding motif, RXXpS. Thus, through binding to 14-3-3 proteins, Odin may play a role in various biological process not limited to RTK signaling.
RAS protein activator like 2 (RASAL2) and DAB2 interacting protein (DAB2IP) have a similar domain structure and contain a RAS GTPase activating domain (RASGAP), a Pleckstrin homology domain (PH) and a Protein kinase C conserved region 2. As RASGAP-containing proteins, they are thought to accelerate the intrinsic RAS GTPase activity after RAS activation, thus switching off RAS activity and negatively regulating RAS signaling. Previous studies have implicated DAB2IP as a putative prostate tumor suppressor gene in aggressive prostate cancer [39, 40]. Functional analyses in cell lines have shown that loss of DAB2IP initiates epithelial-to-mesenchymal transition [41] and that overexpressed DAB2IP is a potent growth inhibitor and proapoptotic in response to stress [42]. Although the roles of these molecules in RTK signaling have not been explored fully, the identification of their interactions with Odin may expand our understanding of Odin as well as these molecules. Previous proteomic studies have also identified RASAL2 as an interactor of 14-3-3 proteins (indicated by a black line in Figure 6) [28, 31], further demonstrating the success of our proteomic strategy in identifying the relevant members of the Odin protein complex.
Other proteins that were identified as members of the Odin protein complex were: talin 2, uveal antoantigen, GART, and mortalin (also known as heat shock 70 kDa protein 9). Talin 2 is a cytoskeleton binding protein. Uveal autoantigen is a less characterized protein that also contains ankyrin repeats domains and may also serve as an adapter protein in signal transduction. The protein encoded by GART is a trifunctional enzyme that has phosphoribosylglycinamide formyltransferase, phosphoribosylglycinamide synthetase, and phosphoribosylaminoimidazole synthetase activities which are required for de novo purine biosynthesis. This protein is also highly conserved across the vertebrates. Mortalin is a heat-shock cognate protein and localized in mitochondria, the endoplasmic reticulum, the plasma membrane and cytoplasmic vesicles. This protein also interacts with 14-3-3γ [31].
3.4. Validation of novel interactions in the Odin protein complex
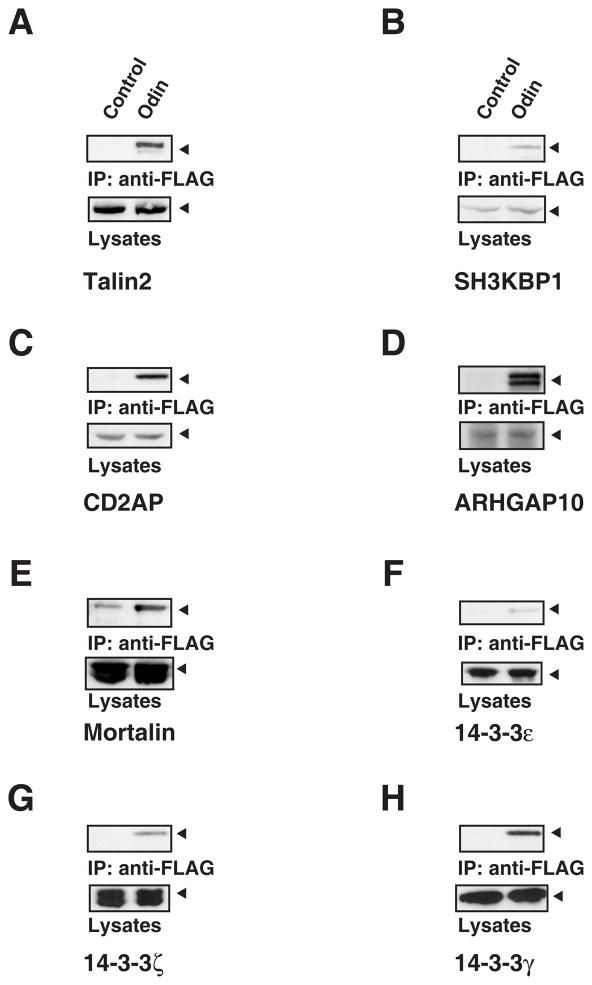
Eight interactions in the Odin protein complex identified in this study were selected based on the availability of antibodies and analyzed further for their ability to form a protein complex with Odin. Protein-protein interactions were examined by co-immunoprecipitation and Western blotting experiments. After affinity purification of Odin, immunoprecipitates were probed by Western blotting with antibodies against the identified proteins. As expected, CD2AP, SH3KBP1, talin 2, mortalin, ARHGAP10, 14-3-3γ, ε and ζ can be detected only in the immunoprecipitates containing Odin (Figure 7). As shown in Figure 7, Western blotting results confirmed the equal loading of proteins. These results further confirmed our identification of these proteins that interact with Odin.
