Abstract
Single-molecule manipulation methods have opened a new vista on the study of molecular motors. Here we describe the use of magnetic traps for the investigation of the mechanism of DNA based motors, in particular helicases and translocases.
1. Introduction
Single-molecule micromanipulation experiments have brought a new approach to DNA protein interactions (Keller and Bustamante, 2000). They allow one to monitor in real time the activity of single proteins on single DNA molecules and hence reduce the complexity of the system under study. In principle, this leads to a simpler data interpretation. While these experiments were initially challenging because of their novel technical aspects, they are becoming more popular and widespread with the availability of commercial instruments (optical tweezers, atomic force microscopes (AFM), and magnetic traps (MT)).
The principle of magnetic traps is quite simple. It uses the fact that the interaction between a magnetic dipole and a magnetic field produces both a force and a torque on the dipole. Everyone has experienced both effects. A small magnet sticks to the wall of a fridge, because it induces in its magnetic material a dipole of opposite polarity that attracts the magnet to the wall. Similarly, the Earth’s magnetic field applies a torque on the magnetized needle of a compass that forces it to align with the field lines and point to the magnetic north pole (which is close to the geographic one). MT uses these common effects to pull on and rotate a micron-sized magnetic bead tethered to a surface by a DNA molecule, through the field of strong and small permanent magnets. The magnetic field applies a force and a torque on the magnetic bead and thus on the DNA molecule. One can thus stretch and coil a DNA molecule (Strick et al., 1996).
Quite often the interaction of proteins with a DNA molecule under tension results in a change of the molecule’s extension. This can be due to DNA bending, looping, denaturation, supercoiling, etc. As a consequence, following the molecule’s extension as function of time allows for real time monitoring of the DNA/protein interaction dynamics. The applied force is not only a way to stretch the molecule in order to monitor its extension, but it is also a way to affect the DNA/protein interaction. It is thus an alternative tool to temperature, pH, and other standard biochemical parameters that alter chemical equilibria.
In comparison with other popular methods to manipulate single molecules, such as optical tweezers (Svoboda et al., 1994) or AFM cantilevers (Engel et al., 1999), MTs are force clamps. They allow one to set the force applied to a molecule rather than fixing its extension. MT also provide an easy way to fix the orientational angle of the magnetic bead (not the applied torque) and thus the DNA’s degree of supercoiling. These properties have a number of experimental consequences. Because MTs are force clamps, their exact positioning is not crucial (the force is not affected significantly if the position of the magnets changes by a few micrometers), which is not the case for optical tweezers or AFM cantilevers. This is quite convenient as it implies that the apparatus does not require particular vibrational isolation or periodic calibrations of the force. It makes an MT setup quite easy to use.
In the present chapter, we are going to first introduce the MT in a more concrete and precise way. We will then give the protocols required to perform an experiment and illustrate some applications in the study of FtsK, a DNA translocase, and gp41, a helicase. These examples will exemplify the use of different DNA substrates in a MT setup.
2. Experimental Setup
A schematic representation of a MT setup is shown in Fig. 13.1A. A DNA molecule (hairpin or double-stranded (ds) DNA) is tethered by its extremities to a glass surface and a magnetic bead. Small magnets positioned above the sample generate a strong magnetic field gradient that pulls the magnetic bead up with constant force. By rotating the magnets one can also coil the molecule. An inverted microscope and a CCD camera are used to image the sample illuminated by the monochromatic focused beam of a LED. The image of the bead displays diffraction rings that are used to estimate its 3-D position as explained elsewhere (Gosse and Croquette, 2002). From the fluctuating positions of the bead both the mean elongation of the molecule and the force applied to it can be deduced (Strick et al., 1996).
Figure 13.1.
Experimental design. (A) Schematic of a magnetic trap setup. (B) DNA hairpin construction used to study gp41 helicase.
Proteins that interact with DNA generally induce distortions in the DNA conformation. These conformational distortions are often translated into changes in the DNA’s extension. For example, a helicase that unwinds a DNA hairpin under tension adds two bases of stretched single-stranded (ss) DNA for each unwound base pair (see below). In this way, DNA manipulation techniques allow for the investigation of DNA/protein interactions in real time. In the following, we will demonstrate the use of MT in the study of translocases and helicases.
3. Methods and Protocols
In order to carry out an MT experiment one needs (in addition to a MT setup) a DNA construct and appropriately functionalized magnetic beads (streptavidin-coated beads are often used) and surfaces. In the following, we present the protocols for preparing surfaces and DNA/bead constructs.
3.1. Surface preparation
Different protocols can be used for preparing the chamber and precoating of the surface. Here we describe one of them (see Lionnet et al., 2008; Revyakin et al., 2003, for alternative protocols).
Work in a clean room.
Put 60 × 40 mm coverslip on a spin coater.
Set acceleration to 4000 rpm2 and speed to 4000 rpm.
Put 200 µL of Teflon AF1600 diluted to 1% in FC72 at the center of the coverslip.
Rotate for 10 s.
Store Teflon-coated coverslips in a clean container to avoid dust contamination.
Remark: The coated surface is highly hydrophobic.
3.2. Chamber preparation
Use a 60 × 40 mm glass coverslip and with a sandblaster make two 1 mm holes in the glass at 50 mm distance.
Cut a 60 × 40 mm piece of parafilm. In the center, cut out a band of dimensions 1 × 52 mm.
Make a sandwich with the parafilm between a Teflon-coated coverslip (made as above) and the perforated coverslip.
Heat the sandwich to 120 °C in order to melt the parafilm and seal the chamber.
Remark: Parafilm can be replaced with double-sided tape.
3.3. Surface coating
Fill the chamber (~10 µL) with a 100 µg/mL solution of polyclonal digoxigenin antibody in phosphate buffer saline (PBS).
Incubate for a few hours at 37 °C or overnight at room temperature.
Rinse with passivation buffer (PBS + 1 mM EDTA, 1 mg/mL BSA, 0.1% (w/v) pluronic F127, and 10 mM azide).
Surfaces can be used after 3 h of incubation at 4 °C.
Chambers can be kept for a few weeks in a humid sealed box at 4 °C.
3.4. DNA preparation
In this section, we present two protocols for preparing different DNA constructs: a dsDNA substrate with specific labels at its extremities and a DNA hairpin with labeled tails.
3.4.1. DNA hairpin
Here we present the protocol for preparing a 6.8-kbp-long DNA hairpin that is used to characterize the loading of the gp41 helicase (see below). The protocol for synthesizing the 1.2 kbp hairpin used in the gp41 unwinding experiments (see below) is presented elsewhere (Manosas et al., 2009). A DNA hairpin, 6.8 kbp long, is synthesized by extending a 5′-biotinylated tailed primer annealed to circular single-stranded M13 DNA with a DNA polymerase. The resulting rolling circle product is linearized by digestion with a restriction enzyme and gel purified. A short 5′-phosphorylated oligonucleotide is then ligated to both strands of the 5′-overhang ahead of the fork structure to form a DNA hairpin. The 3′-end of the template strand is labeled in a two-step process using exonuclease digestion followed by 5′-overhang filling with appropriate DNA polymerases and digoxigenin-labeled dUTP. The completed DNA substrate resembles a DNA replication fork with biotin and digoxigenin labels suitable for manipulation using magnetic tweezers (Fig. 13.1).
Procedure
-
1
The 5′-biotinylated primer (5′-biotin-TTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTTGCGCTTAATGCGCCGCTACAGGGCGCGTAC-3′) may be obtained from numerous companies that provide custom oligonucleotide synthesis. While optional, it is recommended that a primer of this length be purified prior to use. Full-length biotinylated primer may be purified using Pierce Monomeric Avidin UltraLink Resin (Thermo Fisher Scientific) per the manufacturer’s instructions.
-
2
Circular M13 ssDNA is prepared using the large-scale preparation protocol found in Sambrook and Russell (2001).
-
3
Anneal the 5′-biotinylated tailed primer to positions 5485–5514 of circular M13 ssDNA by heating a mixture of 100 nM M13 ssDNA with 200 nM primer in 1.5 mL of ddH2O to 80 °C for 3 min. Allow the mixture to cool slowly to room temperature over several hours by placing into a water bath that is turned off.
