Abstract
In silico analyses have established that transcripts from some genes can be processed into RNAs with rearranged exon order relative to genomic structure (post-transcriptional exon shuffling, or PTES). Although known to contribute to transcriptome diversity in some species, to date the structure, distribution, abundance, and functional significance of human PTES transcripts remains largely unknown. Here, using high-throughput transcriptome sequencing, we identify 205 putative human PTES products from 176 genes. We validate 72 out of 112 products analyzed using RT-PCR, and identify additional PTES products structurally related to 61% of validated targets. Sequencing of these additional products reveals GT-AG dinucleotides at >95% of the splice junctions, confirming that they are processed by the spliceosome. We show that most PTES transcripts are expressed in a wide variety of human tissues, that they can be polyadenylated, and that some are conserved in mouse. We also show that they can extend into 5′ and 3′ UTRs, consistent with formation via trans-splicing of independent pre-mRNA molecules. Finally, we use real-time PCR to compare the abundance of PTES exon junctions relative to canonical exon junctions within the transcripts from seven genes. PTES exon junctions are present at <0.01% to >90% of the levels of canonical junctions, with transcripts from MAN1A2, PHC3, TLE4, and CDK13 exhibiting the highest levels. This is the first systematic experimental analysis of PTES in human, and it suggests both that the phenomenon is much more widespread than previously thought and that some PTES transcripts could be functional.
The pre-mRNAs of multi-exon eukaryotic genes undergo splicing during maturation, with introns being precisely removed by the spliceosomal complex (Rino and Carmo-Fonseca 2009; Hallegger et al. 2010). The vast majority of mammalian genes are also subject to alternative splicing (Johnson et al. 2003; Kampa et al. 2004), which can generate multiple mRNAs and protein isoforms from individual loci. Exons present in mature mRNAs exhibit co-linearity with genomic DNA. However, a growing number of mammalian genes have been shown to also generate transcripts with altered exon order relative to genomic DNA in the absence of underlying genomic rearrangements (Horiuchi and Aigaki 2006). Depending on the context or specific transcript structure, this form of RNA processing has been referred to as exon scrambling (Nigro et al. 1991), mis-splicing (Cocquerelle et al. 1993), exon repetition (Frantz et al. 1999), rearrangement or repetition of exon order (RREO) (Dixon et al. 2005), trans-splicing (Caudevilla et al. 1998; Akopian et al. 1999; Flouriot et al. 2002), alternative trans-splicing (Horiuchi and Aigaki 2006), or homotypic trans-splicing (Takahara et al. 2000). Here, we use the term post-transcriptional exon shuffling (PTES) to specifically refer to rearranged transcripts from a single gene where the defining features are rearrangement at the RNA level and the presence of intact exon junctions at the point where co-linearity with genomic DNA is disrupted. This term effectively excludes transcripts where changes in exon order are due to genomic structural alterations (Patthy 1999; Zhang et al. 2009), excludes transcripts where splice junctions do not coincide with intron/exon boundaries, and excludes fusion transcripts that involve two loci (Li et al. 2009). This term also avoids possible confusion with splice leader (SL) trans-splicing, which is common in some eukaryotes (Blumenthal 1995; Hastings 2005).
PTES transcripts have been interpreted as rare by-products of an error-prone alternative splicing mechanism or as the processed products of lariat intermediates generated during exon skipping (Nigro et al. 1991; Cocquerelle et al. 1993; Zaphiropoulos 1997). Evidence that some transcripts are unpolyadenylated and circular, together with a correlation between the structure of PTES transcripts and exon skipping products in some genes (Zaphiropoulos 1997; Surono et al. 1999), is consistent with this interpretation. Splicing between independent pre-mRNAs has also been proposed as a possible mechanism as some PTES transcripts are both full length and polyadenlyated (Flouriot et al. 2002); some consist of single exon duplications that cannot be generated from lariats produced by exon skipping (Dixon et al. 2005); and the protein product associated with a rearranged transcript from the rat Crot (also known as COT1) gene has been identified (Caudevilla et al. 1998). Consistent with this latter mechanism, short regions of high sequence identity are enriched both within introns downstream from PTES donor exons and within introns upstream of PTES acceptor exons (Dixon et al. 2007). These could influence splice donor and acceptor choice by inducing ectopic base pairing between pre-mRNAs.
In silico analyses of expressed sequence data suggest that PTES transcripts are generated by >1% of human genes (Dixon et al. 2005). However, they only account for ∼20% of all human chimeric ESTs present in public databases as the fusion points of most chimeric ESTs do not correspond to known splice junctions (Li et al. 2009). Although chimeric transcripts generated from two independent loci or from two nonoverlapping transcripts from the same locus have been shown to be of functional importance in Drosophila (Mongelard et al. 2002; Horiuchi et al. 2003) and human (Li et al. 2008; Rickman et al. 2009), very few human PTES events have been verified experimentally or analyzed in detail. To our knowledge, the only human PTES transcripts that have been shown to be both full length and polyadenlyated are derived from the estrogen receptor-α gene, ESR1 (Flouriot et al. 2002). Furthermore only one PTES transcript, from the DCC gene, has been accurately quantified, and it is expressed at about 1/1000th of the level of primary transcripts, consistent with rare errors of the splicing machinery (Nigro et al. 1991). As a result, the contribution of PTES to the human RNA landscape remains poorly defined.
Here, we describe 205 putative human PTES products discovered using high-throughput transcriptome sequencing, and present the first large-scale experimental analysis of such transcripts in human. We confirm their RNA origin and show that they can extend into 5′ and 3′ UTRs, be polyadenylated, and be conserved in mouse. We also show that, within transcripts, PTES exon junctions can be present at levels comparable to un-rearranged (canonical) exon junctions from the same loci. These results suggest that human PTES transcripts may be more abundant than previously thought and appear incompatible with such transcripts being due solely to an error-prone RNA processing system.
Results
Identification of novel human PTES transcripts
As part of a pilot screen for novel splice variants and fusion genes within cancer genomes, a total of 9.7 Gb of Illumina and 454 Life Sciences (Roche) FLX sequence data was generated from seven human pediatric tumor samples and one cell line and was mapped to the RefSeq gene set (see Methods). The data were then filtered to identify reads with two contiguous but independent high-quality hits to RefSeq genes where the junctions between hits terminate at exon boundaries. The reads identified fell into three distinct structural classes: putative splice variants, where the two independent hits were to the same RefSeq entry and in the same order as in RefSeq; putative fusion genes, where the hits were to two RefSeq entries from different genes; and putative PTES events, where the hits were to the same RefSeq entry but in an inverted order with respect to RefSeq (see Methods). After further screening to remove repetitive or suboptimal matches, a total of 205 putative PTES transcripts were identified. The PTES transcripts, including the sequence reads that define them, are presented in Supplemental Table S1. The putative fusion genes and novel splice variants identified by this pipeline are not presented here.
The 205 PTES structures, represented by 378 reads, are derived from 176 different genes. Of these structures, 64 retain an open reading frame and 93 possess frame shifts, and in 48 cases, the donor or acceptor exon is within a 5′ UTR. The number of in-frame structures is not significantly enriched compared to random assortment of coding exons within the sample (data not shown). More than one putative PTES transcript was identified for 17 genes, often involving the same acceptor exon, and these are listed in Table 1A. For example, all four of the structures identified for MIB1 involve exon 2 as the acceptor. In addition, 12 structures from nine genes were identified in more than one sample (Table 1B), with exon 2 or 3 being the acceptor exon in 11 of 12 cases. When the positions of all donor and acceptor exons within genes were plotted, it was found that ∼45% of PTES structures possess exon 2 as acceptor (Supplemental Fig. S1). This accounts for the large number of PTES splices involving UTRs. Furthermore, the first and last exons of genes are not used as donor or acceptor exons. Both these results are consistent with a previous in silico analysis of exon rearrangements identified within EST data (Dixon et al. 2005). In addition, a PTES product from the TLE4 gene was identified serendipitously during RT-PCR analysis of this gene (Liu 2009), and this was included in all subsequent analyses.
