Abstract
Heterochromatin is believed to be associated with increased levels of cytosine methylation. With the recent availability of genome-wide, high-resolution molecular data reflecting chromatin organization and methylation, such relationships can be explored systematically. As well-defined surrogates for heterochromatin, we tested the relationship between DNA replication timing and DNase hypersensitivity with cytosine methylation in two human cell types, unexpectedly finding the later-replicating, more heterochromatic regions to be less methylated than early replicating regions. When we integrated gene-expression data into the study, we found that regions of increased gene expression were earlier replicating, as previously identified, and that transcription-targeted cytosine methylation in gene bodies contributes to the positive correlation with early replication. A self-organizing map (SOM) approach was able to identify genomic regions with early replication and increased methylation, but lacking annotated transcripts, loci missed in simple two variable analyses, possibly encoding unrecognized intergenic transcripts. We conclude that the relationship of cytosine methylation with heterochromatin is not simple and depends on whether the genomic context is tandemly repetitive sequences often found near centromeres, which are known to be heterochromatic and methylated, or the remaining majority of the genome, where cytosine methylation is targeted preferentially to the transcriptionally active, euchromatic compartment of the genome.
The original definition of heterochromatin was wholly derived from cytological studies, identifying it as unusually compacted nuclear material as opposed to the less-condensed euchromatin. Heterochromatin in the eukaryotic genome is subclassified as facultative and constitutive. Whereas sites of constitutive heterochromatin are present in all cell types (e.g., centromeres, G bands) (Holmquist 1989), facultative heterochromatin can be present at different loci in different cell types (e.g., X inactivation in female mammalian cells). Both types of heterochromatin are usually associated with transcriptional silencing, although there is a minority of genes that transcribes preferentially in a heterochromatic context (Vogel et al. 2006). Apart from transcriptional repression, heterochromatin has other functional properties, including associations with centromeres and telomeres and a role in sister chromatid cohesion (Gartenberg 2009). Decades of cytological studies have also characterized heterochromatin by its late-replication timing within the cell cycle (Gilbert 2002). From a molecular point of view, certain proteins (e.g., HP1) (Fanti and Pimpinelli 2008) or post-translational variants (e.g., H3K9me3) (Krauss 2008) characterize heterochromatin. Methyl-binding domain proteins have also been found to accumulate in pericentromeric satellite DNA sequences in mouse (Hendrich and Bird 1998), the type of sequences at which the effects of DNA methyltransferase 3B (DNMT3B) mutations cause loss of cytosine methylation and local decondensation of the heterochromatin (Hansen et al. 1999).
It is believed that the DNA within heterochromatin is highly methylated, the cytosine methylation acting synergistically with chromatin modifications characteristic of heterochromatin, and a repository for transposons maintained in a silent state (Henikoff 2000). However, there is reason to question the association of cytosine methylation with facultative heterochromatin formation, as it has been recognized for some time that the inactive X chromosome in females is globally hypomethylated (Bernardino-Sgherri et al. 2002), possibly related to the decreased methylation in bodies of genes (Hellman and Chess 2007) silenced as part of the X inactivation process.
Now that we have genome-wide and high-resolution maps of DNA-replication timing and chromatin constituents characteristic of heterochromatin, we can study the relationships of heterochromatin with other genomic properties more quantitatively. To study the relationship of cytosine methylation with heterochromatin, we chose to use DNA replication timing (Gilbert 2002) and DNase hypersensitivity as well-characterized indicators of heterochromatin. Our studies of two human cell lines revealed a paradoxical relationship between early DNA replication or increased DNase hypersensitivity, defining euchromatic DNA and increased cytosine methylation, attributable in part to the targeting of cytosine methylation to actively transcribed gene bodies. We conclude that the highly repetitive juxtacentromeric sequences of the human genome represent a special situation, and that in the remainder of the human genome, heterochromatin is less methylated than euchromatin.
Results
Cytosine methylation and replication timing
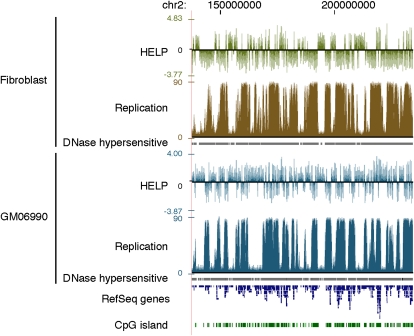
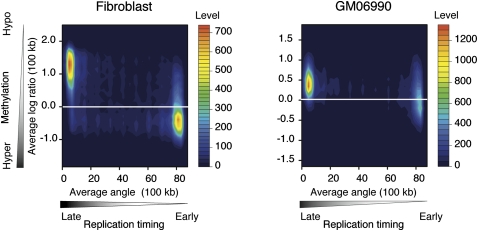
In Figure 1, we show a graphical representation of cytosine methylation status and replication timing in both cell types studied. Visually, the hypomethylated areas appear to be undergoing late replication, whereas the relatively hypermethylated, more gene-rich areas are early replicating. This observation is the opposite of what we initially expected, and motivated us to analyze systematically the correlation between cytosine methylation and replication timing. Figure 2 shows a contour plot illustrating the relationship between cytosine methylation and replication timing in 100-kb windows. The data for both fibroblast and GM06990 cells show a correlation between cytosine hypomethylation and late replication, and vice versa (R2 = −0.574 and R2 = −0.334, respectively). The weaker correlation observed in the GM06990 lymphoblastoid cell line may be explained by greater global hypomethylation in these cells compared with the fibroblast cell line.