-
4
To the annealing reaction, add 100 nM T4 DNA polymerase deficient in exonuclease activity (gp43exo−), 250 µM dNTPs, 5% (v/v) DMSO, 20 mM Tris–Ac (pH 7.5), 150 mM KOAc, 10 mM Mg (OAc)2, and ddH2O up to 3 mL (concentrations given are final concentrations). Extend the primer around the M13 ssDNA by incubating the reaction mixture for 1 h at 37 °C. Heat inactivate the polymerase at 65 °C for 20 min.
Note: Another DNA polymerase may be substituted for gp43exo−; however, the substituted DNA polymerase should lack significant strand displacement activity.
Tip: DMSO is included in the synthesis reaction to disrupt any secondary structure of the M13 ssDNA, thereby increasing the complete extension of the primer around the circular M13 single-stranded template. T4 ssDNA-binding protein (gp32) may be used for this purpose instead of DMSO. The efficiency of the extension reaction appears to vary with each batch of purified M13 ssDNA and enzyme. It is recommended that the DNA synthesis be optimized with small test reactions in which the DMSO and gp32 concentrations are varied between 2.5 and 5% (v/v) and 2.5 and 5 µM, respectively. The efficiency of primer extension may be analyzed on a 0.8% agarose gel containing 0.1 µg/mL ethidium bromide run in 0.5× TBE (Tris–Borate–EDTA); the rolling circle product will migrate at an apparently larger size than the M13 ssDNA.
-
5
Linearize the rolling circle product at position 5976 by adding 60 µL BsmBI restriction enzyme (10,000 U/mL) and incubate for 1.5 h at 55 °C. Quench the digestion reaction with 120 µL of 500 mM EDTA.
-
6
Purify the linearized forked-DNA product by adding 636 µL of 6× DNA gel loading dye and 0.1% (w/v) SDS (final concentration) to the digestion reaction. Load 18.5 µg DNA (~100 µL) per well of a 0.8 % agarose gel containing 0.1 µg/mL ethidium bromide and run in 0.5× TBE until the rolling circle and linearized forked-DNA product bands are separated, approximately 2 h at 100 V for a 12-cm long gel. Cut out the band that migrates at 7.25 kbp (as judged by comparison with a dsDNA ladder) corresponding to the linearized forked-DNA product.
-
7
The DNA is extracted from the gel slices by electroelution in 1× TBE for 16 h at 50 V. Complete elution of the DNA may be confirmed by restaining the gel slices with ethidium bromide; electroelution is continued on any gel slices still containing DNA. The eluted DNA is precipitated by adding 1/10 volume of 3 M NaOAc (pH 5.2) and 2 volumes of ice cold 95% (v/v) ethanol. Incubate on ice for 1 h. Collect the precipitated DNA by centrifugation at 12,000g for 20 min at 4 °C. Allow the DNA pellet to air dry overnight. Dissolve the DNA in 10 mM Tris–HCl (pH 8.0) with a final DNA concentration around 100 nM. Store at −20 °C.
Tip: Minimize the volume of buffer that the DNA is eluted into to increase the amount of DNA recovered by the ethanol precipitation.
Note: Alternatively, the DNA may be extracted from the gel slices using various gel-extraction kits commercially available per the manufacturer’s instructions; however, we find the yield of recovered DNA to be generally lower from these methods.
-
8
The short oligonucleotide (5′-CCAGGTCAGATGCGTTTTCGCATCTGAC-3′), which forms a hairpin structure with a 4-base loop, 10-base stem, and 4-base overhang complementary to the 5′-cohesive end of the forked-DNA product, may be obtained from numerous companies that provide custom oligonucleotide synthesis. The oligonucleotide may be purchased phosphorylated at the 5′-end or alternatively, may be phosphorylated at the 5′-end with T4 polynucleotide kinase and ATP per the manufacturer’s instructions. Following the phosphorylation reaction, the kinase should be heat inactivated at 65 °C for 20 min, but there is no need for further purification.
-
9
Ligate the 5′-phosphorylated oligonucleotide to the forked-DNA product at 4 °C for a minimum of 24 h in a reaction mixture containing approximately 100 nM forked-DNA, 10-fold excess oligonucleotide, 1× ligation buffer, and T4 ligase per the manufacturer’s instructions.
-
10
Excess oligonucleotide may be removed from the large DNA hairpin product using various PCR cleanup kits commercially available, per the manufacturer’s instructions.
Tip: The recovery of the DNA hairpin may often be increased by performing a second DNA elution step and warming the elution buffer to 65 °C.
-
11
The 5′-overhang of the primer/template end of the forked DNA is increased by exonuclease digestion with T4 DNA polymerase (wild-type gp43). Reactions contain a twofold excess of gp43 over DNA substrate and 1 mM dATP in 20 mM Tris–Ac (pH 7.5), 150 mM KOAc, 10 mM MgOAc2. Incubate the reaction mixture for 20 min at 37 °C. Heat inactivate the polymerase at 65 °C for 20 min.
Note: The extent of exonuclease digestion is limited by the presence of dATP in the reaction causing the polymerase to idle or cycle repeatedly between removing and incorporating dATP when it encounters the first dATP in the template strand.
-
12
Multiple digoxigenin labels are incorporated as the 5′-overhang is filled-in with T4 DNA polymerase (gp43exo−) by adding a twofold excess of gp43exo− over the DNA substrate, 250 µM each dGTP and dCTP, and 50 µM digoxigenin-labeled dUTP directly to the previous reaction. Incubate the reaction mixture for 20 min at 37 °C. Heat inactivate the polymerase at 65 °C for 20 min. Remove protein and excess nucleotides from the DNA hairpin product using various PCR clean-up kits commercially available, per the manufacturer’s instructions. Store at −20 °C.
Note: The primer of the forked-DNA substrate is not extended by the gp43exo− polymerase due to its lack of strand displacement activity.
3.4.2. dsDNA construct
The dsDNA construct has been used in FtsK experiments (see below). The protocol must be adapted for the particular DNA sequence being used. Shorter (down to 2 kbps) or longer DNA constructs (typically λ-DNA) may be used.
The guideline is to digest the desired DNA with two restriction enzymes leaving cohesive ends. These ends are then used to bind approximately few hundreds bps PCR products obtained through the incorporation of digoxigenin or biotin modified nucleotides (with a modified/unmodified ratio of about 1/5 or 1/10) as follows (see tables given below)
The PCR products are purified with Microspin SR-400 columns (GE Healthcare): protocol according to the manufacturer’s protocol.
| Labeling DNA anchoring fragments | ||
|---|---|---|
| Reagent | Concentration | Volume (µL) |
| pBluescriptKS | 250 ng/µL | 1 |
| Primer A (CTAAATTGTAAGCGTTAATATTTTGTTAAA) | 100 µM | 1 |
| Primer B, (TATCTTTATAGTCCTGTCGGGTTTCGCCAC) | 100 µM | 1 |
| dNTPs | 10 mM | 1.5 |
| Mg2+ | 25 mM | 2 |
| Taq buffer without Mg2+ | 10× | 5 |
| Taq polymerase | Manufacturer stock (NEB) | 1 |
| Digoxigenin-11-dUTP or biotin-16-dUTP (Roche) | 1 mM | 1.5 |
| DI H2O | 36.5 | |
| PCR program for DNA labeling | |||
|---|---|---|---|
| Step | T (°C) | Duration (min) | Number of cycles |
| 1 | 94 | 5 | 1 |
| 94 | 0.5 | 30 | |
| 2 | 54 | 1 | |
| 72 | 1 | ||
| 3 | 72 | 5 | 1 |
| Central fragment digestion | |
|---|---|
| Reagent | Volume (µL) |
| pFX355 (10 kbp at 100 ng/µL) | 10 |
| XhoI | 1 |
| AatII | 1 |
| Eco109I | 1 |
| NEB 4 | 3 |
| H2O | 14 |
| 37 °C for 1 h + 65 °C for 20 min | |
| DIG-labeled fragment digestion | |
|---|---|
| Reagent | Volume (µL) |
| DIG fragment (~1 kbp at 50 ng/µL) | 10 |
| XhoI | 2 |
| NEB buffer 4 | 4 |
| H2O | 4 |
| 37 °C for 1 h + 65 °C for 20 min | |
| Biotin-labeled fragment digestion | |
|---|---|
| Reagent | Volume (µL) |
| Biotin fragment (~1 kbp at 50 ng/µL) | 10 |
| AatII | 2 |
| NEB buffer 4 | 4 |
| H2O | 4 |
| 37 °C for 1 h + 65 °C for 20 min | |
| Ligation of anchoring fragments to DNA central sequence | |
|---|---|
| Reagent | Volume (µL) |
| Digested central fragment | 2 |
| Digested DIG fragment | 15 |
| Digested Biotin fragment | 15 |
| Ligase buffer 10× | 10 |
| T4 DNA ligase (Fermentas) | 4 |
| H2O | 52 |
| 16 °C for 2 h then 65 °C for 20 min | |
3.5. Bead/DNA preparation
Once the DNA construct has been prepared, mix the DNA with coated magnetic beads at ~1:10 ratio.
