Abstract
Introduction:
A lot of attention has been generated in recent years by digital pathology and telepathology. Multiple reasons for and barriers to effective adoption are discussed in the current literature. Digital slides are the most promising medium at this time. The goal of our study was to evaluate whether the change in the methodology, particularly utilizing the so-called high-definition hematoxylin and eosin (H and E) slides, enhanced the quality of the final digital slide, and whether pathologists who tested the results perceived this as a difference in quality.
Methods:
The study was a blinded comparison of digital slides prepared using two methods: standard H&E batch staining and automated individual “high definition” HD HE staining. Four pathologists have compared 80 cases stained with each method.
Results:
The results discussed in this study show potential promise that the utilization of protocol(s) adapted for tissue and for imaging might be preferable for digital pathology in at least some of the pathology subspecialties. In particular, the protocol evaluated here was capable of turning out digital slides that had more contrast and detail, and therefore were perceived to provide enhanced diagnostically significant information for the pathologist.
Keywords: Digital pathology, digital slides, histochemistry, histology, imaging, staining methods, staining protocols, telepathology
INTRODUCTION
Digital imaging has the potential to revolutionize anatomic pathology, improve workflow, reduce administrative costs, and allow easy sharing of cases and information. However, to reach that potential, image quality must provide the pathologist with a sufficient amount of cellular detail to accurately analyze the tissue being examined. Until recently, this was not technically feasible.[1–5]
High-speed, automated whole slide imaging (WSI) systems (also called digital slide systems, virtual microscopes, or virtual slide imagers) are becoming increasingly capable and robust.[6–8] The images created (also known as digital slides or DS), while not yet equal to viewing slides through high-end microscopes, are approaching a high level of reliability for microscopic diagnosis.[5,9]
This technology is capable of rapid, high throughput digitalization of glass microscope slides, allowing for sharing of diagnostic information between two geographically different locations. Besides making telepathology possible, digitalization opens a new field of cost-efficient preanalytic processing. Unlike glass slides, digital slides can be stored and retrieved electronically, with minimal time and effort, significantly increasing productivity for pathologists and laboratory support staff.
Perhaps most importantly, digital slides are likely to increase diagnostic accuracy and allow for a greater breadth of case workup. Because digital slides can be linked electronically with other patient test results, pathologists will potentially have convenient and timely access to all relevant clinical data for a patient. These additional data allow the pathologist to offer a better turnaround time and greater clinico-pathologic integration of a given diagnosis. Furthermore, digital imaging makes it possible to quickly access images over long distance, providing near-instantaneous access to high-level diagnostic services for patients in rural areas where there are few subspecialists.[10–13] Electronic handling of images eliminates the need for couriers and other expensive means of transferring slides to pathologists at remote locations, virtually eliminating delays associated with more antiquated systems. Thus, digital pathology systems hold the promise for real, long-term cost savings and improvements in access to information, while contributing to improvements in diagnostic accuracy.
Although the articles about the effect of digital manipulation and image optimization were previously published,[14–16] the effect of staining variation per se on image quality was not previously studied in the literature. It is obvious that the method of tissue preparation and glass slide staining can and does inject a variable into the process that directly affects the quality of a glass slide, and consequently, of the final digital slide. A common complaint among pathologists using telepathology systems is that the images lack sufficient contrast required for making an accurate diagnosis;[17,18] furthermore a significant work has been already done in terms of standardization of digital slides.[19] Controlling the staining of tissue to enhance the contrast of the final digital slide may have a significant effect on its usefulness for accurate diagnosis. Individual staining of slides offers the ability to control the quality of the reagents and staining process, thereby enhancing the contrast and quality of the digital slide.
Currently, most anatomic pathology laboratories utilize equipment that implements large-batch processes, where large numbers of slides are immersed in dye baths all at one time. Hematoxylin and eosin (H&E) staining is not standardized; different quantities and qualities of H&E are used, and staining consistency can vary depending on the protocol used by the individual lab and the freshness of the reagents. This inconsistency in staining can create variability in contrast that is an impediment to effective digital imaging. The large-batch method is extremely efficient, but it does not provide the many advantages of individual slide staining, which was previously achievable only by hand. However, as the demands for decreased turnaround times and improved efficiency are ever increasing, individual hand staining is not a viable option in most circumstances. Moreover, processing the slides in multiple or several small batches (to allow for more optimal staining) would decrease efficiency through the addition of labor and markedly increase turnaround time. The variability of staining, due to aging and breakdown of the reagents would still be present, as with all linear staining processes.
