Abstract
Photochemical uncaging techniques use light to release active molecules from otherwise inert compounds. Here we expand this class of techniques by demonstrating the mechanical uncaging of a reactive species within a single protein. We prove this novel technique by capturing the regiospecific reaction between a thiol and a vicinal disulfide bond. We designed a protein that includes a caged cysteine and a buried disulfide. The mechanical unfolding of this protein in the presence of an external nucleophile frees the single reactive cysteine residue, which now can cleave the target disulfide via a nucleophilic attack on either one of its two sulfur atoms. This produces two different and competing reaction pathways. We use single molecule force spectroscopy to monitor the cleavage of the disulfides, which extends the polypeptide by a magnitude unambiguously associated with each reaction pathway. This allowed us to measure, for the first time, the kinetics of disulfide bond isomerization in a protein.
Disulfides, formed by the oxidation of the sulfhydryl groups of two cysteine residues, are the most widespread cross-links in proteins. Differently to other covalent bonds, disulfides can be highly dynamic in physiological conditions as a consequence of a set of reactions known as thiol/disulfide exchange1. In these reactions, electrons are reshuffled between a thiolate (R1-S−) and a disulfide bond (R2-S-S-R3) via an SN2 mechanism that produces a different disulfide and a new thiolate2 (Figure 1a). The relative rate of reactions (1) and (2) in Figure 1a is referred to as the regiospecificity of the reaction, a concept of the highest importance in thiol/disulfide exchanges in biology. Crucial cellular processes, such as the acquisition of native disulfide bonds of proteins in the endoplasmic reticulum, involve regiospecific thiol/disulfide exchange reactions3,4. Although the interaction between redox agents and proteins containing only one disulfide bond is relatively straightforward, a much more complex scenario is predicted for the ~19% of proteins with multiple disulfides. For example, when a disulfide in a protein with more than one disulfide bond is reduced, reactive cysteine residues are generated that can react with the remaining disulfides, leading to disulfide isomerization. Because of their general inability to differentiate isomers with the same oxidation state, current bulk technologies have serious limitations to discriminate parallel intramolecular reaction pathways. These bulk approaches usually rely on quenching agents to freeze thiol/disulfide exchange reactions at specific times. The reaction mixtures are then analyzed by means of techniques such as HPLC, electrophoresis or NMR1,4. Even though smart experimental protocols and careful analysis of the results have provided average rates for intramolecular thiol/disulfide exchange, the values obtained span several orders of magnitude5–7. In addition, divergent interpretations are frequently given to the experimental results8,9. Furthermore, when studying the kinetics of a thiol/disulfide exchange, bulk techniques have to deal with the unavoidable interference coming from the reverse reaction. Hence, the prevalence of disulfide isomerization reactions in proteins remains to be unambiguously quantified. To this end, it is necessary to obtain unequivocal and regiospecific rates for discrete thiol/disulfide exchange reactions occurring within proteins.
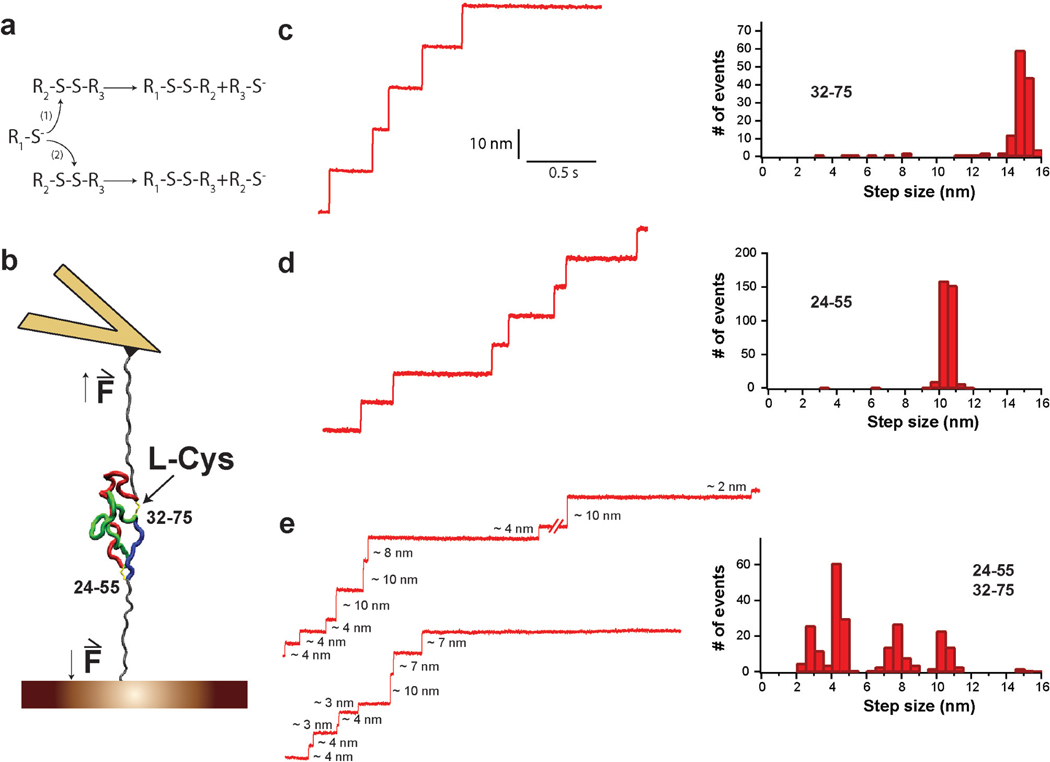
Figure 1. Increased complexity in the reduction of a protein with two disulfides.
