Abstract
Background:
Cytology poses different obstacles in whole slide imaging compared to surgical pathology slides. A single focal plane suffices for most of the latter, but cytology slides are thicker, potentially requiring multiple focal planes for adequate diagnostic information. Multiple focal planes adversely impact scanning time per slide, evaluation times, and file sizes. In this pilot study, we evaluated and compared the multilayer stack method to the extended focus algorithm as an alternative which collapses multiple focal planes into a single image, retaining only focused areas from each plane.
Materials and Methods:
10 SurePath® cervical cytology slides were scanned at three thickness settings: 18, 24, and 30 μm. Three scanners were used: (1) Hamamatsu Nanozoomer 2.0-HT, (2) 3DHISTECH Mirax scan, and (3) Bioimagene iScan Coreo Au. The Nanozoomer and iScan utilized multilayer stacking, while the Mirax files were composited by extended focus. Scan times and file sizes were recorded, and image quality compared.
Results:
The Nanozoomer stacks averaged 1.58 gb and around 25 min for each slide, while the iScan stacks ranged from 6.23 to 9.3 gb and took 34-50 min to scan. The Mirax images averaged 210 mb and took 13-20 min to scan. Multilayer stack image quality from both Nanozoomer and iScan was fairly comparable. The iScan revealed significant mechanical issues that did not correspond to user settings. The Mirax images showed worrisome loss of crisp focus detail, worsening with increasing focal planes and impacting assessment of nuclear contours and chromatin detail.
Conclusions:
The optimal number of focal planes remains unknown for cytology. Multilayer stacks require excessive scanning time, network bandwidth, and file storage. Extended focus was evaluated as an alternative, but significant image quality issues were revealed. Further large-scale studies are needed to assess their clinical impact.
Keywords: Cytology, cytopathology, digital, pathology, whole slide imaging
INTRODUCTION
Whole slide imaging (WSI) is a long awaited, exciting prospect to pathologists, promising value-added computer-aided diagnosis and capability for remote consultation. However, WSI is currently limited by technological limitations, including slow, clinically unviable scanning speeds, focusing errors that compromise diagnostic quality, and most importantly, unprecedented demands for tremendous storage space. Currently, many WSI slides in the surgical pathology setting are scanned on one focal plane that is usually sufficient for diagnosis, with file sizes ranging upwards of 1.5+ gigabytes in compressed file size.
In contrast to surgical pathology, cytopathology has different WSI needs than typical surgical specimen slides [Figure 1]. First, cytology slides are thicker: liquid-based preparation slides (LBP) (e.g. SurePath® ) can range upwards of 30 μm from glass to coverslip (W. Gray, BD Diagnostics, personal communication, March 11, 2010), while the vast majority of surgical paraffin blocks are cut at between 4 and 6 μm in thickness. Second, one focal plane for biopsy and resection surgical slides is usually sufficient to capture diagnostic quality. Third, by definition, tissue architectural features are not present in cytology and therefore the capability to provide fine focusing for every cell and cell cluster within the slide, from glass to coverslip, may be important for adequate evaluation of the diagnostic material to provide the best care to the patient.
Figure 1.
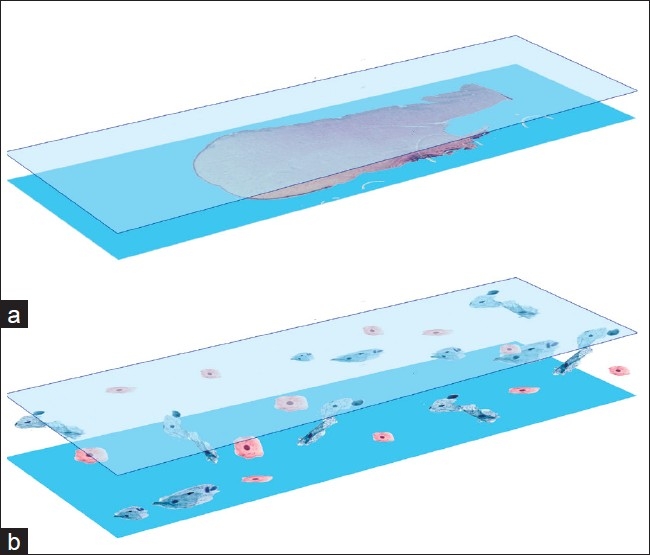
Schematic illustrating differences between (a) surgical pathology slides which are usually 4-6 μm in thickness and (b) cytology slides (bottom diagram) which can range upwards of 30 μm from glass to coverslip. Cells can be positioned anywhere from glass to coverslip in cytology
In the clinical setting, these three issues make WSI problematic as multiple focal planes become a requirement for each slide, a method known as a multilayer stack. Further, the optimal number of focal planes to use per cytology slide is not known, as well as the spacing between these focal planes. Although we can theoretically approximate any glass slide on traditional microscopy by scanning over 100 focal planes at very small intervals (as low as 0.2 μm on some scanners), this is not viable with current technology as of 2011 due to limitations in processing power, memory, networking, and storage space. Each additional focal plane increases both scanning time and file size per slide. Additionally, the true thickness of each individual slide down to the level of the micron is unknown, with the high probability that slide thicknesses vary from one to another.
There is an obvious tradeoff in obtaining an optimal number of focal planes per slide versus the total scan time, speed of evaluating the slide, storage space for the digital slides, and computer memory usage. Adding focal planes directly increases total scanning time and file size per slide. A recent paper used 21 focal planes per slide at 1.5-μm intervals on SurePath liquid-based preparation slides, taking hours to scan each slide and resulting in file sizes of 7.3 gb per slide.[1] Many frustrations were encountered, such as difficulty with manipulating the virtual slides and their 21 focal planes, slow computer response, computer lockups, and network congestion when multiple participants accessed the same digital slide simultaneously were encountered. These issues were directly attributed to the sheer size of the datasets from these slides. These findings parallel those in an earlier paper involving cytology slides involving 10 focal planes each, where the majority of survey participants answered “usable, but requires effort”.[2] Therefore, there clearly is interest in alternative solutions for WSI cytology other than multilayer stacking.