Another option, however, is to employ an automated solution where the specimens are stained individually, allowing for greater control over the staining process, contributing to increased consistency and overall stain quality. Due to recent advances in technology and an increasingly demanding pathology environment, instrumentation capable of automated individual staining is now available. As this is a novel technology, these instruments have incorporated staining protocols that enhance visualization of critical features in tissue samples, possibly enhancing the process of digital imaging. Chief difference is that the on-board software matches a predetermined stain protocol associated with a slide tray, and individually applies fresh reagents to each individual slide present in the tray. Although the effects on diminishing the floaters on the slides via this technology have been studied,[20] the effects of this technology on digital imaging was not investigated.
In this study, the effect of preparing slides through traditional batch staining versus individual staining was evaluated, in the context of the process and the impact these had on digital imaging.
STUDY GOAL
The goal of this study was to determine if high-definition H&E (HDHE) slides enhanced the quality of the final digital slide, and whether pathologists who tested the results perceived this as a difference in quality. In particular, we compared HDHE slides processed using the Ventana Symphony™ staining system (also referred to as “individual slide staining”) to slides prepared by usual means in the histology laboratory (i.e., standard linear H&E batch staining) to determine if the HDHE slides provided a better medium for performance of digital anatomic pathology.
STUDY DESIGN, MATERIALS AND METHODS
The study was a blinded comparison of digital slides prepared using two methods: standard H&E batch staining and automated individual HDHE staining. Review of the current methodologies in histology and histology laboratory are beyond the scope of this article, but can be found in the article published in 2007.[21]
Four pathologists from the Departments of Dermatology and Pathology at University of Pittsburgh Medical Center (UPMC) Presbyterian Shadyside, an academic teaching hospital, were recruited to perform a diagnostic analysis of 20 cases each (each case contained one slide for the purpose of this study) from one of the four tissue types included in the study (for total of 80 digital slides). To ensure that the digital slide focus was maintained, the participants did not review and compare the corresponding glass slides. All four participating pathologists are board certified and have additional subspecialty expertise or additional board certification in their area. The pathologists were compensated for their time while participating in the study, but none held a financial interest of any type with either Ventana Medical Systems, Inc. (“Ventana”) (manufacturer of the Symphony equipment) or Trestle Technologies (which provided the MedMicro software and scanning hardware). None of the four have any previous or existing consulting role of any type with Ventana or Trestle. The study was financed via a Grant between University of Pittsburgh and Ventana, and approved by University of Pittsburgh Institutional Review Board (IRB) approval (exempt study # PRO07050152).
No demographic, diagnostic or clinical data were presented with the slides, as they were deidentified by a University of Pittsburgh “honest broker,” in accordance with University of Pittsburgh IRB procedures.
During this research project the participants completed a side-by-side comparison of digital slides created using batch-stained slides and slides stained using the standard Symphony staining. The study director selected for analysis 80 biopsy cases from the departments of dermatology and pathology at the UPMC Presbyterian Shadyside, after they were diagnosed/signed out. In each of the cases selected, H&E staining sufficed with regard to the final diagnosis (i.e., no additional staining, such as histochemical or immunohistochemical stains, was required), and each case was represented by a single slide (one slide per case).
Twenty (20) cases from each of the following four areas were selected: dermatopathology, pulmonary (thoracic) pathology, genitourinary (GU) pathology, and gastrointestinal (GI) pathology, for a total of 80 cases.
For each case, two unstained slides were cut. One slide from each case was stained using standard batch/linear
H and E staining on Leica instruments (SN:876052004, MN: Autostainer XL, November 2004) installed at UPMC Presbyterian Shadyside, using the accepted and validated clinical protocols in place at UPMC laboratories.
The second slide from each case was stained on the Symphony™ (Stainer SN:910032 , Part Number:2200000, April 2007) instrument installed at UPMC Presbyterian Shadyside, using standard protocols supplied by Ventana. This, in turn, reduces contamination1[20] or degradation over many uses. The detailed description of the differences and methods of the staining instrument itself are beyond the scope of this article, but can be found at: http://www.ventanamed.com/product/page?view=symphony
All work was completed by the four selected pathologists at their regular workstations, using the standard UPMC issued computer hardware and the viewer software provided by Trestle corporation software. The Med Microscopy™ platform, produced by Trestle Corporation, Inc. from 1999 to 2007, utilized an Olympus upright microscope (BX41 frame), robotic focus, nosepiece for objective selection and motorized stage combined with proprietary software to enable full robotic control, imaging and sharing to both local and remote users. A 10× uplan Apo objective was used for scanning, making a “20×” equivalent scanned image, which translates to roughly 0.5 micron/pixel resolution in the images.