a, Generic thiol-disulfide exchange reactions. b, Schematic experimental setup. Polyproteins containing several I27 modules with one or two disulfide bonds are unfolded by force-clamp AFM. For the sake of simplicity, only a single I27 module is shown. The depicted I27 module contains two disulfides linking positions 24–55 and 32–75; I272S-S. After mechanical unfolding, disulfides are rendered solvent accessible and can be cleaved by L-Cys. c, (Left) Experimental trace showing reduction events obtained for (I2732–75)8 polyprotein in the presence of L-Cys at 250 pN. (Right) The reduction events found for (I2732–75)8 are characterized by a single population of 14.5 nm steps. d, (Left) Experimental trace showing reduction events obtained for (I2724–55)8 polyprotein in the presence of L-Cys at 250 pN. (Right) The step sizes for the reduction events found for (I2724–55)8 fall into a single population centered at 10nm. e, (Left) Two experimental traces showing reduction events obtained for (I272S-S)4 polyprotein in the presence of L-Cys at 250 pN. (Right) The distribution of step sizes for the reduction events found for (I272S-S)4 is not a simple combination of the populations found for the proteins with only one disulfide.
In this work, we report the first direct observation of disulfide isomerization in a protein. With that aim, we caged a reactive cysteine residue in a protein disulfide. Using single-molecule force-clamp spectroscopy, we freed the reactive cysteine and studied its regiospecific reactivity towards a second disulfide. Different reaction pathways were distinguished by their different associated extensions of the polypeptide chain. Our experimental approach has allowed the first unambiguous description of the kinetics of intramolecular isomerization of disulfides in proteins. Thus, using our new single-molecule methodology, it becomes possible to dissect the increased complexity in thiol/disulfide exchange reactions in polypeptides with more than one disulfide.
RESULTS
Increased complexity in the reduction of a protein with two disulfides
In previous reports, we have shown that single-molecule force-clamp spectroscopy by atomic force microscopy (AFM) can monitor the reduction of single disulfides in real time10,11. When proteins containing a disulfide are unfolded under forces of few hundreds of piconewtons, they extend up to the disulfide (Figure 1b). Similar to most other covalent bonds, disulfides cannot be broken by forces below ~1 nN10,12. Only if a reducing agent is present in solution can the disulfide be cleaved, which is detected as an additional extension of the polypeptide chain. Importantly, the magnitude of this extension is determined solely by the number of amino acids trapped behind the disulfide13 (Supplementary Text). Furthermore, the reaction is irreversible when studied under force, as both cysteines become separated after the extension of the polypeptide. Thus, the kinetics of the reaction can be monitored with no interference from the reverse reaction. In Figures 1c and 1d, we show experimentally obtained reduction events from two polyproteins (i.e. single polypeptide chains composed of several modules of the same protein molecule) consisting of I27 domains with a single disulfide bond, (I2732–75)8 and (I2724–55)8, in the presence of L-Cys and at a force of 250 pN. A single population of reduction events is detected at 14.5 nm for I2732–75, or 10 nm for I2724–55 (Figures 1c, d). As expected, both experimental values match the predicted ones (Supplementary Text). To study the increase in complexity in the reduction of a protein with multiple disulfides, we performed the same experiment on a polyprotein containing I27 modules with both disulfides 24–55 and 32–75; (I272S-S)4 (Figure 1b, Supplementary Figures S1, S2, Supplementary Text). The results we obtained for (I272S-S)4 cannot be explained by the simple combination of step sizes found for the single-disulfide I27 variants (Figures 1c, d, e). On the contrary, the presence of both disulfide bonds in I272S-S generates a much more complex distribution of step sizes, with at least four different populations centered at 2–3, 4, 7–8, and 10 nm (Figure 1e).
Force-clamp spectroscopy detects specific disulfide isomerization reactions
We hypothesized that the increase in complexity found in the reduction by L-Cys of I272S-S could arise from intramolecular thiol/disulfide exchange reactions. To quantify the kinetics of these isomerization reactions, it was first necessary to assign the experimental reduction steps to specific thiol/disulfide exchange reactions. With this aim, we took advantage of the fact that the step sizes of disulfide reduction detected in force-clamp can be predicted from the number of amino acids released after cleavage of the disulfide13 (Supplementary Text).
The most prevalent step found in the reduction of I272S-S is characterized by an increase in length of 4 nm. We observed that these steps tended to happen before other events, suggesting that they mark the initial event in the reduction of an I272S-S domain (Figures 1e, 2). The reduction of disulfide 32–75 in I272S-S would release 12 amino acids corresponding to a 4-nm elongation of the polypeptide chain (Supplementary Text, Supplementary Figure S3). In contrast, the initial reduction of 24–55 could never produce a 4-nm step (Supplementary Text, Supplementary Figure S4). Therefore, we assigned the 4-nm steps to the reduction of disulfide 32–75 in I272S-S.
