Abstract
The availability and robustness of methods to analyze phosphorylated proteins has greatly expanded our knowledge of phosphorylation-based cell signaling. A key ingredient to the success of these studies is the ability to enrich phosphopeptides using antibodies or other chemical approaches. Most other post-translational modifications, such as lysine acetylation, are still poorly characterized because of the lack of availability of such enrichment methods. Recently, some groups have reported identification of acetylation sites in a global fashion by enriching acetylated peptides with a polyclonal antibody from a single source that was raised against pan-acetylated lysine. Instead of using this polyclonal antibody, we used a cocktail of monoclonal antibodies where each was directed against acetylated lysine in different contexts. Using high resolution Fourier transform mass spectrometry, we observed that the majority of acetylated lysine residues identified using the monoclonal antibody cocktail were distinct from those enriched by the polyclonal antibody used by the other groups. Our study demonstrates that immunoaffinity enrichment of acetylated peptides is somewhat limited by substrate specificity and that an optimal yield of enrichment can be achieved by employing a broader array of affinity reagents.
Post-translational modifications (PTMs) of proteins are integral to the regulation of cellular processes but have been tedious to characterize and quantify in a global fashion until recently. The greatest advances have been made in the field of phosphorylation for which tens of thousands of unique sites have now been described by enriching phosphopeptides based on their chemical characteristics and through immunoaffinity approaches for the low abundance phosphotyrosine peptides(1). Enrichment techniques for other PTMs, such as acetylation, have also been pursued with some success. These approaches have used isoelectric focusing(2) or immunoaffinity(3, 4, 5, 6, 7, 8) based methods to identify acetylated peptides or proteins, and one study identified 3,600 acetylation sites in human cancers using both methods in tandem(9). The analysis described here examines the potential to further broaden the number of acetylation sites identified in an experiment by using a diverse set of antibodies for enrichment of acetylated peptides.
The enzymatic regulation of lysine acetylation of histones has been known for some time(10). Mounting evidence suggests that lysine acetylation of non-histone proteins plays a major role in protein stability, localization and cellular signaling(11, 12, 13). Acetylation results in dynamic changes in protein function by neutralizing the positive charge of lysine residues and through the interplay of competing lysine modifications such as methylation, sumoylation and ubiquitination(13). Early studies suggesting that non-histone proteins are regulated by lysine acetylation examined the regulation of p53 and other proteins of critical importance to diseases such as cancer(11, 11, 14, 15). Similar to the regulation of histone lysine acetylation, non-histone proteins are acetylated by histone acetyltransferases (HAT) and deacetylated by histone deacetylase (HDAC) enzymes, the latter of which are now promising therapeutic targets in the treatment of cancer. The histone deacetylase (HDAC) inhibitor suberoylanilide hydroxamic acid (SAHA) or vorinostat is an FDA-approved drug for the treatment of cutaneous T-cell lymphoma(16, 17) and is currently under investigation for use in breast cancer. Although the rationale for inhibition of HDACs as a cancer therapy was aimed at the targeted re-expression of tumor suppressor genes commonly silenced in cancer progression, the diverse array of non-histone proteins regulated by lysine acetylation underpins the potential pleiotropic effects of HDAC inhibitors as cancer therapies. Identification of these molecules can lead to the development of more targeted therapies and elucidate mechanisms of drug efficacy and resistance.
Recent studies using mass spectrometry to analyze acetylated proteins and peptides enriched using an anti-acetyl lysine polyclonal antibody from a single commercial source have demonstrated the high prevalence of non-histone protein acetylation in human cancers, including breast cancer(4, 6, 9). It should be noted that the immunoprecipitation step in these studies was performed using the rabbit polyclonal pan-acetylated lysine antibody from a single source, which is potentially limited by selectively enriching peptides with a narrow sequence diversity because they bear similarity to the antigen used to generate the antibody. In addition, the supply of the antibody is not unlimited and more difficult to standardize in terms of specificity. To circumvent these limitations, we used a monoclonal antibody cocktail in which the monoclonal antibodies were raised against a variety of antigens containing acetylated lysine (Table 1). Previous studies have demonstrated that pan-acetylated lysine antibodies derived from multiple sources have different affinities for given acetylated peptide sequences (18). Therefore, a cocktail of monoclonal antibodies has the potential to enrich a significant subset of acetylated peptides which might otherwise go unidentified using only the conventional polyclonal antibody.
Table 1.