Extended focusing is an algorithm available from 3DHISTECH Ltd (Hungary) that extracts focused areas from each focal plane and assembles them together into a single, composite image. Although this technique was designed initially for thick slides in cytology and various fluorescent methodologies, its usefulness for cytology has not been studied much. However, a file consisting of only one image, as opposed to 21 image layers as above, is appealing as it presents an alternative solution to the storage, network, and CPU problems that occur when multiple focal planes are utilized.
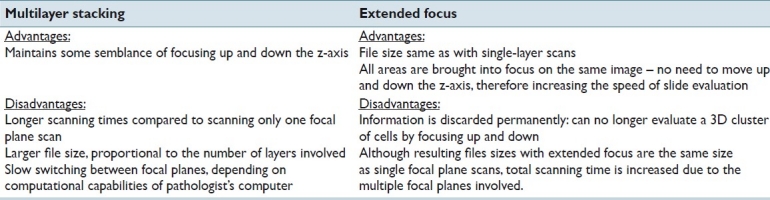
In this paper, we optimize and explore a comparison between multilayered stacking and extended focusing as an alternative to the networking and storage space issues that currently impede the viability of WSI cytology [Table 1].
Table 1.
Advantages and disadvantages unique to each method (multilayer stacking and extended focus algorithm
MATERIALS AND METHODS
After obtaining institutional review board approval from Tufts Medical Center (Boston, MA, USA), 10 cervical pap smear specimens in SurePath® fixative were randomly selected and deidentified, with portions used to produce liquid-based preparation cytology slides (SurePath® ) of similar cellularity, 1 per specimen. The SurePath® system creates a circular field of cells with a surface area of 133 mm2 (13 mm diameter).[3] Diagnoses were restricted to atypical cells of undetermined significance (ASCUS), low grade squamous intraepithelial lesion (LSIL), and high grade squamous intraepithelial lesion (HSIL).
Three scanners were used for this study, all utilizing multiple focal planes on each slide, and connected to different workstations of varying hardware specifications:
Nanozoomer 2.0HT (NDP), Hamamatsu Photonics K.K. (Japan)
Microsoft Windows XP Professional (Service Pack 3), with Intel Xeon 5160 processor (3 Ghz), and 4 gb of RAM
Mirax Scan (Mirax), 3DHISTECH Ltd. (Hungary)
WinXP Professional (SP3), with Intel Xeon E5410 processor (2.33 Ghz), and 3 gb of RAM
iScan Coreo Au (iScan), Bioimagene Inc. (California, USA)
WinXP 64-bit Profession (SP2), with Intel Xeon E5504 (2.0 Ghz), and 12 gb of RAM
-
Three experiments using a different predetermined thickness setting were performed on each scanner, using each of the 10 slides:
- 18 μm
- 24 μm
- 30 μm
Each scanner was used in batch mode with automatic area selection and focus point placement.
Each scanner had its own minimum/maximum settings for number of focal planes and interval levels [Table 2]. We were able to determine and use the correct combination of settings for each scanner in order to perform experiments based on the three specified thickness settings above.
Table 2.
Three-way comparison of flexibility in settings for spacing intervals and number of focal planes. Of note, the Bioimagene scanner only allows multilayer stacking using their proprietary file container and compression scheme (*.bif) instead of JPEG as with the other two scanners
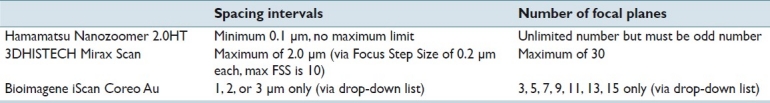
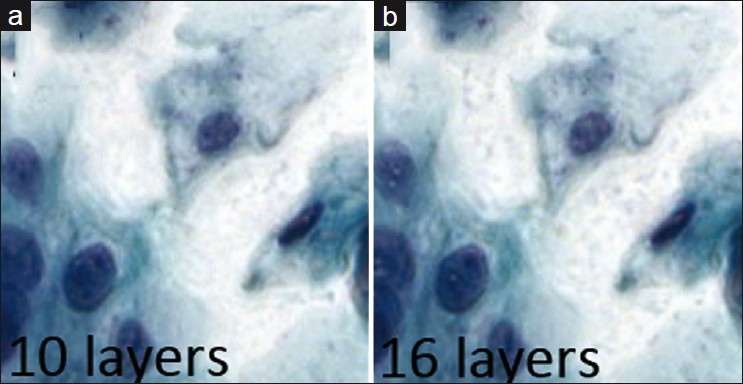
The NDP scanner allowed us to keep the number of focal planes constant at 7, but its three experiments varied by spacing: 3, 4, and 5 μm. The Mirax scanner limits the maximum spacing between focal planes to only 2 μm, and so we scanned 10, 13, and 16 layers in each case. The iScan scanner was more restrictive in the range of focal planes and interval spacing allowed, but matching the three thickness conditions was still possible.
Both the NDP and iScan images were maintained as multilayered stacks consisting of multiple focal planes, while the Mirax scanner utilized the extended focus algorithm.
Total scanning times and file sizes were recorded and visual quality compared between the three systems. In all cases for the NDP and Mirax scanners, the JPEG compression setting was 80 with scanned magnification at 40× (0.25 μm/pixel). The iScan scanner offers three possible compression schemes: JPEG2000, TIFF, and their proprietary Bioimagene image format. However, multilayer stacks on the iScan scanner are restricted to using this file format (*.bif), which uses an unknown compression scheme within a proprietary file container.
Scans for the Mirax and iScan systems were saved over the network to a file server during the scanning process, while the NDP images were saved to the local drive.
For evalutation of image quality and user experience, the images were copied to and viewed off of a local hard drive on a standard Hewlett-Packard desktop computer running 32-bit Windows XP (SP3), with an Intel® Core2™ Duo E8400 @ 3.0 Ghz CPU, with the Intel® Q45/43 Express chipset for graphics, and 4 gb of installed RAM memory.