Figure 7.
Validation of protein-protein interactions by co-immunoprecipitation experiments. The Odin protein complex was harvested by immunoprecipitation with anti-FLAG antibodies. The cell lysates and corresponding immunoprecipitates were resolved by SDS-PAGE and probed with antibodies against talin 2 (A), SH3KBP1 (B), CD2AP (C), ARHGAP10 (D), mortalin (E), 14-3-3ε (F), ζ (G) and γ (H).
4. Conclusions
Adapter proteins contain domains/motifs that mediate protein-protein interactions to form multimeric protein complexes. They play an important role in the regulation of RTK signaling by serving as scaffolds for protein complexes induced by ligand binding to RTK. Odin, previously identified as a tyrosine phosphoprotein, is a negative regulator of growth factor signaling. As an adapter protein, it contains three major domains/motifs - six ankyrin repeat domains, two sterile alpha motifs (SAM) and a phosphotyrosine binding domain (PTB) - that mediate its interaction with other signal molecules in growth factor signaling. Characterization of interactome of Odin may help understand the mechanisms through which signal transduction pathways are negatively regulated. Here, we report a targeted quantitative proteomic analysis to identify interacting proteins of Odin in activated EGFR signaling. Our quantitative proteomic results revealed 18 interacting proteins of Odin. Western blotting validated 8 protein-protein interactions out of 18 that were detected by mass spectrometry. Literature-derived interaction information from HPRD showed that some components in the Odin protein complex were also highly connected via protein-protein interactions. An extensive literature-based functional analysis revealed that the components in the Odin protein complex are involved in various cellular processes. It is known that internalization of activated RTKs and their subsequent delivery to lysosome for degradation play key roles in attenuating RTK-mediated signaling cascades. Among Odin interaction partners, CD2AP and SH3KBP1 are known to play key roles in endocytosis of activated RTKs; ARHGAP10 is an important RhoGTPase in endocytosis and CAPZB and VAPA are important cytoskeletal proteins. Therefore, our studies indicate that Odin may play a role in endocytosis of activated RTK as a negative regulator, which could be directly addressed by experimental studies in the future.
Supplementary Material
Acknowledgments
The work was supported by a contract N01-HV-28180 from the National Heart Lung and Blood Institute, by a grant S10RR023025 from the High End Instrumentation Program of the National Institutes of Health (A.P. and R.N.C.), an NIH Roadmap grant “Technology Center for Networks and Pathways” U54 RR 020839 (A.P.) and by a Breast Cancer Research Program Pre-doctoral Training Award W81XWH-05-1-0304 from the Department of Defense (J. Z.).
Appendix A. Supplementary data
Supplementary data associated with this article can be found, in the online version.
