Abstract
Vertebrate genomes encode large and highly variable numbers of tandem C2H2 zinc finger (tandem ZF) transcription factor proteins. In mammals, most tandem ZF genes also encode a KRAB domain (KZNF proteins). Very little is known about what forces have driven the number and diversity of tandem ZF genes. Recent studies suggest that one role of KZNF proteins is to bind and repress transcription of exogenous retroviruses and their endogenous counterpart LTR retroelements. We report a striking correlation across vertebrate genomes between the number of LTR retroelements and the number of host tandem ZF genes. This correlation is specific to LTR retroelements and ZF genes and was not explained by covariation in other genomic features. We further show that recently active LTR retroelements are correlated with recent tandem ZF gene duplicates across vertebrates. On branches of the primate phylogeny, we find that the appearance of new families of endogenous retroviruses is strongly predictive of the appearance of new duplicate KZNF genes. We hypothesize that retroviral and LTR retroelement burden drives evolution of host tandem ZF genes. This hypothesis is consistent with previously described molecular evolutionary patterns in duplicate ZF genes throughout vertebrates. To further explore these patterns, we investigated 34 duplicate human KZNF gene pairs, all of which underwent an early burst of divergence in the major nucleotide contact residues of their ZF domains, followed by purifying selection in both duplicates. Our results support a host-pathogen model for tandem ZF gene evolution, in which new LTR retroelement challenges drive duplication and divergence of host tandem ZF genes.
Vertebrate genomes contain large and highly variable numbers of tandem C2H2 zinc finger (tandem ZF) transcription factor genes. Outside of mammals, almost nothing is known about the function of these genes. Within mammals, tandem ZF genes are dominated by those with a KRAB domain, which expanded from a KRAB—tandem ZF fusion gene near the root of tetrapod vertebrates (Birtle and Ponting 2006). Most KRAB zinc finger (KZNF) proteins consist of an N-terminal KRAB domain followed by multiple tandem ZF domains (Bellefroid et al. 1991; Huntley et al. 2006). Some KZNF proteins have an additional SCAN protein-interaction domain N-terminal to their KRAB domain (Edelstein and Collins 2005). The KRAB domain represses transcription by binding TRIM28 (also called KAP1), which is part of a large protein complex that modifies histones to promote closed chromatin (e.g., Nielsen et al. 1999; Ryan et al. 1999; Lechner et al. 2000; Schultz et al. 2002; Sripathy et al. 2006). The tandem ZF domains confer DNA-binding specificity in a modular manner, with a turn-helix segment of each ZF domain binding to three nucleotides in target DNA sites (Pavletich and Pabo 1991; Kim and Berg 1996).
Tandem ZF genes have been gained by an ongoing process of lineage-specific duplication and divergence (e.g., Shannon et al. 2003; Emerson and Thomas 2009; Nowick et al. 2010). Throughout vertebrates, tandem ZF gene expansions are characterized by strong positive selection that has changed the number and DNA-binding specificity of zinc fingers, while retaining a conserved KRAB domain (Schmidt and Durrett 2004; Emerson and Thomas 2009). Though most tandem ZF genes outside of mammals lack a KRAB domain, the structure and evolution of their zinc fingers is strikingly similar to that of KZNF genes in mammals (Emerson and Thomas 2009). These evolutionary patterns, combined with remarkably little functional information, have given rise to a set of long-standing puzzles. What are the organismal functions of tandem ZF genes? Why are there so many genes, and why do the gene numbers and their repertoire of DNA-binding sites change so quickly?
Recent work suggests a plausible functional explanation for KRAB tandem ZF gene evolution. First, a series of papers showed that restriction of murine leukemia virus (MLV) in mouse embryonic stem cells results from transcriptional repression by the mouse-specific KZNF gene Zfp809 acting via the TRIM28 complex (Wolf and Goff 2007, 2009). Zfp809 binds integrated MLV DNA at its primer binding site (PBS), which MLV requires to prime reverse transcription via a complementary host tRNA (Wolf and Goff 2009). Shortly thereafter, two papers showed that deletion of TRIM28 (or the TRIM28 effector SETDB1) in mouse embryonic cells causes massive transcriptional derepression of several mouse endogenous retroviruses (ERVs) (Matsui et al. 2010; Rowe et al. 2010). Since TRIM28 and SETDB1 are thought to be shared effectors of all KZNF proteins, this result suggests that a suite of KZNF genes repress transcription of diverse ERVs (for review, see Rowe and Trono 2011). These findings have the potential to explain the large number of evolutionarily volatile KZNF genes based on a host–pathogen interaction. ERVs are the genomic footprints of historical retroviral infections that resulted in viral insertions in the germ line (for review, see de Parseval and Heidmann 2005; Blikstad et al. 2008). When an ERV first appears in a genome, it typically appears as a burst of multiple elements, due either to recurrent viral insertions or to transposition (Belshaw et al. 2004, 2005). This process results in a genomic signature that reflects a sampling of the history of retroviral infections, or at least the subset of infections that successfully established a germ-line copy. Based on the diversity and ages of ERVs present today, both the mouse and human lineages have episodically suffered a large number of infections by diverse retroviruses over the past 80 million years (e.g., de Parseval and Heidmann 2005; Stocking and Kozak 2008). Less extensive analysis suggests that a similar process occurs throughout mammals (e.g., Mouse Genome Sequencing Consortium 2002; Mikkelsen et al. 2007).
Presumably, both new retroviral infections and divergence of endogenous LTR retroelements will drive selection for a host response. One possible host response is the generation of new transcriptional repressors that evolve to target the DNA of the new retrovirus or retrotransposon. The simple modular biochemistry of KZNF transcriptional repression makes KZNF genes particularly suitable for such a role. The existing TRIM28 complex should require only recruitment by a new DNA-binding specificity to result in a repressed chromatin state. This repressed state can spread many kilobases from the DNA-binding site (Groner et al. 2010), so, in principle, binding anywhere in a retroelement could provide effective repression.
Here, we present data that supports the hypothesis that most vertebrate tandem ZF genes evolved to repress retroviral or LTR retroelement activity.
Results
LTR retroelement correlation with ZF domains and genes
Endogenous retroviruses and LTR retrotransposons differ primarily in the presence or absence of an envelope (ENV) coding sequence. These two types of retroelements interconvert by gain or loss of ENV sequences, and the ENV-coding sequence is extremely diverse and rapidly evolving, making it difficult to distinguish these two groups without detailed analysis. We will refer to both types of elements as LTR retroelements, and we did not attempt to distinguish them in our analysis. Our initial goal was to test for correlation between LTR retroelement load and tandem ZF-coding potential across a wide range of vertebrate genomes. Existing genome annotations are uneven, so we implemented genome searches to detect both LTR retroelements and ZF domains in a manner independent of annotation status and phylogeny. In the case of retroelements, this was made possible by the fact that LTR retroelements include ancient protein-coding domains that distinguish them from all other known genomic features (e.g., for review, see Gogvadze and Buzdin 2009). In the case of ZF domains, this was made possible by the fact that the C2H2 ZF domain has the same length and sequence profile throughout animals (Emerson and Thomas 2009). Details of both searches are given in the Methods section and the Supplemental Methods.
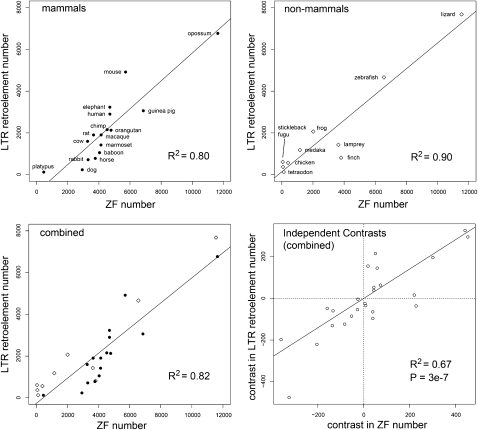
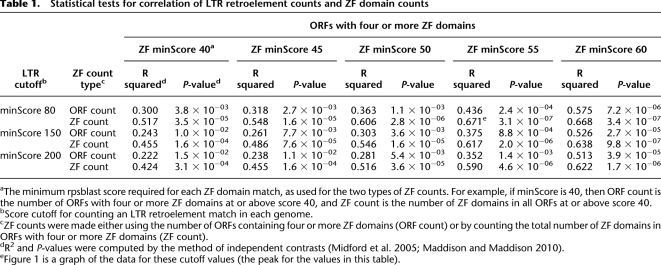
We found a striking correlation across vertebrates between the number of LTR retroelements and the number of ZF domains. This correlation holds within mammals and outside of mammals, and when all 26 taxa are combined (Fig. 1; Supplemental Fig. S1). To account for the fact that shared phylogenetic history probably accounts for some of this correlation, we computed corrected correlations and P-values (Fig. 1; Table1) using the method of independent contrasts (IC) (Felsenstein 1985; Garland et al. 2005). The IC-corrected correlations remained strong and highly significant and were robust to widely different score thresholds for counting LTR retroelements and ZF domains, and to counting the total number ZF domains or the number of putative tandem ZF genes (Table 1; Supplemental Table S1). Given existing evidence that KRAB ZF genes can repress retroviral and LTR retrotransposon transcription, the most obvious inference is that the historical LTR retroelement content of each genome has driven the number of ZF domains, but we considered other possible explanations. First, it seemed possible that each genome has a characteristic rate of segmental duplication or duplicate retention, and that this rate drives both LTR retroelement and ZF domain content. We examined this possibility by testing for IC-corrected correlations of non-LTR (LINE-like) retroelements with ZF domains and of LTR retroelements with other large dynamic protein domain families (olfactory receptor and immunoglobulin C1 and V domains). None of these correlations were statistically significant (Table 2). Second, it seemed possible that unknown constraints on genome size influence the potential for LTR retroelement and ZF domain content in each genome. We tested this possibility by normalizing LTR retroelement content to genome size and testing the normalized correlation to ZF domains. The correlation remained highly significant (Table 2). Given that LTR retroelements are a significant contributor to genome size, it is unsurprising that genome size itself positively correlates with LTR retroelement and ZF domain content, though these correlations were weak and statistically nonsignificant after IC correction (Table 2).