RESULTS
Scanning times and file sizes
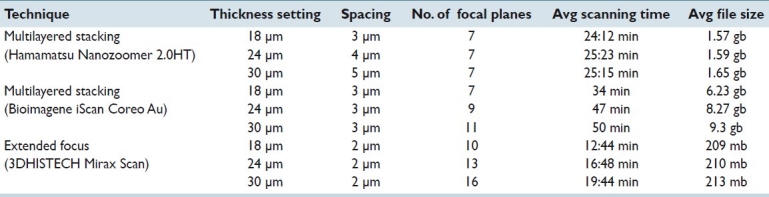
The average scanning times and files sizes between the multilayered stacking of the NDP machine and extended focus algorithm method of Mirax were consistent and predictable after a couple scans [Table 3].
Table 3.
Average scanning times and file sizes. Results are shown for each of the three scanners utilized in this study. All ten slides were scanned for each experiment, each of which is represented by a row in the table
On the NDP machine, the number of focal planes was kept constant at 7 for all three experiments, and the differences in the thickness setting (18, 24, and 30 μm) and spacing between the layers (3, 4, and 5 μm) did not significantly alter scanning times. Average scanning times and file sizes using 7 focal planes were roughly the same amongst the three NDP experiments and appear to be independent of thickness settings and spacing intervals between focal planes.
With the Mirax scanner, the maximum spacing between focal planes was restricted to 2 μm by its scanning software. Therefore, spacing intervals were kept constant at 2 μm and we increased the number of scanned focal planes to match the specified thickness settings used for each scanner′s experiments. As expected, the extended focus algorithm resulted in a single image instead of a multilayer stack, therefore producing file sizes of around 200-215 mb.
Although the average file size of the NDP multilayer stacks were approximately 1.5 gb (7 focal planes), when one compared them on a “per focal plane” basis, the NDP images were comparable in file size to the Mirax files, (230 mb vs. 210 mb, respectively), suggesting similar compression ratios [Table 4]. This was expected because both machines were set to use the same compression scheme (JPEG) at the same compression rate (80).
Table 4.
Summary of scan times and file spaces for each scanner on a per focal plane basis
However, findings for average scanning times between the Mirax and NDP machines were unexpected. First, one would intuitively expect that scanning and saving directly to a local hard drive would be faster than saving to a file server over the network. However, our results revealed opposite results: although the Mirax scans were saved over the network and the NDP files were saved to the local drive instead, scanning times were faster in all cases with the Mirax experiments. Second, although all three Mirax experiments involved more focal planes (10, 13, and 16 planes) than the NDP experiments (7 planes for all three experiments), the average scanning times for the Mirax scanner were all significantly shorter than with the NDP scanner. The average scanning time for Mirax with 16 focal planes was less than 20 min per slide, compared to the roughly average 24-25 min per slide for 7 focal planes on the NDP scanner. A number of reasons can explain this finding (see the Discussion section).
In regards to the Bioimagene scanner, total scanning times were on average, significantly longer than the other two scanners – almost a minute and a half longer than the NDP scanner when compared on a “per focal plane” basis. In terms of comparing its file sizes to the other scanners, the average file size per focal plane was around 887.24 mb, compared to 210-230 mb for NDP and Mirax. It should be noted that this file size comparison is not a fair one because we could not obtain a multilayer stack using the same compression scheme as the others. When creating multilayer stack image, the Bioimagene scanner does not allow JPEG compression, and instead mandates use of its own proprietary file format. This proprietary file format (*.bif) contains an unknown compression scheme.
Multilayer stacking
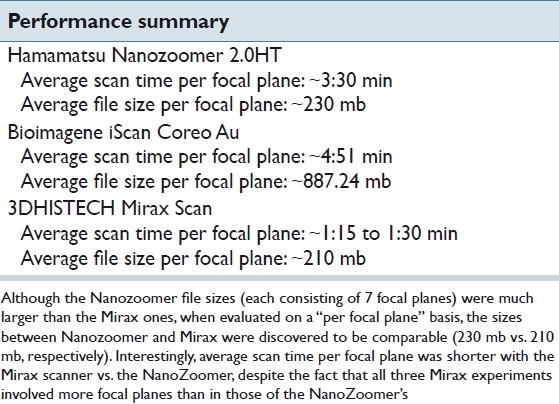
A high power example of the seven NDP focal planes is shown in [Figure 2], corresponding to the same area on the slide as in the Mirax experiments [Figure 3] are shown. Again, as alluded to in the background section, the infinite number of focal planes in traditional microscopy is reduced to only 7 focal planes. These 7 planes simulate focusing up and down on a traditional microscope, and areas that are in focus look as if on a properly focused microscope.
Figure 2.
Multilayer stack from Hamamatsu Nanozoomer (40× view)
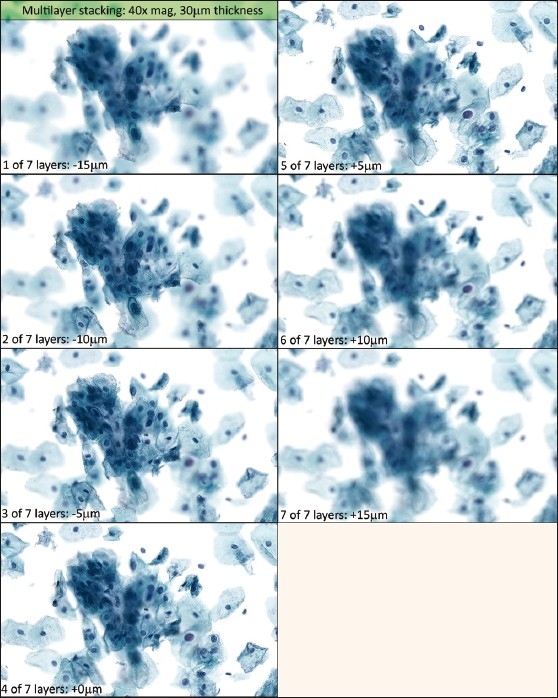
Figure 3.