Figure 1.
Cytosine methylation and replication timing correlate in broad genomic regions. (Top) Fibroblast data; (bottom) GM06690 lymphoblastoid cell line data. Cytosine methylation is shown as the HpaII/MspI log2 intensity ratio from the HELP assay. Positive values indicate relative hypomethylation, and negative values indicate hypermethylation of HpaII sites. DNA replication timing data are generated from raw sequence reads by an arctangent transformation of 1-kb counts comparing early (G1 and S1) and late (S4 and G2) cell samples, as described in the Methods section. Earlier replicated regions have higher values than later replicated regions.
Figure 2.
DNA hypermethylation correlates with early DNA replication timing. Filled contour plots were drawn with two-dimensional histograms. Cytosine methylation data and replication timing data are averaged in 100-kb sliding windows. The cumulative numbers of observations are shown as color-coded levels to generate the contours. Early replicated regions are more methylated in both the fibroblast and lymphoblastoid cell types.
Cytosine methylation and gene expression
Gene body cytosine methylation has previously been correlated with the transcriptional activity of genes (Zhang et al. 2006; Zilberman et al. 2007; Backdahl et al. 2009; Ball et al. 2009). Of these prior studies, Ball et al. (2009) used the same lymphoblastoid cell line that we describe here as well as human fibroblasts. This observation suggests that cytosine hypermethylation associated with early DNA replication may occur within actively transcribed genes. We therefore tested the relationship between cytosine methylation status and gene expression status using our own datasets to see whether we could reproduce the Ball et al. (2009) observations. We tested the cytosine methylation and gene expression values for each RefSeq gene annotated in the human genome, looking separately at promoters and gene bodies. We found the promoter regions to be generally hypomethylated regardless of the gene-expression status, but consistent methylation of gene bodies was found only in actively transcribed genes (Supplemental Fig. 3). We confirmed these genome-wide relationships by locus-specific validation studies, with the results for two representative loci shown in Supplemental Figure 4 and data from further validated loci listed in Supplemental Table 3.
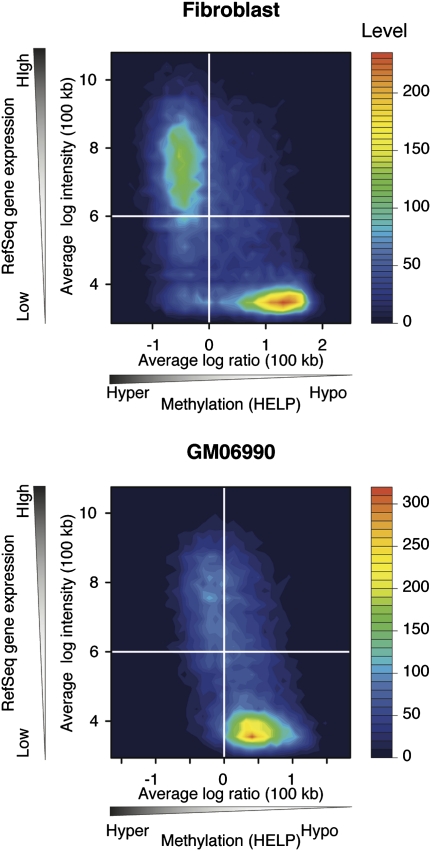
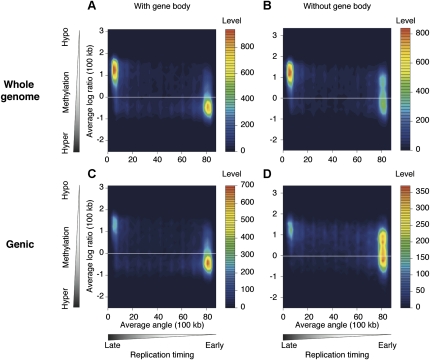
A 100-kb sliding window approach also showed the correlation of gene expression and cytosine methylation, with actively transcribed regions being relatively hypermethylated, and transcriptionally inactive regions being hypomethylated (Fig. 3). We tested to see whether the hypermethylation of regions containing actively transcribed genes is solely due to the targeting of cytosine methylation to active gene bodies, or whether there is also increased cytosine methylation at intergenic loci in these highly transcribed regions. When we excluded the 100-kb genomic windows that do not include annotated genes (∼50% of the windows genome wide) and tested what happened to methylation in the gene-containing windows with the removal of gene body data, we found that the proportion of windows with overall hypomethylation (log2 ratio HpaII/MspI>0) increases from 32.4% to 64.6%. This indicates that the hypermethylation in regions of early replication is substantially, but not solely due to targeting of bodies of annotated, actively transcribed genes (Fig. 4; Supplemental Fig. 5).
