Abstract
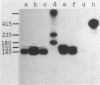
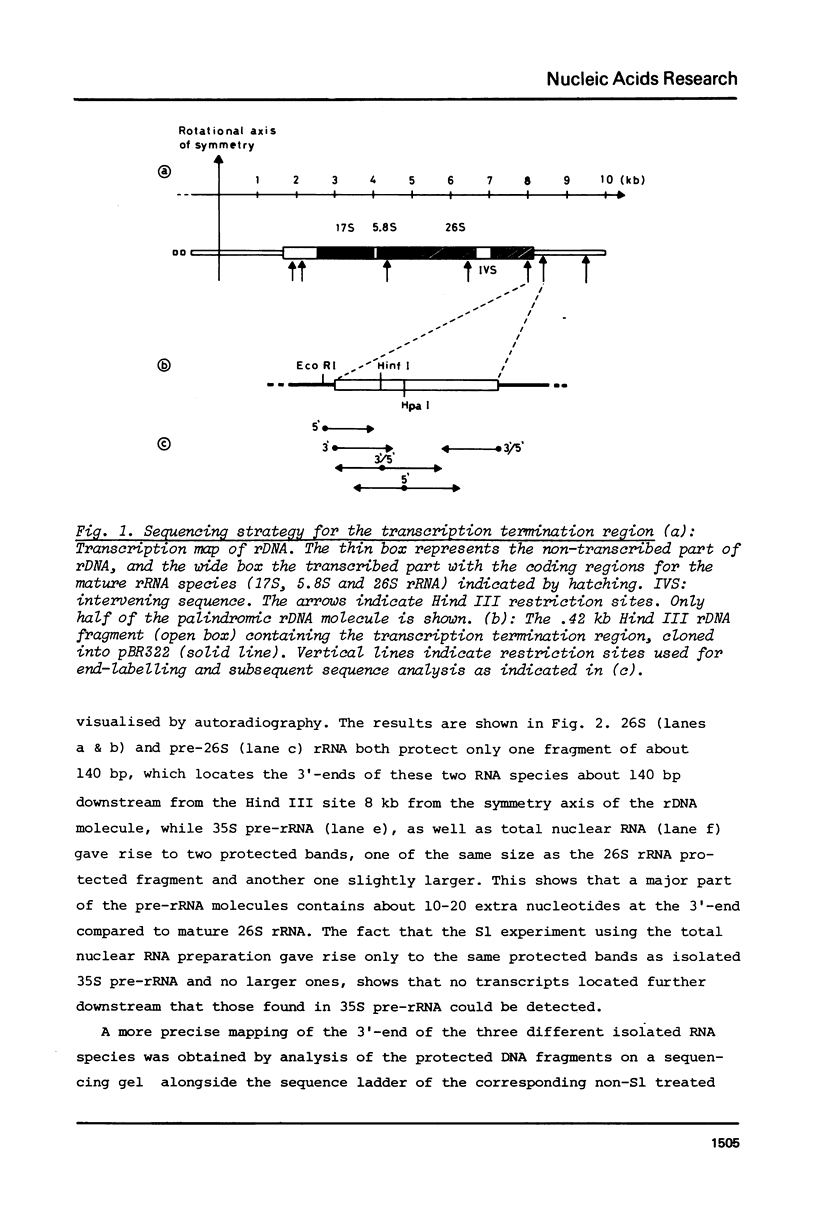
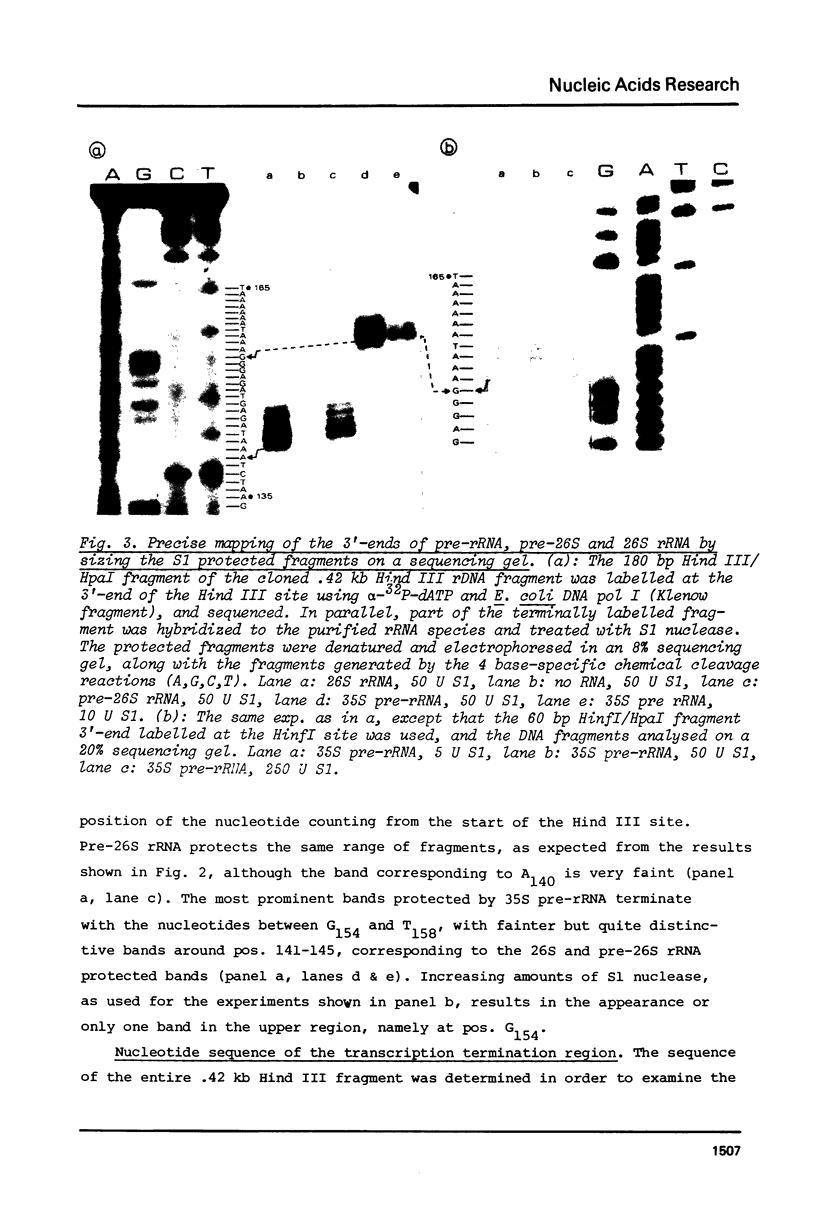
The sequence of 415 nucleotides surrounding the transcription termination site for ribosomal RNA in Tetrahymena thermophila has been determined. The positions of the 3'-ends of mature 26S rRNA, pre-26S rRNA and 35S pre-rRNA were localized within this sequence by hybridization of the purified RNA species to be selected DNA fragments, followed by S1 nuclease treatment of the hybrid and a precise sizing of the RNA-protected DNA fragments on sequencing gels. The 35S pre-rRNA population contained molecules with two distinct 3'-ends, one of which is identical to the end of pre-26S and 26S rRNA, while the other corresponds to a position 15 nucleotides further downstream, which is assumed to be the transcription termination site. The non-coding DNA strand contains a cluster of T's at the putative termination site, and several other T clusters are found further downstream. A short inverted repeat sequence is located near the putative termination site within the transcribed region. The possible role of these structures for transcription termination is discussed.