Table 1.
Genes with multiple PTES transcripts (A) and transcripts in multiple samples (B)
PTES transcripts are widely expressed and polyadenylated
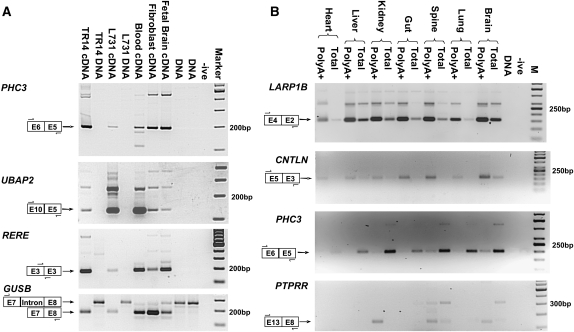
To validate the high-throughput sequence data, the first 112 of the 205 PTES structures identified were analyzed by RT-PCR using the original cDNA sequencing template, independent cDNA templates generated from the same RNAs, and templates from three normal human tissues. Of these 112 structures, 72 were successfully amplified in independent cDNA preparations of the original sequencing templates (listed in Supplemental Table S2), suggesting that approximately two-thirds of the PTES structures represent bona fide transcripts. All 72 also generated amplicons of the expected size in one or more normal human tissue, indicating that none were specific to neoplastic tissue. However in one case (RNF168 E3-E2), the expected product was also amplified from genomic DNA, even under stringent annealing and amplification conditions, suggesting that this transcript may be derived from genomic structures not represented within the current human genome build. Furthermore, in 44 of the 72 validated cases (61%), multiple cDNA-specific amplicons were observed. As examples, results for PHC3 E6-E5, UBAP2 E10-E5, and RERE E3-E3 are shown in Figure 1A. In all three cases, cDNA-specific amplicons of the expected size are generated together with additional products (450 bp for PHC3; 270 bp for UBAP2; and 260 bp, 300 bp, and 750bp for RERE), suggesting that further PTES transcripts from these genes may exist. Results for the GUSB control is also shown, with the expected spliced and unspliced products being observed in cDNAs and genomic DNA samples, respectively.
Figure 1.
Expression of human PTES transcripts. PTES structures, approximate primer location, and expected amplicon size are indicated for each panel. (A) Validation of human PTES transcripts. Amplification of products from the PHC3, UBAP2, and RERE genes are shown. TR14 is a neuroblastoma cell line (Rupniak et al. 1984), and L731 is one of the templates used for HTG sequencing (see Methods). GUSB is a control for template quality (see text). (-ive) No template negative control; (Marker) 100-bp ladder. (B) Polyadenylation and tissue specificity of transcripts. Amplification products from the LARP1B, CNTLN, PHC3, and PTPRR genes are shown. All templates are cDNAs generated from total or PolyA+ RNAs extracted from human fetal tissues (see Methods). (-ive) No template negative control; (M) 50-bp ladder (panels 1–3) and 100-bp ladder (panel 4).
As these transcripts were initially identified in RNA derived from neoplastic tissues, we then used a panel of tissue-specific human cDNA templates generated from both total RNA and polyA+ RNA to investigate expression patterns in normal tissues in more detail by RT-PCR (Fig. 1B; Supplemental Table S2). The results indicate that most PTES transcripts are ubiquitously expressed and that most are polyadenylated to some degree, consistent with the oligo dT primed/purified nature of the sequencing libraries (see Methods). However, significant variation in the extent of polyadenylation is observed, suggesting that the structure of PTES transcripts may be heterogeneous. For example, the LARP1B E4-E2 and CNTLN E5-E3 products are enriched within cDNAs derived from polyA+ RNA, whereas the PHC3 E6-E5 transcript is more abundant within cDNA derived from the total RNA fraction of all the tissues analyzed (Fig. 1B). A minority of transcripts also exhibit tissue-specific expression. For example, PTPRR expression is only observed in kidney and neuronal tissues (bottom panel). Interestingly, the E13-E8 PTPRR transcript is enriched within neuronal PolyA+ samples, whereas the larger transcript (∼320 bp) is not, suggesting that PTES products from the same gene can exhibit different levels of polyadenylation. Canonical transcripts were also analyzed for all genes shown, with the exception of RERE, and were expressed in all cDNAs (data not shown).
Most genes that exhibit PTES produce multiple products
The high-throughput sequencing identified 17 genes with multiple PTES transcripts (Table 1A), and during the RT-PCR validation, 44 primer pairs produced multiple cDNA-specific amplicons (Fig. 1; Supplemental Table S2). This suggests that many genes that generate PTES transcripts may produce multiple products. To confirm this, multiple amplification products recovered from 12 genes were subjected to FLX amplicon sequencing or were gel extracted and Sanger sequenced (see Methods). In total, 23 novel products were sequenced, 22 of which proved to be additional PTES transcripts with exon combinations distinct from those targeted by the original RT-PCR assays. These results are summarized in Table 2. In 18 cases, one or both exons present at the PTES junctions are RefSeq exons distinct from those targeted (e.g., MLL5 E11-E5 recovered as an E10-E11-E5-E6 amplicon using primers designed to amplify the E10-E6 junction). One, HDGFRP3 E6-E2, has a junction that falls within exon 6 and utilizes a novel splice site. A further five have intronic or nongenic flanking DNA at their junctions. Despite this, the dinucleotides flanking all of these additional sequence tracts within genomic DNA are AG and GT, consistent with spliceosomal processing.
Table 2.
Additional PTES products identified by Sanger or FLX amplicon sequencing
As examples, the additional PTES products sequenced from two genes are shown in Figure 2. RT-PCR of C19orf2 E10-E3 and HDGFRP3 E5-E2 generates the expected amplicons (110 bp and 105 bp, respectively) in addition to several larger products that are only observed in cDNA templates (left-hand panels). The sequence corresponding to each of these is spliced relative to genomic DNA (center panels); putative GT-AG splice donor and acceptor sites are present at the termini of all introns (shown in red, center panels); and the sequence traces spanning inferred splice junctions show no evidence of sequence heterogeneity (right-hand panels).
Figure 2.
Additional PTES products identified from RT-PCR amplicons. Amplification products from the C19orf2 E10-E3 and HDGFRP3 E5-E2 RT-PCR validation are shown. Structures, genomic splice junctions, and associated cDNA sequence traces are shown. Splice junctions are indicated using dotted lines in both DNA and cDNA sequences. Terminal gt-ag dinucleotides of inferred introns within genomic sequence are shown in red. Sequence internal to RefSeq exons is shown in upper case. (E) Exonic; (I) intronic; and (3′) novel exonic sequence derived from 3′ of the annotated C19orf2 gene.
One of the 23 sequences, however, has a structure inconsistent with spliceosomal processing. CCDC66 I10-I4, co-amplified during the validation of the CCDC66 E10-E5 transcript, comprises the sequence from intron 10 of CCDC66 upstream of sequence from intron 4. However, the junction between I10 and I4 sequence lies within an 18- to 21-bp region of high sequence identity between two AluY elements (Supplemental Fig. S2). Short regions of sequence identity have been shown to promote template switching during reverse transcription (Cocquet et al. 2006; Houseley and Tollervey 2010), suggesting that this product may be an artifact created during cDNA generation. Consistent with this interpretation, no canonical splice sites are present at the I10-I4 junction. Despite this apparent artifact, >95% of the additional RT-PCR amplicons sequenced (22 out of 23) possess novel rearrangements defined by canonical splice junctions, providing independent evidence that PTES transcripts are processed and suggesting that ∼50% of genes that exhibit this phenomenon generate multiple PTES transcripts.