Pipette 10 µL of MyOne C1 (Invitrogen) streptavidin beads. Clean them according to the manufacturer’s protocol. Resuspend in 10 µL PBS.
Add typically 1 ng of DNA.
After 1 min dilute in 80 µL passivation buffer.
After 30 min beads can be used.
Beads should be kept on a rotator (10 rpm) at room temperature (to prevent their sedimentation) and can be used for weeks.
3.6. Bead injection
Once the previous steps are complete, inject the DNA/bead construct into the chamber for incubation.
Lift the magnets as far away as possible from the sample.
With a syringe pump apply a flow and inject 5 µL of the DNA/bead construct. Stop the flow when a large number of beads can be seen.
Let the beads sediment for about 5 min.
Apply a flow of buffer strong enough to remove the unbound beads, but not so strong as to tear away those that are attached through a DNA molecule to the surface. Stop the flow when no more free beads are passing through the field of view.
3.7. Selection of beads tethered by a single DNA hairpin
Before adding proteins to start the experiment, one needs to identify beads of interest, which are attached to the surface by a single DNA molecule. In order to select suitable beads, we use the previously characterized mechanical properties of the DNA construct.
For the hairpin substrate, the extension remains almost constant below ~15 pN and abruptly increases when the hairpin is unzipped above ~15 pN (see below). If the bead is tethered by two DNA hairpins, the force needed to unzip them is twice as large (~30 pN). Based on these results, we have developed the following protocol for selecting beads with a single DNA hairpin:
Change the position of the magnets so that the applied force varies from low force ~1–5 pN to high force ~20pN.
Measure the difference in DNA extension (Δz) between the two applied forces.
If Δz is consistent with the length of the unfolded hairpin (typically 1 nm for 1 bp unwound, for example, ~1.2 µm for the 1.2 kbp hairpin or 7 µm for the 6.8 kbp hairpin) the bead is selected for the experiment.
If Δz is ~0, the bead is either nonspecifically bound to the surface or is tethered by two or more DNA molecules. In either case, the bead is ignored.
3.8. Selection of beads tethered by a single-nicked DNA molecule
The simplest way to select a single-nicked dsDNA bead with the magnetic tweezers is to rotate the magnets by a large number of positive turns, thereby strongly supercoiling unnicked DNA molecules or braiding the DNA molecules if a bead is tethered by more than one DNA. These supercoils and braids significantly reduce the extension of the molecules. Therefore, only beads tethered by a single-nicked DNA molecule will remain at a fixed distance from the surface.
3.9. Selection of beads tethered by a coilable DNA molecule
Selection of beads attached with a single-unnicked DNA molecule takes advantage of the fact that negatively supercoiled DNA molecules melt if pulled with a force F > Fc ~ 0.5 pN. At forces F < Fc the molecule forms plectonemes that strongly reduce its extension. At larger forces, the molecule does not form plectonemes, but instead denatures (melts) which only slightly affects its extension. To select a bead bound by a single DNA coilable molecule, the idea is to rotate the magnets clockwise by a large number of turns (imposing large negative supercoiling in the DNA). One then tests for a strong change in the bead to surface distance (i.e., the DNA’s extension) as the force is varied between values above and below Fc. This type of behavior is not observed if the bead is bound by a nicked DNA or by two or more braided molecules.
Protocol
Rotate the magnets clockwise to reach a degree of negative supercoiling of σ ~ −0.1 (the number of rotations should be about 10% of the number of helical turns in the DNA).
Scan the sample while moving the magnets vertically in order to modulate the force around Fc (typically between 0.3 and 1 pN). Beads exhibiting large variations in their distance to the surface are good candidates.
At ~1 pN (in passivation buffer) force, rotate the magnets counterclockwise (to reach a positive degree of supercoiling of σ ~ 0.1). The beads of interest should recoil to the surface as positive supercoils are generated.
At this point, it is still possible, though unlikely, that the bead is attached by two molecules, at least one of which is unnicked. To remove that possibility one has two choices.
Rotate the magnets clockwise at F ~ 1 pN to reach σ ~ −0.2. If the bead is bound by two molecules, their braiding should be visible by a decrease in the bead’s extension.
One can also investigate the change in extension for rotations between −1 and +1 turns. If two DNA molecules tether the bead, a strong decrease in extension should appear for the first ± 1/2 turn, before the two molecules cross (Charvin et al., 2004).
3.10. Force determination
The measurement of the force is based on the analysis of the Brownian fluctuations of the tethered bead, which is equivalent to a damped pendulum of length l = 〈z〉 pulled by a force F. That force gives rise to a transverse restoring force given by F = kBT〈z〉/〈δx2〉, where 〈δx2〉 is the mean transverse fluctuations of the bead, kB is Boltzmann’s constant, T is the temperature (Strick et al., 1996).
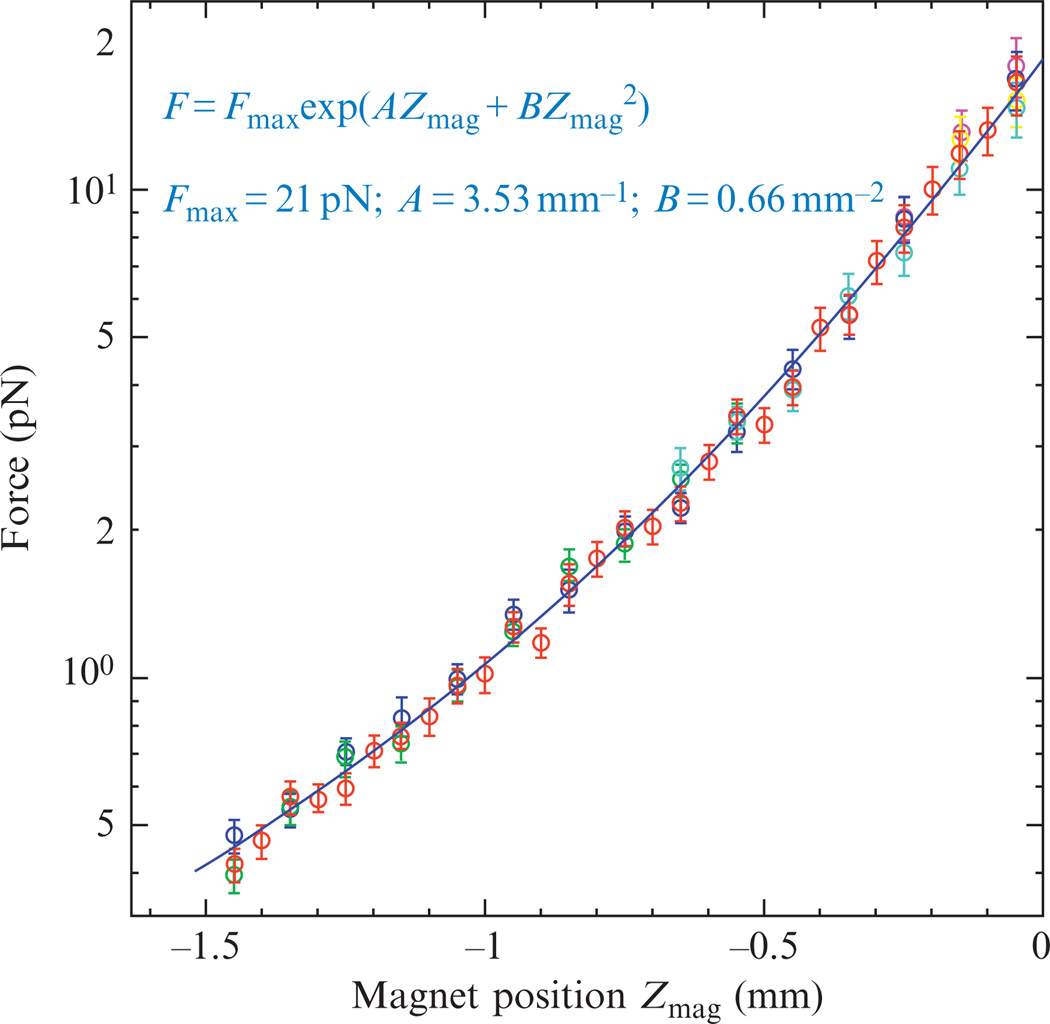
By using a long dsDNA construct (~50 kbp dsDNA molecule obtained from λ-DNA), we have measured the force as a function of the position of the magnets, F(Zmag), for several beads (Fig. 13.2). Typically, there is a 10% variability in force from bead to bead, probably due to small differences in the magnetic properties of the commercial beads. The calibration curve F(Zmag) can be used to estimate the force F given the position of the magnets (Fig. 13.2).