Extended focusing algorithm. This picture corresponds to the same area on the same slide as shown in Figure 2′s multilayer stack. All areas are brought into improved focus; however, extracellular debris are accentuated (worse with increasing focal planes), and crisp detail seen in the multilayer stack is lost
Comparing performance speed of the viewer programs, at high power (40×) the switching between focal planes was fast enough to be usable, although nowhere as fast as turning the focus knob on a traditional microscope. For the two scanners utilizing multilayer stacking (NDP and Bioimagene), switching focal planes at low power (2×) was excessively slow on the standard desktop, on the order of 2-3 s between planes, because the viewing workstation needs to work with the large surface area of the gigapixel-sized image. At high power (40×), zooming in and out of the image using the mouse wheel (the equivalent of changing objective power on a microscope) caused a 0.5 to 1.0 s delay before the actual change.
An evaluation of the multilayer stack of the Bioimagene scanner at the same thickness setting of 30 μm was performed, similar to that of the NDPs in Figure 2. The scanner was not capable of proper focus along the entire z-axis length of 30 μm. Despite an interval setting of 3 μm and 11 focal planes scanned (in contrast to the NDP machine′s settings of 5 μm and 7 focal planes), only 2 out of the 11 focal planes had areas in sharp focus, and the other 9 planes were completely out of focus. Possible explanations follow in the Discussion section.
Nonetheless, it was still possible to compare the iScan′s image quality of the in-focus planes to those of the NDP′s. The two were very comparable in quality, in terms of focused areas, sharpness, and color fidelity. Small issues with the iScan included grainy cytoplasmic textures in the unfocused areas which were not seen in the correlating areas with the NDP counterpart nor on the microscope. Another small issue was that of slight over-white balancing with the default brightness/contrast settings, which resulted in a slight “washed-out” appearance to cell cytoplasm and borders, and loss of sharp detail.
Extended focus
Figure 3 shows an extended focus image of the corresponding area of the slide (scanned by the Mirax machine) from Figure 2. All the cells that were blurry at some point in the multilayer stack were brought into focus. However, the algorithm is also applied to extracellular debris throughout the thickness of the z-stack, resulting in a denser, “dirtier” appearance compared to multilayer stacking or traditional microscopy. The particular example shown in Figure 3 is composed of 16 focal planes, covering a thickness setting of 30 μm. Extracellular debris appears to worsen with increasing numbers of focal planes [Figure 4].
Figure 4.
Extended focus. Extracellular debris appears to become accentuated with increasing layers
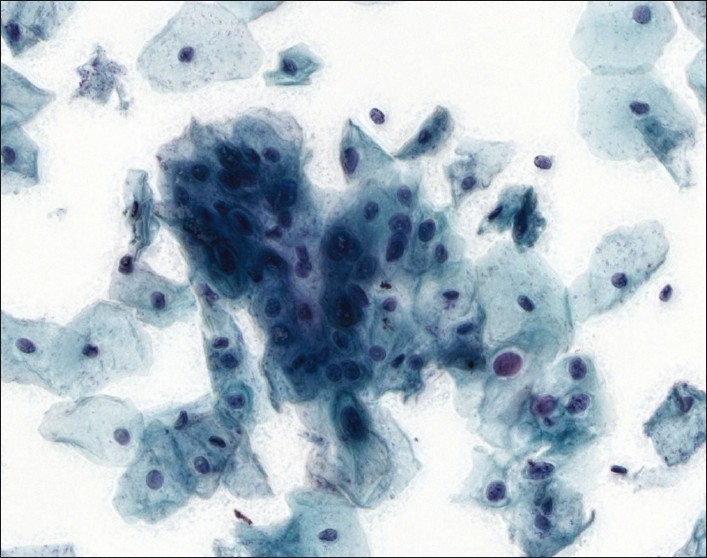
The number of optimal focal planes for extended focus in cytology could not be determined in this study. There was no appreciable improvement in visual quality between 10, 13, and 16 focal planes [Figure 5]. All three focal plane settings were subject to the same visual quality problems (see the next section).
Figure 5.
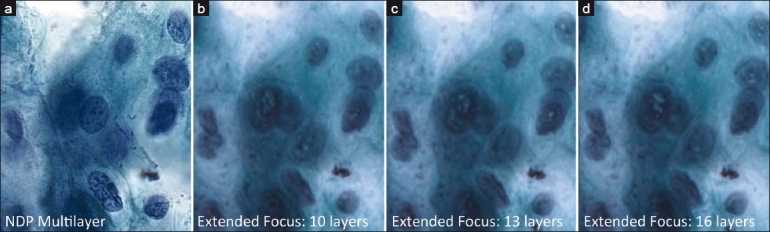
Sequential comparison of extended focus images varying in number of focal planes, with a Multilayer stack image for comparison. With extended focus, intranuclear digital artifacts become more prominent with more focal planes. In all cases, sharp detail is significantly less than with multilayer stack method. (a) Multilayer stack image for comparison, (b) extended focus using 10 focal planes, (c) extended focus using 13 focal planes, (d) extended focus using 16 focal planes
The Mirax viewer application (extended focus) was subjectively much faster than the NDP viewer application (multilayer stack) in terms of navigation and zooming due to less I/O requirements – providing a smoother and lag less navigation experience for the user.
Direct image quality comparison between multilayer stacking and extended focus
Side-by-side comparisons were made between corresponding areas in an extended focus image and in-focus area within the multilayer stack at both high and low power [Figure 6].
Figure 6.