Full text
PDF








Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Azad A. A., Deacon N. J. The 3'-terminal primary structure of five eukaryotic 18S rRNAs determined by the direct chemical method of sequencing. The highly conserved sequences include an invariant region complementary to eukaryotic 5S rRNA. Nucleic Acids Res. 1980 Oct 10;8(19):4365–4376. doi: 10.1093/nar/8.19.4365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogenhagen D. F., Sakonju S., Brown D. D. A control region in the center of the 5S RNA gene directs specific initiation of transcription: II. The 3' border of the region. Cell. 1980 Jan;19(1):27–35. doi: 10.1016/0092-8674(80)90385-2. [DOI] [PubMed] [Google Scholar]
- Bram R. J., Young R. A., Steitz J. A. The ribonuclease III site flanking 23S sequences in the 30S ribosomal precursor RNA of E. coli. Cell. 1980 Feb;19(2):393–401. doi: 10.1016/0092-8674(80)90513-9. [DOI] [PubMed] [Google Scholar]
- Brown D. D., Gurdon J. B. High-fidelity transcription of 5S DNA injected into Xenopus oocytes. Proc Natl Acad Sci U S A. 1977 May;74(5):2064–2068. doi: 10.1073/pnas.74.5.2064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cech T. R., Rio D. C. Localization of transcribed regions on extrachromosomal ribosomal RNA genes of Tetrahymena thermophila by R-loop mapping. Proc Natl Acad Sci U S A. 1979 Oct;76(10):5051–5055. doi: 10.1073/pnas.76.10.5051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clewell D. B., Helinski D. R. Supercoiled circular DNA-protein complex in Escherichia coli: purification and induced conversion to an opern circular DNA form. Proc Natl Acad Sci U S A. 1969 Apr;62(4):1159–1166. doi: 10.1073/pnas.62.4.1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Din N., Engberg J. Extrachromosomal ribosomal RNA genes in Tetrahymena: structure and evolution. J Mol Biol. 1979 Nov 5;134(3):555–574. doi: 10.1016/0022-2836(79)90367-x. [DOI] [PubMed] [Google Scholar]
- Din N., Engberg J., Kaffenberger W., Eckert W. A. The intervening sequence in the 26S rRNA coding region of T. thermophila is transcribed within the largest stable precursor for rRNA. Cell. 1979 Oct;18(2):525–532. doi: 10.1016/0092-8674(79)90069-2. [DOI] [PubMed] [Google Scholar]
- Eckert W. A., Kaffenberger W., Krohne G., Franke W. W. Introduction of hidden breaks during rRNA maturation and ageing in Tetrahymena pyriformis. Eur J Biochem. 1978 Jul 3;87(3):607–616. doi: 10.1111/j.1432-1033.1978.tb12413.x. [DOI] [PubMed] [Google Scholar]
- Engberg J., Andersson P., Leick V., Collins J. Free ribosomal DNA molecules from Tetrahymena pyriformis GL are giant palindromes. J Mol Biol. 1976 Jun 25;104(2):455–470. doi: 10.1016/0022-2836(76)90281-3. [DOI] [PubMed] [Google Scholar]
- Engberg J., Nasir-ud-Din, Eckert W. A., Kaffenberger W., Pearlman R. E. Detailed transcription map of the extrachromosomal ribosomal RNA genes in Tetrahymena thermophila. J Mol Biol. 1980 Sep 25;142(3):289–313. doi: 10.1016/0022-2836(80)90274-0. [DOI] [PubMed] [Google Scholar]
- Galas D. J., Schmitz A. DNAse footprinting: a simple method for the detection of protein-DNA binding specificity. Nucleic Acids Res. 1978 Sep;5(9):3157–3170. doi: 10.1093/nar/5.9.3157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamada H., Kominami R., Muramatsu M. 3'-terminal processing of ribosomal RNA precursors in mammalian cells. Nucleic Acids Res. 1980 Feb 25;8(4):889–903. [PMC free article] [PubMed] [Google Scholar]