Comparison of pan-acetyl lysine antibodies
| Immunogen | Clone | Antibody Class | Supplier |
|---|---|---|---|
| Acetylated peptide coupled KLH | Not applicable | Rabbit Polyclonal | ImmuneChem |
| Acetylated KLH | AKL5C1 | Mouse Mono IgG1 | Santa Cruz |
| KLH coupled XXX(Kac)XXX | Ac-K-103 | Mouse Mono IgG2a | Cell Signaling |
| Acetylated protein mixture | 15G10 | Mouse Mono IgG2b | Novus |
| Synthetic Ac K9 of Histone H3 | 1C6 | Mouse Mono IgG | Pierce |
EXPERIMENTAL SECTION
Materials
Acetonitrile was obtained from Fisher Scientific. All other chemicals were obtained from Sigma unless stated otherwise. Antibodies were obtained from the suppliers listed in Table 1.
Cell Culture and Sample Preparation
MDA-MB-231 cells were obtained from the American Tissue Culture Collection (ATCC) and grown in Dulbecco’s Modified Eagle Medium with 5% FBS at 37°C and 5% CO2. Cells were plated in 15cm culture dishes, grown to 80% confluency and treated with 10 μM suberoylanilide hydroxamic acid (SAHA, Cayman Chemical) for 24 hours prior to harvesting by scraping in lysis buffer (9.0 M Urea, 20 mM Hepes pH 8.0, 1 mM sodium orthovanadate, 2.5 mM sodium pyrophosphate, 1 mM β-glycerophosphate and 0.5 μM trichostatin A), yielding roughly 3 to 5 mg of protein lysate per culture dish. Cells were lysed by sonication and centrifuged at 20,000 × g for 15 minutes at 15°C, followed by reduction in 5 mM DTT and alkylation of cysteines by incubation in 10 mM iodoacetamide in the dark. The protein lysate was then digested using 1mg of trypsin (Worthington) per 50mg of protein after diluting 5-fold with 20 mM Hepes pH 8.0. The peptide mixture was acidified using trifluoroacetic acid (TFA) at final concentration of 1% and purified by C18 Sep-Pak filtration (Waters) followed by elution with 40% acetonitrile, 0.1% TFA and lyophylization at −60° C.
Immunoprecipitation
Dried peptides were resuspended in immunoprecipitation buffer (50 mM MOPS pH 7.2, 10 mM sodium phosphate and 50 mM NaCl) and incubated with either polyclonal antibody (ImmuneChem) or a monoclonal cocktail (four monoclonal antibodies combined in equal ratio) coupled to protein G agarose beads according to the manufacturer’s instructions (Roche). Immunoaffinity enrichment was carried out by incubating 100 μg of total antibody with 10 mg of peptide with constant mixing at 4°C overnight. Peptides were washed 3 times with immunoprecipitation buffer and then eluted with 0.1% TFA and purified using the Stage-tip method as previously described(19).
Mass Spectrometry
Peptides were analyzed by direct online injection into an LTQ-Orbitrap XL ETD mass spectrometer (Thermo) from an Agilent 1100 autosampler coupled to an Eksigent nanopump, or an LTQ-Orbitrap Velos mass spectrometer (Thermo) with a NanoAcquity UPLC (Waters). Chromatographic separation was performed over a gradient from 9–55% acetonitrile containing 0.1% formic acid over 120 minutes on a 50 μm × 15 cm analytical column packed in-house (Magic 5.0 μm, 300Å pore size, Michrom). Using optimized resolution parameters(20), survey scans were acquired in the Orbitrap from a range of m/z 350–1800 at 60,000 resolution with polydimethylcyclosiloxane from ambient air as a lock mass (m/z 445.120025)(21). In each cycle, the 8 most intense ions were fragmented by collisionally induced dissociation (CID) prior to ion trap detection using the LTQ-Orbitrap XL and the 10 most intense ions were fragmented by high energy collision dissociation (HCD) prior to Orbitrap detection at 7500 resolution using the LTQ-Orbitrap Velos. Ions were isolated at a target of 10,000 ions or 150 ms maximum injection time before being excluded for the following 30 seconds.
Data Processing
Spectra were processed using Proteome Discoverer Version 1.2 (Thermo) to merge search results from Mascot (v.2.2, Matrix Science), with and without deconvolution, and Sequest (Thermo), and subjected to a strict cutoff of 1% false discovery rate (FDR). Spectra were searched against the RefSeq45 human protein database using a 20 ppm mass cutoff for MS and either a 0.5 Da (CID) or 0.05 Da cutoff for MS/MS spectra. Carbamidomethyl cysteine was searched as a fixed modification while methionine oxidation, pyroglutamine, protein N-terminal acetylation and lysine acetylation were used as variable modifications. Full details of search results and analysis are available as part of the Supporting Information (Table S1–S4). Sequence logos for acetylation sites were generated using enoLOGOS(22). The parameters were used such that weight type was ‘alignment counts’ and LOGO plot method was based on frequency.