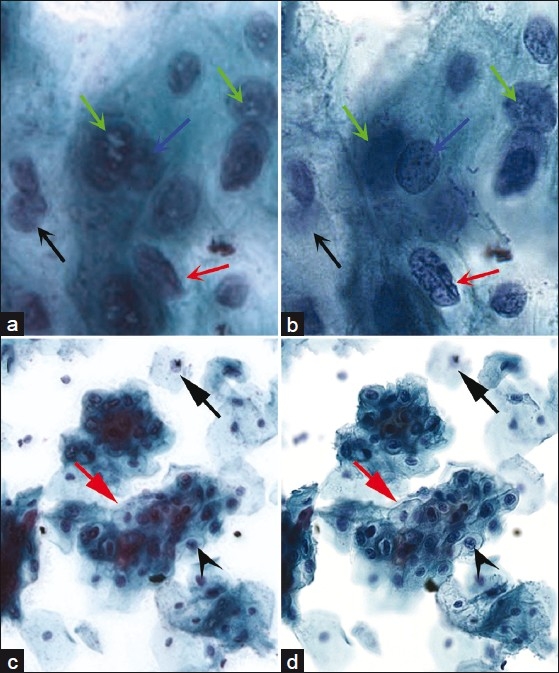
Side by side comparisons between extended focus and multilayer stacking. At high power (40×), (a) extended focus improves focus (black arrow) for areas out of the plane of focus but sharpness and detail appears to be adversely affected. Nuclear contours (red arrow) and chromatin detail (blue arrow) are harder to assess, and white intranuclear digital artifacts are seen only in extended focus (green arrows). 16 focus planes were used in this example. (b) Multilayer stack counterpart to (a). At low power (10×), (c) loss of sharp detail can still be seen in extended focus compared to its multilayer stack counterpart. Nuclear detail is blurred compared to the sharply focused, corresponding area within the multilayer stack version (black arrowhead), and cell borders are difficult to assess (red arrow). (d) Multilayer stack counterpart to (c).
Although the extended focus algorithm generally improved otherwise blurry areas closer to focus, there appeared to be significant degradation in sharpness and crisp detail in the extended focus image compared to the NDP multilayer stack versions. At low power, the intercellular borders in the extended focus image became difficult to distinguish. At high power, chromatin detail and nuclear contours became very difficult to assess, presenting a critical problem for cytology. A “digital pseudo-intranuclear inclusion”, appearing as white-colored areas within the nucleus, was apparent only in the extended focus version and never in the multilayered stack.
An experiment utilizing one focal plane from the Mirax scanner revealed excellent nuclear contours and chromatin detail. However, a direct comparison of multilayer stacking methods between the Mirax and NDP was not possible because with multiple focal planes, the Mirax is not capable of producing multilayer stacks - only extended focus is permitted.
DISCUSSION
Evaluation of the Bioimagene iScan coreo Au for cytology
The Bioimagene iScan machine is a relatively new entrant to WSI hardware. In comparison, the NDP scanner has already been used and described in many telecytology literature studies.
Our initial evaluation of the Bioimagene machine′s image quality for cytology use revealed four issues: (1) questionable interval spacing/focusing, (2) limited user choices for selection of number of focal planes and intervals, (3) grainy texture, and (4) overuse of white-balancing.
The first issue is the most worrisome, because the scanner most likely did not perform at user settings for interval spacing. Its multilayer stack of 11 focal planes, with an interval distance of 3 μm, had only 2 planes in focus, and the rest were completely out of focus. In comparison, the multilayer stack of the NDP (previously seen in Figure 2), which used 7 planes and an interval distance of 5 μm, provided a nice progression of different focusing while traversing the focal planes [Figure 2]. This unexpected finding of 9 (out of 11) unusable focal planes in the Bioimagene experiment is worrisome because although it could reflect scanner miscalibration, it could also suggest that software settings do not correlate with actual hardware operation. In other words, if the interval setting is set to 2 μm but the machine instead scans at 5 μm intervals without notifying the user, then a silent, potentially significant discrepancy arises. If this were the case, then a pathology laboratory cannot trust such a machine to do what is requested.
In addition, the Bioimagene scanning software′s options for selecting the number of focal planes and intervals between them was extremely limiting in comparison to those of the NDP and Mirax scanners. In the setting of thicker slides such as in cytology, these limitations can have a large impact on scanning result. The Bioimagene scanning software utilizes drop-down lists of limited choices for both variables. For spacing intervals, there were only three possible choices: 1, 2, or 3 μm. In comparison, the NDP scanning software is capable of intervals as low as 0.1 μm, and the user specifies the intervals in a text field as opposed to a drop-down list with fixed choices. The number of focal planes allowed with the Bioimagene software was limited to only 3, 5, 7, 9, 11, 13, and 15. Additionally, the Mirax scanning software allows any number of focal planes from 1 to 30, and the NDP is capable of unlimited numbers of focal planes though limited to odd numbers only.
The minor issues involving grainy textures and over-white balancing would likely not have any impact on diagnosis. Although the in-focus areas on the Bioimagene iScan images appeared sharp, the grainy texture seen especially in the out-of-focus areas suggests an over-application of a sharpening filter in software, and does not accurately reflect what is seen with a glass counterpart. Overuse of white balancing under default settings resulted in some loss of image quality; however, this issue is easily corrected via the Bioimagene image viewer′s settings to adjust brightness and contrast.
Comparison between multilayer stack and extended focus methods
Multilayer stacks obtained from these cytology slides provided superior image quality over that of extended focus as discussed earlier. Images were sharper and chromatin detail could be assessed easily as with glass slides. Although the chances of obtaining perfectly focused areas increases with more focal planes (thereby closer approximating the original glass slide), the drawbacks include larger file sizes, higher network congestion, and slower pathologist workstation response. It is still unknown how to determine the optimal balance between too little and too many focal planes. Current limitations of storage space, networking speeds, workstation memory capacity, and workstation processing power prevent multilayer stacks from becoming a viable solution for WSI cytology in 2011. However, due to rapid developments in technology, these issues will likely be solved in the near future.
Extended focus was used in the context of cytology as a possible alternative method to obviate the multilayer stack method′s limitations. Because there is only one image to transmit through the network and store, this method of WSI is potentially cost and time saving: storage requirements are markedly less, and during slide examination the pathologist only needs to work in two dimensions – the z-axis is not involved at all. However, this method′s drawbacks were discovered during direct comparison to the multilayer stack: assessment of nuclear contours and chromatin detail are some of most basic principles in cytology, and become very difficult if not impossible with these image degradations. Additionally, collapsing multiple focal planes into a single image may mislead a pathologist into misinterpreting the true amount of extracellular debris. Debris is accentuated with increasing focal planes because the extended focus algorithm will retain the focused areas of debris in the composite image. Lastly, the introduction of digital artifacts, especially within nuclei, is not acceptable.