PTES products can extend into 5′ and 3′ UTRs
Previous analyses have provided evidence that some PTES transcripts are not polyadenylated and may be circular (Nigro et al. 1991; Cocquerelle et al. 1993; Zaphiropoulos 1997), while others are linear, resembling two mRNAs fused at the PTES junction (Caudevilla et al. 1998; Flouriot et al. 2002). The transcripts identified here were originally sequenced from PolyA+ purified or oligo dT primed templates, and some are enriched within polyA+ purified material (see Methods; Fig. 1B; Supplemental Table S2). We therefore used primers spanning the PTES junctions of four genes (C19orf2 E10-E2, UBAP2 E10-E5, LARP1B E4-E2, and PHC3 E6-E5) to try to amplify in the 5′ and 3′ directions, to establish if these PTES exon junctions can be present in RNAs that contain the known 5′ or 3′ UTRs. In each case, the PTES junction primers produced amplicons in combination with primers from both the 5′ and 3′ UTRs. Furthermore, the amplicon expected if all intervening exons were present was recovered in each case. This exon organization is inconsistent with formation by lariat processing during alternative splicing. Critically, in three cases (C19orf2, UBAP2, and LARP1B) template specificity of the PTES junction primers could be confirmed using publicly available cDNA clones as control templates.
As an example, the results for C19orf2 E10-E2 are presented in Figure 3. Primers specific for the E10-E2 exon junction used in conjunction with primers from the 5′ and 3′ UTRs (primers 1 + 2 and 3 + 4) yield products of ∼1.3 kb and ∼1.8 kb from the NB3 cDNA template (left-hand panel). These are the sizes expected from transcripts containing all intervening exons; that is, E1-E10 followed by E2 for the 5′ amplicon and E10 followed by E2-E11 for the 3′ amplicon (shown schematically in right-hand panel). Additional, smaller products are also observed in the 3′ amplification, which may represent splice variants. No products were generated using these primer pairs with the un-rearranged, sequence validated, positive control C19orf2 cDNA template (AK292170). This confirms that the E10-E2 junction primers do not anneal to the canonical transcript and are PTES specific. In contrast, when the 5′ UTR and 3′ UTR primers are used together (primers 1 + 4) a product of 1.85 kb is obtained from both NB3 and AK292170 templates, corresponding to the full-length, un-rearranged, C19orf2 transcript. Comparable results were obtained for both LARP1B and UBAP2 (Supplemental Fig. S3). While these experiments indicate that PTES splice junctions are connected in some molecules to either the 5′ or 3′ ends of transcripts, no full-length PTES structures were co-amplified with canonical structures from cDNA templates using the 5′ and 3′ UTR primers (primers 1 and 4). This means that the entirely full-length molecules are present at a low frequency relative to the canonical transcript, are outcompeted by the smaller product during PCR amplification, or are entirely absent.
Figure 3.
Identification of extended PTES products using junction-specific primers. Amplicons generated using primers specific for C19orf2 PTES E10-E2 splice junction are shown. Primers 1 and 2 amplify from the 5′ UTR to the E10-E2 breakpoint. Primers 3 and 4 amplify from the E10-E2 breakpoint to the 3′ UTR. Templates are as follows: (NB3) cDNA where the E10-E2 PTES product was originally identified; (+ive) unrearranged (canonical) C19orf2 cDNA clone (AK292170); and (-ive) no template. The exon organization of the inferred E10-E2 spliced PTES RNAs, the full-length C19orf2 gene from RefSeq (canonical), and the AK292170 +ive control is also shown, together with the position of primers used and the expected amplicon sizes. Individual exons are shown as boxes, with coding regions shown in gray and UTRs in white. The sizes of the PTES amplicons are given relative to the E10-E2 junction. Additional products of ∼0.4–0.65 kb and ∼1.2 kb are seen when the NB3 template is amplified using primers 1 and 2, suggesting that shorter C19orf2 PTES isoforms also exist. For all primers, see Supplemental Table S6.
Conservation of PTES structures in mouse
The conservation of exonic structures in diverse species is routinely used to infer possible function, and although several PTES structures analyzed previously have been shown to be present in both human and mouse (Nigro et al. 1991; Dixon et al. 2005), it is not clear if these results are representative. To establish the extent of conservation, we designed murine primers to search for orthologs of 41 randomly chosen validated human PTES structures in a panel of adult mouse tissues. Amplicons of the expected size were observed in one or more tissues for orthologs of seven human genes (MAN1A2, TLE4, ICA1, MLL5, CDK13, VRK1, and ZNF236), and results for three murine genes are shown in Figure 4. Man1a2 and TLE4 show similar expression patterns to that found in human, with the Man1a2 E5-E2 PTES junction being found in all tissues analyzed, and Tle4 E8-E5 being expressed in the brain and, to a lesser extent, testis. In contrast, Cdk13 E5-E2, which is widely expressed in human, is only found in the bladder and lung in the mouse. As in human, additional amplicons are observed in many cases, including an amplicon of the size expected for Man1a2 E4-E2. These results suggest that orthologs of ∼17% of human PTES events are conserved in the mouse (90% CI 7.4%–26.7%), although tissue distribution can differ between the two species.
Figure 4.
RT-PCR amplification of murine PTES products corresponding to known human structures. The amplicon corresponding to the expected PTES structure is highlighted in each case. The additional amplicons seen in panels 1 and 3 are the expected size for murine orthologs of additional human products (MAN1A2 E6-E2, Supplemental Table S6; TLE4 E8-E5, GenBank accession no. HQ283388). For Tle4, a Gapdh loading control was included. For details of primers and amplicons, see Supplemental Table S6.
PTES transcripts can be highly expressed relative to canonical transcripts
To date, very few human PTES transcripts have been quantified due, partly, to the lack of unique priming sites relative to canonical transcripts (e.g., Fig. 3). We therefore used both bioinformatic and experimental approaches to investigate the frequency of PTES and canonical junctions within cDNA as a measure of relative transcript abundance. It is important to note that while the structure (and heterogeneity) of PTES transcripts remains to be fully investigated, it is clear that some can contain both PTES junctions and related canonical splice junctions (e.g., transcript structures in Figs. 3, 4). As a result, the relative frequency of PTES to canonical junctions provides a conservative estimate of the proportion of PTES transcripts relative to canonical transcripts for each gene.
Within our HTGS data we compared the frequency of reads that spanned the PTES junctions (defined here as E(X) − E(N)), with the frequency of reads spanning the two associated canonical splice junctions (E(X) − E(X+1) and E(N−1) − E(N)). As FLX sequencing has not been validated for mRNA quantitation, we confined this analysis to Illumina reads from the acute lymphoblastic leukemia (ALL) samples, and to minimize ascertainment bias, we only analyzed PTES structures identified in neuroblastomas (NBs; see Methods). The results of this analysis are summarized in Table 3 and presented in full in Supplemental Table S3. Of 52 PTES transcripts analyzed, 41 were expressed at a high enough level in one or more ALL samples to be informative, with an average read depth of over 30 and an average coverage of over 90% (Supplemental Table S3). Of these, 25 gave no reads spanning PTES junctions, suggesting that they are not present in these samples or they occur at a low frequency relative to the canonical junctions in polyA+ RNA (Supplemental Table S3). From the remaining 16 genes, a total of 64 E(X) − E(N) PTES reads were recovered compared to 2011 reads from all proximal canonical junctions (E(N−1) − E(N)) and 3456 reads from distal canonical junctions (E(X) − E(X+1)). The higher number of reads recovered from distal canonical junctions is consistent with more efficient recovery of 3′ sequence from oligo-dT captured RNA (Edery et al. 1995). Taking the average of these values suggests that in the PolyA+ transcripts of genes that exhibit this phenomenon, PTES exon junctions are present at an average frequency of 2.3% compared with the levels of related canonical junctions. However, the frequency of E(X) − E(N) junctions varied extensively, ranging from ∼0.5% of the average number of canonical junctions (LUC7L2) to over 50% (MAN1A2 E5-E2), with reads containing all three MAN1A2 PTES junctions being observed at high frequency. These results suggest that for most genes PTES junctions are present at low levels but that transcripts from a small number of genes can contain a high frequency of PTES junctions.