Figure 1.
Correlation of genomic LTR retroelement and ZF domain content: Three panels show the number of detected LTR retroelements plotted against the number of detected ZF domains in different vertebrate groups or all groups combined. The lines show the linear least-squares best fit with its squared correlation coefficient R2. The fourth panel (lower right) shows the combined data after correction for phylogenetic relatedness by the method of independent contrasts. The line is the linear least-squares best fit forced to go through the origin and its associated R2 and P-value. The summed score cutoff for LTR retroelements was 50 (see Supplemental Methods). The ZF number was determined from all genomic open reading frames with four or more ZF domain matches with a minimum average score of 55. These counting criteria gave the maximum correlation for combined data, but a wide variety of other counting criteria also gave highly significant correlations (Supplemental Table S1).
Table 1.
Statistical tests for correlation of LTR retroelement counts and ZF domain counts
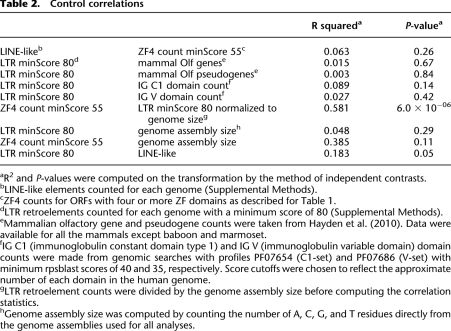
Table 2.
Control correlations
Other features of these data can be explained by a model in which LTR retroelements drive tandem ZF gene evolution. First, testing various score cutoffs for counting ZF domains showed that the correlation to LTR retroelements is strongest very near the score that best distinguishes human ZF domains in known genes from those in pseudogenes (Table 1; Supplemental Fig. S2). This result suggests that the correlation is stronger for functional ZF genes than for pseudogenes. Second, LTR retroelement correlation to total ZF domain number was slightly stronger than to the number of putative tandem ZF genes (Table 1). This result suggests that the total DNA-binding potential of ZF genes is more important than the number of genes. Finally, the most prominent correlation outlier in mammals is mouse, which has fewer ZF domains than predicted by its LTR retroelement content (Fig. 1). The mouse genome is known to have several groups of recently and currently active ERVs (Stocking and Kozak 2008), suggesting the possibility that the host ZF response is lagging behind a recent burst of LTR retroelement activity. Alternative explanations of this mouse result are considered in the Discussion.
If the major function of ZF genes is to transcriptionally repress LTR retroelements, then the sequence diversity of LTR retroelements should be an important factor in driving ZF number. We estimated the relative sequence diversity of LTR retroelements in each species by extracting their reverse-transcriptase coding regions and measuring their total protein tree length. Unsurprisingly, we found that retroelement diversity correlates strongly with retroelement number, so it was difficult to distinguish the influence of copy number and copy diversity. As expected given this result, retroelement diversity also strongly correlated with ZF number, though not quite as strongly as did retroelement copy number (Supplemental Table S1).
The 26 genome assemblies analyzed above are all based on more than fivefold sequence read coverage, but they vary in read coverage and in the degree of assembly finishing (Supplemental Fig. S1). This variation could affect the apparent retroelement and ZF gene content differentially, for example, by collapsing recent ZF gene duplicates into apparent single genes. Such variation in assembly quality is difficult to detect and control for, but we made one simple test by repeating the analysis only on the 16 published genomes (higher than average read coverage and finishing effort). Using the LTR retroelement and ZF domain cutoffs that gave the best correlation across all species, the IC-corrected correlation for published genomes was higher than for all genomes (R2 0.71 vs. R2 0.67) and the correlation remained highly significant despite the smaller data set (P-value 7.2 × 10−5). This result suggests that improved genome assemblies will most likely improve the observed correlations.
In addition to LTR retroelements containing part or all of their internal sequences, vertebrate genomes contain large numbers of solo LTR sequences that arise by recombination between flanking LTRs (Copeland et al. 1983). It is very difficult to obtain unbiased counts of solo LTRs, because they have no generally shared sequence features. In addition, solo LTRs are more abundant for older retroelements, because they have had more time to recombine since their original insertion. Nevertheless, we assessed correlation between ZF sequences and total annotated LTR retroelement sequence content (including solo LTRs) for the 16 published vertebrate genomes, since they have the best annotated general repeat content. The IC-corrected correlation was significant (R2 0.36, P = 0.018), though weaker than for elements with internal sequence. This lower correlation could result from uneven annotation of solo LTR sequences, the expected skew toward older retroelements that are less reflective of recent selective pressure on ZF genes, or other unknown factors.
Though mammalian tandem ZF genes are dominated by those encoding a KRAB domain, this domain association is less common in other tetrapods and is absent in fish (Supplemental Table S1; Looman et al. 2002; Birtle and Ponting 2006; Emerson and Thomas 2009). The fact that ZF domain content correlates strongly with LTR retroelement content throughout all of these groups suggests that the function of non-KRAB ZF genes in other tetrapods and in fish is related to the function of KRAB ZF genes in mammals. This inference is also supported by similarities in sequence evolution of tandem ZF genes in each of these groups (see below; Emerson and Thomas 2009). We speculate that other domains in these taxa play a role analogous to the KRAB domain in mammals, or that tandem ZF proteins bound to DNA can directly repress transcription.
Recent LTR retroelement activity
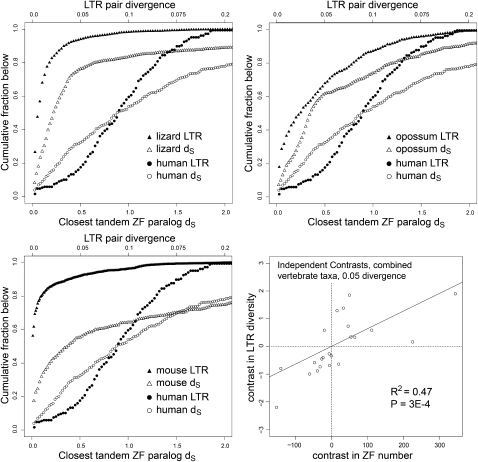
The data above reflect an historical aggregate of LTR retroelement activity and tandem ZF gene duplications. To test whether these characters are temporally correlated, we estimated the age of LTR retroelement insertions based on divergence between the two long terminal repeats of each retroelement (Johnson and Coffin 1999) and the age of ZF gene duplicates based on synonymous site divergence (dS). Though mutation rates surely vary among the species, this variation should not affect relative divergence rates of ZF genes and LTR retroelements within a species. Using 5% and 10% divergence cutoffs, we found significant IC-corrected correlations between recent LTR retroelement activity and recent tandem ZF gene duplications across the combined taxa (Supplemental Table S3). The highest correlation was between sequence diversity among recently active LTR retroelements and the number of ZF domains in recent tandem ZF gene duplicates (Fig. 2), but comparisons of the numbers of LTR retroelements and ZF gene duplicates were also highly significant (Supplemental Table S3). Mouse, opossum, and lizard show evidence of especially high recent LTR retroelement activity, and all three species have correspondingly high numbers of recent tandem ZF gene duplicates (Fig. 2, compare the steepness of the curves near the origin). In addition, all three species show possible evidence of an earlier period of relatively quiescent LTR retroelement activity associated with fewer tandem ZF gene duplicates, as evidenced by the plateaus on each curve. Alternatively, these plateaus could result from a higher rate of deletion removing older LTR retroelements. Except for a very recent drop in LTR retroelement activity, the patterns on the human lineage suggest relatively slow and constant rates of LTR retrotransposition and ZF gene duplication. Other genomes with relatively low recent LTR retroelement activity (e.g., horse, elephant, and medaka) have curves broadly similar to that of human (data not shown).