Extended focus was never intended to be the perfect solution to multilayer stacking′s limitations, but rather as a workaround to current IT limitations until multilayer stacking becomes more feasible in the future. Despite the issues this study raises about its image quality, the extended focus method still provides important benefits that would optimize pathology workflow: faster scan times and slide navigation, less file storage consumption, and overall decreased slide review time.
If the extended focus algorithm were to be improved such that these loss of quality and introduction of digital artifact issues were solved, then the matter would become moot. Thus, the important question to ask is why crisp detail is being lost with this algorithm.
That said, cytology expertise is a key factor, and cytologists with many years of experience would still be able to interpret the image adequately despite lesser sharpness compared to the multilayer stack counterpart. In contrast, a pathology resident might have difficulty interpreting digital cytology slides with these quality issues. This begs the question of what is good enough for diagnosis, as opposed to education.[4]
Performance-wise, as noted earlier, both the Mirax viewer and scanning applications were much faster than those of the two other scanner machines, despite the involvement of more focal planes and saving the image files to a network file server as opposed to the local drive. One possible (and likely) explanation for this improvement in speed is likely due to inclusion of MMX instruction sets in their software engineering, which they stated on their website to improve performance by 50% over previous versions. It is unknown whether Hamamatsu or Bioimagene utilize any special instruction sets in their software code.
The optimal number of focal planes for cytology is still unknown. Although larger numbers of focal planes result in artifacts such as excess extracellular debris, the “less is more” approach could be valid. Recently, it has been suggested that 3 focal planes may be the optimal number.[5] Today′s non-WSI, FDA approved, automated LBP cytology screeners utilize 3 planes during their process, determine the best focused field of view out of the three, and then use it for analysis. Although one may argue that a mere three focal planes is insufficient to ensure adequate capturing of information involving three-dimensional clusters of cells, there are already published studies suggesting adequacy.[6,7] Nevertheless, investigation into the optimal number of focal planes is ongoing.
Lack of information and manual control of z-axis
A major problem with current scanners and their handling of the z-axis is that their chosen focal planes may or may not be representative of equal distribution along the entire z-axis, and we cannot confirm whether they are clustered or biased towards the glass slide or coverslip.
A frequent cause of user frustration occurs when the z-axis is only partially captured instead of its full range by multiple focal planes from glass to coverslip. At the first and last focal planes, the so-called boundary planes, the user can encounter a suspicious lesion just starting to come into focus, but cannot proceed beyond these boundary focal planes because the focal planes beyond them were never scanned. One can never be confident how much un-scanned spatial volume remains between the bottom boundary layer of the multilayer stack and glass slide, and between the upper boundary layer and the cover slip.
Full capability to evaluate the entire volume of space along the z-axis requires full coverage from glass to coverslip, potentially covering 30 μm (or thicker). However, due to the lack in certain hardware capabilities in these WSI scanners, full z-axis coverage is either impossible or we are never informed by the scanning software which portions of the z-axis were covered. Patient safety could be severely compromised if critical planes are missed by the scanner, and therefore the pathologist cannot trust these focal planes on blind faith.
Subsequently, it is also important to realize that, the extended focus algorithm is susceptible to this problem as well. With extended focus, it becomes less obvious to realize that crucial focal planes still may not be represented in the composite image.
Further, it is unknown if digitizing a physical slide containing a limited number of focal planes is sufficient enough to prevent misdiagnosis. Red blood cells have a diameter of 5-7 μm, which is approximately the same size as the nucleus of the intermediate squamous cell of the cervix. By Bethesda criteria, low-grade squamous intraepithelial lesion (LSIL) cells have nuclei that are at least three times that of a normal intermediate cell′s. Assuming a slide thickness of 30 μm, and a LSIL nucleus range of roughly 15-21 μm, there is still potential of a focal plane completely missing the nucleus of that cell. All dysplastic cells must be captured, and none missed, during the scanning process.
Secretory cells from the endocervix and superior to it are often found in three-dimensional clusters. Fine adjustment of the focus knob on the microscope by “wiggling back and forth” is often used to identify ciliated cells in this setting. Within a given cluster of cells, there may be multiple findings at different focal planes.
The traditional microscope provides a continuous range of an infinite number of focal planes along the entire z-axis, from glass to coverslip, with focusing errors easily corrected in real time. In contrast, because current WSI methods reduce the data set to a discrete number of focal planes, pathologists are unaware of missing diagnostic information between focal planes. A pathologist would not want to miss a critical cluster of cells that could potentially lead to misdiagnosis, especially when it is present on the original glass slide.
Additionally, a sparse slide that still fulfills Bethesda criteria under the microscope could be wrongly called inadequate for diagnosis due to the scanner not properly focusing on enough ciliated endocervical or squamous metaplastic cells within its focal planes.
The microscope, a technology hundreds of years old, remains immune to these z-axis issues.
Possible solutions
In many cases, the performance of viewing software is likely not optimized for speed. For wide adoption among pathologists, the user experience should be lagless, smooth, and responsive while navigating. It is believed that a minimum of around 50 frames per second (fps) is required for the human eye to perceive fluid motion without flicker or motion blur.[8] With this frame rate, navigation would become quick and smooth, as if gliding across the entire virtual slide.
Also, switching between focal planes needs to be as fast as moving up and down while on a microscope. While this may not be yet possible with today′s common desktop computer, it can be possible with a high-end workstation with a modern CPU (such as the Intel i7), with 16 gb of RAM, and a higher end accelerated 3d video card. Viewing programs can be written take advantage of the multiple core architecture in modern video cards, especially if a high-performance workstation equipped with a high-end consumer graphics card to help relieve CPU load. This is a solution possible today, and already implemented in various areas of science, especially in bioinformatics.[9–12] In addition, if the file is viewed on a local computer as opposed to over a network, then undesirable elements such as network latency and congestion are completely avoided.