Statistical Analysis
The proportion of acetylation sites common between two replicate runs using the same affinity reagent (either the monoclonal antibody cocktail or the polyclonal antibody) or between runs using two different affintity reagents (i.e. monoclonal versus polyclonal) were calculated as the number of common sites divided by the average number of sites identified in the two runs. To assess whether the two different enrichment approaches contributed to a difference in the acetylation sites identified, a z-test for two independent proportions was performed. We tested the null hypothesis that the proportion of common sites identified between two runs of different enrichment approaches was no different than what would be observed between two replicate runs of the same enrichment approach. The results of analysis is summarized in Table S5.
RESULTS AND DISCUSSION
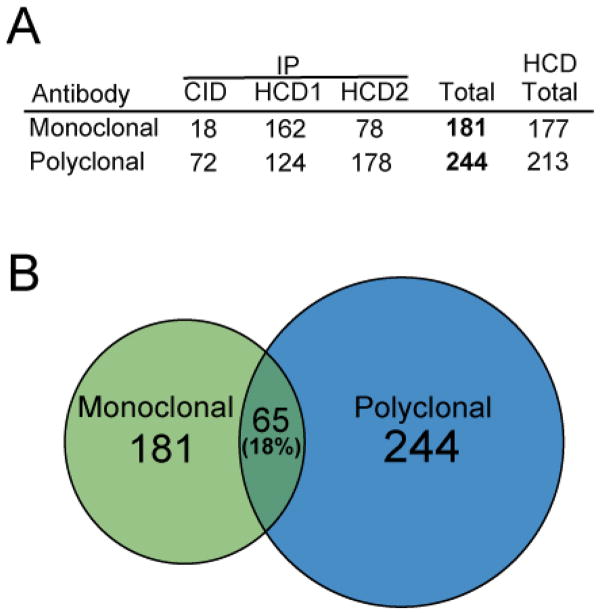
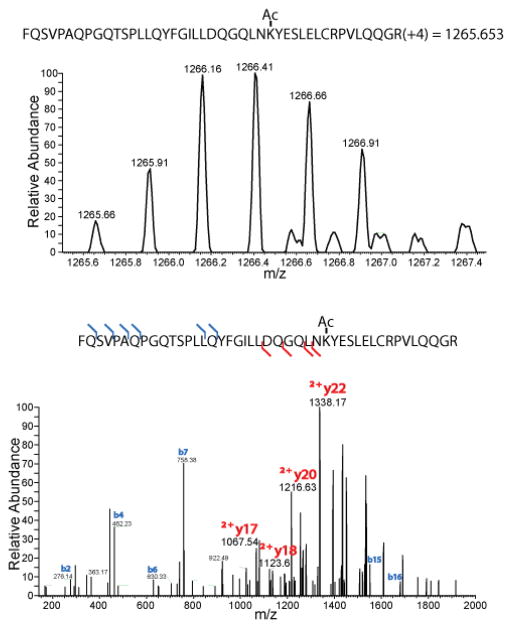
Three independent peptide immunopreciptions were performed using the monoclonal cocktail or the polyclonal antibody, for which one sample was analyzed by ion trap detection of CID fragments and the other two were analyzed by Orbitrap detection of MS/MS ions from HCD fragmentation. In total, the three biological replicates led to identification of 181 and 244 acetylation sites from enrichment with the monoclonal antibody cocktail and the polyclonal antibody, respectively (Figure 1). Both immunoprecipitation procedures yielded acetylated lysines on histones and a variety of non-histone proteins, suggesting that both reagents possess binding affinity for a broad spectrum of acetylated peptide sequences. Sample analysis with the high resolution MS/MS detection of HCD fragments dramatically outperformed the ion trap CID method, yielding considerably more acetylation sites identified in each HCD experiment compared to the corresponding CID experiment (Figure 1A). The enhancement of acetylation sites identified in the HCD analysis relative to CID analysis is due to multiple factors, including differences in ion accumulation and scan speed between the two instruments used. In particular, high resolution detection of HCD fragments is critical to the identification of large, highly charged acetylated peptides as seen in Figure 2. In addition to accurate mass measurement, large highly charged peptides can be confidently identified by several doubly charged fragments when using high resolution detection of MS/MS fragments, that would not be identified by ion trap detection.
Figure 1.
The number of sites identified from each immunoprecipitation experiment using various fragmention schemes as indicated is listed in (A). The relative overlap of the datasets is displayed as a Venn diagram with the circles drawn to scale in (B).