Figure 2.
ZF gene duplicate and LTR divergence time courses. Three panels show cumulative histograms of LTR nucleotide divergence and closest ZF paralog dS for the indicated species. The axes have been scaled to best display the full curve for both data sets. The human data are included in all three panels for comparison. The fourth panel (lower right) shows statistical analysis at or below one divergence point (0.05 LTR divergence/0.05 paralog dS) for all species combined after correction by independent contrasts (see Supplemental Table S3). The line is the linear least-squares best fit forced to go through the origin and its associated R2 and P-value.
Primate LTR retroelements and tandem ZF gene duplicates
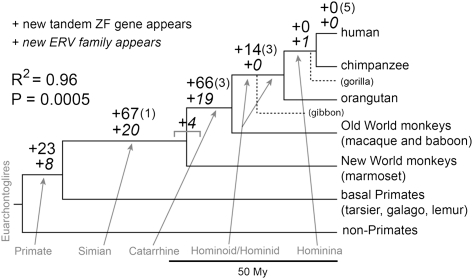
Among existing vertebrate genome sequences, primates provide the densest phylogeny and the best annotation of LTR retroelements. Using RepeatMasker annotations and tandem ZF gene annotations in humans as a starting point, we investigated in detail the appearance of new LTR retroelements and tandem ZF genes on the primate lineage (Supplemental Methods). We could divide the primate lineage into six distinct branches based on available sequences: a basal primate branch (before the divergence of basal primates), a Simian branch (before the divergence of New World from Old World monkeys), a Catarrhine branch (before the divergence of Old World monkeys from apes), a Hominoid/Hominid branch (before the divergence of orangutan from human; this branch is bisected by gibbons, which currently lack a whole-genome assembly), a Hominina branch (before the divergence of chimpanzee from human), and a human-specific branch. Using a combination of insertion-site analysis and sequence trees of retroelement internal sequences, we defined the branch on which each of 48 primate-specific LTR retroelement families first appeared (Fig. 3; Supplemental Table S4). Another four retroelement families were imperfectly resolved, appearing just before or just after the divergence of New World monkeys. Starting with annotated human tandem ZF genes, we used a combination of genome sequence searches, maximum-likelihood trees, and synteny to determine the branch on which each new tandem ZF gene duplicate appeared. Among ZF gene duplicates, we distinguished between those that diverged by at least 5% in amino acid sequence in an attempt to distinguish between selected duplicates and possibly neutral copy-number variation. Since events on each branch of the phylogeny are statistically independent, we analyzed correlations without using independent contrasts. Correlation between appearance of new LTR retroelement families and new tandem ZF genes was remarkably strong and statistically significant regardless of whether or not ZF genes with low divergence were included and regardless of branch assignment of the four ambiguous retroelement families (Supplemental Table S7). The correlation was highest when ZF genes with low divergence were excluded and the ambiguous retroelement families were split equally on the two possible branches (Fig. 3, statistics). These results are consistent with our global analysis of vertebrate correlations, suggesting that many or most tandem ZF genes in primates arose in response to the appearance of new families of endogenous retroviruses. The most prominent deviation from perfect correlation is on the Hominoid/Hominid branch, where no new LTR retroelement families, but 14 new tandem ZF genes appeared. The immediately preceding Catarrhine branch was subject to a particularly intense burst of new LTR retroelements (Fig. 3); we speculate that some of the 14 Hominoid/Hominid ZF genes arose in response to this slightly earlier burst. In contrast to the human-specific branch, new LTR retroelement families have arisen on the chimpanzee- and macaque-specific branches (Jern et al. 2006; Polavarapu et al. 2006; Rhesus Macaque Genome Sequencing and Analysis Consortium 2007), but the number of families is small, and we lack statistical power to test for tandem ZF gene response.
Figure 3.
Primate phylogeny with the appearance of new endogenous retroviral families and new tandem ZF genes. Data were derived by tracking the first appearance of human ERV families and tandem ZF genes (Supplemental Methods). On each branch leading to the human, the top number indicates the number of tandem ZF gene duplicates (with additional duplicates that diverged by <5% in amino acid sequence in parentheses) and the bottom number indicates the number of new ERV families. Four families of ERVs could not be confidently assigned to a specific branch and are shown straddling the Simian/Catarrhine branch point. The R2 and ANOVA P-value shown are for the peak correlation based on various criteria for partitioning the data (see Supplemental Table S7); all other partitions were also significant. The gorilla genome is low coverage and was not systematically analyzed, but the single ERV (HERV-Fc1) that appears on the branch leading to human and chimpanzee is clearly present in gorilla (data not shown). The Hominoid and Hominid branches are split by the gibbons, for which there are currently no genome assemblies.
Predictions for duplicate gene divergence
The hypothesis that most tandem ZF genes function to repress LTR retroelements predicts certain patterns of molecular evolution driven by the epidemiology of retroelements. First, each host genome should acquire distinct expansions of tandem ZF genes in response to lineage-specific retroelement challenges. Second, the duplicate genes that comprise these expansions should be subject to positive selection to modify their DNA-binding specificity as they adapt to new retroelements. These two predictions have already been confirmed for several of the genomes we analyzed here, including human, mouse, cow, frog, fugu, and zebrafish (Emerson and Thomas 2009). We extended these analyses to several additional species, namely, rat, horse, elephant, opossum, lizard, tetraodon, medaka, and lamprey. In all cases, we found large species-specific clades of tandem ZF genes with overwhelming evidence of positive selection affecting predominantly nucleotide contact residues of ZF domains (Supplemental Table S8; Supplemental Fig. S6).
A final prediction of our hypothesis is that often when a new divergent duplicate tandem ZF gene pair arises, one of the duplicates will retain the ancestral DNA-binding specificity, while the other duplicate acquires a new or modified DNA-binding specificity that targets a new retroelement. After optimizing its new DNA-binding function, the divergent duplicate should be subject to purifying selection. These patterns are expected in cases in which repression of an ancestrally targeted retroelement remains selectively significant, so that only one copy of a duplicate gene pair is free to alter specificity to protect against a new challenge. These patterns are also predicted when the ancestral tandem ZF gene has been exapted for host transcriptional regulation and by some other hypotheses for ZF gene evolution (see Discussion). As described in the next sections, we explored these predictions among human duplicate KZNF genes, since they are best annotated and there exist multiple closely related primate genomes that help resolve the time of duplication and ancestral gene identity.
Tracing the origins of human KZNF genes
We identified 34 cases in which two human KZNF genes were clearly closest relatives (see Methods); each pair is assumed to have arisen by gene duplication from an ancestral gene at some time during tetrapod evolution. In order to trace the evolutionary history of each pair of genes, we used the two human proteins to find all closely related sequences in a set of increasingly divergent mammalian genomes. Because of the patterns of conservation and divergence detailed below, these searches were remarkably effective in unambiguously tracing the ancestry of each gene.
Considering one duplicate pair, one common result was as follows. In one or more of the most closely related species, one clear copy of each gene was found, indicating that both genes were present in the last common ancestor of human and those species. On deeper branches in the tree, each species had only one gene closely related to the two human genes, suggesting that their last common ancestral species had one copy of the gene, which later duplicated and diverged on the human lineage. The other common result was that clear copies of both genes were found back to some point on the phylogenetic tree, but deeper in the tree no specific ancestral genes were found (more accurately, many possible ancestral genes were found, but it was unclear which of them was the true ancestor). Examples of trees reflecting these assignments are given in Supplemental Fig. S8. Below, we consider the latter case first, in which the precise origin of two closely related human genes is unclear, but the pattern of divergence of the two copies from each other can nevertheless be analyzed.