Figure 3.
Broad correlation exists between DNA hypermethylation and actively transcribed gene regions. Extending the analysis of Supplemental Figure 3 to a 100-kb sliding window representation continues to show the relationship between increased gene expression and hypermethylation of DNA. A two-dimensional histogram of the averaged HpaII/MspI log2 ratio in 100-kb windows and averaged signal intensities of the genes are represented by filled contour plots.
Figure 4.
The hypermethylation of early-replicating regions is predominantly due to gene-body hypermethylation. We tested how gene-body methylation could be contributing to the patterns shown in Figure 2, reproducing the fibroblast plot to facilitate comparison in A. C shows the results when 100-kb windows that do not contain genes are removed, with a decrease in the late-replicating/hypomethylated population of signals. Excluding gene bodies, to study only intergenic methylation, generates a shift in signal distribution toward hypomethylated DNA (B), especially when the analysis is restricted to the gene-containing regions of the genome (D). These results show that a substantial proportion of the correlation of cytosine methylation with early replication is due to the methylation targeting transcribed sequences.
Gene expression and replication timing
The preceding results indicate that we should expect to see a positive correlation between gene expression and early replication. We tested this formally, showing the results in Supplemental Figure 6. Again, using the 100-kb sliding window approach, we show that the actively transcribed regions are replicated earlier, and that a small subset of early replicating regions contain relatively inactive loci (Supplemental Fig. 6). We also show that late-replicating regions are largely inactive in this 100-kb context. This result is concordant with previous observations in organisms from insects (Schubeler et al. 2002) to mammals (Desprat et al. 2009).
DNase hypersensitivity and replication timing
Although the replication timing data were processed differently in this study, we confirmed our previous correlation (Hansen et al. 2010) of increased DNase hypersensitivity with earlier DNA replication (Supplemental Fig. 7).
Self-organizing map analysis
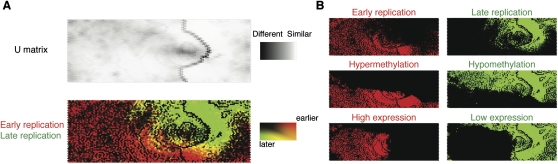
While global correlations are indicative of relationships of genomic processes, these kinds of analyses may fail to reveal the presence of a subset of genomic regions diverging from overall genome-wide relationships. We therefore applied a self-organizing map (SOM) analysis, wherein we were able to define distinct subsets of loci that posses similar properties. The SOM is a lossless unbiased clustering method that projects high-dimensional data onto a two-dimensional map, while at the same time preserving the topology of the data. To perform an unbiased test of the relationship between replication timing and other vector elements, we excluded the replication timing data from the training process. Following the construction of the SOM, the replication timing data were tagged on all vectors and overlaid to visualize whether the variables used to build the SOM predicted replication timing. For example, as a negative control experiment, we examined vectors comprising only the number of RefSeq genes and HAFs in 100-kb windows and were unable to observe any discernible clustering in the U-matrix or separation of replication timing data, as expected (Supplemental Fig. 8). However, adding gene expression, CpG island number, and cytosine methylation data to the vectors provided sufficient information to enable the data to separate into two distinct clusters enriched in vectors tagged to show either early or late replication (data not shown). We illustrate the performance of all five variables in Figure 5, which shows two distinct clusters of loci exhibiting alternative replication timing patterns. These two clusters mainly consist of loci with early replication/hypermethylation/high gene expression or late replication/hypomethylation/low expression (Fig. 5B), as would be predicted by our previous analyses. However, the SOM analysis was also able to identify a new group of loci, where early replication and hypermethylation were found in regions with unexpectedly low levels of gene expression (Fig. 6A). These low-expression loci consisted not only of regions containing genes expressed at low levels, but also regions lacking any annotated RefSeq genes, Gencode genes (Harrow et al. 2006), or expressed sequence tags (ESTs, ≤5) (Fig. 6B). Adding DNase hypersensitivity data (Hansen et al. 2010) revealed a substantial proportion of these regions to be nuclease accessible, indicating these to be euchromatically organized, but retaining the increased methylation pattern of the remainder of the early replicating regions (Fig. 6B).