While the extended focus algorithm appears to significantly degrade sharp detail, thereby questioning correct diagnosis, it could still be useful for quick screening, perhaps via automated image analysis to identify areas on the slide that should be examined, then evaluated by a pathologist using a multilayer stack version of the same slide where it could be assessed with more accuracy. More studies are required to assess where/when use of the extended focus algorithm provides efficiency along the workflow process.
Regarding file sizes, compression schemes may improve in the future. JPEG2000[13] is currently available and has been shown to be a superior at compressing files than standard JPEG, with upwards of 33% more compression. However, JPEG2000 is not currently viable on many standard desktop computers because it is more computationally intensive than JPEG, and most current low-cost desktop computers lack the computational horsepower to provide 50 fps rendering with the JPEG2000 codec .
As for the z-axis problem, as users, we are forced to work with unknown distributions of focal planes along the z-axis. This is undesirable because complete coverage from glass to coverslip is required in order to avoid missing important lesions and to ensure optimal evaluation of the patient′s slides. Affecting both WSI methods (multilayer stacking and extended focusing), this problem could be a new source of misdiagnoses, representing new medico-legal implications. The solution to this problem is heavily dependent on pathologists working closely with vendors and their engineers to help them understand these crucial medico-legal implications and to develop solutions in providing full z-axis information, coverage, and mechanical control.
Aside from ensuring full coverage of the z-axis from glass to coverslip, another argument for mechanical z-axis control on the scanner is that of focusing. No pathology slide is truly flat at the microscopic level – there is a microscopic topography which presents autofocusing challenges.
Although all scanners provide autofocusing, their algorithms are not perfect, and still often fail resulting in full slide blurriness. In these cases, additional algorithms can be developed to address and negate some possible causes of focusing failure, such as tissue folds in histology slides and physical annotation by traditional dotting pens.
Another possible solution when focusing failure occurs is to attempt repeat scanning using a different vendor′s scanner. However, this approach is unfeasible because (1) it assumes that the histology lab possesses more than one scanner, whether a different model or from a different vendor, and (2) it is no guarantee of obtaining a well-focused image because the second attempt may still result in total focusing failure [Figure 7]. In these cases, because there is no manual z-axis control for the scanner, there is no other viable way to view the tissue other than reverting back to the actual glass slide on a traditional microscope.
Figure 7.
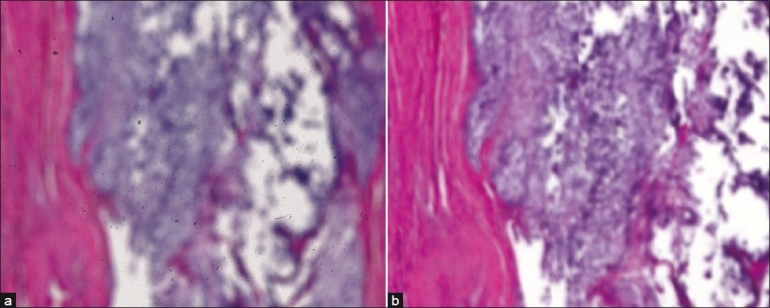
Example of total autofocus failure on a chondrosarcoma slide with two attempts using different scanners. A third attempt using a third scanner is unlikely to fix the problem. Both images are low power (5×) views, and both unusable for neither education nor diagnosis. (a) NanoZoomer, (b) Mirax
It should be noted that if autofocus algorithms were to successfully produce perfectly focused images for any tissue type, then many of these problems could be solved for surgical slides.
However, in the realm of cytology, many three-dimensional structures such as clusters of cells can be positioned anywhere along the z-axis, and it is doubtful that a viable autofocus solution is possible because there would never be a single focus point that would suffice for these structures within a slide of that thickness. Depth of field issues would prevent such three-dimensional structures from perfect focus throughout its entire volume. Using the traditional microscope, pathologists evaluate these structures by manually focusing up and down through them, revealing different cells and findings as they do so. As they focus up and down the z-axis, focused areas become blurry, and previously blurry areas come into focus. Usually, these clusters are large enough that there will always be out-of-focus areas when evaluating them. We argue that the solution to this issue is not exclusively through autofocusing, but also through improved focal plane capture. Theoretically, if a machine can accurately identify a cell cluster′s center through autofocus, then it can intelligently determine adjacent focal planes with greater accuracy.
Either manual, mechanical z-axis control or assurance of full z-axis coverage would provide the advantage of determining precise slide thickness on a slide-by-slide basis, since they all vary at the sub-micron level. Therefore, instead of generically estimating slide thickness, the scanner itself can determine and report actual thickness between glass and coverslip, which would be extremely helpful in future studies towards finding optimal values for parameters such as number of focal plane layers and distance between focal planes.
CONCLUSION
In this pilot study, we evaluated and compared different methods to improve cytology WSI and encountered a serious issue involving the z-axis affecting both methods. We identified the pros and cons of each method. It is not clear which method is more optimal in today′s setting and technological limitations, but it is important to note that diagnosis was not the endpoint of this study, and that further studies are needed to assess the impact of our findings on functionality and diagnosis.
Extended focusing was investigated as a promising alternative to multilayer stacking because it addresses crucial infrastructural aspects such as insufficient networking bandwidth and storage space, which translates into superior scan times, slide navigation and overall review time. However, worrisome image quality concerns found here requires further study on diagnostic impact.
Future directions could include large-scale studies between multilayered stacking versus extended focus. We would want to compare the average time for a pathologist to evaluate slides from each method, and to evaluate the diagnostic concordance between them. In addition, it would be interesting to see which method the majority of pathologists prefer.
Alternatively, a large-scale study comparing extended focus to conventional glass would be useful in evaluating the diagnostic concordance between glass and extended focusing in the cytology setting. Similar to the above-mentioned comparison, it would also be interesting to evaluate the average length of time to review an extended focus cytology image compared to conventional glass.