Footnotes
Conflict of interest: The authors declare no financial or commercial conflict of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Soubeyran P, Kowanetz K, Szymkiewicz I, Langdon WY, Dikic I. Cbl-CIN85-endophilin complex mediates ligand-induced downregulation of EGF receptors. Nature. 2002;416:183–7. doi: 10.1038/416183a. [DOI] [PubMed] [Google Scholar]
- 2.Szymkiewicz I, Kowanetz K, Soubeyran P, Dinarina A, Lipkowitz S, Dikic I. CIN85 participates in Cbl-b-mediated down-regulation of receptor tyrosine kinases. J Biol Chem. 2002;277:39666–72. doi: 10.1074/jbc.M205535200. [DOI] [PubMed] [Google Scholar]
- 3.Petrelli A, Gilestro GF, Lanzardo S, Comoglio PM, Migone N, Giordano S. The endophilin-CIN85-Cbl complex mediates ligand-dependent downregulation of c-Met. Nature. 2002;416:187–90. doi: 10.1038/416187a. [DOI] [PubMed] [Google Scholar]
- 4.Steen H, Kuster B, Fernandez M, Pandey A, Mann M. Tyrosine phosphorylation mapping of the epidermal growth factor receptor signaling pathway. J Biol Chem. 2002;277:1031–9. doi: 10.1074/jbc.M109992200. [DOI] [PubMed] [Google Scholar]
- 5.Pandey A, Blagoev B, Kratchmarova I, Fernandez M, Nielsen M, Kristiansen TZ, et al. Cloning of a novel phosphotyrosine binding domain containing molecule, Odin, involved in signaling by receptor tyrosine kinases. Oncogene. 2002;21:8029–36. doi: 10.1038/sj.onc.1205988. [DOI] [PubMed] [Google Scholar]
- 6.Pandey A, Podtelejnikov AV, Blagoev B, Bustelo XR, Mann M, Lodish HF. Analysis of receptor signaling pathways by mass spectrometry: identification of vav-2 as a substrate of the epidermal and platelet-derived growth factor receptors. Proc Natl Acad Sci U S A. 2000;97:179–84. doi: 10.1073/pnas.97.1.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kristiansen TZ, Nielsen MM, Blagoev B, Pandey A, Mann M. Mouse embryonic fibroblasts derived from Odin deficient mice display a hyperproliiferative phenotype. DNA Res. 2004;11:285–92. [PubMed] [Google Scholar]
- 8.Kim J, Lee H, Kim Y, Yoo S, Park E, Park S. The SAM domains of Anks family proteins are critically involved in modulating the degradation of EphA receptors. Mol Cell Biol. 2010;30:1582–92. doi: 10.1128/MCB.01605-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shin J, Gu C, Park E, Park S. Identification of phosphotyrosine binding domain-containing proteins as novel downstream targets of the EphA8 signaling function. Mol Cell Biol. 2007;27:8113–26. doi: 10.1128/MCB.00794-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ong SE, Blagoev B, Kratchmarova I, Kristensen DB, Steen H, Pandey A, et al. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol Cell Proteomics. 2002;1:376–86. doi: 10.1074/mcp.m200025-mcp200. [DOI] [PubMed] [Google Scholar]
- 11.Blagoev B, Kratchmarova I, Ong SE, Nielsen M, Foster LJ, Mann M. A proteomics strategy to elucidate functional protein-protein interactions applied to EGF signaling. Nat Biotechnol. 2003;21:315–8. doi: 10.1038/nbt790. [DOI] [PubMed] [Google Scholar]
- 12.Schulze WX, Mann M. A novel proteomic screen for peptide-protein interactions. J Biol Chem. 2004;279:10756–64. doi: 10.1074/jbc.M309909200. [DOI] [PubMed] [Google Scholar]