Figure 13.2.
Force calibration. Force as a function of the magnets position (Zmag) measured for several micron-sized beads (Myone Invitrogen) attached by a single λ-DNA molecule (each color corresponds to a separate bead). Note that the position Zmag is measured with respect to the sample chamber (e.g., Zmag = 0 when the magnets touch the upper surface of the chamber and when the magnets are 1 mm above the chamber Zmag = −1). The blue line corresponds to a fit of the data to the function , which yields Fmax = 21 pN, A = 3.53 mm−1 and B = 0.66 mm−2. This curve can be used to estimate the force on the bead at a known position of the magnets.
4. Application to the Study of FtsK
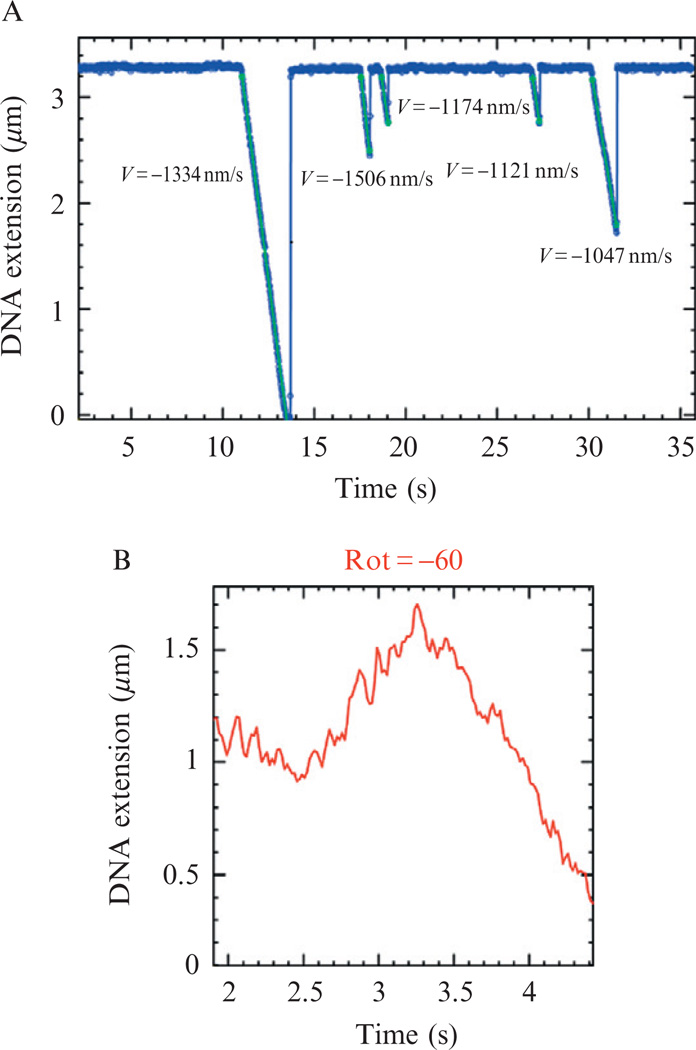
Recombination events in bacteria with circular chromosomes may lead to chromosome dimer formations that are deleterious to cell division. In order to resolve these dimers, bacteria use site-specific recombinases, which in the case of Escherichia coli are called XerC and D. These recombinases bind to a specific DNA sequence called dif. E. coli chromosome has a single dif site. However, in the case of unresolved dimers two dif sites appear in the dimeric chromosome. XerC/D bind to both sites forming a DNA loop and resolve the chromosome dimer into two monomers by a single recombination event. For this to occur, the two dif sites need to be brought into proximity of each other on a timescale that is shorter than cell division (~20 min for E. coli). FtsK is the molecular motor responsible for rapidly translocating the chromosomal DNA and bringing the dif sites into proximity. FtsK is a protein that is bound to the membrane septum via its N-terminus and whose C-terminus is an ATPase catalyzing DNA translocation. Additionally, FtsK catalyses the recombination reaction by interacting with XerD. Until recently, only the C-terminal part of the motor has been studied in vitro. Initial studies showed that the FtsK motor forms transient loops of DNA (Aussel et al., 2002), which could be more adequately investigated by real-time measurements. In the following, we shall see how MT have helped us address this issue.
4.1. FtsK activity
Protocol
Once a DNA molecule is characterized (nicked or not) and the force is set as desired:
Exchange buffer by flowing FtsK buffer (5 mM ATP unless otherwise specified, 10 mM Mg2+, 10 mM Tris (pH 7.6) and 100 mM NaCl) into the chamber.
Starting from 10 nM introduce increasing protein concentrations until protein activity is observed. Wait several minutes between protein injections to see if translocation events occur.
Protein activity results in rapid shortening of the DNA extension. Figure 13.3A shows a typical burst of activity. Such events can be recorded for a few hours as long as there are enough active proteins in solution. For data analysis one should make sure that the protein concentration is low enough to observe well separated events to ensure that only one motor is active at any time. Data analysis is relatively simple. Each event (such as the ones shown in Fig. 13.3A) is characterized by the slope, the duration, and the extension (height). These values are related to the motor’s speed, activity, and processivity, respectively, and may vary with the force, ATP, or salt concentration. In order to relate the enzymatic rate and processivity to the number of base pairs translocated, one must translate the change in the DNA extension at a given force into the DNA contour length. To give an example: at 0.1 pN the DNA extension is only 1/2 of its crystallographic length. Therefore, a decrease in DNA extension at a rate of 1 µm/s corresponds to the motor’s actual rate of 2 × 1 µm/s.The relation giving the DNA extension l as a function of the force F is provided by the Worm Like Chain model (Bustamante et al., 1994). A useful approximate formula is (Bouchiat et al., 1999):
where a2 = −0.5164228, a3 = −2.737418, a4 = −16.07497, a5 = −38.87607, a6 = −39.49944, and a7 = −14.17718. Here l0 denotes the molecular contour length and ξ is the DNA persistence length, which under physiological salt conditions is ~50 nm. This relation gives the relative DNA extension z = l/l0 at each force from which the measured change in DNA extension (δl) can be translated into a change in contour length δl0 = δl/z.
Figure 13.3.
Detecting FtsK translocation activity on a DNA molecule by MT (A) FtsK activity events. The DNA molecule shortens as FtsK translocates and forms a DNA loop. The total change in DNA extension indicates the processivity of the enzyme, while the change in extension with time yields the translocation rate for each separate event; the average of these values would be measured in bulk experiments. (B) When negative supercoiling is introduced, one observes an initial increase in the DNA extension due to supercoil removal by FtsK as it twists and translocates on DNA to form a coiled DNA loop.