Figure 5.
A self-organizing map analysis correlates DNA replication with methylation and transcription patterns. In this self-organizing map (SOM) representation of the multivariate data set, the top panel shows a U matrix representation of the map derived from genome-wide DNA methylation log2 ratios, RefSeq gene expression, RefSeq gene number, CpG island number, and HpaII-amplifiable fragment number in each 100-kb window. Each node is shaded using a linear grayscale that represents the mean Euclidean distance of that node vector relative to its immediate neighbors on the map ([white] most similar; [black] least similar). Overlaying loci with information about late (green) and early (red) replication shows that the parameters tested are predictive of replication timing, as evidenced by the clear separation of the red and green regions (A). We break out some of the variables used in generating the SOM (cytosine methylation, gene expression) to illustrate their overall correlations with DNA replication (B).
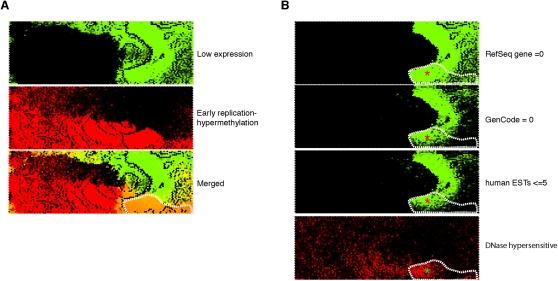
Figure 6.
Identification of a genomic compartment where early replication and cytosine hypermethylation occur at nongenic regions. To highlight the loci where gene expression appeared to be behaving discordantly from the overall relationship with DNA replication and methylation, we represented early replicating and hypermethylated loci in red and low-expressing loci in green to illustrate these loci in the merged plot as orange (outlined in A). In B we show that a substantial proportion of these loci (area marked with red asterisks) have neither RefSeq nor Gencode genes annotated, nor even the lowest quintile of EST densities annotated for the UCSC Genome Browser. These loci are not only lacking any measurable gene expression, they do not even have any evidence for any transcriptional potential, regions usually referred to as gene deserts but with DNA-replication characteristics and DNase hypersensitivity (bottom right, green asterisk) that may indicate noncoding, nonprocessed transcription.
Discussion
To explore the relationship between heterochromatin and cytosine methylation, we integrated the results of three genome-wide assays and a number of genomic sequence feature annotations. The Repli-seq assay maps DNA-replication timing (Hansen et al. 2010), while cytosine methylation was measured using the HELP assay (Khulan et al. 2006; Oda et al. 2009) and gene expression by microarray studies in two human cell lines. Our results show a strong correlation between cytosine methylation and DNA replication timing genome wide.
However, this correlation is the opposite of what might have been expected, as the late-replicating compartment of the genome, which functionally defines heterochromatin, is less methylated than the early replicating, euchromatic compartment. Our prior studies showing regions of increased DNase hypersensitivity correlating well with early replication (Hansen et al. 2010) supports the link between euchromatin and early replication. The finding of increased methylation associated with euchromatin is paradoxical for a number of reasons, including the fact that cytosine methylation is a known repressive mark in the context of gene promoters, and heterochromatin is likewise a repressive environment for gene transcription, while loci such as pericentromeric satellite DNA (Gopalakrishnan et al. 2009) are classic examples of heterochromatically organized DNA at cytological resolution (Plohl et al. 2008) known to be hypermethylated in vivo (Hassan et al. 2001). However, any assumption that increased cytosine methylation is a universal feature of heterochromatic DNA may be questionable based on prior studies. In the case of mammalian X chromosome inactivation, one copy of the two X chromosomes is inactivated in female cells. During S phase, the active X chromosome replicates early and the inactive X chromosome late (Gribnau et al. 2005), consistent with the heterochromatic organization of the inactive X (Chow and Brown 2003). The inactivated X chromosome has been reported to be globally hypomethylated (Viegas-Pequignot et al. 1988), and the gene bodies on inactive X chromosomes have been found to be hypomethylated (Hellman and Chess 2007) despite the increased cytosine methylation at promoters causing gene inactivation of the inactive X (Zeschnigk et al. 2009). As the physical amount of sequence occupied by gene bodies vastly exceeds promoters, the bulk effect of gene-body hypomethylation on the inactive X chromosome is consistent with the observation of global hypomethylation of the inactive X using methylation-sensitive restriction enzymes (Viegas-Pequignot et al. 1988). Extending this approach to the whole genome, a comparison of cytogenetic patterns obtained from the digestion of human chromosomes in situ with methylation-insensitive MspI and methylation-sensitive HpaII revealed that R bands (gene-rich, euchromatic) are relatively methylated, whereas heterochromatic blocks of sequence can be strikingly unmethylated (Fernandez-Peralta et al. 1994). Our results associating increased methylation with earlier replication are therefore not inconsistent with prior observations.