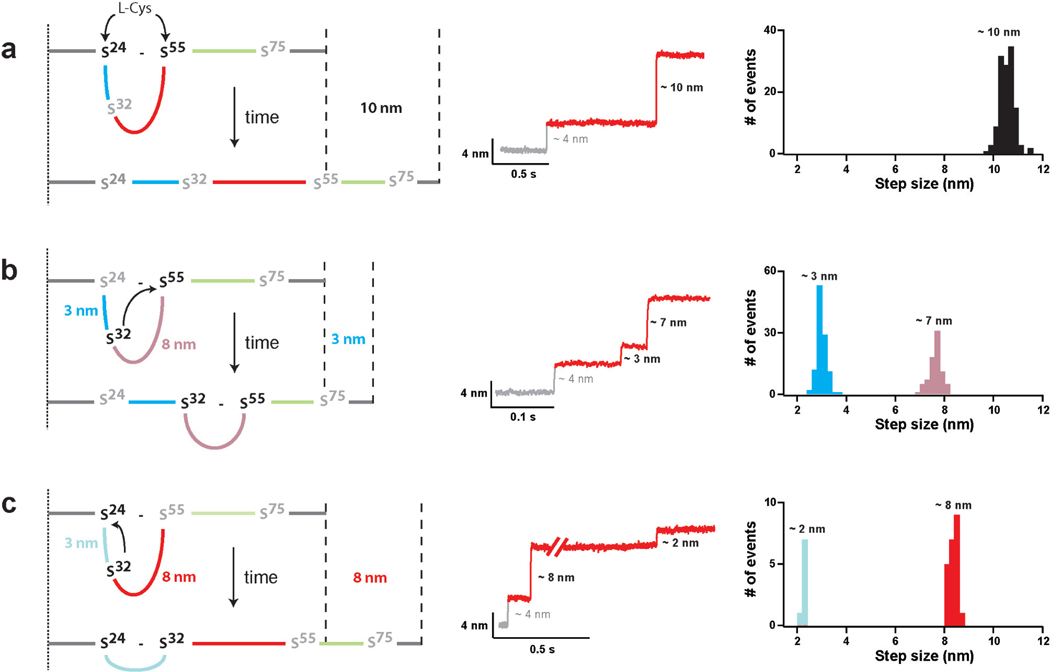
Figure 2. Fingerprints of the pathways available for the reduction of disulfide 24–55 after the reduction of disulfide 32–75.
In ~95% of the events, disulfide 32–75 in I272S-S is reduced before disulfide 24–55 producing a step of 4 nm (Supplementary Text, Supplementary Figure S3). Following the initial reduction of disulfide 32–75, disulfide 24–55 can be cleaved following three different reaction pathways. a, Cleavage of disulfide 24–55 by L-Cys. b, Cleavage of disulfide 24–55 by the attack of Cys32 on position 55. c, Cleavage of disulfide 24–55 by the attack of Cys32 on position 24. The figure shows (left) diagrams that illustrate the reaction pathways, (center) representative experimental traces, and (right) computed histograms of the step sizes associated with each one of the thiol/disulfide exchanges. The step sizes for different reactions rarely overlap. Hence, the different step sizes together with the order in which the events are detected can be used to assign reaction pathways. In the diagrams on the left, the colored lines between residues 24–32 (blue), 32–55 (red) and 55–75 (green) represent the polypeptide chain linking the various cysteine residues. The remaining polypeptide chain is depicted in gray. The same color code is used in Figures 1b and 4a.
Immediately after cleavage of disulfide 32–75, Cys75 is pulled away by force, while Cys32 remains in the vicinity of disulfide 24–55 (Figure 2, Supplementary Figure S3). Thus, disulfide 24–55 can be cleaved following three different reaction pathways (Figure 2). As demonstrated above for I2724–55, the straightforward reduction of disulfide 24–55 by an external L-Cys molecule extends the protein by 10 nm (Figures 1d, 2a). However, if instead of L-Cys, Cys32 attacks disulfide 24–55 at Cys55, the protein is extended by 3 nm (predicted extension 2.8 nm). This reaction generates a new disulfide bond, 32–55, not present in I272S-S (Figure 2b). Alternatively, if Cys32 reacts with the sulfur at Cys24, an 8-nm increase in length is observed (predicted extension 8.1 nm). In this case, the reaction renders a disulfide between residues 24–32 (Figure 2c). Whenever the reduction of disulfide 24–55 is mediated by Cys32, a second step adding up to the 10-nm total extension follows the initial 3 or 8-nm steps, indicating the cleavage of the newly isomerized disulfide by an external L-Cys molecule (Figures 2b, c). Thus, the different regiospecific thiol/disulfide exchange reactions described in Figure 2 explain the different populations of step sizes found in the reduction of I272S-S (Figure 1e). The three pathways start with the cleavage of disulfide 32–75 rendering a 4-nm step. The events detected afterwards are a signature of the particular reduction pathway. Two of the reactions are intramolecular thiol/disulfide exchanges, which had not been directly observed before. To unambiguously assign the different step sizes to any of the possible reaction pathways, both the size of the steps and the order in which they appear in a trace were employed (Figures 1e, 2).