This platform utilizes the same mechanism and hardware to create digital (virtual) slides, ranging from submicron per pixel values to macro imagery, facilitating rapid panning and zooming. The linkage of the various resolution layers is typically encoded in a pyramid image structure, and the data optimized such that the transition from layer to layer is highly optimized for speed.
The MedMicroscopy Digital Slide Module (DSM) viewer (Trestle) is provided for viewing images produced on this scanning platform technology. The software, developed for the PC platform, enables rapid panning and zooming of digital slide files stored on a standard hard drive or network share.
Minimum requirements for workstations (hardware) of test pathologists were those of a monitor with 1600 × 1200 resolution; 21-inch diagonal size; 0.26 dot pitch; video cardwith 256 MB of video RAM, Pentium IV , 512 MB of RAM (1 GB preferred); Internet Explorer 7 provided by UPMC was utilized. Each machine used was running Microsoft Windows XP Professional.
Each of the pathologists that participated in the study was familiar with use of digital slides, which they actually used in everyday practice and teaching; therefore, it was deemed that no further training was necessary to participate in the study.
During the slide scanning phase of the study, light intensity, focus, and all other pertinent microscope settings (e.g., condenser, iris, filters) and telepathology camera settings were monitored to ensure that they remain constant.
The pathologists viewed the slides from each case side by side (HDHE and regular H&E) while being blinded as to which is which, and were asked to choose one slide over the other according to the following parameters:
Contrast
Nuclear detail
Diagnostic information/clinical utility
Pathologists also answered the following questions regarding each slide:
Are you comfortable diagnosing from this slide (diagnostic quality of the specimen)?
Is the staining quality ideal?
RESULTS
The HDHE slides were rated as more acceptable than the traditional H&E batch-stained slides for the genitourinary (GU) samples and the gastrointestinal (GI) samples. The thoracic (THX) samples were rated as being approximately equal. The dermatopathology bench results were mixed. The dermatopathologist rated the samples equal on some characteristics, while on one characteristic the HDHE slides rated higher. On another characteristic, the batch-stained slide was rated higher.
Genitourinary pathology results
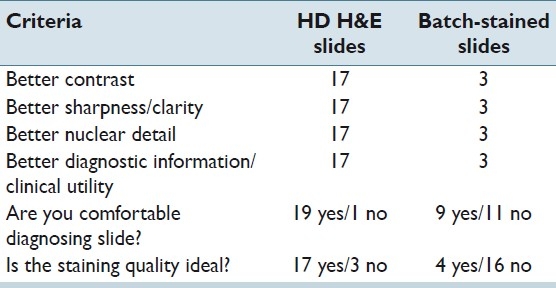
The pathologist examined 20 cases (slide pairs) for the study. Overall, the HDHE slides were rated more acceptable by a ratio of 17:3. The results are tabulated in Table 1.
Table 1.
GU pathology results
In the Figure 1, we contrast the tinctorial properties of the prostatic glands imaged at approximately 20× magnification.
Figure 1.
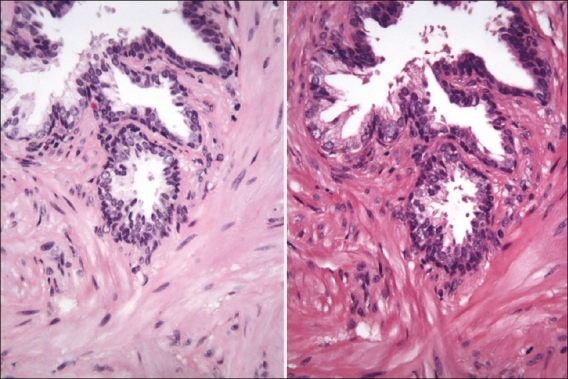
Prostate biopsy stained with the linear-batch method on the left and the HDHE method on the right. One can observe differences especially in the collagen area (screenshot of the Trestle MedMicro interface, approximate magnification 20×)
Gastrointestinal pathology results
The pathologist examined 20 cases (slide pairs) for the study. The HDHE stained slides were rated as more acceptable by a ratio of 16:4 [Table 2]. The results were rather similar to the results obtained from the GU bench. The two benches have similar needs in terms of contrast and nuclear detail, and for the most part, evaluate glandular tissue where eosin carries more weight in diagnosis than hematoxylin [Figure 2]; thus, the quality of the eosin stain is extremely important - it allows for a precise visualization and localization of features such as inclusions, H. pylori microorganisms, secretions, membrane color, crystals and eosinophils. The impression was that because the protocol used for HDHE slides produced a stain that was rich in red, it was particularly well suited to these slides.