Figure 2.
The high resolution MS spectrum of a large peptide with a +4 charge (top) is unambiguously identified by high resolution detection of both singly and doubly charged MS/MS fragments (bottom).
Interestingly, the overlap of identified acetylated residues was limited to 65 lysines belonging to only 37 different proteins, indicating distinctive antigen specificities between the different reagents. Statistical analysis of the replicate HCD datasets shows that the proportion of common acetylation sites identified between the monoclonal antibody cocktail and polyclonal antibody ranged from 28.8% to 37.6%, while the proportions between replicate runs of the same enrichment approach were 52.5% for monoclonal antibody cocktail and 58.9% for the polyclonal antibody. The difference between the proportion of common sites from runs of the same approach is significantly higher than that from runs of different approaches, with p-values ranging between 0.038 to <0.001. This indicates that the monoclonal antibody cocktail enrichment identified a unique subset of acetylation sites compared to the sites identified using the polyclonal antibody with statistical significance, even when adjusting for the run-to-run variation. (Table S5).
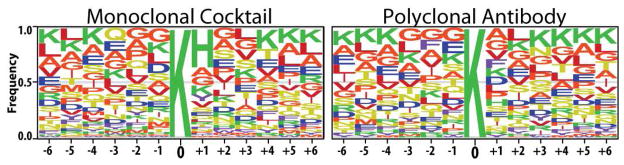
One of the reasons why the monoclonal cocktail led to identification of acetylated lysines not identified by the polyclonal antibody is likely due to the specificity of the monoclonal antibodies for a distinct subset of peptides. To assess this possibility, we compared the peptide sequences surrounding the acetylation sites identified by each method (Figure 3). The only major sequence motif which stands out in this data is the occurrence of histidine (H) at the +1 position at a 35% frequency in the monoclonal dataset but with only a 6% frequency in the polyclonal dataset. This motif difference has not been described by other dataset analyses searching for acetylation motifs (23, 24), suggesting that it is specific to the peptide population enriched by the monoclonal cocktail. Additionally, it is possible that subsets of more complex motifs are hidden in these datasets. The significant number of acetylation sites unique to the monoclonal-derived dataset, together with the identification of a new acetylation motif that is exclusive to the monoclonal cocktail enrichment, is consistent with the possibility that a unique subset of acetylated peptides was enriched using the monoclonal antibody cocktail.
Figure 3.
Sequence alignment of the regions six amino acids from either side of the acetylated lysine residues identified using the monoclonal cocktail (left) and the polyclonal antibody (right).
CONCLUSIONS
Although current methods using a polyclonal antibody from a single supplier have been used to identify a considerable number of acetylation sites in the human proteome, this work demonstrates that a cocktail of multiple monoclonal antibodies has the potential to complement the breadth of acetylated peptides enriched for this type of analysis. Clearly, further biological insights will be necessary to determine the full benefit of additional acetylation sites identified using the monoclonal cocktail.
Supplementary Material
Acknowledgments
The authors acknowledge Vikram Katju and Jyoti Sharma of the Institute of Bioinformatics for their assistance in the data analysis and Robert O’Meally and Robert Cole of the Johns Hopkins Mass Spectrometry Core Facility for help with sample analysis. This work was supported by grants from the National Institutes of Health (CA88843, S10RR023025, U54 RR 020839, P30 ES03819), the Breast Cancer Research Foundation and by a Breast Cancer Research Program Era of Hope Award (W81XWH-06-1-0428) from the Department of Defense.