Divergence of gene pairs of uncertain origin
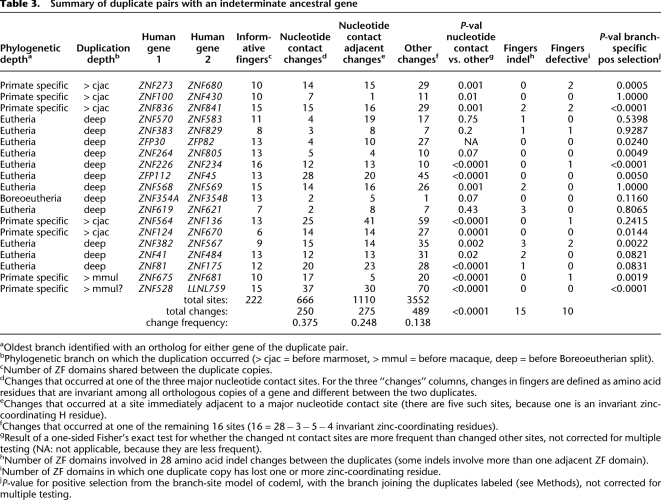
We could unambiguously identify and analyze 19 human duplicate gene pairs with two copies in a number of species but no clear specific ancestor. Table 3 summarizes key features of these duplicates and Supplemental Table S10 gives additional details. The phylogenetic depth of the traceable ancestry of the two genes varied from early in the primate lineage to early in the placental mammalian lineage. It may be presumed that the two genes arose by one or more rounds of duplication and divergence from some specific ancestral gene, but the identity and sequence of the ancestral gene is indeterminate. For example, clear copies of both ZNF273 and ZNF680 were identified from all five primate species, but no species outside of primates. A detailed example of sequence divergence patterns for one pair of genes is shown in Supplemental Figure S7.
Table 3.
Summary of duplicate pairs with an indeterminate ancestral gene
Three patterns were apparent in most or all cases:
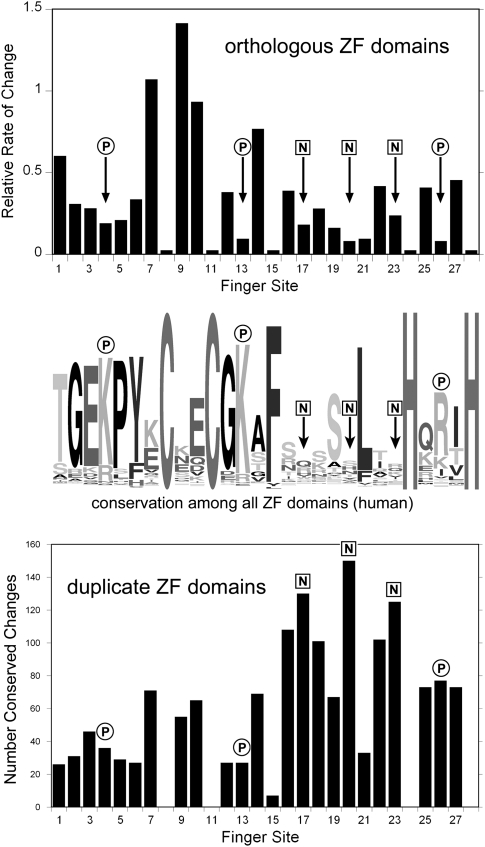
Orthologs of each gene were subject to purifying selection across the entire set of DNA-binding domains: Both the number of ZF domains and the amino acid sequence of each ZF domain are highly conserved. Averaged across 280 ZF domains from 22 genes randomly selected from these duplicates, the nucleotide and phosphate contact residues are among the slowest evolving (Fig. 4). The most plausible explanation for this pattern is that each orthologous ZF domain is subject to purifying selection to retain its DNA-binding specificity.
A second pattern is evident when comparing two duplicate genes to each other: Amino acid changes are more abundant in major nucleotide contact residues than elsewhere (18 of 19 duplicate pairs). When the divergence between the duplicates was relatively low, this difference did not reach statistical significance, but in 12 of 19 duplicates, changes were enriched in major nucleotide contact residues with P < 0.01 (Fisher's exact test). Summed over all 222 testable ZF domains from all 19 duplicate pairs, changes between paralogs that are conserved among orthologs occurred in 250 of 666 major nucleotide contact residues (37.5%), but only 489 of 3552 other residues (13.8%), a highly significant enrichment. This pattern is summarized graphically in Figure 4.
A third pattern is that entire ZF domains were often lost or gained in one duplicate gene relative to the other, consistent with previous observations (Looman et al. 2002; Huntley et al. 2006). When such a difference was observed it was strongly conserved among orthologs of each of the two genes, suggesting that these domain arrangements are also subject to purifying selection. Some such events involved insertion or deletion of ZF domains and others involved point mutations that disrupt the canonical finger structure (Table 3). These results suggest that finger gain and loss contribute to changes in DNA-binding specificity between duplicate genes.
Figure 4.
Changes in orthologous zinc fingers and duplicate zinc fingers compared with diversity among all fingers. (Top) The averaged relative rates of divergence in 280 orthologous ZF domains from 22 randomly chosen KZNF genes (see Methods). (Middle) The diversity among all ZF domains from human KZNF genes as a logo plot (http://weblogo.berkeley.edu). (Bottom) The number of conserved changes observed among the 390 testable zinc fingers in all 34 duplicate gene pairs analyzed. Circles labeled P indicate residues that make major phosphate contacts and squares labeled N indicate residues that make the major nucleotide contacts.
Duplicate divergence is asymmetric
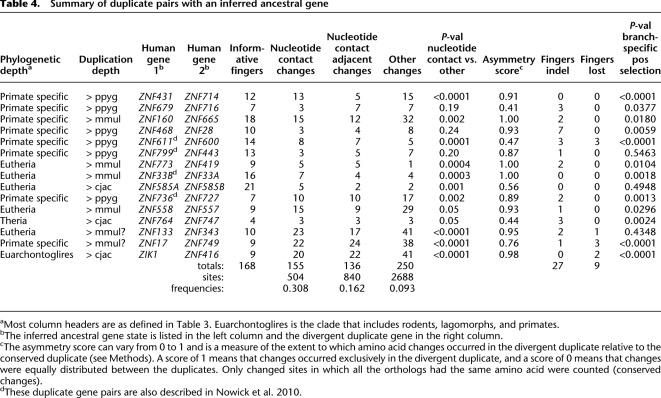
In the 19 cases discussed above, the absence of an identified ancestral gene in the outgroup species precluded analysis of the symmetry of divergence following duplication. In the other 15 duplicate cases, the ancestral gene state could be identified based on the pattern of gene number and gene type in various species (Table 4; Supplemental Table S10). For example, copies of both human ZNF557 and ZNF558 were clearly identified in chimpanzee and orangutan, but only one related gene was found in macaque, marmoset, cow, dog, horse, and rodents, suggesting that a single ancestral gene duplicated on the branch leading to great apes. By comparison of the two duplicates with the single gene from outgroup species, we could address whether divergence occurred in one or both duplicate copies. As shown in Table 4, the results usually indicated highly asymmetric divergence of the duplicate genes. Alignments for two examples are shown in Supplemental Figure S7. Amino acid changes following duplication were strongly biased toward nucleotide contact residues: In total, conserved changes occurred in 155 of 504 major nucleotide contact residues (30.8%), but in only 250 of 2688 other residues (9.3%). After an initial period of divergence, the divergent copy became subject to purifying selection, since its copies in all descendant species are very similar in amino acid sequence. These patterns suggest that one duplicate retains the ancestral DNA-binding specificity, whereas the other duplicate acquires a new or modified DNA-binding specificity.
Table 4.
Summary of duplicate pairs with an inferred ancestral gene
Apparent divergence asymmetry could result from genome assembly artifacts or repeated gene loss in specific lineages. A combination of measuring assembly completeness, dating duplicates by synonymous site divergence, and parsimony analysis of loss events, indicates that these artifacts cannot account for the asymmetry results (Supplemental Methods).
Positive selection following duplication
If new duplicate KZNF genes are subject to selection to acquire new DNA-binding specificities, codon-based methods for analyzing selection might be able to detect branch-specific positive selection. We used the branch-site maximum-likelihood models implemented in codeml to test this possibility. For 17 of the 34 duplicates, highly significant evidence (P < 0.01) was obtained for positive selection on the branch connecting the two duplicate copies (Table 4). In many cases, the number of available sequences and their total tree length are well below the optimum for detection of positive selection by this method (Anisimova et al. 2001), so it is possible that divergence of all of the duplicates involved positive selection, but reached statistical significance only in the strongest cases. As expected, the specific residues implicated in positive selection are strongly enriched in the major nucleotide contact residues (data not shown). These results directly support the idea that initial duplicate divergence is driven by selection to acquire new DNA-binding specificity.
Stable genes
Though duplication is common in the KZNF family, some genes are old and highly conserved. Marsupial mammals diverged from placental mammals about 180 Mya (Kumar and Hedges 1998; Mikkelsen et al. 2007). Using systematic genome searches, we identified 20 human KZNF genes that have clear orthologs in the marsupialopossum genome and are present in all or nearly all placental mammalian genomes (ZKSCAN1, ZNF3, ZNF18, ZNF192, ZNF202, ZNF205, ZNF212, ZNF213, ZNF263, ZNF282, ZNF398, ZNF436, ZNF446, ZNF496, ZNF641, ZNF746, ZNF764, ZNF777, ZNF783, and ZNF786). Each gene was present in single copy in opossum and throughout placental mammals, and their ZF domains were invariant in number and highly conserved. These results indicate that some KZNF genes adopted stable functional roles early in mammalian evolution and that they have subsequently retained the same DNA-binding specificity. Explanations that reconcile this finding with the retroelement hypothesis are given in the Discussion.