With MT one also has the ability to investigate the coupling of translocation and rotation as the enzyme moves forming a DNA loop. At forces below 0.5 pN (Strick et al., 1996), supercoils form on unnicked DNA molecules when the bead is rotated. As a result of the formation of plectonemes, the molecule DNA extension decreases by ~40 nm for every turn added. If the motor’s step size does not fit the helical pitch of DNA, then it will swivel around the DNA as it proceeds along. If the motor forms a DNA loop, by remaining attached to the DNA molecule at one point while continuing to translocate along the DNA, then the DNA in the loop will be coiled. Since the total linking number of DNA is a topological constant, the amount of coil in the loop has to be compensated for opposite supercoiling in the remaining DNA.
MTs allow one to easily control the degree of DNA supercoiling and as such they are ideally suited to investigate the coupling between translocation and rotation. For that purpose, one studies the motor’s activity on a DNA molecule that has been supercoiled through rotation of the magnets. Suppose that the motor’s step size is such that it creates supercoils outside the formed DNA loop of opposite sign to the supercoils generated in the stretched DNA. In this case, as the motor proceeds along, the DNA molecule’s supercoils are absorbed in twisting of the loop associated with the motor. The observed change in extension of the DNA molecule due to loop formation is therefore smaller than for a nicked (uncoiled) DNA (each adsorbed supercoil lengthens the molecule by about 40 nm). Experimentally, a stretched negatively supercoiled DNA actually lengthens as the FtsK motor translocates, indicating that FtsK generates positive supercoils as it moves along DNA (Fig. 13.3B; Saleh et al., 2005). The experimental protocol is exactly the same as for a nicked DNA except that the molecule is negatively coiled after protein injection.
5. Application to the Study of the GP41 Helicase
DNA helicases are ATP-dependent enzymes capable of unwinding dsDNA to provide the ssDNA template required in many biological processes such as DNA replication, repair, and recombination (Delagoutte and Hippel, 2002, 2003; Hippel and Delagoutte, 2001). Generally, a helicase operating in isolation is difficult to assay as the ssDNA intermediates of the unwinding reaction are transient and may reanneal in the wake of the enzyme. Bulk assays measuring helicase activity use DNA traps such as proteins (e.g., single strand binding (SSB) proteins) or enzymes (e.g., single strand nucleases or the cell replication machinery) that trap or process the ssDNA generated by the helicase activity. In MT experiments, the applied force prevents the DNA from reannealing and allows one to follow the activity of helicases in real time in the absence of DNA trap molecules. Moreover, as discussed below, varying the applied force can help determine the unwinding mechanism of helicases. MT experiments enables one to directly measure the unwinding rate (how many base pairs are opened per second), ssDNA translocation rate (how many nucleotides are translocated per second), and processivity (how many base pairs are unwound before the enzyme dissociates from its substrate) of helicases. In order to illustrate how MT can be used to characterize the behavior of DNA helicases, we next present results on the T4 gp41 replicative helicase working on a DNA hairpin substrate.
5.1. Force–extension curve
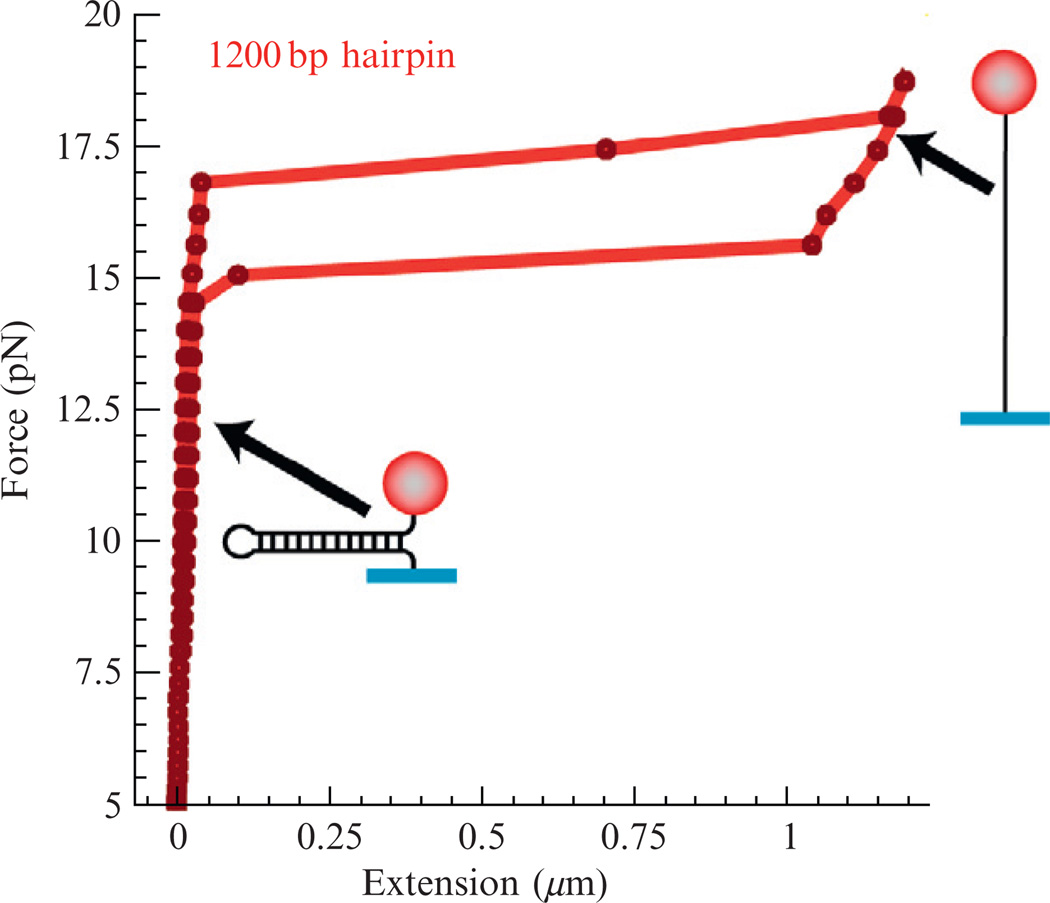
First, we characterize the mechanical stability of the DNA hairpin by measuring the extension of the substrate as a function of the pulling force along a force-cycle in which the force is first increased and then relaxed. The force–extension curve for a 1.2 kbp hairpin is shown in Fig. 13.4. As the force is increased, the hairpin remains annealed at a constant extension until the force reaches Fu = 16 ± 1 pN when the extension abruptly increases due to the mechanical unfolding of the hairpin. As the force is decreased below 14 ± 1 pN (Fr), the hairpin reanneals, returning to its initial extension. At forces F < Fr, the extension of the DNA molecule remains constant at the level corresponding to the folded hairpin. Thus, in that force range and in the presence of helicase, any unfolding observed results from its unwinding activity.
Figure 13.4.
Force–extension curve for a 1.2 kbp hairpin. At low forces the hairpin is annealed and displays a constant extension. As the force is increased above ~16 pN, the hairpin is mechanically unzipped and its extension abruptly increases. The force needs to be reduced below 15 pN for the hairpin to fully reanneal; a certain hysteresis is observed (due to the nucleation of a dsDNA seed in the vicinity of the hairpin loop).
Force–extension protocol
Move the position of the magnets with respect to the top surface of the flow chamber from Zmag = −1 mm (F ~ 1 pN) to Zmag = 0 mm (F ~ 17 pN) and back to Zmag = −1 mm in small steps.
Use the calibrated force versus magnet position curve to estimate the force (Fig. 13.2).
For each position of the magnets measure the DNA extension.