Table 2.
GI pathology results
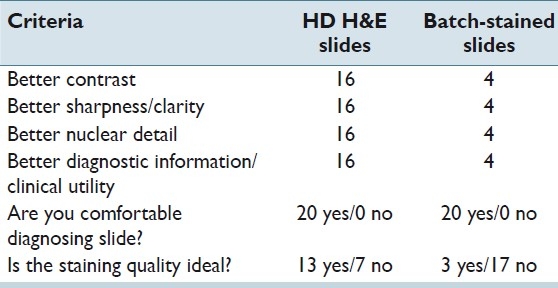
Figure 2.
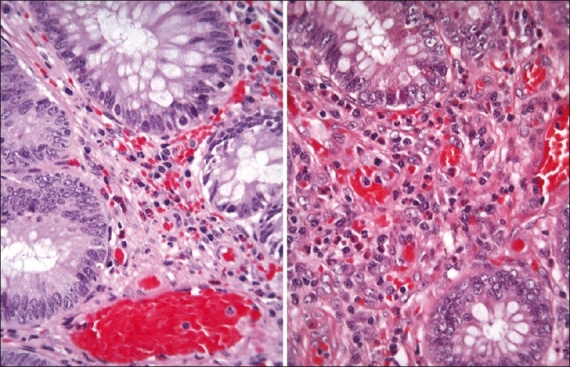
Contrast the biopsy stained with the linear-batch method on the left and the HDHE method on the right. Eosinophils are more readily distinguished with the HDHE method (screenshot of the Trestle MedMicro interface, approximate magnification 20×)
Thoracic pathology results
The thoracic pathologist examined 19 cases (slide pairs) for the study (the pathologist omitted one case with no explanation given) – results are tabulated in Table 3. Overall, both sets of slides were rated approximately equal on the test parameters. Both sets of slides were well accepted by the pathologist, with little difference in quality or diagnostic comfort. One can contrast the tinctorial slide properties of lung tissue in the Figure 3.
Table 3.
Thx pathology results
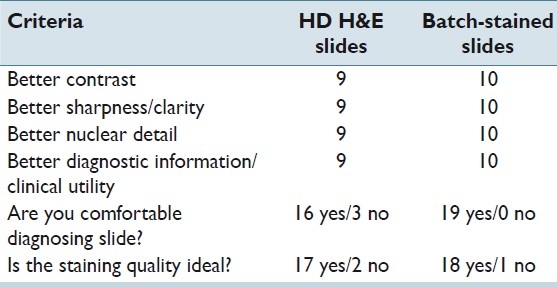
Figure 3.
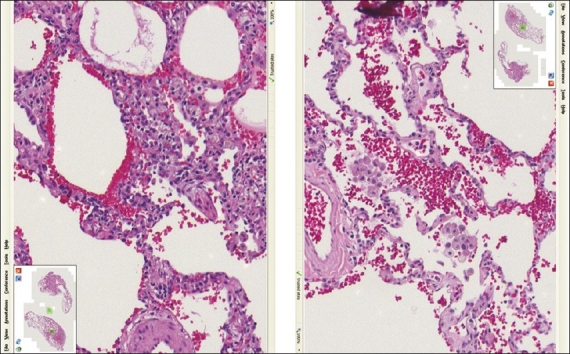
Lung tissue stained with the linear method (left) compared with the HDHE method (right) - (screenshot of the Trestle MedMicro interface, approximate magnification 20×)
Dermatopathology results
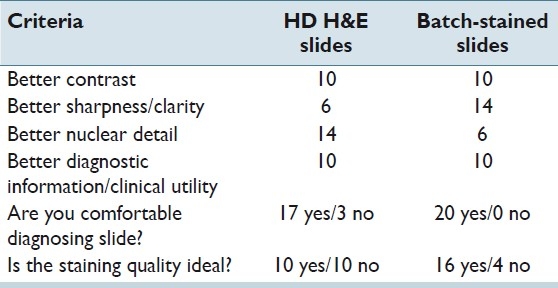
The dermatopathologist examined 20 cases (slide pairs) for the study. Results were mixed, with both conventional H&E slides and the HDHE showing marked advantage on at least one parameter [Table 4]. Neither method showed a clear advantage over the other on the total test parameters. Worth noting is that the HDHE slides were preferred in diagnosis of cellular features that are usually examined in detail at higher powers (10× and 20×) with ratio 14:6 in the area of nuclear detail; a comment from the study dermatopathologist was that the nucleoli were clearly visible and the nuclear membrane was distinct.