Kinetic characterization of the intramolecular isomerization of disulfides
To get information about the kinetics of the reactions, we developed a model accounting for all the possible reaction pathways for the reduction of I272S-S by L-Cys, including the regiospecificity of two of the reactions (Figure 3a, for a full description, see Supplementary Text). Our model predicts that the frequency of appearance of the different steps is governed both by the corresponding rate constants and the concentration of L-Cys. For example, the first-order constants k1 and k7 at a given concentration of L-Cys determine the rate of appearance of the 4-nm steps. However, the appearance of the 10-, 3- and 8-nm steps is a much more complex scenario. First, they only happen after a 4-nm step, so their frequency depends on the rate of occurrence of the 4-nm steps. In addition, as they are the signature of competing reaction pathways, their frequency does not only depend on their characteristic rate constants, but also on the competing constants. Despite this complexity, the model implies a simple expected behavior: at high concentrations of L-Cys the frequency of the 10-nm steps should increase at the expense of the 3- and 8-nm steps. This is a direct consequence of the competition between inter- and intramolecular thiol/disulfide exchange reactions, with the former being accelerated by increasing concentrations of external L-Cys (Figure 3a, top). In addition, the relative frequency of the 3- and 8-nm steps should be independent of the concentration of L-Cys, as both pathways are purely intramolecular. Thus, to test the validity of the kinetic model, we monitored the reduction of I272S-S in the presence of increasing concentrations of L-Cys. We computed the frequency of appearance of the 10-, 3- and 8-nm steps. As predicted by our model, at high concentrations of L-Cys the 10-nm steps become more prevalent, while the 3- and 8-nm steps appear less frequently (Figures 3b, c). At the same time, the relative frequency of the 3- and 8-nm steps is independent of the concentration of L-Cys (Figure 3d).
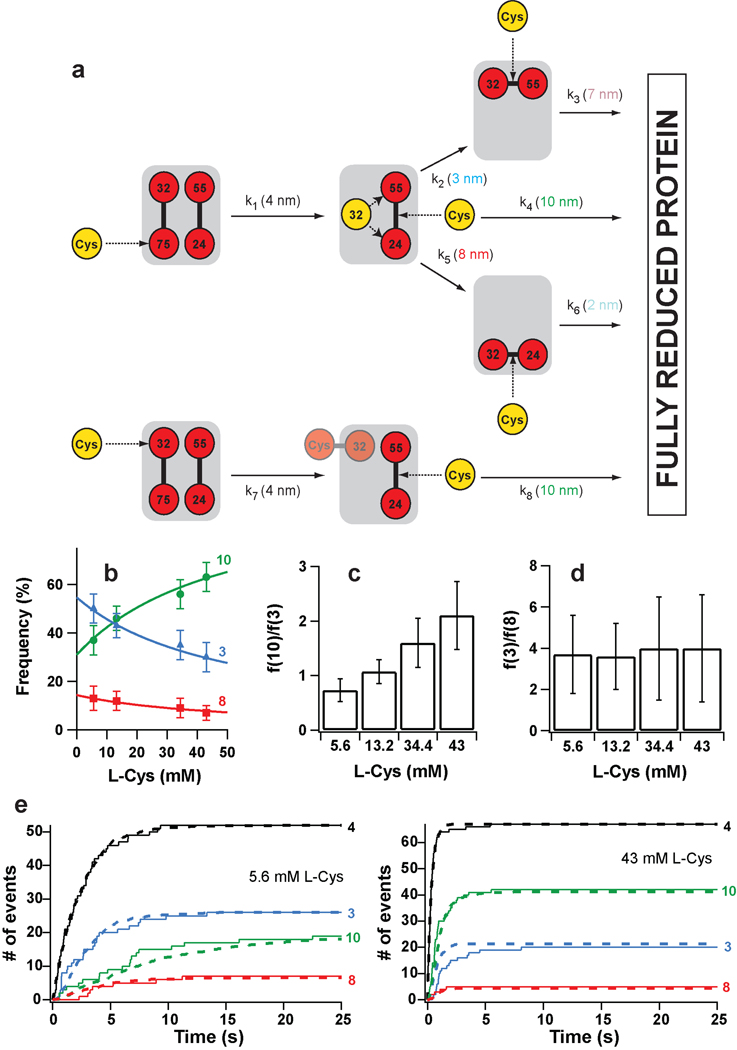
Figure 3. Kinetic model of complete protein reduction by L-Cys following the initial reduction of disulfide 32–75.
a, L-Cys can react with either sulfur in disulfide 32–75. (Top) Reaction at position 75 generates a free thiolate at position 32, which can then react intramolecularly with disulfide 24–55. This attack can happen at either sulfur atom, and competes with the intermolecular reaction with L-Cys. The intramolecular reactions generate new disulfides that can only be cleaved by L-Cys. (Bottom) If the initial reaction happens at position 32, the intramolecular reactions are blocked by the formation of a mixed disulfide between L-Cys and Cys32. In this case, a free thiol group is generated at position 75, but it is pulled away by force and can not participate in intramolecular exchanges with disulfide 24–55. The rate constants governing the reactions and their characteristic step sizes are indicated. b, The frequency of appearance of the step sizes depends on the concentration of L-Cys. The solid lines are the plots of Supplementary Equations S4–S6 using the rate constants derived from the downhill simplex method. c, With increasing concentrations of L-Cys, the relative frequency of appearance of the 10- and 3-nm increases. d, The relative frequency of the 3- and 8-nm steps is independent of L-Cys concentration. e, (Solid lines) Time course for the appearance of the different step sizes at two L-Cys concentrations. (Dashed lines) Theoretical curves obtained from the rate constants determined using the downhill simplex method. In panels b and e, traces are identified by the magnitude of the steps.
In order to quantify the rate of the reactions, we computed the dwell times associated with every type of event (Supplementary Figure S2). Time courses of appearance of the different step sizes were generated by plotting the cumulative number of events at each particular dwell time (Figure 3e, Supplementary Figure S5). As expected from the end-point results discussed above, we found that the kinetics of appearance of the steps is also highly dependent on the concentration of L-Cys (Figure 3e). To find the rate constants that best fit our experimental data, we solved the coupled differential equations governing the time evolution of our model system (i.e. the probability of the system occupying each of the states depicted in Figure 3a). We then used a downhill simplex method to find the optimal parameters that minimized the discrepancy between the predicted outcome and the experimental data (Supplementary Text). Using this procedure, we determined a set of rate constants that predict kinetic traces highly similar to the experimental results (Figure 3e, Supplementary Figure S5). As a further test, we used the same rate constants to fit the frequency of the different step sizes as predicted by our model (Supplementary Text). The fits of Supplementary Equations S4–S6 to the frequencies of appearance of the different step sizes are remarkably accurate (Figure 3b).