5.2. Detecting helicase activity
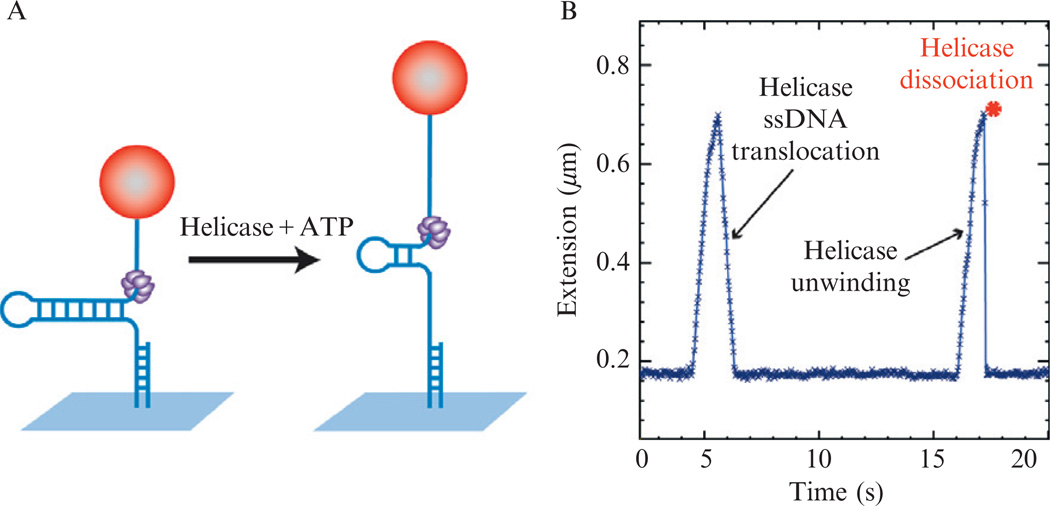
When helicase and ATP are added to the chamber, bursts of helicase activity are observed (Lionnet et al., 2007). Unwinding of the hairpin by a single helicase results in an increase in the end-to-end distance of the DNA molecule observed as a change in the distance between the bead and the surface. Complete unwinding is followed by either the instantaneous rehybridization of the hairpin after enzyme dissociation or by hairpin reannealing in the wake of the helicase as it moves along the ssDNA until the extension of the folded hairpin is recovered (Fig. 13.5B).
Figure 13.5.
Detecting helicase activity on hairpin substrate by MT. (A) Schematic showing increase in DNA extension as a result of helicase activity. (B) Trace showing helicase activity on a 600 bp hairpin (1.2 kbp DNA substrate with a blocking oligo that generate a 600 bp hairpin with 600 nts tails, see below Fig. 13.6B). Two bursts of helicase activity are observed. The first burst corresponds to a full unwinding of the hairpin and the slow hairpin reannealing following the translocation of the helicase on ssDNA, whereas the second burst corresponds to the partial unwinding of the hairpin followed by enzyme dissociation and abrupt hairpin rehybridization.
Materials
T4 gp41 helicase (20–100 nM)
T4 reaction buffer (25 mM Tris–Ac (pH 7.5), 150 mM KOAc, 10 mM Mg(OAc)2, and 1 mM DTT) and 5 mM ATP.
Flow cell with DNA/bead construct.
Protocols
Maintain the force constant at the desired value.
Wait until an extension increase is detected, indicating the initiation of an helicase burst.
Record data during the desired period of time.
5.3. Optimizing helicase loading
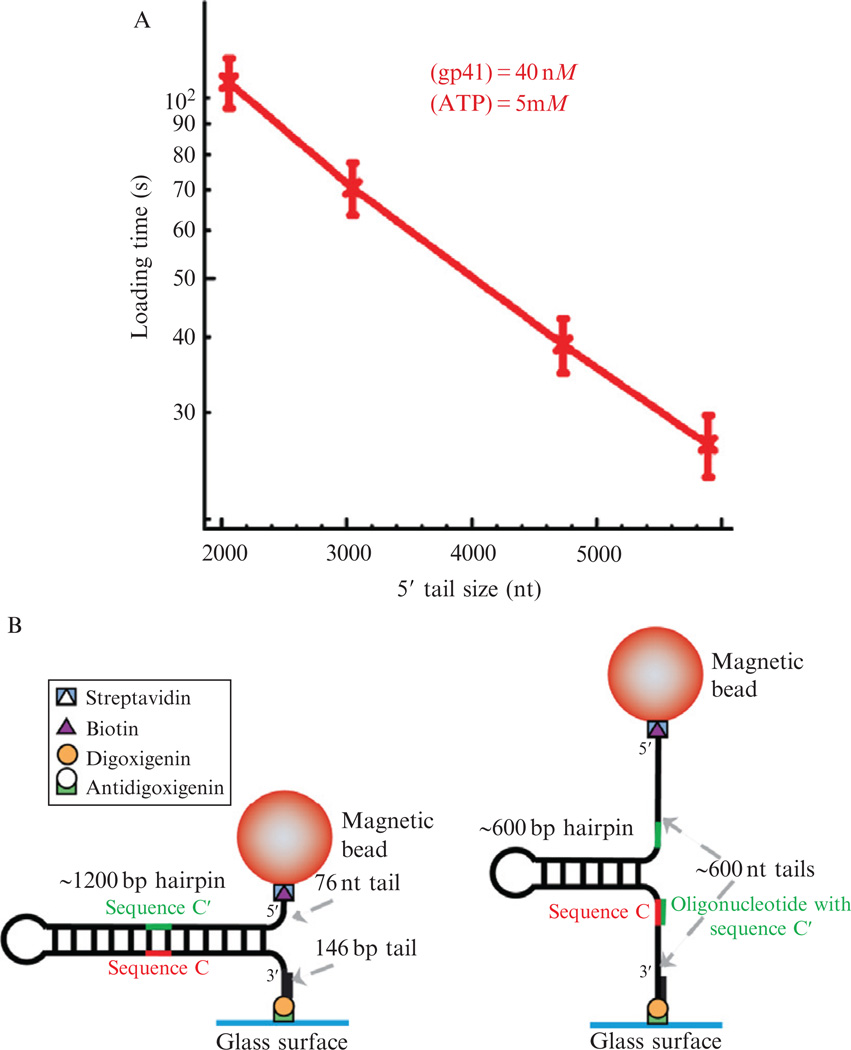
The time required for gp41 helicase to load and start unwinding the DNA hairpin decreases as the length of the 5′ ssDNA tail increases (Fig. 13.6A). As the hairpin is unwound by the helicase, the length of the 5′ ssDNA tail increases. Therefore, a second helicase may bind more rapidly as the substrate is unwound, possibly leading to multiple enzymes binding to the substrate. To ensure single-enzyme conditions, the helicase concentration must be well below 100 nM; however, this results in long initial enzyme loading times.
Figure 13.6.
Optimizing helicase loading time with annealed oligo. (A) The mean helicase loading time as a function of the length of the 5′ ssDNA tail. Assays measured the loading of 40 nM gp41 at saturating ATP concentration on a 6.8 kbp hairpin substrate. Four oligonucleotides complementary to different sequences along the hairpin were used to obtain 5′ ssDNA tails of approximately 2000, 3000, 4500, and 6000 nt. (B) Schematic representation of the DNA hairpin substrate consisting of a 1239 bp hairpin with a 4-nt loop, a 76-nt 5′-biotinylated ssDNA tail, and a 146-bp 3′-digoxigenin labeled dsDNA tail (Manosas et al., 2009), and the half-hairpin substrate created with a complementary 50-mer oligonucleotide (grey) used to reduce the length of the hairpin and increase the length of the 5′ ssDNA tail.
In order to optimize the conditions for helicase loading on the 1.2 kbp hairpin substrate, we have used a complementary 50-mer oligonucleotide that binds near the middle of the hairpin to increase the length of the 5′ ssDNA tail (Fig. 13.6B; Manosas et al., 2009). Before starting an experiment, the oligonucleotide is introduced into the chamber at a high concentration, 1 µM. Then a force large enough to unfold the hairpin (F > 16 pN) is applied for a few seconds allowing the oligonucleotide to hybridize to its complementary sequence in the hairpin. When the force is reduced to low values, the hairpin reanneals up to the position of the oligonucleotide resulting in a substrate with an ~600 bp hairpin, and long 5′ and 3′ ssDNA tails of ~600 nucleotides (nt).