- 13.Keshava Prasad TS, Goel R, Kandasamy K, Keerthikumar S, Kumar S, Mathivanan S, et al. Human Protein Reference Database--2009 update. Nucleic Acids Res. 2009;37:D767–72. doi: 10.1093/nar/gkn892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Peri S, Navarro JD, Kristiansen TZ, Amanchy R, Surendranath V, Muthusamy B, et al. Human protein reference database as a discovery resource for proteomics. Nucleic Acids Res. 2004;32:D497–501. doi: 10.1093/nar/gkh070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ren XR, Du QS, Huang YZ, Ao SZ, Mei L, Xiong WC. Regulation of CDC42 GTPase by proline-rich tyrosine kinase 2 interacting with PSGAP, a novel pleckstrin homology and Src homology 3 domain containing rhoGAP protein. J Cell Biol. 2001;152:971–84. doi: 10.1083/jcb.152.5.971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Amanchy R, Kalume DE, Iwahori A, Zhong J, Pandey A. Phosphoproteome analysis of HeLa cells using stable isotope labeling with amino acids in cell culture (SILAC) J Proteome Res. 2005;4:1661–71. doi: 10.1021/pr050134h. [DOI] [PubMed] [Google Scholar]
- 17.Mortensen P, Gouw JW, Olsen JV, Ong SE, Rigbolt KT, Bunkenborg J, et al. MSQuant, an open source platform for mass spectrometry-based quantitative proteomics. J Proteome Res. 2010;9:393–403. doi: 10.1021/pr900721e. [DOI] [PubMed] [Google Scholar]
- 18.Ballif BA, Villen J, Beausoleil SA, Schwartz D, Gygi SP. Phosphoproteomic analysis of the developing mouse brain. Mol Cell Proteomics. 2004;3:1093–101. doi: 10.1074/mcp.M400085-MCP200. [DOI] [PubMed] [Google Scholar]
- 19.Beausoleil SA, Jedrychowski M, Schwartz D, Elias JE, Villen J, Li J, et al. Large-scale characterization of HeLa cell nuclear phosphoproteins. Proc Natl Acad Sci U S A. 2004;101:12130–5. doi: 10.1073/pnas.0404720101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dustin ML, Olszowy MW, Holdorf AD, Li J, Bromley S, Desai N, et al. A novel adaptor protein orchestrates receptor patterning and cytoskeletal polarity in T-cell contacts. Cell. 1998;94:667–77. doi: 10.1016/s0092-8674(00)81608-6. [DOI] [PubMed] [Google Scholar]
- 21.Gout I, Middleton G, Adu J, Ninkina NN, Drobot LB, Filonenko V, et al. Negative regulation of PI 3-kinase by Ruk, a novel adaptor protein. Embo J. 2000;19:4015–25. doi: 10.1093/emboj/19.15.4015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hutchings NJ, Clarkson N, Chalkley R, Barclay AN, Brown MH. Linking the T cell surface protein CD2 to the actin-capping protein CAPZ via CMS and CIN85. J Biol Chem. 2003;278:22396–403. doi: 10.1074/jbc.M302540200. [DOI] [PubMed] [Google Scholar]
- 23.Weir ML, Klip A, Trimble WS. Identification of a human homologue of the vesicle-associated membrane protein (VAMP)-associated protein of 33 kDa (VAP-33): a broadly expressed protein that binds to VAMP. Biochem J. 1998;333 (Pt 2):247–51. doi: 10.1042/bj3330247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Foster LJ, Weir ML, Lim DY, Liu Z, Trimble WS, Klip A. A functional role for VAP-33 in insulin-stimulated GLUT4 traffic. Traffic. 2000;1:512–21. doi: 10.1034/j.1600-0854.2000.010609.x. [DOI] [PubMed] [Google Scholar]
- 25.Wyles JP, McMaster CR, Ridgway ND. Vesicle-associated membrane protein-associated protein-A (VAP-A) interacts with the oxysterol-binding protein to modify export from the endoplasmic reticulum. J Biol Chem. 2002;277:29908–18. doi: 10.1074/jbc.M201191200. [DOI] [PubMed] [Google Scholar]
- 26.Dubois T, Paleotti O, Mironov AA, Fraisier V, Stradal TE, De Matteis MA, et al. Golgi-localized GAP for Cdc42 functions downstream of ARF1 to control Arp2/3 complex and F-actin dynamics. Nat Cell Biol. 2005;7:353–64. doi: 10.1038/ncb1244. [DOI] [PubMed] [Google Scholar]
- 27.Kumari S, Mayor S. ARF1 is directly involved in dynamin-independent endocytosis. Nat Cell Biol. 2008;10:30–41. doi: 10.1038/ncb1666. [DOI] [PubMed] [Google Scholar]
- 28.Meek SE, Lane WS, Piwnica-Worms H. Comprehensive proteomic analysis of interphase and mitotic 14–3–3-binding proteins. J Biol Chem. 2004;279:32046–54. doi: 10.1074/jbc.M403044200. [DOI] [PubMed] [Google Scholar]