Discussion
Based on the striking correlations between LTR retroelement content and C2H2 ZF domain content throughout vertebrates and over time, we propose that most tandem ZF genes originate to repress transcription of LTR retrotransposons or retroviruses. The linear-regression lines for the raw correlations pass close to 0 (the origin) LTR retroelements and ZF domains (Fig. 1), suggesting that most or all tandem ZF genes are involved. Consistent with this hypothesis, recent publications show that a mouse KZNF gene represses murine leukemia retrovirus (Wolf and Goff 2009) and that an unknown suite of KZNF genes probably repress a wide variety of IAP and MusD LTR retroelements (Matsui et al. 2010; Rowe et al. 2010). The vast majority of tandem ZF genes have no experimentally determined organismal function, a situation fully compatible with retroelement repression because this function should be difficult to ascertain. Nevertheless, a handful of tandem ZF genes are implicated in other processes, including sex-limited gene expression, imprinting, and mouse embryonic development (Krebs et al. 2005; García-García et al. 2008; Li et al. 2008; Mackay et al. 2008). Similarly, some KZNF genes arose early in mammalian evolution and have been retained throughout Therian mammals with nearly invariant DNA-binding domains. These genes are unlikely to have current-day retroelement-related functions, since there is no evidence for such widely shared retroelements. One plausible explanation is simply that some tandem ZF genes evolved directly to fulfill other host functions and that they were never involved in retroelement repression. Alternatively, it is well established that transcriptional promoters and enhancers present in retroelements are sometimes exapted for host transcription, following chance integration in an appropriate location to confer useful transcriptional regulation on a host gene (for review, see Cohen et al. 2009). A few studies provide indirect evidence that host exaptation of retroelement regulatory sequences may be extremely common (Lowe et al. 2007; Conley et al. 2008). In addition, in at least two cases a retroviral gene itself appears to have been adopted for a host function (Best et al. 1996; Dupressoir et al. 2009). Thus, it is possible that tandem ZF genes that now function as host transcription factors evolved initially to repress LTR retroelements and were later retained on the basis of their regulatory role for a host gene.
In mammals, transposition competence of new ERV families is usually relatively transient, decaying over a period of several million years; after transposition-specific ERV sequences evolve neutrally and eventually lose protein-coding function and transcription competence (e.g., de Parseval and Heidmann 2005; Stocking and Kozak 2008). During this transition, selection to retain specifically protective ZF genes will attenuate. Unless they are exapted for a distinct host function, most such ZF genes should eventually become pseudogenes or be deleted from the genome. This predicted pattern has not been analyzed in detail, but the general expectation of abundant ZF pseudogenes is clearly met in the human genome and probably in other genomes (Supplemental Fig. S2).
The state of ERVs and tandem ZF genes in the mouse is of particular interest, because further experimental tests of our hypothesis are most feasible there. Our data show that the mouse reference genome assembly has an unusually high number and diversity of ERVs relative to tandem ZF genes. One possible explanation is that tandem ZF gene response in mouse lags behind the recent high ERV activity that is known in mouse (Stocking and Kozak 2008), but alternative explanations are possible. First, mouse has a higher rate of genomic deletion than human (Mouse Genome Sequencing Consortium 2002), which should remove older ERVs more quickly, potentially freeing ZF genes involved in their defense for directional positive selection to protect against new ERV challenges. This possibility may be testable by a focused analysis of the patterns of duplication and positive selection among mouse ZF genes. Second, a strong recent evolutionary ZF response to high ERV activity is expected to result in an abundance of unfixed ZF gene duplicates, causing heterogeneity in the number of ZF genes in mouse populations. This possibility predicts that sequences from other wild Mus musculus isolates will vary in ZF gene content, with some isolates having more or fewer ZF genes than the reference genome. Finally, though the mouse genome assembly is one of the highest quality that we analyzed, it is possible that a recent burst of ZF gene duplication would be obscured by assembly collapse of multiple similar paralogs, resulting in an underestimate of ZF gene number.
In mammals, the large majority of LTR retroelements are clearly ERVs, as indicated by the presence of a viral envelope gene or close relatedness to a known retrovirus. Outside of mammals, this relationship is less clear. Fish appear to have relatively few ERVs and a large burden of LTR retrotransposons with no clear retroviral connection (Basta et al. 2007, 2009). In chicken and finch, most retroelements are classified by RepeatFinder as ERVs, but detailed analysis is lacking, and in lizard no analysis of LTR retroelements is available. Retroviruses have repeatedly evolved from vertically transmitted retrotransposons by acquisition of an envelope gene (e.g., Doolittle and Feng 1992; Laten et al. 1998; Malik et al. 2000). Conversely, integrated retroviruses can readily convert to vertically transmitted transposons by loss of the envelope gene (Ribet et al. 2008). It is possible that tandem ZF genes repress exogenous retroviruses, endogenous retroviruses, and LTR retrotransposons, but the balance of activities for these groups remains unclear and may vary in different species.
Implications for retroviral repression in mammals
Judging from patterns of endogenized retroviral sequences, mammals have been subject to an ongoing barrage of retroviral infections of diverse types (de Parseval and Heidmann 2005; Blikstad et al. 2008). New retroviral infections in a particular species can arise by a shift or expansion of host range by a retrovirus that infects another species (e.g., Benveniste and Todaro 1974; Chen et al. 1996; Gao et al. 1999; Martin et al. 1999). The consequence for the new host is the occasional appearance of an unpredictable new retroviral challenge. If a retrovirus successfully integrates in the germ cell lineage, it may also result in the spread of a new deleterious ERV in the host genome. It is well established that mammals combat retroviral infection in multiple ways, including attacking viral RNA with APOBEC cytidine deaminases and ZAP, interfering with viral capsid with Fv1 and TRIM5alpha, and preventing viral particle release with Tetherin (for review, see Wolf and Goff 2008). The pervasiveness of retroviruses in vertebrates and the multiple layers of viral restriction by the host support the idea that there should also be strong selection on the host to repress retroviral transcription. The size, diversity, and rapid evolution of the tandem ZF gene family suggests that these genes fill this role.
The sequence divergence patterns of new duplicate genes suggests the following model for the contribution of KZNF genes to host response to a new retroviral infection in mammals. Starting from either a new duplicate gene or a pre-existing copy-number polymorphism, a KZNF gene with significant, even minor, off-target binding to a new retroviral sequence is driven to fixation and starts to evolve improved target recognition by changes in amino acid sequence and changes in ZF number. This pattern of duplicate evolution corresponds in many ways to that proposed for bacterial genes (Bergthorsson et al. 2007). The initial off-target binding to a new retrovirus may arise purely by chance or may result from sequence relatedness of the new retrovirus to a previously encountered retrovirus for which the host has already evolved a cognate KZNF gene. If the previously encountered retrovirus (or its endogenized copies) remain selectively significant for the host, there will be pressure for one copy of the KZNF gene to retain its ancestral DNA-binding specificity and for adaptation to the new retrovirus to act on the other copy. If the previously encountered retrovirus is no longer selectively significant for the host, targeting a new retrovirus could be achieved by directional selection on an ancestral KZNF gene without gene duplication, though we didn't observe any clear instances of this pattern.
Other possible evolutionary drivers
A number of other potential drivers of tandem ZF gene duplication and divergence have been suggested and probably apply in specific cases. Based on the expansion and diversification of the KZNF gene sequence and expression patterns on the primate lineage, it has been suggested that these genes underlie the evolution of novel primate traits, including an enlarged brain (Hamilton et al. 2003; Nowick et al. 2009, 2010). Based on the expansion of genes in a cluster of KZNF genes in mouse that includes two genes that modify sex-limited expression of other genes, it has been suggested that KZNF genes play a role in speciation via modification of sex-specific traits (Krebs et al. 2005). One KZNF gene with an ortholog in mouse (Zfp57) and human (ZFP57) has been shown to be required for genomic imprinting at several loci (Li et al. 2008; Mackay et al. 2008). Since imprinting involves maternal-zygotic conflict (e.g., Smith et al. 2006), this process has the potential to drive KZNF duplication and diversification. Finally, the PRDM9 tandem ZF gene is strongly implicated in specification of recombination hotspots (Baudat et al. 2010; Myers et al. 2010; Parvanov et al. 2010). Though recombination hotspots evolve rapidly, the domain structure and evolution of PRDM9 are clearly different from all other tandem ZF genes (Oliver et al. 2009; Thomas et al. 2009), and it has not been subject to the expansion seen in the genes described here. None of these explanations alone suffice to explain the general correlations between genomic LTR retroelement content and tandem ZF coding potential. In contrast, the established potential for host exaptation of retroviral regulatory elements provides a plausible mechanism by which tandem ZF genes initially selected for retroelement repression could, over time, adopt a variety of other host functions.