Table 3.
Frequency of Illumina reads spanning PTES and canonical splice junctions
This in silico analysis is limited by the modest number of sequence reads that span canonical splice junctions in many genes (Supplemental Table S3). To analyze junction frequencies in more detail, PTES and canonical junction real-time PCR assays were designed for 10 genes, including MAN1A2, PHC3, UBAP2, and LARP2 (Table 3) and those conserved in mouse (Fig. 4). Five genes (RTN4, CDK13, PHC3, TLE4, and KTN1) gave comparable amplification efficiencies in both their PTES and canonical junction assays so were used to assess relative junction levels within cDNAs generated from a variety of adult and fetal human tissues using the Δ − Ct method. (Supplemental Fig. S4). Threshold values obtained for canonical junctions from all five genes were five to 10 cycles lower than the average of the three control genes used for normalization (GAPDH, ACTB, and PPIA; see Methods), indicating modest levels of expression for all genes analyzed. For RTN4 and KTN1, PTES junctions were much rarer than canonical in virtually all tissues tested with PTES thresholds appearing three to five cycles later than canonical. However, for PHC3, TLE4, and CDK13, the PTES thresholds were only zero to two cycles later than the canonical thresholds in some tissues, suggesting that PTES junctions in these three genes could be present at anything from 20%–100% of canonical levels (Supplemental Fig. S4).
To confirm the results obtained using the Δ − Ct method and to control for variation in the efficiency of reverse transcription between templates, PTES and canonical junctions from genes exhibiting the highest PTES frequencies (PHC3, CDK13, TLE4, MAN1A2 E5-E2, and MAN1A2 E4-E2) were then cloned into plasmid expression vectors and transcribed, and the resulting RNAs were used to generate standard curves (SCs). Estimates of junction abundance for these genes using both the Δ − Ct and SC methods are shown in Figure 5, and SCs are shown in Supplemental Figure S5. The SC results were comparable to those obtained with the Δ − Ct method and identify high frequencies of PTES junctions in one or more tissue for each gene. For example, the TLE4 E8-E5 junction is present at ∼15% (Δ − Ct) − 50% (SC) of canonical levels in adult cerebellum (panel A), PHC3 E6-E5 is present at ∼60% (Δ − Ct) − 90% (SC) of canonical levels in fetal heart (panel B), CDK13 E5-E2 is present at ∼43% (Δ − Ct) − 57% (SC) of canonical levels in fetal thalamus, while MAN1A2 E5-E2 is present at ∼47% (Δ − Ct) − 96% (SC) of canonical levels in fetal spine. In all tissues and for all genes, canonical junctions are more abundant than individual PTES junctions. However, in the fetal spine, all MAN1A2 PTES junctions (i.e., E5-E2 and E4-E2 combined) are more abundant than the related canonical structures.
Figure 5.
Identification of abundant PTES products using real-time PCR. (A) TLE4; (B) PHC3; (C) CDK13; and (D) MAN1A2. Each panel shows PTES and canonical transcript abundance in four human tissues estimated using both standard curves (upper bars) and the Δ − Ct method (lower bars) As all genes are expressed at a lower level than control genes, −Δ − Ct values are plotted to facilitate comparison with data from standard curves. (Thal) Thalamus; (Cereb) cerebellum. Additional data are presented in Supplemental Figures S4 and S5. For all primers, see Supplemental Table S6.
While there is some variation in the results obtained with the different methods used to estimate junction frequency within transcripts, the quantitative PCR analyses are consistent. The SC analysis of MAN1A2 also confirms the high proportion of PTES junctions identified in the in silico analysis. Collectively they provide strong evidence that PTES exon junctions can be present at levels comparable to canonical junctions within transcripts from a small number of human genes.
Genomic rearrangement cannot account for PTES
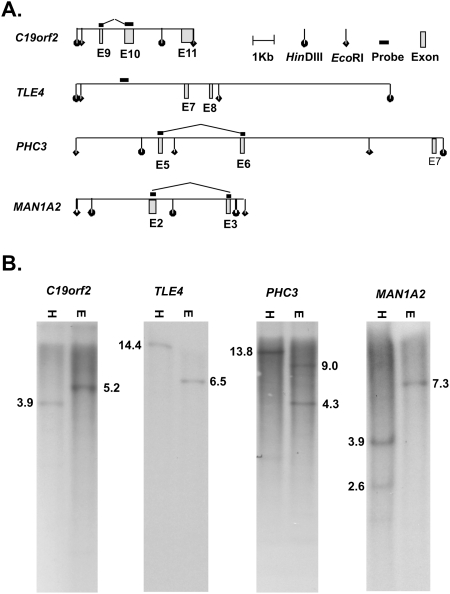
Finally, naturally occurring exon duplication at the genomic level, or errors in the current genome build, could result in transcripts with apparently rearranged exon order relative to RefSeq entries. While these are unlikely to account for the large numbers of PTES transcripts identified here, they could account for the high expression levels of a small number of PTES transcripts relative to canonical isoforms. To exclude this possibility, Southern analyses were used to investigate the integrity of restriction maps surrounding the donor and/or acceptor exons of six genes, and results from four of these are presented in Figure 6. In all cases only genomic fragments of the expected sizes are observed, excluding genomic rearrangement or build errors as possible explanations for the high frequency of PTES junctions within transcripts from genes such as MAN1A2, TLE4, and PHC3.
Figure 6.
Genomic Southern analysis of PTES exons. (A) Position of exons flanking PTES junctions in four genes relative to genomic HinDIII and EcoR1 sites (Build GRCh37/19). (B) Southern blots of human genomic DNA using the probe and enzyme combinations shown in A ([H] HinDIII; [E] EcoR1). The expected product sizes are shown in each case. For details, see Methods and Supplemental Table S6.
Discussion
This is the first systematic experimental analysis of PTES in humans. We demonstrate for the first time that most of the genes that exhibit this phenomenon generate multiple transcripts, that these transcripts can be both polyadenylated and can extend into 5′ and 3′ UTRs, and that they are present in a wide variety of normal human tissues. We also provide the first experimental evidence that some human PTES transcripts can be expressed at high levels relative to canonical (un-rearranged) transcripts and that they can also be conserved in the mouse. Collectively, these data indicate that the transcriptional output of many human genes is even more diverse than previously thought and that shuffled transcripts could make a significant contribution to human total and polyA+ RNA.
Although specific examples of this phenomenon have been well characterized in other species, the few human PTES transcripts characterized experimentally to date have been identified by chance (Nigro et al. 1991; Zaphiropoulos 1997; Takahara et al. 2000; Flouriot et al. 2002). The first indication that large numbers of human genes might generate PTES transcripts was provided by an in silico analysis of ESTs (Dixon et al. 2005) that identified 263 putative PTES transcripts from 178 human genes and concluded that ∼1% of human genes may exhibit this phenomenon. A bias toward the use of 5′ exons with large upstream introns as PTES acceptor exons was also identified, and this is empirically confirmed here. Our analysis has identified a comparable number of PTES transcripts from a similar number of genes (205/176) and has subsequently validated ∼64% of the PTES structures experimentally. The structures that could not be validated are likely to be present at very low levels within cDNAs or to represent artifacts of cDNA generation or subsequent PCR amplification. Importantly, only 16 genes (∼9%) and seven PTES structures (∼3.4%) identified here are also identified by the Dixon analysis (Supplemental Table S4), and in both studies, the majority of PTES transcripts are defined by a small number of ESTs/sequence reads. This suggests that a much larger proportion of human genes may be capable of generating PTES transcripts than have been identified to date and that sampling at a much greater depth will be required to characterize the full extent of this phenomenon.