- 29.Peri S, Navarro JD, Amanchy R, Kristiansen TZ, Jonnalagadda CK, Surendranath V, et al. Development of human protein reference database as an initial platform for approaching systems biology in humans. Genome Res. 2003;13:2363–71. doi: 10.1101/gr.1680803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Benzinger A, Muster N, Koch HB, Yates JR, 3rd, Hermeking H. Targeted proteomic analysis of 14–3–3 sigma, a p53 effector commonly silenced in cancer. Mol Cell Proteomics. 2005;4:785–95. doi: 10.1074/mcp.M500021-MCP200. [DOI] [PubMed] [Google Scholar]
- 31.Jin J, Smith FD, Stark C, Wells CD, Fawcett JP, Kulkarni S, et al. Proteomic, functional, and domain-based analysis of in vivo 14–3–3 binding proteins involved in cytoskeletal regulation and cellular organization. Curr Biol. 2004;14:1436–50. doi: 10.1016/j.cub.2004.07.051. [DOI] [PubMed] [Google Scholar]
- 32.Pozuelo Rubio M, Geraghty KM, Wong BH, Wood NT, Campbell DG, Morrice N, et al. 14–3–3-affinity purification of over 200 human phosphoproteins reveals new links to regulation of cellular metabolism, proliferation and trafficking. Biochem J. 2004;379:395–408. doi: 10.1042/BJ20031797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Muslin AJ, Tanner JW, Allen PM, Shaw AS. Interaction of 14–3–3 with signaling proteins is mediated by the recognition of phosphoserine. Cell. 1996;84:889–97. doi: 10.1016/s0092-8674(00)81067-3. [DOI] [PubMed] [Google Scholar]
- 34.Yaffe MB, Rittinger K, Volinia S, Caron PR, Aitken A, Leffers H, et al. The structural basis for 14–3–3:phosphopeptide binding specificity. Cell. 1997;91:961–71. doi: 10.1016/s0092-8674(00)80487-0. [DOI] [PubMed] [Google Scholar]
- 35.Ganguly S, Weller JL, Ho A, Chemineau P, Malpaux B, Klein DC. Melatonin synthesis: 14–3–3-dependent activation and inhibition of arylalkylamine N-acetyltransferase mediated by phosphoserine-205. Proc Natl Acad Sci U S A. 2005;102:1222–7. doi: 10.1073/pnas.0406871102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ku NO, Liao J, Omary MB. Phosphorylation of human keratin 18 serine 33 regulates binding to 14–3–3 proteins. Embo J. 1998;17:1892–906. doi: 10.1093/emboj/17.7.1892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang SH, Kobayashi R, Graves PR, Piwnica-Worms H, Tonks NK. Serine phosphorylation-dependent association of the band 4. 1-related protein-tyrosine phosphatase PTPH1 with 14–3–3beta protein. J Biol Chem. 1997;272:27281–7. doi: 10.1074/jbc.272.43.27281. [DOI] [PubMed] [Google Scholar]
- 38.Amanchy R, Periaswamy B, Mathivanan S, Reddy R, Tattikota SG, Pandey A. A curated compendium of phosphorylation motifs. Nat Biotechnol. 2007;25:285–6. doi: 10.1038/nbt0307-285. [DOI] [PubMed] [Google Scholar]
- 39.Duggan D, Zheng SL, Knowlton M, Benitez D, Dimitrov L, Wiklund F, et al. Two genome-wide association studies of aggressive prostate cancer implicate putative prostate tumor suppressor gene DAB2IP. J Natl Cancer Inst. 2007;99:1836–44. doi: 10.1093/jnci/djm250. [DOI] [PubMed] [Google Scholar]
- 40.Hsieh JT, Karam JA, Min W. Genetic and biologic evidence that implicates a gene in aggressive prostate cancer. J Natl Cancer Inst. 2007;99:1823–4. doi: 10.1093/jnci/djm263. [DOI] [PubMed] [Google Scholar]
- 41.Xie D, Gore C, Liu J, Pong RC, Mason R, Hao G, et al. Role of DAB2IP in modulating epithelial-to-mesenchymal transition and prostate cancer metastasis. Proc Natl Acad Sci U S A. 2010;107:2485–90. doi: 10.1073/pnas.0908133107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xie D, Gore C, Zhou J, Pong RC, Zhang H, Yu L, et al. DAB2IP coordinates both PI3K-Akt and ASK1 pathways for cell survival and apoptosis. Proc Natl Acad Sci U S A. 2009;106:19878–83. doi: 10.1073/pnas.0908458106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.