Methods
Species key
A key for species abbreviations, common names, and genome assemblies is provided in the Supplemental Methods.
Retroelement counts
RepeatMasker data were unavailable for several genomes of interest and misleading for others, apparently because some genomes contain abundant retroelement sequences that do not yet appear in the RepBase sequences used as queries by RepeatMasker (AFA Smit, R Hubley, P Green. 1996–2010. RepeatMasker Open-3.0, http://www.repeatmasker.org). To make counts of retroelements in a manner independent of repeat annotation status and species phylogeny, we used the fact that LTR retroelements are distinguished from all other known sequences by the appearance of a characteristic pattern of conserved coding elements, namely protease, reverse transcriptase, RNaseH, and integrase domains (many retroelements also encode gag and env proteins, but these are poorly conserved across the broad phylogenetic space we wished to analyze). We used patterns of genomic matches to Pfam profiles for these domains to identify LTR retroelements as detailed in the Supplemental Methods.
ZF domain searches
We performed a search for zinc finger domains on selected genomes using the program rpsblast with the –p F option (6-frame translation of DNA query). The search profile consisted of a 28 amino acid weight matrix profile of the ZF domain (including the 7-amino acid linker region upstream of the 21-amino acid ZF core). This profile was generated from the set of functional human tandem ZF proteins using the psiblast program as directed in the NCBI blast documentation. Subsequent analysis showed that this profile is nearly identical to profiles derived from tandem ZF proteins from other species (examples shown in Supplemental Fig. S5). Genome searches were carried out in two forms: one search of the entire genome assembly and a second search of all open reading frame (ORF) segments of 100 codons or longer. From ORF searches, we counted: (1) the number of ORFs with one or more ZF match above some score cutoff, and (2) the total number of ZF matches above some score cutoff. Counts from all searches with various score cutoffs are reported in Supplemental Table S1. To be sure that the ZF domain matches reflect bona fide tandem ZF genes, we determined the ZF domain profile and the number of tandem ZF domains for each genome with large numbers of ZF genes (examples shown in Supplemental Fig. S5). To determine the ZF domain score distribution expected for genes and pseudogenes, all human ZF domain matches were divided into those in known RefSeq genes (plus a few probable genes not yet appearing in RefSeq) (Huntley et al. 2006) and those outside genes (which are likely to belong to pseudogenes and gene fragments). We made a density histogram of the rpsblast scores for each group (representing how well each hit matches the ZF profile) and superimposed the histograms (Supplemental Fig. S2). The pseudogene scores presumably reflect a distribution of times of neutral evolution since pseudogenization. The crossover point where the hit density for genes first exceeds the hit density for pseudogenes occurs at about rpsblast score 57. This crossover point is close to the peak correlations of LTR retroelements and ZF domains (Supplemental Table S1).
Phylogenetic correction by independent contrasts
Comparing two characters in a scatter plot assumes statistical independence, an assumption that is violated when related species are used as data points (Felsenstein 1985; Garland et al. 2005). This can create spurious correlations across broad phylogenies (Whitney and Garland 2010). To account for such phylogenetic concerns we tested our data using Felsenstein's independent contrasts method implemented in the PDAP package (Midford et al. 2005) of Mesquite (Maddison and Maddison 2010). Specifically, we used the positivized x vs. y contrasts (mode 9 in PDAP) to measure the correlation between respective zinc finger metrics and ERV metrics. The tree used in the analysis was based on best estimates for species divergence times derived largely from TimeTree (http://www.timetree.org/; Hedges et al. 2006). Pearson correlations were forced to go through the origin.
Identification of new LTR retroelement families and tandem ZF genes on primate branches
Phylogenetic branches on which new LTR retroelement families were added were determined using a combination of RepeatMasker annotations, shared insertion site analysis, and DNA trees of retroelement internal sequences (Supplemental Methods; Supplemental Fig. S4; Supplemental Table S4). Branches on which new tandem ZF genes were added were determined by identifying primate orthologs of human tandem ZF genes and determining the first appearance of each gene (Supplemental Methods; Supplemental Table S5).
Identification of closest human gene pairs and their orthologs in other species
Pairs of human duplicate KZNF genes were identified as reciprocal best blastp matches or as neighbors on a pairwise distance tree among all human KZNF genes, with further tests to eliminate unclear gene pairs (Supplemental Methods). For each pair of human duplicate genes, a TBLASTN pipeline was used to identify all close relatives in other genome assemblies (Supplemental Methods). Final analysis of candidate sequences was based on maximum-likelihood trees using protein sequence (Supplemental Methods). Most analysis was carried out on partial genes corresponding to the human ZF exon, and thus lacking the KRAB domain. Many identified gene candidates did not correspond to “UCSC known”, RefSeq, or Ensembl predicted genes. To be sure that the orthologs identified in nonhuman genomes are bona fide candidate KZNF genes, we used a form of chained TBLASTN search for all coding exons (Supplemental Methods).
Rate of change in orthologous fingers
We measured the relative rates of change of specific amino acid residues in orthologous KZNF domains as follows. A total of 11 KZNF gene pairs were arbitrarily selected from among the duplicate pairs with an indeterminate ancestral gene. For each pair, all available orthologs were gathered from the five primates: cow, dog, and horse. The 22 genes encoded a total of 280 ZF domains. Each orthologous finger protein set was “aligned” (these are gap-free alignments since all fingers conformed to the standard 28-amino acid finger domain), and PhyML 3.0 was used to estimate rates of change at each site (JTT matrix, 20 rate categories, gamma parameter 1.0) (Guindon and Gascuel 2003; Guindon et al. 2009). The “lk” output file from PhyML gives the likelihood that each site belongs in each of the 20 estimated rate categories (Anisimova and Gascuel 2006). The peak likelihood rate value was extracted for each position in each ZF domain. These rates were averaged across all 280 orthologous finger groups. Note that this method does not measure the absolute divergence of the sequences, which varied from gene to gene depending on available orthologs. Since all 28 positions were present in all aligned finger sets, this method does produce an average estimate of the relative rates of change at each ZF site, as plotted in Figure 4.
Asymmetry calculation
For duplicate KZNF genes with an identified ancestral gene, asymmetry of divergence between the two duplicates was computed as follows. For each aligned site, the ancestral state was inferred when all copies of the ancestral gene (the single-copy gene present in early branching species) encoded the same amino acid. At such sites, when all copies of each of the two duplicate genes encoded the same amino acid and at least one gene diverged from the ancestral state (i.e., the site changed and was conserved among orthologs), the site was counted as informative. When duplicate copy 1 (the copy overall most similar to the ancestral state) was divergent, the site received a score of −1; when duplicate copy 2 was divergent, the site received a score of +1; when both were divergent, the site received a score of 0. When averaged across all informative sites, the expected score is 0 if divergence is perfectly symmetric and the expected score is 1 if divergence is perfectly asymmetric.
Tests for positive selection
For species-specific expansion analysis, clades of species-specific tandem ZF exons were collected and analyzed by site models 7, 8, and 8A implemented in codeml PAML 3.15 (Yang et al. 2005; Zhang et al. 2005). Additional details are described in Emerson and Thomas (2009). Results and statistical tests are shown in Supplemental Table S8. Strong evidence of positive selection was detected in 30 of 35 clades. To determine the types of protein sites subject to positive selection, ZF sites with Bayes-Empirical-Bayes P-values of 0.98 or higher were counted, summing over all the clades. Counts for each of the 28 classes of ZF sites are shown in Supplemental Figure S6. For each human duplicate gene pair, branch-site models implemented in codeml were applied to test for branch-specific positive selection (Yang et al. 2005; Zhang et al. 2005). For the control model A1, the foreground dN/dS was constrained to be 1.0 on all branches of the tree, whereas for selection model A, the foreground dN/dS was allowed to differ on the branch joining the duplicate copies. Statistical analysis was based on a χ2 test of twice the difference in log likelihoods between the two models with one degree of freedom. Specific codeml output details and statistics are shown in Supplemental Table S9.
Acknowledgments
We thank Ryan Emerson and members of the Swanson lab for discussions of the results. This work received no external funding.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.121749.111.