All of the PTES events identified using our bioinformatics pipeline are associated with canonical GT-AG splice junctions. In contrast, an in silico analysis of EST data by Li et al. (2009) identified 31,005 human chimeric ESTs from 11,645 genes, over 80% of which showed no evidence of spliceosomal processing. However, the majority of these spliced ESTs involved two loci (putative fusion genes). Short regions of sequence identity (<10 bp) were also found at the fusion points of ∼20% of chimeric transcripts, and a transcription slippage model, which hypothesizes that RNA polymerase II switches from one template to another in the same transcription factories, was proposed to accommodate this observation. This high proportion of putative fusion genes contrasts sharply with our results, where only 13 were identified, 12 of which were subsequently found to be PCR artifacts (H. Al-Balool, unpubl.). The high proportion of putative fusion genes identified by Li et al. (2009) may be due to such transcripts being created by a mechanism that does not involve spliceosomal processing (Zhang et al. 2003) but it may also be due, at least in part, to the high representation of transcripts from neoplastic tissues within human EST databases (Qiu et al. 2004), including many recovered during targeted searches for fusion genes using techniques such as panhandle PCR (Jones and Winistorfer 1992; Robinson and Felix 2009). In addition, homology-dependent template switching by reverse transcriptase during cDNA generation has been shown to be responsible for some noncanonical splice variants within human EST data (Cocquet et al. 2006), raising the possibility that the chimeric ESTs with short tracts of sequence identity at the putative fusion points observed by Li et al. (2009) may be artifactual.
Template switching has also recently been shown to generate PTES like structures from artificial templates derived from the intronless Saccharomyces cerevisiae SPT7 gene (Houseley and Tollervey 2010). However, no evidence of template switching has been found among spliced human ESTs with canonical splice junctions (Cocquet et al. 2006), and it cannot account for the PTES structures defined here. Of 23 products co-amplified and sequenced during our validation procedure, 22 proved to be spliced PTES products related in structure to the transcripts targeted for validation, and each contained a novel PTES junction with canonical splice donor and acceptor sites (Fig. 2; Table 2). This result is incompatible with template switching. Furthermore, sequences generated by template switching have been shown to be heterogeneous, reflecting subtle variation in template switch position (Houseley and Tollervey 2010), whereas the PTES splice junctions sequenced here show no such heterogeneity (e.g., Fig. 2).
PTES structures have now been reported and verified in a wide variety of eukaryotes. Despite this, the precise mechanism (or mechanisms) that underpins this phenomenon remains to be formally defined. The processing of lariat intermediates generated during alternative splicing has been invoked to account for some structures (Cocquerelle et al. 1993; Zaphiropoulos 1997; Surono et al. 1999). However, this has been excluded in specific instances through the identification of full-length transcripts (e.g., Flouriot et al. 2002; Rigatti et al. 2004), suggesting that in some cases splicing between two pre-mRNA molecules is responsible (Flouriot et al. 2002; Dixon et al. 2005). Detailed analysis of alleles of the rat Crot and Sa genes that exhibit variable levels of PTES expression have established that the mechanism is determined in cis (Rigatti et al. 2004), and in silico analyses have identified a common 21-bp motif present in inverted orientation within 79% of PTES donor and acceptor introns (Dixon et al. 2007), further suggesting that the mechanism is homology dependent. This is consistent with the extensive experimental evidence that hairpins formed by base pairing between different regions of the same pre-mRNA can influence exon choice during normal splicing (Solnick 1985; Miriami et al. 2003; Lev-Maor et al. 2008; Warf and Berglund 2010). Within the data presented here, the enrichment of specific PTES transcripts in polyA+ RNA (Fig. 1; Supplemental Table S2), the amplification of 5′ and 3′ UTRs using primers specific for PTES exon junctions (Fig. 3), and the identification of PTES structures containing multiple copies of the same exon (GenBank accession nos. HQ234317 and HQ283388) are all consistent with exon splicing between two pre-mRNA molecules, providing further support for this mechanism as one source of PTES transcripts. However, the possibility that a proportion of PTES transcripts may be derived from lariat intermediates cannot be ruled out.
Evidence for the functional importance of alternative splicing is incontrovertible, with specific events having clear impact upon a wide variety of phenotypes, including human disease states (for review, see Tazi et al. 2009). Currently, however, there is no evidence for the function of PTES transcripts. Our in silico and real-time PCR analyses of transcript abundance are, therefore, of interest as they are consistent with possible function. Most PTES junctions analyzed here are observed at frequencies <5% of related canonical exon junctions, higher than the previous quantitative analysis of DCC transcripts (Nigro et al. 1991) but still relatively low. In contrast, PHC3, TLE4, CDK13, and MAN1A2 PTES junctions are expressed at anything from 15%–90% of the levels of canonical junctions in some tissues. While high expression levels have been reported for a rat Crot transcript containing two copies of exon 2 (Rigatti et al. 2004), this is the first evidence that human PTES transcripts can be expressed at such high levels.
Protein products derived from PTES transcripts have been identified in other species (Caudevilla et al. 1998), and the PTES transcripts detected for PHC3 and TLE4 do not disrupt the reading frame, suggesting that they could be translated. However, both of these transcripts are highly enriched in total RNA compared with polyA+ RNA (Fig. 1B; Supplemental Table S2), making this unlikely. Furthermore, the majority of PTES transcripts identified here and elsewhere (Dixon et al. 2005) result in frame shifts or involve acceptor exons upstream of the translation start site, suggesting that the vast majority of these transcripts do not contribute to the proteome. Thus, if these transcripts are functional, a regulatory role seems more plausible given the growing number of functional noncoding RNAs being identified (Ponting et al. 2009; Wilusz et al. 2009). Our discovery that 17% (±9.7%) of human PTES junctions are conserved in mouse is also of interest as this level of conservation is comparable to levels seen for standard alternative splicing events (Modrek and Lee 2003; Pan et al. 2005).
We have confirmed that some PTES junctions can be linked to both 5′ and 3′ terminal exons. However, the extensive variation in polyadenylation observed, as well as the multiple products generated from many loci, indicates that PTES transcripts are heterogeneous in structure as well as abundance. As a result, it is also possible that PTES transcripts are noise within the complex and dynamic transcriptional system (Graveley 2001; Kan et al. 2002; Melamud and Moult 2009) and that the genes identified here represent extremes of this noise. For instance, it is possible that, for reasons currently unknown, MAN1A2, PHC3, and TLE4 are particularly prone to PTES generation or that the PTES products from these genes are unusually stable and not efficiently degraded by the nonsense mediated decay pathway. If this is the case, the underlying reasons for this stability, and how it is controlled, will be of interest. Furthermore, as there is evidence for circular (Cocquerelle et al. 1993; Zaphiropoulos 1997; Surono et al. 1999) and linear PTES RNAs (Caudevilla et al. 1998; Flouriot et al. 2002; this study), the possibility that both could be generated from the same loci must be considered. Lariats from spliced isoforms that may be more abundant than full-length transcripts could, for instance, contribute to PTES structures generated by the genes identified here. Detailed experimental analyses of all transcripts from these genes, and of the processes that create them, are therefore now warranted.
Finally, the evidence that RNA secondary structure can be involved both in exon skipping during normal spliceosomal processing (Solnick 1985; Miriami et al. 2003; Lev-Maor et al. 2008; Warf and Berglund 2010) and in PTES (Dixon et al. 2007), raises the further possibility that a proportion of co-linear spliced products (which exhibit no disruption of exon order relative to genomic DNA) could also be generated from more than one pre-mRNA. Since the discovery of splicing (Berget et al. 1977; Chow et al. 1977), it has been implicitly assumed that spliced mRNA isoforms are generated from single pre-mRNA molecules with looping out of intervening introns and exons. However, there is no a priori reason why a proportion of some spliced mRNAs could not be generated via the interaction of two pre-mRNAs. This could be particularly relevant to splices involving the proximal and distal exons of genes spanning large genomic regions. Rates of RNA polymerase II transcription in humans have been estimated to be ∼2.4–3.8 kb/min, and splicing can occur co-transcriptionally (Roberts et al. 1998), being observed as early as 10 min after synthesis (Tennyson et al. 1995; Singh and Padgett 2009). As a result, the 5′ introns of many genes may be processed before 3′ introns are transcribed, yet transcripts with alternative splices requiring removal of ∼800 kb of transcribed genomic DNA have been reported (Surono et al. 1999). While the generation of such transcripts could utilize secondary structure and/or protein interactions to prevent intervening exons from being spliced (Roberts et al. 1998; Warf and Berglund 2010), they could also be generated through the splicing of two independent pre-mRNAs. Interestingly, both increased intron length and transcriptional pausing can increase the level of PTES transcripts generated from transgenic Sp1 constructs (Takahara et al. 2005). Thus, while the data presented here indicate that PTES transcripts are generated by a much larger number of human genes than previously thought, that they can be conserved, and that they can be expressed at a high level relative to canonical transcripts, the data also suggest that the fundamental assumption that human mRNA processing is overwhelmingly co-linear may need to be reassessed.