Footnotes
SUPPORTING INFORMATION AVAILABLE
Additional information as noted in text. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Grimsrud PA, Swaney DL, Wenger CD, Beauchene NA, Coon JJ. ACS Chem Biol. 2010;5:105–119. doi: 10.1021/cb900277e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Xie H, Bandhakavi S, Roe MR, Griffin TJ. J Proteome Res. 2007;6:2019–2026. doi: 10.1021/pr060691j. [DOI] [PubMed] [Google Scholar]
- 3.Iwabata H, Yoshida M, Komatsu Y. Proteomics. 2005;5:4653–4664. doi: 10.1002/pmic.200500042. [DOI] [PubMed] [Google Scholar]
- 4.Kim SC, Sprung R, Chen Y, Xu Y, Ball H, Pei J, Cheng T, Kho Y, Xiao H, Xiao L, Grishin NV, White M, Yang XJ, Zhao Y. Mol Cell. 2006;23:607–618. doi: 10.1016/j.molcel.2006.06.026. [DOI] [PubMed] [Google Scholar]
- 5.Zhang J, Sprung R, Pei J, Tan X, Kim S, Zhu H, Liu CF, Grishin NV, Zhao Y. Mol Cell Proteomics. 2009;8:215–225. doi: 10.1074/mcp.M800187-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhou Q, Chaerkady R, Shaw PG, Kensler TW, Pandey A, Davidson NE. Proteomics. 2010;10:1029–1039. doi: 10.1002/pmic.200900602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang Q, Zhang Y, Yang C, Xiong H, Lin Y, Yao J, Li H, Xie L, Zhao W, Yao Y, Ning ZB, Zeng R, Xiong Y, Guan KL, Zhao S, Zhao GP. Science. 2010;327:1004–1007. doi: 10.1126/science.1179687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhao S, Xu W, Jiang W, Yu W, Lin Y, Zhang T, Yao J, Zhou L, Zeng Y, Li H, Li Y, Shi J, An W, Hancock SM, He F, Qin L, Chin J, Yang P, Chen X, Lei Q, Xiong Y, Guan KL. Science. 2010;327:1000–1004. doi: 10.1126/science.1179689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Choudhary C, Kumar C, Gnad F, Nielsen ML, Rehman M, Walther TC, Olsen JV, Mann M. Science. 2009;325:834–840. doi: 10.1126/science.1175371. [DOI] [PubMed] [Google Scholar]
- 10.Inoue A, Fujimoto D. Biochem Biophys Res Commun. 1969;36:146–150. doi: 10.1016/0006-291x(69)90661-5. [DOI] [PubMed] [Google Scholar]
- 11.Kouzarides T. EMBO J. 2000;19:1176–1179. doi: 10.1093/emboj/19.6.1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang XJ, Seto E. Mol Cell. 2008;31:449–461. doi: 10.1016/j.molcel.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Spange S, Wagner T, Heinzel T, Kramer OH. Int J Biochem Cell Biol. 2009;41:185–198. doi: 10.1016/j.biocel.2008.08.027. [DOI] [PubMed] [Google Scholar]
- 14.Ito A, Lai CH, Zhao X, Saito S, Hamilton MH, Appella E, Yao TP. EMBO J. 2001;20:1331–1340. doi: 10.1093/emboj/20.6.1331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hubbert C, Guardiola A, Shao R, Kawaguchi Y, Ito A, Nixon A, Yoshida M, Wang XF, Yao TP. Nature. 2002;417:455–458. doi: 10.1038/417455a. [DOI] [PubMed] [Google Scholar]
- 16.Olsen EA, Kim YH, Kuzel TM, Pacheco TR, Foss FM, Parker S, Frankel SR, Chen C, Ricker JL, Arduino JM, Duvic M. J Clin Oncol. 2007;25:3109–3115. doi: 10.1200/JCO.2006.10.2434. [DOI] [PubMed] [Google Scholar]
- 17.Mann BS, Johnson JR, Cohen MH, Justice R, Pazdur R. Oncologist. 2007;12:1247–1252. doi: 10.1634/theoncologist.12-10-1247. [DOI] [PubMed] [Google Scholar]
- 18.Komatsu Y, Yukutake Y, Yoshida M. J Immunol Methods. 2003;272:161–175. doi: 10.1016/s0022-1759(02)00500-8. [DOI] [PubMed] [Google Scholar]
- 19.Rappsilber J, Mann M, Ishihama Y. Nat Protoc. 2007;2:1896–1906. doi: 10.1038/nprot.2007.261. [DOI] [PubMed] [Google Scholar]
- 20.Kim MS, Kandasamy K, Chaerkady R, Pandey A. J Am Soc Mass Spectrom. 2010 doi: 10.1016/j.jasms.2010.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Olsen JV, de Godoy LM, Li G, Macek B, Mortensen P, Pesch R, Makarov A, Lange O, Horning S, Mann M. Mol Cell Proteomics. 2005;4:2010–2021. doi: 10.1074/mcp.T500030-MCP200. [DOI] [PubMed] [Google Scholar]
- 22.Workman CT, Yin Y, Corcoran DL, Ideker T, Stormo GD, Benos PV. Nucleic Acids Res. 2005;33:W389–92. doi: 10.1093/nar/gki439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schwartz D, Chou MF, Church GM. Mol Cell Proteomics. 2009;8:365–379. doi: 10.1074/mcp.M800332-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Basu A, Rose KL, Zhang J, Beavis RC, Ueberheide B, Garcia BA, Chait B, Zhao Y, Hunt DF, Segal E, Allis CD, Hake SB. Proc Natl Acad Sci U S A. 2009;106:13785–13790. doi: 10.1073/pnas.0906801106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.