The paradoxical relationship between increased cytosine methylation and early DNA replication is consistent with another recent report (Aran et al. 2011). Both studies concur that increased cytosine methylation in early replicating regions is substantially but not solely attributable to transcription-targeted cytosine methylation, in our case shown by the depletion of methylated loci in early replicating regions when we remove RefSeq gene bodies from the analysis (Fig. 4), and in Supplemental Figure 5C relative correlation coefficient values when genes are included or excluded from the analysis in a manner similar to Aran et al. (2011). It is apparent from these analyses that the exclusion of gene bodies does not remove all loci with increased methylation in early replicating regions, raising the question as to why these supposedly euchromatic genomic compartments have methylation also targeted to intergenic regions. Our use of the SOM approach revealed that some of the early replicating, highly methylated loci in the genome are devoid of annotated genes and at the lowest quintile of EST density, but retain the DNase hypersensitivity of euchromatin (Fig. 6). These SOM-defined regions, which would have been difficult to identify through the preceding two-variable comparisons, are candidates for being transcribed as nonannotated, noncoding RNAs in these cell types, causing targeting of cytosine methylation and associated with early replication of DNA. Based on these SOM findings, it is possible that a similar phenomenon of nonannotated transcription occurs in gene-containing regions and helps to account for the remaining cytosine methylation when annotated gene bodies are removed from early replicating regions (Fig. 4).
A question that arises is whether cytosine methylation has a possible role in helping to define the choice of replication origins in the genome. DNA replication is initiated from sites in the genome called origins of replication. In mammalian cells, replication is organized into discrete zones of similar replication timing, which consist of multiple replication origins. The zones are heterogeneous in size (30–450 kb, with the most frequent sizes in the range of 75–150 kb) (Berezney et al. 2000). Since later replication timing is correlated with closed chromatin, a logical conclusion would be that the repressive cytosine methylation mark should be enriched in regions of later replication timing. With the identification of specific origins of replication in mammals, direct testing of these loci could be performed to test their methylation status. A consistent observation has been that nascent strands or defined replication origins are derived from or located at CpG islands (Tasheva and Roufa 1995; Rein et al. 1997; Delgado et al. 1998; Sequeira-Mendes et al. 2009) and, intriguingly, that these CpG islands may be characterized by being methylated (Tasheva and Roufa 1995; Rein et al. 1997), a characteristic that defines only a small subset of these genomic elements (Glass et al. 2007). Those methylated CpG islands are listed in Supplemental Tables 4 and 5.
Our appreciation of the relationship between cytosine methylation and heterochromatin needs to be refined in terms of genomic context—the positive association between cytosine methylation at tandemly repetitive sequences (such as those found in paracentromeric regions) and heterochromatin is well-established, and as such repetitive sequences are not tested by assays using the microarrays or massively parallel sequencing of the current project, our data do nothing to challenge this established relationship. The heterochromatically organized DNA in the remaining majority of the genome represents a distinct genomic context where the relationship with cytosine methylation is the opposite to that of the tandemly repetitive sequences. This has implications for the mechanism of drugs such as DNMT inhibitors, which may promote demethylation and chromosomal instability primarily in tandemly repetitive DNA, but may have different effects in the context of actively transcribed euchromatic regions. The other intriguing implication is a context-dependent association of cytosine methylation with regulators of post-translational modifications of histones. As the ENCODE project tested the GM06990 cell line in its pilot phase, we were able to correlate a number of histone modifications with replication timing for the 1% of the genome surveyed in the pilot phase of the ENCODE project (The ENCODE Project Consortium 2007). We show these results in Supplemental Figure 9. We were able to observe one histone modification in particular to be strongly correlated with late replication, histone H3 lysine 9 trimethylation (H3K9me3). This result indicates that histone modifications rather than cytosine methylation are likely to be responsible for the heterochromatic organization associated with late replication, but it also indicates that enriched cytosine methylation and H3K9me3 are not necessarily colocalized in the genome, despite biochemical (Rottach et al. 2009), genetic, and cytological (Peng and Karpen 2007) data that link the two epigenetic regulatory processes in eukaryotes. The function of histone methyltransferases to influence the targeting of cytosine methylation may thus be subject to the DNA sequence composition of specific genomic contexts rather than acting in the same manner throughout the genome. Overall, we conclude that heterochromatin is inherently heterogeneous, and that rules that determine relationships within this compartment may not be universal, but have genomic context dependencies for epigenetic regulators such as cytosine methylation.