DISCUSSION
In vivo, thiol/disulfide exchange reactions are highly dynamic and regiospecific, and often involve the participation of transient disulfides14–17. These properties generate complex behaviors in proteins with multiple disulfide bonds, such as the intramolecular isomerization of disulfides described in this report. However, so far it has been impossible to predict the extent of these ubiquitous isomerization reactions because no accurate estimates existed for the rate of intramolecular thiol/disulfide exchange occurring in proteins. This is a consequence of the lack of experimental methods able to dissect parallel reaction pathways while retaining kinetic information1,4,18.
Our experimental approach bears a resemblance to the photochemical uncaging experiments frequently employed to study biological processes such as neurotransmitter physiology or Ca2+-mediated cell signaling19,20. In our case, we caged Cys32 in the form of an inert disulfide with Cys75 in I272S-S. After the forced unfolding of I272S-S in the presence of L-Cys, the reactive Cys32 is uncaged and situated in the vicinity of disulfide 24–55. This allowed us to monitor the three different competing reaction pathways available for cleavage of disulfide 24–55 in a synchronized manner (Figure 4a). Using an analytical optimization procedure, we obtained the rates for each one of the discrete competing pathways (Figure 4b). Thus, for the first time, we have directly measured the rate of intramolecular thiol/disulfide exchange reactions in a protein.
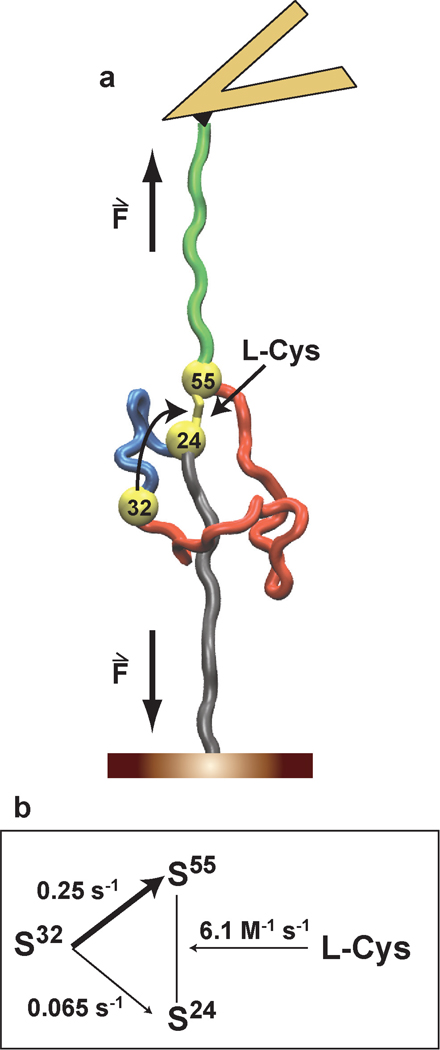
Figure 4. The uncaging of a single cysteine residue in a protein allows calculating the rate of spontaneous disulfide isomerization.
a, In the presence of external nucleophiles, a single reactive cysteine, Cys32, is uncaged after the mechanical unfolding of I272S-S. Cys32 remains in the vicinity of disulfide 24–55, which can be cleaved by the reaction of any of its sulfur atoms with Cys32 or by the attack of an external L-Cys molecule. b, Using our experimental approach, we determine the rates of two intramolecular thiol/disulfide exchange reactions, i.e. the attack of Cys32 to Cys24 or Cys55.
Noticeably, our results provide a regiospecific description of two different thiol/disulfide exchange reactions. Regarding the intermolecular cleavage of disulfide 32–75 by L-Cys, we found that Cys75 is attacked 2.2 times more frequently than Cys32 (compare k1 and k7 in Supplementary Text, Figure 3a). We also observed that the intramolecular attack of Cys32 on position 55 is 3.8 times more frequent than on position 24 (Figure 4b). Thus, both thiol/disulfide exchange reactions showed a considerable regiospecificity, the origin of which is intriguing. Steric effects should not have a strong influence on the reactivity towards an unfolded polypeptide. From a chemical point of view, it has been proposed that the quality of a thiolate as a leaving group is related to its pKa, with lower pKa values being associated with better leaving groups2. The presence of positive residues next to Cys32 and Cys55 would suggest that these two cysteines are better leaving groups than their counterparts Cys75 and Cys24 (Supplementary Table S1)2. We found that Cys32 is the preferred leaving group when cleaving disulfide 32–75. However, Cys24, and not Cys55, is the most frequent leaving group in the rupture of disulfide 24–55. Therefore, our results demand new theoretical developments that can account for the observed experimental regiospecificities.