Table 4.
DP results
For the cellular features usually evaluated using mid to low power on the microscope, it was the impression of the study dermatopathologist that some of the HDHE slides lacked clarity - the cell borders looked indistinct [Figure 4]. This is represented in the fact that the linear-stained slides were rated higher on this parameter (ratio 6:14). We interpreted this as a concern, because the dermatopathology bench generally uses low-power magnification in diagnosing lesions [http://www.derm101.com/abstract/32672] and it was felt that a protocol adjustment will be necessary for further evaluation.)
Figure 4.
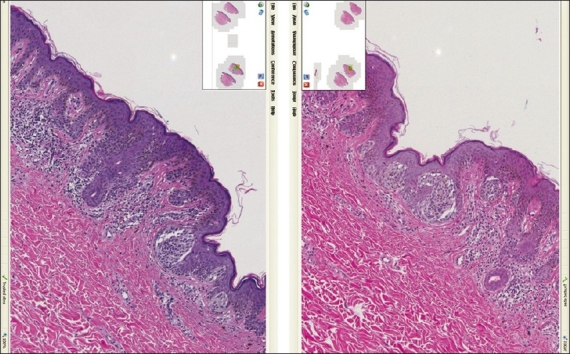
Skin tissue (compound melanocytic nevus) stained with the linear method on the left vs. the HDHE method on the right. In this figure, as well as previous ones, the small differences in the tissue outline arise from the fact that these are two adjacent (or nearly adjacent) tissue levels and are thus not identical (screenshot of the Trestle MedMicro interface, approximate magnification 20×)
DISCUSSION
The results of this study show that the utilization of HDHE protocol(s), as opposed to standard batch-stained slides might be preferable for digital pathology in some of the pathology subspecialties, notably GU and GI. It appears that the HDHE protocol is capable of turning out digital slides that have more contrast and detail, and therefore could offer more diagnostically significant information for the pathologist.
However, more work needs to be done to test protocols to find the optimal stain methodology for additional tissue types and various viewing modes; in particular, this entertains the notion that a tissue-specific staining protocols are needed – or at least that the tissue staining requirements in dermatopathology differ from the tissue staining requirements in “glandular” pathology (such as GU and GI pathology).
Thus, as more staining protocols are developed and fine-tuned to produce the best results for varying tissue types and viewing modes, this technology should yield greater utility for digital diagnostic pathology. It might be interesting to further evaluate the staining with HDHE protocol by the method described by Tadrous[22] or a similar color separation method.
Considering the advantages of digital slide technology for individually staining glass slides and the possibility that it can produce higher quality digital slides could play a decisive role to advancing the transition to a laboratory that is dominated by digital pathology.
ACKNOWLEDGMENTS
The authors thank Karen Lynn Branz for editorial assistance in preparing this article.