Methods
Cell reagents
We used the GM06990 cell line and a human foreskin fibroblast cell line for these studies. GM06990 is a karyotypically normal lymphoblastoid cell line available from the Coriell repository (http://www.coriell.org/) that has been used by the ENCODE consortium for a number of studies (The ENCODE Project Consortium 2007), and was used for our recent genome-wide analysis of DNA replication timing (Hansen et al. 2010). The GM06690 cells were cultured as recommended by the Coriell repository. The human fibroblast cells were grown in Dulbecco's modified Eagle medium (DMEM) supplemented with 10% Fetal Bovine Serum, 2 mM Glutamine and Penicillin–Streptomycin (Invitrogen) in a 37°C incubator with 5% CO2. The cells were harvested at 80%–90% confluence in 150-cm2 flasks by trypsin-EDTA dissociation. DNA and RNA were extracted from the cells using standard protocols.
Microarray design
We used our previously published HELP microarray design representing >1.32 million loci genome wide, representing each HpaII-amplifiable fragment from 50 to 2000 bp with one to two oligonucleotides encoding unique sequence at each locus (Oda et al. 2009). The human gene expression microarray was a standard Roche-NimbleGen design (2006-08-03_HG18_60mer_expr).
Microarray sample preparation and hybridization
We performed the HELP assay as previously described (Oda et al. 2009). For expression studies, we converted mRNA to dsDNA using the SuperScript Double-Strand cDNA Synthesis kit (Invitrogen) with T7-Oligo(dT)24 (5′-GGCCAGTGAATTGTAATACGACTCACTATAGGGAGGCGGTTTTTTTTTTTTTTTTTTTTTTTT-3′). DNA labeling and hybridization to the microarrays were performed using a published technique (Selzer et al. 2005).
Single-locus quantitative validation assays
Bisulphite conversion and MassArray (Sequenom) were performed using the same sample of DNA used for the high-throughput assays above. Bisulphite conversion was performed with the EZ DNA Methylation kit. Bisulphite primers were designed using MethPrimer (http://www.urogene.org/methprimer/) with the following parameters: product length (250–450 bp), primer length (23–29 bp) and primer Tm (56–62°C). PCR was performed in the following conditions with FastStart High Fidelity Taq polymerase (Roche): 95°C for 10 min and 42 cycles of 95°C for 30 sec, primer-specific Tm for 30 sec and 72°C for 1 min, followed by 72°C for 10 min for the final extension. Primer-specific Tms and primer sequences are provided in Supplemental Table 1. Bisulphite MassArray assays were performed by the Einstein's Genomics Core Facility.
Quantitative RT–PCR
Complementary DNA (cDNA) was generated from 2 μg of total RNA with Superscript III reverse transcriptase (Invitrogen) using oligo(dT)20. RT–PCR primers were designed with Primer3 software (http://frodo.wi.mit.edu/primer3/input.htm). The primer sequences that we used in this study are provided in Supplemental Table 2. The quantitative PCR was performed using SYBR Green (Power SYBR Green PCR Master mix [Applied Biosystems]).
Analysis of cytosine methylation (HELP) assays
HELP assay data analysis was performed using our published pipeline (Thompson et al. 2008), an open source resource available through BioConductor (HELP package).
Analysis of gene expression microarray data
Gene expression microarrays performed on the GM06990 and fibroblast cells were analyzed using NimbleScan2.3 (NimbleGen) and R version 2.9.2 (http://www.R-project.org). We selected loci for validation that appeared to be expressed in one or both cell types and performed real-time RT–PCR with the ABI7500. The expression status was normalized using the human GAPDH expression level. We show the correlation between the microarray expression intensity and the real-time PCR validation data in Supplemental Figure 1. The expression microarrays and RT–PCR results had high correlation values (R = 0.97). Using the RT–PCR validation we were able to define a threshold for highly expressed genes as a log intensity of ≥6.
Timing of replication analysis
The original massively parallel sequencing-based data measuring timing of replication and DNase hypersensitivity used in this study have been published previously (Hansen et al. 2010). For the DNA replication timing, in cell cycle phases G1, S1, S2, S3, S4, and G2, newly replicated DNA positions were analyzed by massively parallel sequencing (Hansen et al. 2010). The newly replicated sequences were counted in windows of 1-kb size. In their correlation of replication timing with DNase hypersensitivity, they calculated the sum of read numbers for (G1 + S1) and divided this by the sum of read numbers for (S4 + G2) to get a single value for each 1-kb window. Rather than dividing values, we used an approach we recently described for our HELP-tagging assay to study cytosine methylation (Suzuki et al. 2010), transforming the read depth as shown in Supplemental Figure 2, comparing the (G1 + S1) with the (S4 + G2) sequence read counts by measuring the inverse tangent (arctangent) for each data point.