Nevertheless, if one were in a setting where it is possible to smoothly manipulate, navigate, and work with 10 gb files, or even 100 gb files, and perhaps transfer the entire file completely within 1-2 s, then this setting could favor multilayer stacking over extended focus. The 100 Gb Ethernet standard was ratified in July of 2010,[14] and file transfer at these speeds would be capable of transferring a 10 gb file within 1 s under optimal network conditions. Based on prior generation cycles for Ethernet,[15] we may see this speed as the common standard by 2025, available on all low-cost desktop computers. In addition a 14 TiB capacity laptop-sized hard drive is projected to be a common, mass-produced capacity by 2020 as well.[16]
Therefore, with this increase in storage capacities processing power, and networking bandwidth, eventually it would be entirely feasible in cytology to utilize a multilayer stack consisting of tens to hundreds of layers, with enough computational power to allow seamless navigation up and down the z-axis, at any magnification, compared to today′s unacceptable 2-3 s when switching to different focal planes at low power. Advanced compression schemes such as JPEG2000 would likely be in widespread use as well in 2020.
A multilayer stack consisting of that many focal planes could facilitate three-dimensional reconstruction and manipulation, similar to what radiology is capable of today: instead of a flat, two-dimensional plane, one would now possess a high-resolution volume of spatial data, allowing views of different planes at different angles.
While extended focus very likely could still be extremely valuable in many other settings such as FISH, today′s technological limitations in computational power and networking speed will have diminished by then, and this algorithm′s value in cytology could be lessened as an alternative to the multilayer stack method.
ACKNOWLEDGEMENTS
The authors wish to thank David C. Wilbur, MD, for his gracious and valuable input for this manuscript.
Footnotes
Available FREE in open access from: http://www.jpathinformatics.org/text.asp?2011/2/1/46/86285
REFERENCES
- 1.Evered A, Dudding N. Accuracy and perceptions of virtual microscopy compared with glass slide microscopy in cervical cytology. Cytopathology. 2010 May 17; doi: 10.1111/j.1365-2303.2010.00758.x. DOI: 101111/j1365-2303201000758x. [DOI] [PubMed] [Google Scholar]
- 2.Mori I, Nunobiki O, Ozaki T, Taniguchi E, Kakudo K. Issues for application of virtual microscopy to cytoscreening, perspectives based on questionnaire to Japanese cytotechnologists. Diagn Pathol. 2008;3(Suppl 1):S15. doi: 10.1186/1746-1596-3-S1-S15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Solomon D, Nayar R. The Bethesda System for Reporting Cervical Cytology. 2nd ed. New York: Springer Science + Business Media LLC; 2001. p. 4. [Google Scholar]
- 4.Wilbur DC. Digital cytology: current State of the art and prospects for the future. Acta Cytol. 2011;55:227–38. doi: 10.1159/000324734. [DOI] [PubMed] [Google Scholar]
- 5.Qayyum S, Yagi Y, Wilbur DC. Optimization of Whole Slide Imaging Parameters for Liquid-Based Cervical Cytology Slides. Presented at 57th Annual Scientific Meeting of the American Society of Cytopathology in 2009 [Google Scholar]
- 6.Eichhorn JH, Brauns TA, Gelfand JA, Crothers BA, Wilbur DC. A novel automated screening and interpretation process for cervical cytology using the internet transmission of low-resolution images: a feasibility study. Cancer. 2005;105:199–206. doi: 10.1002/cncr.21098. [DOI] [PubMed] [Google Scholar]
- 7.Eichhorn JH, Buckner L, Buckner SB, Beech DP, Harris KA, McClure DJ, et al. Internet-based gynecologic telecytology with remote automated image selection: results of a first-phase developmental trial. Am J Clin Pathol. 2008;129:686–96. doi: 10.1309/GRAV16QP8JR5XTPF. [DOI] [PubMed] [Google Scholar]
- 8.Wikipedia contributors. Frame Rate. Wikipedia, The Free Encyclopedia. 2002. Feb 25, [Last retrieved on 2011 Feb 5]. Last revised 16 Jan 2011. Available from: http://en.wikipedia.org/wiki/Frame_rate .
- 9.Wirawan A, Kwoh CK, Hieu NT, Schmidt B. CBESW: sequence alignment on the Playstation 3. BMC Bioinformatics. 2008;9:377. doi: 10.1186/1471-2105-9-377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Scharfe M, Pielot R, Schreiber F. Fast multi-core based multimodal registration of 2D cross-sections and 3D datasets. BMC Bioinformatics. 2010;11:20. doi: 10.1186/1471-2105-11-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pinto N, Doukhan D, DiCarlo JJ, Cox DD. A high-throughput screening approach to discovering good forms of biologically inspired visual representation. PLoS Comput Biol. 2009;5:e1000579. doi: 10.1371/journal.pcbi.1000579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wilson J, Dai M, Jakupovic E, Watson S, Meng F. Supercomputing with toys: harnessing the power of NVIDIA 8800GTX and playstation 3 for bioinformatics problem. Comput Syst Bioinformatics Conf. 2007;6:387–90. [PubMed] [Google Scholar]
- 13.Tuominen VJ, Isola J. The application of JPEG2000 in virtual microscopy. J Digit Imaging. 2009;22:250–8. doi: 10.1007/s10278-007-9090-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wikipedia contributors. 100 Gigabit Ethernet. Wikipedia, the Free Encyclopedia. 2006. Dec 3, [Last retrieved 2011 Feb 5]. Last revised 4 Feb 2011. Available from: http://en.wikipedia.org/wiki/100_Gigabit_Ethernet .
- 15.Wikipedia contributors. IEEE 802.3. Wikipedia, the Free Encyclopedia. 2001. Nov 5, [Last retrieved 2011 Feb 5]. Last revised 4 Feb 2011. Available from: http://en.wikipedia.org/wiki/IEEE_802.3 .
- 16.Kryder MH, Kim CS. After Hard Drives—What Comes Next? IEEE Trans Magn. 2009;45:3406–13. [Google Scholar]