References
- Anisimova M, Gascuel O 2006. Approximate likelihood-ratio test for branches: A fast, accurate, and powerful alternative. Syst Biol 55: 539–552 [DOI] [PubMed] [Google Scholar]
- Anisimova M, Bielawski JP, Yang Z 2001. Accuracy and power of the likelihood ratio test in detecting adaptive molecular evolution. Mol Biol Evol 18: 1585–1592 [DOI] [PubMed] [Google Scholar]
- Basta HA, Buzak AJ, McClure MA 2007. Identification of novel retroid agents in Danio rerio, Oryzias latipes, Gasterosteus aculeatus and Tetraodon nigroviridis. Evol Bioinform Online 3: 179–195 [PMC free article] [PubMed] [Google Scholar]
- Basta HA, Cleveland SB, Clinton RA, Dimitrov AG, McClure MA 2009. Evolution of teleost fish retroviruses: characterization of new retroviruses with cellular genes. J Virol 83: 10152–10162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudat F, Buard J, Grey C, Fledel-Alon A, Ober C, Przeworski M, Coop G, de Massy B 2010. PRDM9 is a major determinant of meiotic recombination hotspots in humans and mice. Science 327: 836–840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellefroid EJ, Poncelet DA, Lecocq PJ, Revelant O, Martial JA 1991. The evolutionarily conserved Kruppel-associated box domain defines a subfamily of eukaryotic multifingered proteins. Proc Natl Acad Sci 88: 3608–3612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belshaw R, Pereira V, Katzourakis A, Talbot G, Paces J, Burt A, Tristem M 2004. Long-term reinfection of the human genome by endogenous retroviruses. Proc Natl Acad Sci 101: 4894–4899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belshaw R, Katzourakis A, Paces J, Burt A, Tristem M 2005. High copy number in human endogenous retrovirus families is associated with copying mechanisms in addition to reinfection. Mol Biol Evol 22: 814–817 [DOI] [PubMed] [Google Scholar]
- Benveniste RE, Todaro GJ 1974. Evolution of C-type viral genes: inheritance of exogenously acquired viral genes. Nature 252: 456–459 [DOI] [PubMed] [Google Scholar]
- Bergthorsson U, Andersson DI, Roth JR 2007. Ohno's dilemma: evolution of new genes under continuous selection. Proc Natl Acad Sci 104: 17004–17009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Best S, Le Tissier P, Towers G, Stoye JP 1996. Positional cloning of the mouse retrovirus restriction gene Fv1. Nature 382: 826–829 [DOI] [PubMed] [Google Scholar]
- Birtle Z, Ponting CP 2006. Meisetz and the birth of the KRAB motif. Bioinformatics 22: 2841–2845 [DOI] [PubMed] [Google Scholar]
- Blikstad V, Benachenhou F, Sperber GO, Blomberg J 2008. Evolution of human endogenous retroviral sequences: a conceptual account. Cell Mol Life Sci 65: 3348–3365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z, Telfer P, Gettie A, Reed P, Zhang L, Ho D, Marx PA 1996. Genetic characterization of new West African simian immunodeficiency virus SIVsm: geographic clustering of household-derived SIV strains with human immunodeficiency virus type 2 subtypes and genetically diverse viruses from a single feral sooty mangabey troop. J Virol 70: 3617–3627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen CJ, Lock WM, Mager DL 2009. Endogenous retroviral LTRs as promoters for human genes: a critical assessment. Gene 448: 105–114 [DOI] [PubMed] [Google Scholar]
- Conley AB, Piriyapongsa J, Jordan IK 2008. Retroviral promoters in the human genome. Bioinformatics 15: 1563–1567 [DOI] [PubMed] [Google Scholar]
- Copeland NG, Hutchison KW, Jenkins NA 1983. Excision of the DBA ecotropic provirus in dilute coat-color revertants of mice occurs by homologous recombination involving the viral LTRs. Cell 33: 379–387 [DOI] [PubMed] [Google Scholar]
- de Parseval N, Heidmann T 2005. Human endogenous retroviruses: from infectious elements to human genes. Cytogenet Genome Res 110: 318–332 [DOI] [PubMed] [Google Scholar]
- Doolittle RF, Feng DF 1992. Tracing the origin of retroviruses. Curr Top Microbiol Immunol 176: 195–211 [DOI] [PubMed] [Google Scholar]
- Dupressoir A, Vernochet C, Bawa O, Harper F, Pierron G, Opolon P, Heidmann T 2009. Syncytin-A knockout mice demonstrate the critical role in placentation of a fusogenic, endogenous retrovirus-derived, envelope gene. Proc Natl Acad Sci 106: 12127–12132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edelstein LC, Collins T 2005. The SCAN domain family of zinc finger transcription factors. Gene 359: 1–17 [DOI] [PubMed] [Google Scholar]
- Emerson RO, Thomas JH 2009. Adaptive evolution in zinc finger transcription factors. PLoS Genet 5: e1000325 doi: 10.1371/journal.pgen.1000325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J 1985. Phylogenies and the comparative method. Am Nat 125: 1–15 [Google Scholar]
- Gao F, Bailes E, Robertson DL, Chen Y, Rodenburg CM, Michael SF, Cummins LB, Arthur LO, Peeters M, Shaw GM, et al. 1999. Origin of HIV-1 in the chimpanzee Pan troglodytes troglodytes. Nature 397: 436–441 [DOI] [PubMed] [Google Scholar]
- García-García MJ, Shibata M, Anderson KV 2008. Chato, a KRAB zinc-finger protein, regulates convergent extension in the mouse embryo. Development 135: 3053–3062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garland T Jr, Bennett AF, Rezende EL 2005. Phylogenetic approaches in comparative physiology. J Exp Biol 208: 3015–3035 [DOI] [PubMed] [Google Scholar]
- Gogvadze E, Buzdin A 2009. Retroelements and their impact on genome evolution and functioning. Cell Mol Life Sci 66: 3727–3742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groner AC, Meylan S, Ciuffi A, Zangger N, Ambrosini G, Dénervaud N, Bucher P, Trono D 2010. KRAB-zinc finger proteins and KAP1 can mediate long-range transcriptional repression through heterochromatin spreading. PLoS Genet 6: e1000869 doi: 10.1371/journal.pgen.1000869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S, Gascuel O 2003. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol 52: 696–704 [DOI] [PubMed] [Google Scholar]
- Guindon S, Delsuc F, Dufayard JF, Gascuel O 2009. Estimating maximum likelihood phylogenies with PhyML. Methods Mol Biol 537: 113–137 [DOI] [PubMed] [Google Scholar]
- Hamilton AT, Huntley S, Kim J, Branscomb E, Stubbs L 2003. Lineage-specific expansion of KRAB zinc-finger transcription factor genes: implications for the evolution of vertebrate regulatory networks. Cold Spring Harb Symp Quant Biol 68: 131–140 [DOI] [PubMed] [Google Scholar]
- Hayden S, Bekaert M, Crider TA, Mariani S, Murphy WJ, Teeling EC 2010. Ecological adaptation determines functional mammalian olfactory subgenomes. Genome Res 20:1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedges SB, Dudley J, Kumar S 2006. TimeTree: a public knowledge-base of divergence times among organisms. Bioinformatics 22: 2971–2972 [DOI] [PubMed] [Google Scholar]
- Huntley S, Baggott DM, Hamilton AT, Tran-Gyamfi M, Yang S, Kim J, Gordon L, Branscomb E, Stubbs L 2006. A comprehensive catalog of human KRAB-associated zinc finger genes: insights into the evolutionary history of a large family of transcriptional repressors. Genome Res 16: 669–677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jern P, Sperber GO, Blomberg J 2006. Divergent patterns of recent retroviral integrations in the human and chimpanzee genomes: probable transmissions between other primates and chimpanzees. J Virol 80: 1367–1375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson WE, Coffin JM 1999. Constructing primate phylogenies from ancient retrovirus sequences. Proc Natl Acad Sci 96: 10254–10260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim CA, Berg JM 1996. A 2.2 A resolution crystal structure of a designed zinc finger protein bound to DNA. Nat Struct Biol 3: 940–945 [DOI] [PubMed] [Google Scholar]
- Krebs CJ, Larkins LK, Khan SM, Robins DM 2005. Expansion and diversification of KRAB zinc-finger genes within a cluster including Regulator of sex-limitation 1 and 2. Genomics 85: 752–761 [DOI] [PubMed] [Google Scholar]
- Kumar S, Hedges SB 1998. A molecular timescale for vertebrate evolution. Nature 392: 917–920 [DOI] [PubMed] [Google Scholar]