Protocol for optimizing helicase loading using a blocking oligonucleotide
Inject the 50-mer oligonucleotide at 1 µM diluted in the T4 buffer (25 mM Tris–Ac (pH 7.5), 150 mM KOAc, 10 mM Mg(OAc)2, and 1 mM DTT).
Increase the force to ~16 pN in order to denaturate the hairpin.
Wait for few seconds to allow the oligonucleotide to hybridize to the complementary sequence.
Decrease the force to the initial value.
5.4. Measuring unwinding and ssDNA translocation activities
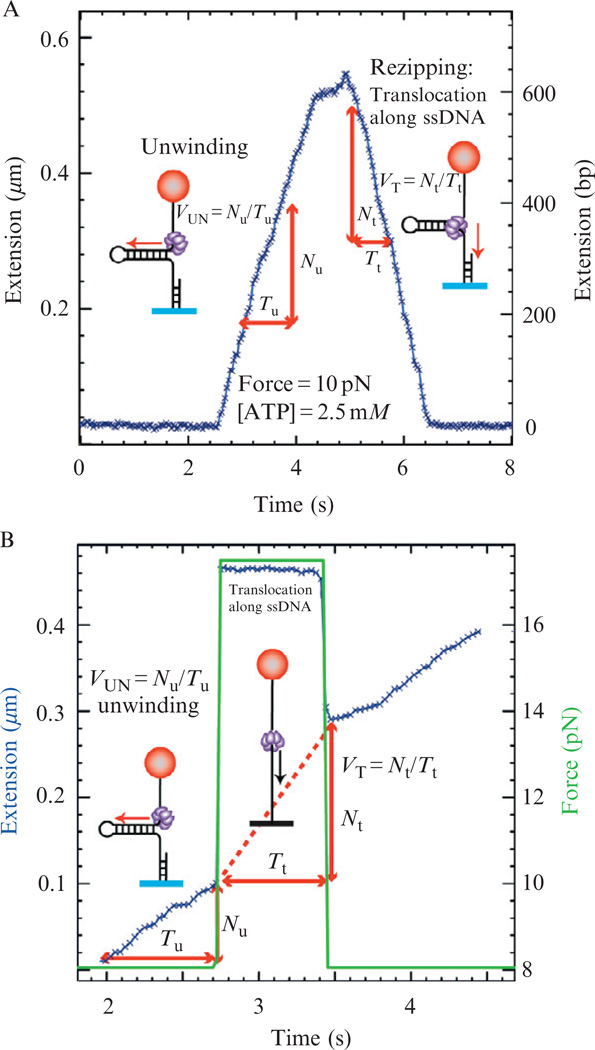
On DNA hairpin substrates, helicase activity is composed of two phases: the unwinding phase (rising edge) corresponding to the release of two nucleotides of ssDNA for each base pair unwound and the rezipping phase (falling edge) corresponding to the slow reannealing of the hairpin following helicase translocation on ssDNA (Lionnet et al., 2007). Conversion of the measured change in DNA extension into the number of base pairs unwound can easily be done by either assigning the maximum DNA extension of the unwinding events to the full length of the unwound hairpin, or by using the previously measured elasticity of ssDNA. The unwinding and ssDNA translocation rates, VUN and VT, can then be directly computed from the slopes of the unwinding and rezipping phases, respectively (Fig. 13.7A). Alternatively, VT can be deduced from an experiment where the force is transiently increased to a value of F > Fu in order to denaturate the hairpin during an unwinding event (Fig. 13.7B). After reducing the force to its initial value (F < Fr) the hairpin reforms and is shorter by Nt base pairs, corresponding to the distance traveled by the helicase on ssDNA.
Figure 13.7.
Measuring unwinding and ssDNa translocation activities. (A) Experimental trace corresponding to the gp41 helicase activity on the 600 bp hairpin (generated from a 1.2 kbp DNA substrate, Fig. 13.6B). Extension in µm (left axis) is converted to number of base pairs unwound (right axis) by assigning to the maximum length of the unwinding events to the full length of the DNA hairpin. The trace shows the unwinding phase (rising edge) and the rezipping phase (falling edge) in which the enzyme translocates on the ssDNA and the hairpin reanneals in its wake. (B) Experimental trace corresponding to the gp41 helicase activity on the 600 bp hairpin. The applied force (grey) is transiently increased during DNA unwinding by the helicase in order to measure the translocation on ssDNA.
Force jump protocol
Wait until a burst of helicase activity is observed.
Increase the force to ~16 pN in order to denaturate the hairpin.
Wait for a few seconds (Δt) to allow the helicase to advance along the ssDNA.
Decrease the force to the initial value.
Calculate the translocation rate by measuring the number of bases the helicase has advanced during Δt.
5.5. Using force to investigate the helicase mechanism
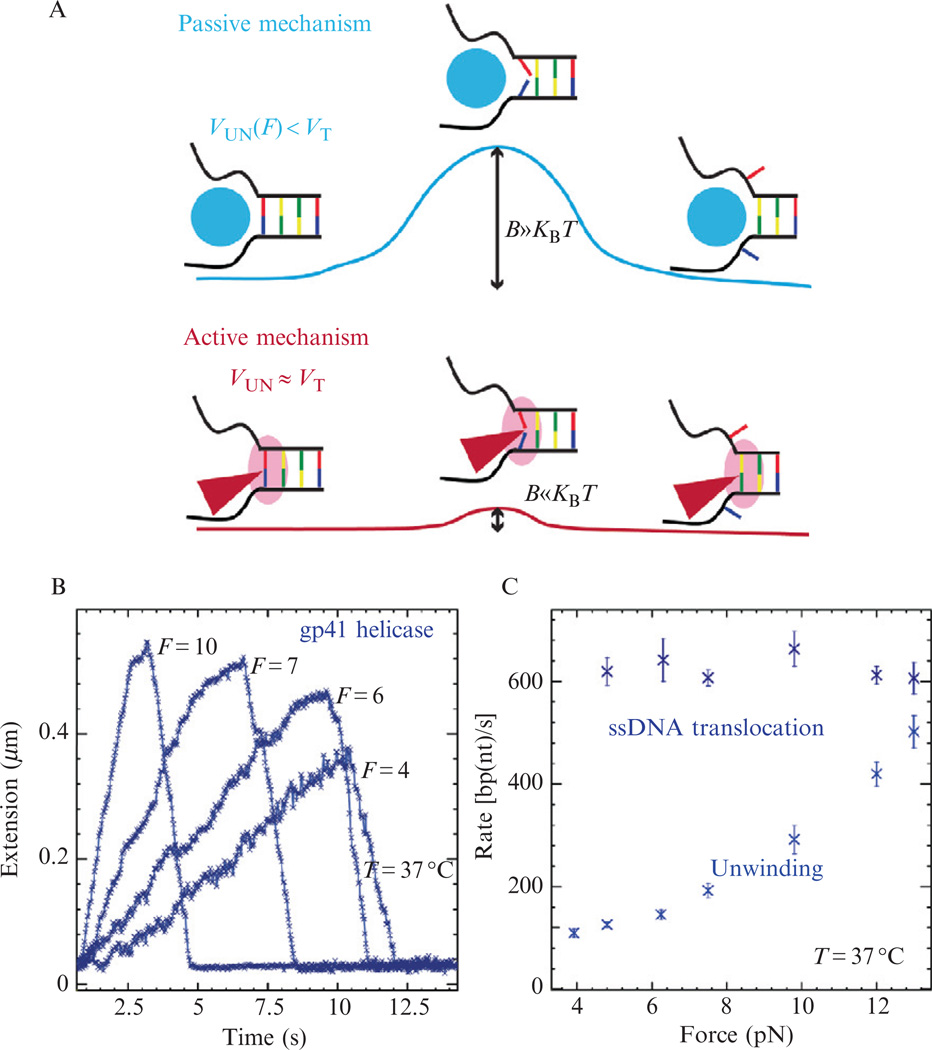
From a mechanistic point of view, the most important issue concerning the function of helicases is the coupling between translocation and DNA unwinding. Two mechanisms for helicase unwinding have been proposed. In the passive model the helicase is a ssDNA translocase simply trapping the transient opening fluctuations of the dsDNA; while in the active model the interaction of the helicase with the dsDNA is sufficient to destabilize the double helix removing it as a block to the forward progression of the enzyme. In general, helicases are enzymes that act by lowering the activation barrier of the reaction they catalyze, that is, DNA unwinding. From that point of view, the difference between an active and a passive helicase rests on the size of the activation energy B (Fig. 13.8A). Therefore, an active helicase is one that is able to lower the activation energy significantly below the base pairing energy (3.4kBT for a GC base pair, where T is the temperature and kB the Boltzman’s constant), that is B ≪ kBT. In contrast, a passive helicase is one that is unable to lower the activation energy resulting in DNA melting being the rate limiting step to DNA unwinding (Fig. 13.8A). In the case of a passive helicase, the application of force on the DNA fork is expected to reduce the activation barrier and result in an increase in the enzyme’s unwinding rate at increasing force. In the case of an active helicase, the effect of the force may be negligible as expected for an inch-worm active model (Lohman and Bjornson, 1996) or counterproductive (i.e., slowing down the enzyme) as expected for an active rolling model (Lohman and Bjornson, 1996). Therefore, single-molecule measurements of the rate of a single helicase unwinding a DNA fork under a given tension can yield insight into the mechanism of the studied helicase. Results obtained for gp41 demonstrate that its unwinding rate is extremely sensitive to the applied force, revealing that this helicase is a predominantly passive helicase (Lionnet et al., 2007) (Fig. 13.8B and C). Similar studies have been performed on other helicases (Cheng et al., 2007; Johnson et al., 2007; Sun et al., 2008).