Methods
Sample preparation
Total RNA for sequencing was extracted using the RNeasy Micro kit (Qiagen). Other total RNAs were isolated using Trizol (Invitrogen). PolyA+ RNA was isolated using a Dynabeads mRNA purification kit (Invitrogen). DNA was isolated using phenol/chloroform/isoamyl alcohol extraction following proteinase K digestion (Maniatis et al. 1982). The quantity of RNA and DNA was estimated using the NanoDrop ND-1000 spectrophotometer (NanoDrop), and RNA quality was established using the Agilent 2100 Bioanalyser (Agilent Technologies).
Transcriptome sequencing
Sequence from four primary NB tumors (Lastowska et al. 2007), four ALL samples (Case et al. 2008), and the NB cell line IMR32 (Clementi et al. 1986) was generated, originally to develop and assess mutation detection pipelines. Appropriate informed consent for use of all primary material was obtained (Lastowska et al. 2007, Case et al. 2008). Six templates (from L466, L547, L612, L731, NB5, and NB19) were prepared using the Illumina mRNA-seq sample preparation kit (part no. 1004898) according to manufacturer's recommendations, with the exception that 500 ng of total RNA was used for each template with 17 cycles of amplification. Approximately 9.2 Gb of unpaired 76-bp sequence reads was generated on the Illumina Genome Analyzer II platform according to the manufacturer's protocols (Illumina) by Geneservice. Six templates (NB3, IMR32, NB6, NB5, L466, and L612) were also prepared by oligo dT priming using the SMART cDNA Synthesis kit (Clontech). These were used to generate a further ∼0.5 Gb of unpaired sequence data on the GS-FLX 454 platform (Roche) by NewGene using the GS LR70 sequencing kit according to the recommended protocols. Average read lengths varied between templates from 195–252 bp. For all samples, two independent cDNA templates were sequenced to facilitate the subsequent identification of PCR artifacts. All sequence data are available from the Sequence Read Archive under accession no. SRA023629.
In silico identification of PTES transcripts
Illumina reads were converted to FASTA format using the sol2sanger and fq-all2std scripts within MAQ (http://maq.sourceforge.net/maq-man.shtml). All reads were aligned to the human RefSeq gene set build 36.1 using the GS Reference Mapper (Roche). Perl scripts were then used to identify rearranged transcripts as follows: reads within FLX PairAlign files with two or more independent matches to RefSeq, each with >95% identity over >25 bp, where the two matches overlapped by a maximum of 5 bp were identified. Reads where the matches were >10 bp apart within the sequencing read and/or in different orientations within RefSeq were then removed. Reads where both matches were to the same RefSeq entries were further processed to distinguish reads where both matches to RefSeq were in the same order relative to the read (alternatively spliced isoforms) from those where the matches were in an inverted order (rearranged transcripts). All perl scripts are available upon request. The structure of all reads passing these filters were then manually analyzed using BLAT (Kent 2002) to remove reads where the junction between the two independent hits to RefSeq did not correspond precisely to two exon boundaries and to remove reads that mapped to other regions of the genome at a higher identity that to the top RefSeq entries. To compare read counts at PTES and canonical splice junctions, reads were mapped as before to 60-bp sequences consisting of 30 bp on either side of each PTES splice junction (E(x) − E(N)) and equivalent 60-bp sequences from both related canonical junctions (E(X) − E(X+1) and E(N−1) − E(N)). A minimal sequence match of 40 bp was enforced to ensure specificity. Minor differences in read counts relative to other analyses were observed due to the different mapping parameters used.
PCR
Human fetal RNAs and DNAs were extracted from tissue samples obtained from the MRC/Wellcome Trust Human Developmental Biology tissue bank (http://www.hdbr.org/). RNAs from adult tissues were obtained from BioChain Institute Inc. All cDNA and DNA samples for RT-PCR validation experiments were generated using the random priming TransPlex Whole Transcriptome Amplification Kit or Genome Plex Complete Whole Genome Amplification Kit (Sigma Aldrich) using 50–100 ng of template according to the manufacturer's instructions. Templates for subsequent RT-PCR and real-time PCR experiments were generated using the High Capacity cDNA reverse transcription system (Applied Biosystems). Templates for 5′ and 3′ UTR reactions were generated by oligo-dT primed SMART cDNA Synthesis (Clontech). PCR reactions were performed as 20 μL reactions using 0.065 U/μL GoTaq polymerase in 1× buffer (Promega) or HotStar Taq (Qiagen) with 200 μM of each dNTPs (Fermentas), 500 pM forward and reverse primer (Metabion), and 0.5ng/μL cDNA or 2.5 ng/μL DNA, respectively. Amplifications to 5′ and 3′ UTRs were performed using Phusion high-fidelity DNA polymerase (Finnzymes) according to the manufacturer's recommended protocols. Primers were designed using Pimer3 (Rozen and Skaletsky 2000). Thermo cycling was performed using an MJ Research Peltier ThermalCycler (MJ Research) with 5-min denaturation at 95°C, 30 cycles of 30-sec denaturation at 95°C, 30 sec annealing at 58°C–64°C depending on the primers used, and 1-min elongation at 72°C followed by a final 10-min elongation at 72°C. All PCR primer sequences are given in Supplemental Tables S5 and S6.
Sequencing of PCR products
Individual fragments were gel extracted and purified using Qiaquick PCR purification columns (Qiagen), sequenced using Sanger sequencing by GeneService, and deposited in GenBank (accession nos. HQ234305-HQ234314, HQ234317, and HQ283388). PCR reactions containing multiple products from the same gene were subjected to FLX amplicon sequencing, and the data were deposited in the Sequence Read Archive under accession no. SRA023629.
Real-time PCR
All real-time PCR was performed using amplicons of between 90 bp and 166 bp in length using internal probes with a 5′ FAM fluorescent dye and 3′ TAMRA quencher (Metabion). All primer pairs were designed to amplify across at least one exon boundary, and the specificity of amplification in cDNA was confirmed prior to use. PTES and canonical (unrearranged) amplicons from each gene were designed to use the same probe and to be of similar size to minimize amplification bias. The concentration of forward and reverse primers varied from 100–300 pM as primer concentrations for each transcript analyzed was optimized to maximize reaction efficiencies. Only PTES and canonical assay pairs where the amplification efficiencies differed by <3% were subsequently used (see Supplemental Table S6). Reactions were performed in 15 μL volume using 0.6 ng/μL cDNA, 100 pM TaqMan probe, and TaqMan PCR Master Mix (Roche) using the ABI 7900 HT Fast real-time PCR System (Applied Biosystems) with the following cycling parameters: 2-min initial activation of the polymerase at 50°C, 10-min initial denaturation at 95°C, and 45 cycles comprising a 15-sec denaturation step at 95°C and a 1-min combined annealing and elongation step at 60°C. The mean Ct value of three amplifications from each template was normalized against the averaged expression level of the endogenous control genes PPIA (Fischer et al. 2005), ACTB, and GAPDH (Applied Biosystems), each run in triplicate.