Correlative analyses
For our correlative analyses, we calculated the mean representation of the data we generated for cytosine methylation and gene expression, and added our previously published DNase hypersensitivity and DNA replication timing data (Hansen et al. 2010) in 100-kb sliding windows with a 50-kb step size. The number of DNase-hypersensitive sites and the mean arctangent DNA-replication timing values per window were calculated. Two-dimensional histograms were generated as contour plots using R version 2.9.2 and the areas between the contours were filled.
Self-organizing map analysis
To examine the relationships between the variables being tested, we used an artificial neural learning-based approach, the self-organizing map (SOM) (Kohonen 2001). Using 100-kb sliding windows, with a step size of 50 kb, we calculated mean log ratio values from the HELP assay (indicative of DNA methylation levels), mean RefSeq gene expression levels, and the cumulative number of HpaII amplifiable fragments (HAFs), CpG islands, and genes per window. These data were used to generate vectors for analysis. A total of 26 experiments were performed to use each of the five vector elements in all possible two-, three-, four-, and five-element combinations. Vector elements were mean centered, scaled between −1 and 1 based on the range of the data, and vectors were then normalized to unit length. All vectors were tagged as being either late (replication timing angle 0–30), intermediate (31–60), or early (61–90) replicating. These tags were provided with the sole purpose of revealing the whereabouts of vectors of these classes on the maps post-training, and did not provide any assistance to the training process itself. The data were formatted to be compatible with the GACT SOMengine C++ program implementing the SOM algorithm and SOMviewer, the accompanying Java-based SOM visualization software (AS McLellan, AA Golden, in prep.), which were used to perform the analysis. This software produces a SOM analysis using an implementation of the batch map SOM algorithm, featuring accelerated best-matching unit (BMU) finding (Kohonen 2001), and is parallelized with openMP (AS McLellan, AA Golden, in prep.). SOM maps were generated using the entire data set of 58,621 vectors. Each trained SOM map was generated using a 112 × 54 hexagonally arranged grid with random initialization of the codebook vectors (vectors of the same dimensions as the data vectors that represent each node of the grid). A total of 10,000 cycles of training was performed, each time presenting the entire data set to the grid and allowing grid nodes to compete for the vectors in the data set to which they were most similar (using an Euclidean distance metric). The SOM software was run on our local ROCKS/Sun Grid Engine (SGE)-based cluster using all eight processors on a single node for each experiment. After each training cycle, grid vectors were modified to resemble the data vectors “won” by that node with some influence from neighboring nodes. This was achieved using a Gaussian neighborhood function for updating codebook vectors at the end of each cycle and Gaussian neighborhood-radius decay with time. An initial neighborhood-radius of 69 was used. After training, all data was reintroduced to the grid a final time and selected annotations revealed as a dual color intensity graph in order to examine the distribution of features. For example, replication-timing status could be examined by coloring the nodes with a shade and intensity proportional to the number of vectors associated with each label from green (all late replicating) to red (all early replicating). Overall clustering patterns in the data were also examined using a U-matrix representation of the grid, which represents a similarity graph where a linear grayscale is used to indicate how similar a node vector is to its immediate neighbors in vector space.
Data access
Genome-wide molecular data from HELP microarray experiments have been submitted to the NCBI Gene Expression Omnibus (GEO) (http://www.ncbi.nlm.nih.gov/geo), under accession numbers GSM679751 (fibroblast HELP), GSM679750 (lymphoblast HELP), GSM679748 (fibroblast gene expression), and GSM679749 (lymphoblast gene expression).
Acknowledgments
This work is supported by grants from the National Institutes of Health (NIH, R01HG004401, J.M.G.; P30HD02274, R.S.H.; U54HG004592, J.A.S.). We thank Shahina Maqbool PhD, Raul Olea, and Gael Westby of Einstein's Epigenomics Shared Facility for their contributions, and Einstein's Center for Epigenomics.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.116509.110.