The order of magnitude we obtain for the rate of intramolecular thiol/disulfide exchange suggests that the spontaneous intramolecular isomerization of disulfides in proteins can effectively compete with their intermolecular reduction. Indeed, for small reducing agents such as glutathione, the fastest experimental rate constants for protein disulfide reduction are ~102 M−1s−1 21. Considering that the concentration of glutathione inside cells is 10 mM22, a maximum rate of ~1 s−1 is predicted for disulfide reduction by glutathione. A similar figure is obtained for enzyme-catalyzed reduction of disulfides22–24. Thus, with a rate of ~0.3 s−1 (Figure 4b), the isomerization of disulfides may interfere with physiological redox processes. The existence of this unavoidable competition may contribute to explain why different mechanisms have evolved to mitigate the harmful effects coming from undesired intramolecular thiol/disulfide exchange reactions. An interesting example can be found in protein disulfide isomerases25. The malfunction of any such mechanisms could be responsible for the persistent formation of aberrant disulfides that may trigger unfolding and/or aggregation of proteins. Interestingly, some pathologies such as familial amyotrophic lateral sclerosis or cataracts formation have been linked to the formation of incorrect disulfides in proteins26,27.
Our study demonstrates the feasibility of mechanically uncaging reactive thiols that can engage in subsequent thiol/disulfide exchange reactions. We speculate that similar mechanisms may be found as regulatory switches in Biology, since the activity of some proteins is controlled by the oxidation state of specific cysteines28. Interestingly, some of these proteins are also subject to mechanical stress that can trigger thiol/disulfide exchanges. For instance, it has been shown that the adhesion activity of the von Willebrand factor is regulated by shear-induced disulfide formation29.
Specific transfers of electrons between cysteine residues are essential for a myriad of cellular redox systems14,15,17,30 and their relevance in an increasing number of diseases is starting to be unveiled. Additionally, in the emerging field of nanobioelectronics there is a need to develop nanoscopic biomaterials able to transport electrons in a controlled manner31–33. Our results validate a new platform to study the complexity of intramolecular thiol/disulfide exchange reactions in proteins with multiple disulfides. The single-molecule approach that we introduce opens up the possibility to systematic studies on how the environment of cysteine residues in proteins modifies their reactivity34. Such information may be useful both to predict the behavior of cysteines and disulfides in physiological reactions and to engineer proteins with particular electron reshuffling properties. Our methodology relies on the generation of a reactive thiol group from a buried disulfide. A similar experimental design could potentially be employed to study other chemical reactions involving thiol groups. The mechanical uncaging of different reactive groups may broaden the applicability of our methodology to other chemical reactions that result in the cleavage of mechanically resistant covalent bonds.
METHODS
Protein purification
The expression and purification of (I2732–75)8 and (I2724–55)8 has been described before13. In order to produce the cDNA for I272S-S, we applied the QuikChange Multi Site-Directed Mutagenesis kit from Stratagene (La Jolla, CA) using the cDNA coding for I2724–55 as a template. We constructed polyproteins using an iterative process of digestion and ligation of DNA fragments, as described before35,36. BamHI and KpnI sites were used to insert cDNAs into the expression vector pQE80L (Qiagen, Valencia, CA)36. Protein production in Origami B E. coli cells was induced at OD600 ~1.0 with 1 mM IPTG overnight at 23 °C. Soluble proteins were obtained by a combination of sonication and passes through a French press. Polyproteins were purified from the soluble fraction by an affinity chromatography using Talon resin (Mountain View, CA), followed by an FPLC step in a Superdex 200 column (Amersham Biosciences, Pittsburgh, PA). The buffer employed was 10 mM Hepes pH 7.2, 1 mM EDTA, 150 mM NaCl. We observed that polyproteins based on I27 modules solely including cysteine residues at positions 24, 32, 55, and 75 were mainly recovered in the insoluble fraction, which precludes purification. However, we found that the amount soluble proteins could be improved by decreasing the number of repeats in the polyprotein and by mutations affecting the loop between residues 24 and 32. In our experiments, we used a polyprotein with four repetitions of I272S-S that included the following substitutions at positions 26–30: Ser26-Glu-Pro-Asp-Val30 to Asp26-Asp-Asp-Asp-Lys30. The purification procedure applied to this engineered polyprotein rendered homogeneous soluble fractions of (I272S-S)4, as estimated by SDS-PAGE (Supplementary Figure. S1).
Single-molecule force-clamp spectroscopy
1–5 µL of a ~0.2 mg/mL (I272S-S)4 solution were deposited onto an evaporated gold coverslip. Veeco silicon nitride MLCT (Camarillo, CA) cantilevers were used. They were calibrated using the equipartition theorem37. Typical spring constants were in the range of 15 – 25 pN/nm. To pick up single molecules, the cantilever was pushed against the surface at contact forces of ~2 nN for 1–2 seconds, and then retracted. We used a custom-built atomic force microscopy (AFM) setup that allows a strict control of the force based on en electronic feedback with a ~5 ms response time that controls the extension of the piezoelectric actuator38. Experiments were done at room temperature (22 – 25 °C) in 10 mM Tris-HCl pH7.6. The effective concentration of L-Cys was calculated as described in the Supplementary Text.
Traces showing steps that could be assigned to specific thiol/disulfide exchanges were selected. Only traces that showed a pattern of steps that was compatible with the reaction pathways described in Figure 2 and Supplementary Figure S4 (>85% of the selected traces) were included in the analysis. It has been described before that the criteria used to select traces in single-molecule force-clamp experiments can affect the calculated rates39. For instance, including traces with short detachment times can bias the results towards faster rates. Consequently, we only considered traces that showed long times between the 4-nm steps and the detachment event (5.6 mM L-Cys; 20 s; 13.2 mM L-Cys; 10 s, 33.4 mM L-Cys; 7.5 s; 43 mM L-Cys; 5 s). These cut-off values were chosen according to slow reaction events observed for each experimental condition.