Footnotes
Available FREE in open access from: http://www.jpathinformatics.org/text.asp?2011/2/1/45/86284
REFERENCES
- 1.Jara-Lazaro AR, Thamboo TP, Teh M, Tan PH. Digital pathology: exploring its applications in diagnostic surgical pathology practice. Pathology. 2010;42:512–8. doi: 10.3109/00313025.2010.508787. [DOI] [PubMed] [Google Scholar]
- 2.Lele SM. Digital pathology in clinical consultation practice. J Pathol Inform. 2010;1:pii–17. doi: 10.4103/2153-3539.68334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pantanowitz L. Digital images and the future of digital pathology. J Pathol Inform. 2010;1:pii–15. doi: 10.4103/2153-3539.68332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Al-Janabi S, Huisman A, Van Diest PJ. Digital pathology: current status and future perspectives. Histopathology. 2011 doi: 10.1111/j.1365-2559.2011.03814.x. doi: 10.1111/j.1365-2559.2011.03814.x. [DOI] [PubMed] [Google Scholar]
- 5.Jukić DM, Drogowski LM, Martina J, Parwani AV. Clinical examination and validation of primary diagnosis in anatomic pathology using whole slide digital images. Arch Pathol Lab Med. 2011;135:372–8. doi: 10.5858/2009-0678-OA.1. [DOI] [PubMed] [Google Scholar]
- 6.Chen X, Zheng B, Liu H. Optical and digital microscopic imaging techniques and applications in pathology. Anal Cell Pathol (Amst) 2011;34:5–18. doi: 10.3233/ACP-2011-0006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Daniel C, et al. Standardizing the use of whole slide images in digital pathology. Comput Med Imaging Graph. 2011;35:496–505. doi: 10.1016/j.compmedimag.2010.12.004. [DOI] [PubMed] [Google Scholar]
- 8.Velez N, Jukic D, Ho J. Evaluation of 2 whole-slide imaging applications in dermatopathology. Hum Pathol. 2008;39:1341–9. doi: 10.1016/j.humpath.2008.01.006. [DOI] [PubMed] [Google Scholar]
- 9.Gilbertson JR, Ho J, Anthony L, Jukic DM, Yagi Y, Parwani AV. Primary histologic diagnosis using automated whole slide imaging: a validation study. BMC Clin Pathol. 2006;6:4. doi: 10.1186/1472-6890-6-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kldiashvili E, Schrader T, Burduli A, Ghortlishvili G. Application of medical information system for telepathology--Georgian experience. Telemed J E Health. 2010;16:699–704. doi: 10.1089/tmj.2010.0022. [DOI] [PubMed] [Google Scholar]
- 11.Williams S, Henricks WH, Becich MJ, Toscano M, Carter AB. Telepathology for patient care: what am I getting myself into? Adv Anat Pathol. 2010;17:130–49. doi: 10.1097/PAP.0b013e3181cfb788. [DOI] [PubMed] [Google Scholar]
- 12.Ayad E. Virtual telepathology in Egypt, applications of WSI in Cairo University. Diagn Pathol. 2011;6(Suppl 1):S1. doi: 10.1186/1746-1596-6-S1-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hitchcock CL. The future of telepathology for the developing world. Arch Pathol Lab Med. 2011;135:211–4. doi: 10.5858/135.2.211. [DOI] [PubMed] [Google Scholar]
- 14.Bautista PA, Yagi Y. Detection of tissue folds in whole slide images. Conference proceedings: … Annual International Conference of the IEEE Engineering in Medicine and Biology Society. IEEE Engineering in Medicine and Biology Society. Conference, 2009. 2009:3669–72. doi: 10.1109/IEMBS.2009.5334529. [DOI] [PubMed] [Google Scholar]
- 15.Bautista PA, Yagi Y. Improving the visualization and detection of tissue folds in whole slide images through color enhancement. J Pathol Inform. 2010;1:25. doi: 10.4103/2153-3539.73320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yagi Y. Color standardization and optimization in whole slide imaging. Diagn Pathol. 2011;6(Suppl 1):S15. doi: 10.1186/1746-1596-6-S1-S15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ho J, Parwani AV, Jukic DM, Yagi Y, Anthony L, Gilbertson JR. Use of whole slide imaging in surgical pathology quality assurance: design and pilot validation studies. Hum Pathol. 2006;37:322–31. doi: 10.1016/j.humpath.2005.11.005. [DOI] [PubMed] [Google Scholar]
- 18.Weinstein RS, Graham AR, Richter LC, Barker GP, Krupinski EA, Lopez AM, et al. Overview of telepathology, virtual microscopy, and whole slide imaging: prospects for the future. Hum Pathol. 2009;40:1057–69. doi: 10.1016/j.humpath.2009.04.006. [DOI] [PubMed] [Google Scholar]
- 19.Singh R, Chubb L, Pantanowitz L, Parwani A. Standardization in digital pathology: Supplement 145 of the DICOM standards. J Pathol Inform. 2011;2:23. doi: 10.4103/2153-3539.80719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Platt E, Sommer P, McDonald L, Bennett A, Hunt J. Tissue floaters and contaminants in the histology laboratory. Arch Pathol Lab Med. 2009;133:973–8. doi: 10.5858/133.6.973. [DOI] [PubMed] [Google Scholar]
- 21.Buesa RJ. Histology: a unique area of the medical laboratory. Ann Diagn Pathol. 2007;11:137–41. doi: 10.1016/j.anndiagpath.2007.01.002. [DOI] [PubMed] [Google Scholar]
- 22.Tadrous PJ. Digital stain separation for histological images. J Microsc. 2010;240:164–72. doi: 10.1111/j.1365-2818.2010.03390.x. [DOI] [PubMed] [Google Scholar]