In vitro transcription
PTES and canonical splice junctions were amplified using primers containing the BamH1 and Sal1 restriction sites (Supplemental Table S6), ligated into the polylinker of pBluescript KS+ (Strategene) using T4 DNA ligase (New England Biolabs), and electroporated into DH5α cells according to the manufacturer's recommendations. Clones with the desired inserts were identified using Xgal/IPTG color selection followed by PCR and Sanger sequencing. Plasmid DNAs were isolated using Qiagen mini prep columns (Qiagen), and the inserts were amplified using M13 forward and reverse primers. In vitro transcription of 250 ng of each plasmid was performed using T7 MEGAscript kit (Applied Biosystems) according to the manufacturer's recommended protocols. DNase-treated RNA was purified using NucAway Spin Columns (Applied Biosystems), and RNA concentrations were adjusted to 2 × 1012 molecules/μL based on amplicon size. Equal volumes of each RNA were then pooled, and 3 μg of this pool was used to generate a single template that was subject to first-strand cDNA synthesis using a high-capacity cDNA reverse transcription kit (Applied Biosystems). Three independent pools were created and reverse transcribed to give three control templates that were serially diluted for SC generation.
Electrophoresis, Southern transfer, and hybridizations
Digestion with restriction enzymes (New England Biolabs), electrophoresis, and Southern blotting using Hybond-N+ membranes (GE Healthcare) were carried out using standard methods (Maniatis et al. 1982). DNA probes were generated by PCR, cleaned using Qiaquick purification columns (Qiagen), and labeled with α32P-dCTP by random oligonucleotide priming (Feinberg and Vogelstein 1984) using the Megaprime DNA labeling system Kit (GE Healthcare). Filters were prehybridized for 1 h at 65°C in 6× SSC 1%SDS with 0.1 mg/mL denatured sheared salmon sperm DNA and 5× Denhardt's solution (Maniatis et al. 1982), hybridized for 16 h at 65°C in the same solution, washed at high stringency (0.5× SSC 0.1% SDS at 65°C), and exposed to Kodak Biomax MR X-ray film for 24–72 h at −70°C with intensifying screens.
Data access
Sequence data from this study have been submitted to the NCBI Sequence Read Archive (http://trace.ncbi.nlm.nih.gov/Traces/sra/sra.cgi) under accession no. SRA023629 and to GenBank (http://www.ncbi.nlm.nih.gov/genbank/) under accession nos. HQ234305–HQ234314, HQ234317, and HQ283388.
Acknowledgments
The financial support of the Newcastle Healthcare Charity, Tyneside Leukaemia Research Association, BBSRC (grant no. BB/D013917/1), and Wellcome Trust (grant no. WT080368MA) is gratefully acknowledged. Mark Cooper provided technical assistance with the analysis of murine samples. H.H.A.-B. was supported by the Government of Kuwait. Y.L. was partly supported by an ORSAS studentship. We thank Ian Eperon for commenting on an earlier version of this manuscript.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.116442.110.
References
- Akopian AN, Okuse K, Souslova V, England S, Ogata N, Wood JN 1999. Trans-splicing of a voltage-gated sodium channel is regulated by nerve growth factor. FEBS Lett 445: 177–182 [DOI] [PubMed] [Google Scholar]
- Berget SM, Moore C, Sharp PA 1977. Spliced segments at the 5′ terminus of adenovirus 2 late mRNA. Proc Natl Acad Sci 74: 3171–3175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumenthal T 1995. Trans-splicing and polycistronic transcription in Ceanorhabditis elegans. Trends Genet 11: 132–136 [DOI] [PubMed] [Google Scholar]
- Case M, Matheson E, Minto L, Hassan R, Harrison CJ, Bown N, Bailey S, Vormoor J, Hall AG, Irving JAE 2008. Mutation of genes impacting on the RAS pathway are common in childhood acute lymphoblastic leukemia. Cancer Res 68: 6803–6809 [DOI] [PubMed] [Google Scholar]
- Caudevilla C, Serra D, Miliar A, Codony C, Asins G, Bach M, Hegardt FG 1998. Natural trans-splicing in carnitine octanoyltransferase pre-mRNAs in rat liver. Proc Natl Acad Sci 95: 12185–12190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow LT, Gelinas RE, Broker TR, Roberts RJ 1977. An amazing sequence arrangement at the 5′ ends of adenovirus 2 messenger RNA. Cell 12: 1–8 [DOI] [PubMed] [Google Scholar]
- Clementi F, Cabrini D, Gotti C, Sher E 1986. Pharmacological characterization of cholinergic receptors in a human neuroblastoma cell line. J Neurochem 47: 291–297 [DOI] [PubMed] [Google Scholar]
- Cocquerelle C, Mascrez B, Hétuin D, Bailleul B 1993. Mis-splicing yields circular RNA molecules. FASEB J 7: 155–160 [DOI] [PubMed] [Google Scholar]
- Cocquet J, Chong A, Zhang G, Veitia RA 2006. Reverse transcriptase template switching and false alternative transcripts. Genomics 88: 127–131 [DOI] [PubMed] [Google Scholar]
- Dixon RJ, Eperon IC, Hall L, Samani NJ 2005. A genome-wide survey demonstrates widespread non-linear mRNA in expressed sequences from multiple species. Nucleic Acids Res 33: 5904–5913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon RJ, Eperon IC, Samani NJ 2007. Complementary intron sequence motifs associated with human exon repetition: a role for intragenic, inter-transcript interactions in gene expression. Bioinformatics 23: 150–155 [DOI] [PubMed] [Google Scholar]
- Edery I, Chu LL, Sonenberg N, Pelletier J 1995. An efficient strategy to isolate full-length cDNAs based on an mRNA cap retention procedure (CAPture). Mol Cell Biol 15: 3363–3371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg A, Vogelstein B 1984. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Addendum. Anal Biochem 137: 266–267 [DOI] [PubMed] [Google Scholar]
- Fischer M, Skowron M, Berthold F 2005. Reliable transcript quantification by real-time reverse transcriptase-polymerase chain reaction in primary neuroblastoma using normalization averaged expression levels of the control genes HPRT1 and SDHA. J Mol Diagn 7: 89–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flouriot G, Brand H, Seraphin B, Gannon F 2002. Natural trans-spliced mRNAs are generated from the human estrogen receptor-α (hERα) gene. J Biol Chem 277: 26244–26251 [DOI] [PubMed] [Google Scholar]
- Frantz SA, Thiara AS, Lodwick D, Ng LL, Eperon IC, Samani NJ 1999. Exon repetition in mRNA. Proc Natl Acad Sci 96: 5400–5405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graveley BR 2001. Alternative splicing: increasing diversity in the proteomic world. Trends Genet 17: 100–107 [DOI] [PubMed] [Google Scholar]
- Hallegger M, Llorian M, Smith CW 2010. Alternative splicing: global insights. FEBS J 277: 856–866 [DOI] [PubMed] [Google Scholar]
- Hastings KE 2005. SL trans-splicing: easy come or easy go? Trends Genet 21: 240–247 [DOI] [PubMed] [Google Scholar]
- Horiuchi T, Aigaki T 2006. Alternative trans-splicing: a novel mode of pre-mRNA processing. Biol Cell 98: 135–140 [DOI] [PubMed] [Google Scholar]
- Horiuchi T, Giniger E, Aigaki T 2003. Alternative trans-splicing of constant and variable exons of a Drosophila axon guidance gene, lola. Genes Dev 17: 2496–2501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houseley J, Tollervey D 2010. Apparent non-canonical trans-splicing is generated by reverse transcriptase in vitro. PLoS ONE 5: e12271 doi: 10.1371/journal.pone.0012271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson JM, Castle J, Garrett-Engele P, Kan Z, Loerch PM, Armour CD, Santos R, Schadt EE, Stoughton R, Shoemaker DD 2003. Genome-wide survey of human alternative pre-mRNA splicing with exon junction microarrays. Science 302: 2141–2144 [DOI] [PubMed] [Google Scholar]