The error bars in Figures 3b, c, d were estimated by bootstrapping40. To this end, 10,000 artificial data sets including the number of observed events for each step size were randomly generated from the experimental traces at a given L-Cys concentration. Next, the frequency of appearance of each step size was calculated for every simulated data set, resulting in a distribution of frequencies. The error bars in Figure 3b are given by the standard deviation of such distributions. The errors in the relative frequencies shown in Figures 3c and 3d were calculated from the errors in the corresponding absolute frequencies.
Supplementary Material
Acknowledgements
This work has been supported by NIH grants HL66030 and HL61228 to J.M.F. J.A-.C. thanks Fundación Caja Madrid, Fundación Alfonso Martín Escudero (Madrid, Spain) and Fundación Ibercaja (Zaragoza, Spain) for their financial support. J.A.R-.P is the recipient of a fellowship from CONICYT and a MECESUP visiting scholar fellowship (Chile). We thank Jingyuan Li and Bruce J. Berne for their help with SMD simulations. We also thank Sergi Garcia-Manyes for careful reading of the manuscript.
Footnotes
Supplementary information is available as a separate file.
Author Contributions J.A-.C. and J.M.F designed the research project. J.A-.C and J.A.R-P. performed the experiments. J.A-.C., P.K., J.A.R-P. and J.M.F. analyzed the data. J.A-.C., P.K. and J.M.F wrote the paper.
Author Information The authors declare no competing financial interests.
REFERENCES
- 1.Gilbert HF. Thiol/disulfide exchange equilibria and disulfide bond stability. Methods Enzymol. 1995;251:8–28. doi: 10.1016/0076-6879(95)51107-5. [DOI] [PubMed] [Google Scholar]
- 2.Gilbert HF. Molecular and cellular aspects of thiol-disulfide exchange. Adv Enzymol Relat Areas Mol Biol. 1990;63:69–172. doi: 10.1002/9780470123096.ch2. [DOI] [PubMed] [Google Scholar]
- 3.Mamathambika BS, Bardwell JC. Disulfide-linked protein folding pathways. Annu Rev Cell Dev Biol. 2008;24:211–235. doi: 10.1146/annurev.cellbio.24.110707.175333. [DOI] [PubMed] [Google Scholar]
- 4.Wedemeyer WJ, Welker E, Narayan M, Scheraga HA. Disulfide bonds and protein folding. Biochemistry. 2000;39:4207–4216. doi: 10.1021/bi992922o. [DOI] [PubMed] [Google Scholar]
- 5.Creighton TE, Goldenberg DP. Kinetic role of a meta-stable native-like two-disulphide species in the folding transition of bovine pancreatic trypsin inhibitor. J Mol Biol. 1984;179:497–526. doi: 10.1016/0022-2836(84)90077-9. [DOI] [PubMed] [Google Scholar]
- 6.Rothwarf DM, Scheraga HA. Regeneration of bovine pancreatic ribonuclease A. 2. Kinetics of regeneration. Biochemistry. 1993;32:2680–2689. doi: 10.1021/bi00061a028. [DOI] [PubMed] [Google Scholar]
- 7.Weissman JS, Kim PS. Reexamination of the folding of BPTI: predominance of native intermediates. Science. 1991;253:1386–1393. doi: 10.1126/science.1716783. [DOI] [PubMed] [Google Scholar]
- 8.Creighton TE. The disulfide folding pathway of BPTI. Science. 1992;256:111–114. doi: 10.1126/science.1373519. [DOI] [PubMed] [Google Scholar]
- 9.Weissman JS, Kim PS. Response. Science. 1992;256:112–114. doi: 10.1126/science.256.5053.112. [DOI] [PubMed] [Google Scholar]
- 10.Wiita AP, Ainavarapu SR, Huang HH, Fernandez JM. Force-dependent chemical kinetics of disulfide bond reduction observed with single-molecule techniques. Proc Natl Acad Sci U S A. 2006;103:7222–7227. doi: 10.1073/pnas.0511035103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wiita AP, et al. Probing the chemistry of thioredoxin catalysis with force. Nature. 2007;450:124–127. doi: 10.1038/nature06231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Grandbois M, et al. How strong is a covalent bond? Science. 1999;283:1727–1730. doi: 10.1126/science.283.5408.1727. [DOI] [PubMed] [Google Scholar]
- 13.Ainavarapu SR, et al. Contour length and refolding rate of a small protein controlled by engineered disulfide bonds. Biophys J. 2007;92:225–233. doi: 10.1529/biophysj.106.091561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rozhkova A, et al. Structural basis and kinetics of inter- and intramolecular disulfide exchange in the redox catalyst DsbD. EMBO J. 2004;23:1709–1719. doi: 10.1038/sj.emboj.7600178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.van der Neut Kolfschoten M, et al. Anti-inflammatory activity of human IgG4 antibodies by dynamic Fab arm exchange. Science. 2007;317:1554–1557. doi: 10.1126/science.1144603. [DOI] [PubMed] [Google Scholar]
- 16.Wypych J, et al. Human IgG2 antibodies display disulfide-mediated structural isoforms. J Biol Chem. 2008;283:16194–16205. doi: 10.1074/jbc.M709987200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kadokura H, Beckwith J. Four cysteines of the membrane protein DsbB act in concert to oxidize its substrate DsbA. EMBO J. 2002;21:2354–2363. doi: 10.1093/emboj/21.10.2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Messens J, et al. How thioredoxin can reduce a buried disulphide bond. J Mol Biol. 2004;339:527–537. doi: 10.1016/j.jmb.2004.04.016. [DOI] [PubMed] [Google Scholar]