Figure 13.8.
Determining helicase mechanism. (A) Schematic representing passive versus active unwinding. (B) Several helicase activity traces at various applied forces. (C) Force dependence of the helicase translocation rate, VT, and unwinding rate, VUN.
6. Conclusions
In this article we have shown how MTs can be used efficiently to monitor in real time the activity of DNA translocases and helicases. From such data, one can extract enzymatic states (e.g., translocation on dsDNA or ssDNA, forward or backward), unwinding and translocation rates, processivity, and step-size as well as learn about enzymatic mechanisms by studying the variation of these measurables with force and twist on the DNA molecule.
ACKNOWLEDGMENTS
We acknowledge the partial support of grants from ANR, HFSP (RGP0003/2007-C), BioNanoSwitch, IUF and PUF.
REFERENCES
- Aussel L, Barre FX, Aroyo M, Stasiak A, Stasiak AZ, Sherratt D. FtsK is a DNA motor protein that activates chromosome dimer resolution by switching the catalytic state of the XerC and XerD recombinases. Cell. 2002;108(2):195–205. doi: 10.1016/s0092-8674(02)00624-4. [DOI] [PubMed] [Google Scholar]
- Bouchiat C, Wang M, Block SM, Allemand J-F, Strick T, Croquette V. Estimating the persistence length of a worm-like chain molecule from force–extension measurements. Biophys. J. 1999;76:409–413. doi: 10.1016/s0006-3495(99)77207-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustamante C, Marko J, Siggia E, Smith S. Entropic elasticity of λ-phage DNA. Science. 1994;265:1599–1600. doi: 10.1126/science.8079175. [DOI] [PubMed] [Google Scholar]
- Charvin G, Allemand J, Strick T, Bensimon D, Croquette V. Twisting DNA: Single molecule studies. Contemp. Phys. 2004;45(5):383–403. [Google Scholar]
- Cheng W, Dumont S, Tinoco I, Bustamante C. NS3 helicase actively separates RNA strands and senses sequence barriers ahead of the opening fork. Proc. Natl. Acad. Sci. USA. 2007;104(35):13954–13959. doi: 10.1073/pnas.0702315104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delagoutte E, Hippel PHV. Helicase mechanisms and the coupling of helicases within macromolecular machines part I: Structures and properties of isolated helicases. Q. Rev. Biophys. 2002;35:431–478. doi: 10.1017/s0033583502003852. [DOI] [PubMed] [Google Scholar]
- Delagoutte E, Hippel PHV. Helicase mechanisms and the coupling of helicases within macromolecular machines part II: Integration of helicases into cellular processes. Q. Rev. Biophys. 2003;36:1–69. doi: 10.1017/s0033583502003864. [DOI] [PubMed] [Google Scholar]
- Engel A, Gaub H, Müller D. Atomic force microscopy: A forceful way with single molecules. Curr. Biol. 1999;9:R133–R136. doi: 10.1016/s0960-9822(99)80081-5. [DOI] [PubMed] [Google Scholar]
- Gosse C, Croquette V. Magnetic tweezers: Micromanipulation and force measurement at the molecular level. Biophys. J. 2002;82:3314–3329. doi: 10.1016/S0006-3495(02)75672-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hippel PHV, Delagoutte E. A general model for nucleic acid helicases and their “coupling” within macromolecular machines. Cell. 2001;104:177–190. doi: 10.1016/s0092-8674(01)00203-3. [DOI] [PubMed] [Google Scholar]
- Johnson D, Bai L, Smith B, Patel S, Wang M. Asingle-molecule studies reveal dynamics of DNA unwinding by the ring-shaped t7 helicase. Cell. 2007;129(7):1299–1309. doi: 10.1016/j.cell.2007.04.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller D, Bustamante C. The mechanochemistry of molecular motors. Biophys. J. 2000;78:541–556. doi: 10.1016/S0006-3495(00)76615-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lionnet T, Spiering M, Benkovic S, Bensimon D, Croquette V. Real-time observation of bacteriophage t4 gp41 helicase reveals an unwinding mechanism. Proc. Natl. Acad. Sci. USA. 2007;104:19790–19795. doi: 10.1073/pnas.0709793104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lionnet T, Allemand J-F, Andrey Revyakin TRS, Saleh OA, Bensimon D, Croquette V. Single Molecule Techniques A Laboratory Manual. Chapter 19. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2008. [Google Scholar]
- Lohman TM, Bjornson KP. Mechanisms of helicase-catalysed unwinding. Annu. Rev. Biochem. 1996;65:169–214. doi: 10.1146/annurev.bi.65.070196.001125. [DOI] [PubMed] [Google Scholar]
- Manosas M, Spiering MM, Zhuang Z, Benkovic SJ, Croquette V. Coupling DNA unwinding activity with primer synthesis in the bacteriophage T4 primosome. Nat. Chem. Biol. 2009;5(12):904–912. doi: 10.1038/nchembio.236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revyakin A, Allemand J, Croquette V, Ebright R, Strick T. Single-molecule DNA nanomanipulation: Detection of promoter-unwinding events by RNA polymerase. Methods Enzymol. 2003;370:577–598. doi: 10.1016/S0076-6879(03)70049-4. [DOI] [PubMed] [Google Scholar]
- Saleh OA, Bigot S, Barre FX, Allemand JF. Analysis of DNA supercoil induction by FtsK indicates translocation without groove-tracking. Nat. Struct. Mol. Biol. 2005;12(5):436–440. doi: 10.1038/nsmb926. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Russell DW. Molecular Cloning: A Laboratory Manual. 3rd edn. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- Strick TR, Allemand JF, Bensimon D, Bensimon A, Croquette V. The elasticity of a single supercoiled DNA molecule. Science. 1996;271:1835–1837. doi: 10.1126/science.271.5257.1835. [DOI] [PubMed] [Google Scholar]
- Sun B, Wei K, Zhang B, Zhang X, Dou S, Li M, Xi X. Impediment of E. coli UvrD by DNA-destabilizing force reveals a strained-inchworm mechanism of DNA unwinding. EMBO J. 2008;27(24):3279–3287. doi: 10.1038/emboj.2008.240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svoboda K, Mitra PP, Block SM. Fluctuation analysis of motor protein movements and single enzyme kinetics. Proc. Natl. Acad. Sci. USA. 1994;91:11782–11786. doi: 10.1073/pnas.91.25.11782. [DOI] [PMC free article] [PubMed] [Google Scholar]