- Jones DH, Winistorfer SC 1992. Sequence specific generation of a DNA panhandle permits PCR amplification of unknown flanking DNA. Nucleic Acids Res 20: 595–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kampa D, Cheng J, Kapranov P, Yamanaka M, Brubaker S, Cawley S, Drenkow J, Piccolboni A, Bekiranov S, Helt G, et al. 2004. Novel RNAs identified from an in-depth analysis of the transcriptome of human chromosomes 21 and 22. Genome Res 14: 331–342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kan Z, States D, Gish W 2002. Selecting for functional alternative splices in ESTs. Genome Res 12: 1837–1845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kent WJ 2002. BLAT—the BLAST-like alignment tool. Genome Res 12: 656–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lastowska M, Viprey V, Santibanez-Koref M, Wappler I, Peters H, Cullinane C, Roberts P, Hall AG, Tweddle DA, Pearson AD, et al. 2007. Identification of candidate genes involved in neuroblastoma progression by combining genomic and expression microarrays with survival data. Oncogene 26: 7432–7444 [DOI] [PubMed] [Google Scholar]
- Lev-Maor G, Ram O, Kim E, Sela N, Goren A, Levanon EY, Ast G 2008. Intronic Alus influence alternative splicing. PLoS Genet 4: e1000204 doi: 10.1371/journal.pgen.1000204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Wang J, Mor G, Sklar J 2008. A neoplastic gene fusion mimics trans-splicing of RNAs in normal human cells. Science 321: 1357–1361 [DOI] [PubMed] [Google Scholar]
- Li X, Zhao L, Jiang H, Wang W 2009. Short homologous sequences are strongly associated with the generation of chimeric RNAs in eukaryotes. J Mol Evol 68: 56–65 [DOI] [PubMed] [Google Scholar]
- Liu Y 2009. “Identification and functional dissection of physiological targets of the RNA binding proteins RBMY, hnRNP G-T and T-Star.” PhD thesis, Newcastle University, Newcastle upon Tyne, UK [Google Scholar]
- Maniatis T, Fritsch EF, Sambrook J 1982. Molecular cloning: A laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- Melamud E, Moult J 2009. Stochastic noise in splicing machinery. Nucleic Acids Res 37: 4873–4886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miriami E, Margalit H, Sperling R 2003. Conserved sequence elements associated with exon skipping. Nucleic Acids Res 31: 1974–1983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modrek B, Lee C 2003. Alternative splicing in the human, mouse and rat genomes is associated with an increased frequency of exon creation and/or loss. Nat Genet 34: 177. [DOI] [PubMed] [Google Scholar]
- Mongelard F, Labrador M, Baxter EM, Gerasimova TI, Corces VG 2002. Trans-splicing as a novel mechanism to explain interallelic complementation in Drosophila. Genetics 160: 1481–1487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nigro JM, Cho KR, Fearon ER, Kern SE, Ruppert JM, Oliner JD, Kinzler KW, Vogelstein B 1991. Scrambled exons. Cell 64: 607–613 [DOI] [PubMed] [Google Scholar]
- Pan Q, Bakowski MA, Morris Q, Zhang W, Frey BJ, Hughes TR, Blencowe BJ 2005. Alternative splicing of conserved exons is frequently species-specific in human and mouse. Trends Genet 21: 73. [DOI] [PubMed] [Google Scholar]
- Patthy L 1999. Genome evolution and the evolution of exon-shuffling—a review. Gene 238: 103–114 [DOI] [PubMed] [Google Scholar]
- Ponting CP, Oliver PL, Reik W 2009. Evolution and function of long noncoding RNAs. Cell 136: 629–641 [DOI] [PubMed] [Google Scholar]
- Qiu P, Wang L, Kostich M, Ding W, Simon JS, Greene JR 2004. Genome wide in silico SNP-tumor association analysis. BMC Cancer 4: 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rickman DS, Pflueger D, Moss B, VanDoren VE, Chen CX, de la Taille A, Kuefer R, Tewari AK, Setlur SR, Demichelis F, et al. 2009. SLC45A3-ELK4 is a novel and frequent erythroblast transformation-specific fusion transcript in prostate cancer. Cancer Res 69: 2734–2738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rigatti R, Jia JH, Samani NJ, Eperon IC 2004. Exon repetition: a major pathway for processing mRNA of some genes is allele-specific. Nucleic Acids Res 32: 441–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rino J, Carmo-Fonseca M 2009. The spliceosome: a self-organized macromolecular machine in the nucleus? Trends Cell Biol 19: 375–384 [DOI] [PubMed] [Google Scholar]
- Roberts GC, Gooding C, Mak HY, Proudfoot NJ, Smith CW 1998. Co-transcriptional commitment to alternative splice site selection. Nucleic Acids Res 26: 5568–5572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson BW, Felix CA 2009. Panhandle PCR approaches to cloning MLL genomic breakpoint junctions and fusion transcript sequences. Methods Mol Biol 538: 85–114 [DOI] [PubMed] [Google Scholar]
- Rozen S, Skaletsky H 2000. Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol 132: 365–386 [DOI] [PubMed] [Google Scholar]
- Rupniak HT, Rein G, Powell JF, Ryder TA, Carson S, Povey S, Hill BT 1984. Characteristics of a new human neuroblastoma cell line which differentiates in response to cyclic adenosine 3′:5′-monophosphate. Cancer Res 44: 2600–2607 [PubMed] [Google Scholar]
- Singh J, Padgett RA 2009. Rates of in situ transcription and splicing in large human genes. Nat Struct Mol Biol 16: 1128–1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solnick D 1985. Alternative splicing caused by RNA secondary structure. Cell 43: 667–676 [DOI] [PubMed] [Google Scholar]
- Surono A, Takeshima Y, Wibawa T, Ikezawa M, Nonaka I, Matsuo M 1999. Circular dystrophin RNAs consisting of exons that were skipped by alternative splicing. Hum Mol Genet 8: 493–500 [DOI] [PubMed] [Google Scholar]
- Takahara T, Kanazu SI, Yanagisawa S, Akanuma H 2000. Heterogeneous Sp1 mRNAs in human HepG2 cells include a product of homotypic trans-splicing. J Biol Chem 275: 38067–38072 [DOI] [PubMed] [Google Scholar]
- Takahara T, Tasic B, Maniatis T, Akanuma H, Yanagisawa S 2005. Delay in synthesis of the 3′ splice site promotes trans-splicing of the preceding 5′ splice site. Mol Cell 18: 245–251 [DOI] [PubMed] [Google Scholar]
- Tazi J, Bakkour N, Stamm S 2009. Alternative splicing in disease. Biochim Biophys Acta 1792: 14–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tennyson CN, Klamut HJ, Worton RG 1995. The human dystrophin gene requires 16 hours to be transcribed and is cotranscriptionally spliced. Nat Genet 9: 184–190 [DOI] [PubMed] [Google Scholar]
- Warf MB, Berglund JA 2010. Role of RNA structure in regulating pre-mRNA splicing. Trends Biochem Sci 35: 169–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilusz JE, Sunwoo H, Spector DL 2009. Long noncoding RNAs: functional surprises from the RNA world. Genes Dev 23: 1494–1504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaphiropoulos PG 1997. Exon skipping and circular RNA formation in transcripts of the human cytochrome P-450 2C18 gene in epidermis and of the rat androgen binding protein gene in testis. Mol Cell Biol 17: 2985–2993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Xie Y, Martignetti JA, Yeo TT, Massa SM, Longo FM 2003. A candidate chimeric mammalian mRNA transcript is derived from distinct chromosomes and is associated with nonconsensus splice junction motifs. DNA Cell Biol 22: 303–315 [DOI] [PubMed] [Google Scholar]
- Zhang F, Gu W, Hurles ME, Lupski JR 2009. Copy number variation in human health, disease, and evolution. Annu Rev Genomics Hum Genet 10: 451–481 [DOI] [PMC free article] [PubMed] [Google Scholar]