- 19.Mayer G, Heckel A. Biologically active molecules with a "light switch". Angew Chem Int Ed Engl. 2006;45:4900–4921. doi: 10.1002/anie.200600387. [DOI] [PubMed] [Google Scholar]
- 20.Gorostiza P, Isacoff EY. Optical switches for remote and noninvasive control of cell signaling. Science. 2008;322:395–399. doi: 10.1126/science.1166022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shaked Z, Szajewski RP, Whitesides GM. Rates of thiol-disulfide interchange reactions involving proteins and kinetic measurements of thiol pKa values. Biochemistry. 1980;19:4156–4166. doi: 10.1021/bi00559a004. [DOI] [PubMed] [Google Scholar]
- 22.Schafer FQ, Buettner GR. Redox environment of the cell as viewed through the redox state of the glutathione disulfide/glutathione couple. Free Radic Biol Med. 2001;30:1191–1212. doi: 10.1016/s0891-5849(01)00480-4. [DOI] [PubMed] [Google Scholar]
- 23.Holmgren A. Reduction of disulfides by thioredoxin. Exceptional reactivity of insulin and suggested functions of thioredoxin in mechanism of hormone action. J Biol Chem. 1979;254:9113–9119. [PubMed] [Google Scholar]
- 24.Dyson HJ, et al. Effects of buried charged groups on cysteine thiol ionization and reactivity in Escherichia coli thioredoxin: structural and functional characterization of mutants of Asp 26 and Lys 57. Biochemistry. 1997;36:2622–2636. doi: 10.1021/bi961801a. [DOI] [PubMed] [Google Scholar]
- 25.Hatahet F, et al. Protein disulfide isomerase: a critical evaluation of its function in disulfide bond formation. Antioxid Redox Signal. 2009;11:2807–2850. doi: 10.1089/ars.2009.2466. [DOI] [PubMed] [Google Scholar]
- 26.Wang J, Xu G, Borchelt DR. Mapping superoxide dismutase 1 domains of non-native interaction: roles of intra- and intermolecular disulfide bonding in aggregation. J Neurochem. 2006;96:1277–1288. doi: 10.1111/j.1471-4159.2005.03642.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pande A, Gillot D, Pande J. The cataract-associated R14C mutant of human gamma D-crystallin shows a variety of intermolecular disulfide cross-links: a Raman spectroscopic study. Biochemistry. 2009;48:4937–4945. doi: 10.1021/bi9004182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hogg PJ. Disulfide bonds as switches for protein function. Trends Biochem Sci. 2003;28:210–214. doi: 10.1016/S0968-0004(03)00057-4. [DOI] [PubMed] [Google Scholar]
- 29.Choi H, et al. Shear-induced disulfide bond formation regulates adhesion activity of von Willebrand factor. J Biol Chem. 2007;282:35604–35611. doi: 10.1074/jbc.M704047200. [DOI] [PubMed] [Google Scholar]
- 30.Frand AR, Kaiser CA. Two pairs of conserved cysteines are required for the oxidative activity of Ero1p in protein disulfide bond formation in the endoplasmic reticulum. Mol Biol Cell. 2000;11:2833–2843. doi: 10.1091/mbc.11.9.2833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Davis JJ, et al. Molecular bioelectronics. Journal of Materials Chemistry. 2005;15:2160–2174. [Google Scholar]
- 32.Willner I. Tech.Sight. Bioelectronics. Biomaterials for sensors, fuel cells, and circuitry. Science. 2002;298:2407–2408. doi: 10.1126/science.298.5602.2407. [DOI] [PubMed] [Google Scholar]
- 33.Birge RR, et al. Biomolecular electronics: Protein-based associative processors and volumetric memories. Journal of Physical Chemistry B. 1999;103:10746–10766. [Google Scholar]
- 34.Weerapana E, et al. Quantitative reactivity profiling predicts functional cysteines in proteomes. Nature. 2010;468:790–795. doi: 10.1038/nature09472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Carrion-Vazquez M, Marszalek PE, Oberhauser AF, Fernandez JM. Atomic force microscopy captures length phenotypes in single proteins. Proc Natl Acad Sci U S A. 1999;96:11288–11292. doi: 10.1073/pnas.96.20.11288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Alegre-Cebollada J, Badilla CL, Fernandez JM. Isopeptide bonds block the mechanical extension of pili in pathogenic Streptococcus pyogenes. J Biol Chem. 2010;285:11235–11242. doi: 10.1074/jbc.M110.102962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Florin EL, et al. Sensing Specific Molecular-Interactions with the Atomic-Force Microscope. Biosensors & Bioelectronics. 1995;10:895–901. [Google Scholar]
- 38.Schlierf M, Li H, Fernandez JM. The unfolding kinetics of ubiquitin captured with single-molecule force-clamp techniques. Proc Natl Acad Sci U S A. 2004;101:7299–7304. doi: 10.1073/pnas.0400033101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Garcia-Manyes S, Brujic J, Badilla CL, Fernandez JM. Force-clamp spectroscopy of single-protein monomers reveals the individual unfolding and folding pathways of I27 and ubiquitin. Biophys J. 2007;93:2436–2446. doi: 10.1529/biophysj.107.104422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Efron B. The Jackknife, the Bootstrap, and Other Resampling Plans. Philadelphia: SIAM; 1982. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.