- Laten HM, Majumdar A, Gaucher EA 1998. SIRE-1, a copia/Ty1-like retroelement from soybean, encodes a retroviral envelope-like protein. Proc Natl Acad Sci 95: 6897–6902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechner MS, Begg GE, Speicher DW, Rauscher FJ III 2000. Molecular determinants for targeting heterochromatin protein 1-mediated gene silencing: direct chromoshadow domain-KAP-1 corepressor interaction is essential. Mol Cell Biol 20: 6449–6465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Ito M, Zhou F, Youngson N, Zuo X, Leder P, Ferguson-Smith AC 2008. A maternal-zygotic effect gene, Zfp57, maintains both maternal and paternal imprints. Dev Cell 15: 547–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Looman C, Abrink M, Mark C, Hellman L 2002. KRAB zinc finger proteins: an analysis of the molecular mechanisms governing their increase in numbers and complexity during evolution. Mol Biol Evol 19: 2118–2130 [DOI] [PubMed] [Google Scholar]
- Lowe CB, Bejerano G, Haussler D 2007. Thousands of human mobile element fragments undergo strong purifying selection near developmental genes. Proc Natl Acad Sci 104: 8005–8010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay DJ, Callaway JL, Marks SM, White HE, Acerini CL, Boonen SE, Dayanikli P, Firth HV, Goodship JA, Haemers AP, et al. 2008. Hypomethylation of multiple imprinted loci in individuals with transient neonatal diabetes is associated with mutations in ZFP57. Nat Genet 40: 949–951 [DOI] [PubMed] [Google Scholar]
- Maddison WP, Maddison DR 2010. Mesquite: A modular system for evolutionary analysis. Version 1.1. http://mesquiteproject.org
- Malik HS, Henikoff S, Eickbush TH 2000. Poised for contagion: Evolutionary origins of the infectious abilities of invertebrate retroviruses. Genome Res 10: 1307–1318 [DOI] [PubMed] [Google Scholar]
- Martin J, Herniou E, Cook J, O'Neill RW, Tristem M 1999. Interclass transmission and phyletic host tracking in murine leukemia virus-related retroviruses. J Virol 73: 2442–2449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui T, Leung D, Miyashita H, Maksakova IA, Miyachi H, Kimura H, Tachibana M, Lorincz MC, Shinkai Y 2010. Proviral silencing in embryonic stem cells requires the histone methyltransferase ESET. Nature 464: 927–931 [DOI] [PubMed] [Google Scholar]
- Midford PE, Garland T Jr, Maddison WP 2005. PDAP Package of Mesquite. Version 1.07. http://mesquiteproject.org/pdap_mesquite/
- Mikkelsen TS, Wakefield MJ, Aken B, Amemiya CT, Chang JL, Duke S, Garber M, Gentles AJ, Goodstadt L, Heger A, et al. 2007. Genome of the marsupial Monodelphis domestica reveals innovation in non-coding sequences. Nature 447: 167–177 [DOI] [PubMed] [Google Scholar]
- Mouse Genome Sequencing Consortium 2002. Initial sequencing and comparative analysis of the mouse genome. Nature 420: 520–562 [DOI] [PubMed] [Google Scholar]
- Myers S, Bowden R, Tumian A, Bontrop RE, Freeman C, MacFie TS, McVean G, Donnelly P 2010. Drive against hotspot motifs in primates implicates the PRDM9 gene in meiotic recombination. Science 327: 876–879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen AL, Ortiz JA, You J, Oulad-Abdelghani M, Khechumian R, Gansmuller A, Chambon P, Losson R 1999. Interaction with members of the heterochromatin protein 1 (HP1) family and histone deacetylation are differentially involved in transcriptional silencing by members of the TIF1 family. EMBO J 18: 6385–6395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowick K, Gernat T, Almaas E, Stubbs L 2009. Differences in human and chimpanzee gene expression patterns define an evolving network of transcription factors in brain. Proc Natl Acad Sci 106: 22358–22363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowick K, Hamilton AT, Zhang H, Stubbs L 2010. Rapid sequence and expression divergence suggests selection for novel function in primate-specific KRAB-ZNF genes. Mol Biol Evol 27: 2606–2617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver PL, Goodstadt L, Bayes JJ, Birtle Z, Roach KC, Phadnis N, Beatson SA, Lunter G, Malik HS, Ponting CP 2009. Accelerated evolution of the Prdm9 speciation gene across diverse metazoan taxa. PLoS Genet 5: e1000753 doi: 10.1371/journal.pgen.1000753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parvanov ED, Petkov PM, Paigen K 2010. Prdm9 controls activation of mammalian recombination hotspots. Science 327: 835 doi: 10.1126/science.1181495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavletich NP, Pabo CO 1991. Zinc finger-DNA recognition: crystal structure of a Zif268-DNA complex at 2.1 A. Science 252: 809–817 [DOI] [PubMed] [Google Scholar]
- Polavarapu N, Bowen NJ, McDonald JF 2006. Identification, characterization and comparative genomics of chimpanzee endogenous retroviruses. Genome Biol 7: R51 doi: 10.1186/gb-2006-7-6-r51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhesus Macaque Genome Sequencing and Analysis Consortium. 2007. Evolutionary and biomedical insights from the rhesus macaque genome. Science 316: 222–234 [DOI] [PubMed] [Google Scholar]
- Ribet D, Harper F, Dupressoir A, Dewannieux M, Pierron G, Heidmann T 2008. An infectious progenitor for the murine IAP retrotransposon: Emergence of an intracellular genetic parasite from an ancient retrovirus. Genome Res 18: 597–609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowe HM, Trono D 2011. Dynamic control of endogenous retroviruses during development. Virology 411: 273–287 [DOI] [PubMed] [Google Scholar]
- Rowe HM, Jakobsson J, Mesnard D, Rougemont J, Reynard S, Aktas T, Maillard PV, Layard-Liesching H, Verp S, Marquis J, et al. 2010. KAP1 controls endogenous retroviruses in embryonic stem cells. Nature 463: 237–240 [DOI] [PubMed] [Google Scholar]
- Ryan RF, Schultz DC, Ayyanathan K, Singh PB, Friedman JR, Fredericks WJ, Rauscher FJ 3rd 1999. KAP-1 corepressor protein interacts and colocalizes with heterochromatic and euchromatic HP1 proteins: a potential role for Krüppel-associated box-zinc finger proteins in heterochromatin-mediated gene silencing. Mol Cell Biol 19: 4366–4378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt D, Durrett R 2004. Adaptive evolution drives the diversification of zinc-finger binding domains. Mol Biol Evol 21: 2326–2339 [DOI] [PubMed] [Google Scholar]
- Schultz DC, Ayyanathan K, Negorev D, Maul GG, Rauscher FJ 3rd 2002. SETDB1: A novel KAP-1-associated histone H3, lysine 9-specific methyltransferase that contributes to HP1-mediated silencing of euchromatic genes by KRAB zinc-finger proteins. Genes Dev 16: 919–932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon M, Hamilton AT, Gordon L, Branscomb E, Stubbs L 2003. Differential expansion of zinc-finger transcription factor loci in homologous human and mouse gene clusters. Genome Res 13: 1097–1110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith FM, Garfield AS, Ward A 2006. Regulation of growth and metabolism by imprinted genes. Cytogenet Genome Res 113: 279–291 [DOI] [PubMed] [Google Scholar]
- Sripathy SP, Stevens J, Schultz DC 2006. The KAP1 corepressor functions to coordinate the assembly of de novo HP1-demarcated microenvironments of heterochromatin required for KRAB zinc finger protein-mediated transcriptional repression. Mol Cell Biol 26: 8623–8638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stocking C, Kozak CA 2008. Murine endogenous retroviruses. Cell Mol Life Sci 65: 3383–3398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas JH, Emerson RO, Shendure J 2009. Extraordinary molecular evolution in the PRDM9 fertility gene. PLoS ONE 30: e8505 doi: 10.1371/journal.pone.0008505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitney KD, Garland T Jr 2010. Did genetic drift drive increases in genome complexity? PLoS Genet 6: e1001080 doi: 10.1371/journal.pgen.1001080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf D, Goff SP 2007. TRIM28 mediates primer binding site-targeted silencing of murine leukemia virus in embryonic cells. Cell 131: 46–57 [DOI] [PubMed] [Google Scholar]
- Wolf D, Goff SP 2008. Host restriction factors blocking retroviral replication. Annu Rev Genet 42: 143–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf D, Goff SP 2009. Embryonic stem cells use ZFP809 to silence retroviral DNAs. Nature 458: 1201–1204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Wong WS, Nielsen R 2005. Bayes empirical bayes inference of amino acid sites under positive selection. Mol Biol Evol 22: 1107–1118 [DOI] [PubMed] [Google Scholar]
- Zhang J, Nielsen R, Yang Z 2005. Evaluation of an improved branch-site likelihood method for detecting positive selection at the molecular level. Mol Biol Evol 22: 2472–2479 [DOI] [PubMed] [Google Scholar]