- Karrer K. M., Gall J. G. The macronuclear ribosomal DNA of Tetrahymena pyriformis is a palindrome. J Mol Biol. 1976 Jun 25;104(2):421–453. doi: 10.1016/0022-2836(76)90280-1. [DOI] [PubMed] [Google Scholar]
- Kelly J. M., Cox R. A. The nucleotide sequence at the 3'-end of Neurospora crassa 25S-rRNA and the location of a 5.8S-rRNA binding site. Nucleic Acids Res. 1981 Mar 11;9(5):1111–1121. doi: 10.1093/nar/9.5.1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A. Ribosome synthesis in Tetrahymena pyriformis. J Cell Biol. 1970 Jun;45(3):623–634. doi: 10.1083/jcb.45.3.623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leer J. C., Tiryaki D., Westergaard O. Termination of transcription in nucleoli isolated from Tetrahymena. Proc Natl Acad Sci U S A. 1979 Nov;76(11):5563–5566. doi: 10.1073/pnas.76.11.5563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandal R. K., Dawid I. B. The nucleotide sequence at the transcription termination site of ribosomal RNA in Drosophila melanogaster. Nucleic Acids Res. 1981 Apr 24;9(8):1801–1811. doi: 10.1093/nar/9.8.1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Niles E. G. Isolation of a high specific activity 35S ribosomal RNA precursor from Tetrahymena pyriformis and identification of its 5' terminus, pppAp. Biochemistry. 1978 Oct 31;17(22):4839–4844. doi: 10.1021/bi00615a035. [DOI] [PubMed] [Google Scholar]
- Prescott D. M., Bostock C., Gamow E., Lauth M. Characterization of rapidly labeled RNA in Tetrahymena pyriformis. Exp Cell Res. 1971 Jul;67(1):124–128. doi: 10.1016/0014-4827(71)90627-6. [DOI] [PubMed] [Google Scholar]
- Rae P. M., Kohorn B. D., Wade R. P. The 10 kb Drosophila virilis 28S rDNA intervening sequence is flanked by a direct repeat of 14 base pairs of coding sequence. Nucleic Acids Res. 1980 Aug 25;8(16):3491–3504. doi: 10.1093/nar/8.16.3491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg M., Court D. Regulatory sequences involved in the promotion and termination of RNA transcription. Annu Rev Genet. 1979;13:319–353. doi: 10.1146/annurev.ge.13.120179.001535. [DOI] [PubMed] [Google Scholar]
- Samols D. R., Hagenbuchle O., Gage L. P. Homology of the 3' terminal sequences of the 18S rRNA of Bombyx mori and the 16S rRNA of Escherchia coli. Nucleic Acids Res. 1979 Nov 10;7(5):1109–1119. doi: 10.1093/nar/7.5.1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith M., Leung D. W., Gillam S., Astell C. R., Montgomery D. L., Hall B. D. Sequence of the gene for iso-1-cytochrome c in Saccharomyces cerevisiae. Cell. 1979 Apr;16(4):753–761. doi: 10.1016/0092-8674(79)90091-6. [DOI] [PubMed] [Google Scholar]
- Sollner-Webb B., Reeder R. H. The nucleotide sequence of the initiation and termination sites for ribosomal RNA transcription in X. laevis. Cell. 1979 Oct;18(2):485–499. doi: 10.1016/0092-8674(79)90066-7. [DOI] [PubMed] [Google Scholar]
- Veldman G. M., Klootwijk J., de Jonge P., Leer R. J., Planta R. J. The transcription termination site of the ribosomal RNA operon in yeast. Nucleic Acids Res. 1980 Nov 25;8(22):5179–5192. doi: 10.1093/nar/8.22.5179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wieslander L. A simple method to recover intact high molecular weight RNA and DNA after electrophoretic separation in low gelling temperature agarose gels. Anal Biochem. 1979 Oct 1;98(2):305–309. doi: 10.1016/0003-2697(79)90145-3. [DOI] [PubMed] [Google Scholar]