References
- Aran D, Toperoff G, Rosenberg M, Hellman A 2011. Replication timing-related and gene body-specific methylation of active human genes. Hum Mol Genet 20: 670–680 [DOI] [PubMed] [Google Scholar]
- Backdahl L, Herberth M, Wilson G, Tate P, Campos LS, Cortese R, Eckhardt F, Beck S 2009. Gene body methylation of the dimethylarginine dimethylamino-hydrolase 2 (Ddah2) gene is an epigenetic biomarker for neural stem cell differentiation. Epigenetics 4: 248–254 [DOI] [PubMed] [Google Scholar]
- Ball MP, Li JB, Gao Y, Lee JH, LeProust EM, Park IH, Xie B, Daley GQ, Church GM 2009. Targeted and genome-scale strategies reveal gene-body methylation signatures in human cells. Nat Biotechnol 27: 361–368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berezney R, Dubey DD, Huberman JA 2000. Heterogeneity of eukaryotic replicons, replicon clusters, and replication foci. Chromosoma 108: 471–484 [DOI] [PubMed] [Google Scholar]
- Bernardino-Sgherri J, Flagiello D, Dutrillaux B 2002. Overall DNA methylation and chromatin structure of normal and abnormal X chromosomes. Cytogenet Genome Res 99: 85–91 [DOI] [PubMed] [Google Scholar]
- Chow JC, Brown CJ 2003. Forming facultative heterochromatin: silencing of an X chromosome in mammalian females. Cell Mol Life Sci 60: 2586–2603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delgado S, Gomez M, Bird A, Antequera F 1998. Initiation of DNA replication at CpG islands in mammalian chromosomes. EMBO J 17: 2426–2435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desprat R, Thierry-Mieg D, Lailler N, Lajugie J, Schildkraut C, Thierry-Mieg J, Bouhassira EE 2009. Predictable dynamic program of timing of DNA replication in human cells. Genome Res 19: 2288–2299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The ENCODE Project Consortium 2007. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature 447: 799–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fanti L, Pimpinelli S 2008. HP1: a functionally multifaceted protein. Curr Opin Genet Dev 18: 169–174 [DOI] [PubMed] [Google Scholar]
- Fernandez-Peralta AM, Navarro P, Tagarro I, Gonzalez-Aguilera JJ 1994. Digestion of human chromosomes by means of the isoschizomers MspI and HpaII. Genome 37: 770–774 [DOI] [PubMed] [Google Scholar]
- Gartenberg M 2009. Heterochromatin and the cohesion of sister chromatids. Chromosome Res 17: 229–238 [DOI] [PubMed] [Google Scholar]
- Gilbert DM 2002. Replication timing and transcriptional control: beyond cause and effect. Curr Opin Cell Biol 14: 377–383 [DOI] [PubMed] [Google Scholar]
- Glass JL, Thompson RF, Khulan B, Figueroa ME, Olivier EN, Oakley EJ, Van Zant G, Bouhassira EE, Melnick A, Golden A, et al. 2007. CG dinucleotide clustering is a species-specific property of the genome. Nucleic Acids Res 35: 6798–6807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gopalakrishnan S, Sullivan BA, Trazzi S, Della Valle G, Robertson KD 2009. DNMT3B interacts with constitutive centromere protein CENP-C to modulate DNA methylation and the histone code at centromeric regions. Hum Mol Genet 18: 3178–3193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gribnau J, Luikenhuis S, Hochedlinger K, Monkhorst K, Jaenisch R 2005. X chromosome choice occurs independently of asynchronous replication timing. J Cell Biol 168: 365–373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen RS, Wijmenga C, Luo P, Stanek AM, Canfield TK, Weemaes CM, Gartler SM 1999. The DNMT3B DNA methyltransferase gene is mutated in the ICF immunodeficiency syndrome. Proc Natl Acad Sci 96: 14412–14417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen RS, Thomas S, Sandstrom R, Canfield TK, Thurman RE, Weaver M, Dorschner MO, Gartler SM, Stamatoyannopoulos JA 2010. Sequencing newly replicated DNA reveals widespread plasticity in human replication timing. Proc Natl Acad Sci 107: 139–144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrow J, Denoeud F, Frankish A, Reymond A, Chen CK, Chrast J, Lagarde J, Gilbert JG, Storey R, Swarbreck D, et al. 2006. GENCODE: producing a reference annotation for ENCODE. Genome Biol 7 (Suppl 1): S4 doi: 10.1186/gb-2006-7-s1-s4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassan KM, Norwood T, Gimelli G, Gartler SM, Hansen RS 2001. Satellite 2 methylation patterns in normal and ICF syndrome cells and association of hypomethylation with advanced replication. Hum Genet 109: 452–462 [DOI] [PubMed] [Google Scholar]
- Hellman A, Chess A 2007. Gene body-specific methylation on the active X chromosome. Science 315: 1141–1143 [DOI] [PubMed] [Google Scholar]
- Hendrich B, Bird A 1998. Identification and characterization of a family of mammalian methyl-CpG binding proteins. Mol Cell Biol 18: 6538–6547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S 2000. Heterochromatin function in complex genomes. Biochim Biophys Acta 1470: 1–8 [DOI] [PubMed] [Google Scholar]
- Holmquist GP 1989. Evolution of chromosome bands: molecular ecology of noncoding DNA. J Mol Evol 28: 469–486 [DOI] [PubMed] [Google Scholar]
- Khulan B, Thompson RF, Ye K, Fazzari MJ, Suzuki M, Stasiek E, Figueroa ME, Glass JL, Chen Q, Montagna C, et al. 2006. Comparative isoschizomer profiling of cytosine methylation: The HELP assay. Genome Res 16: 1046–1055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohonen T 2001. Self Organizing Maps. Springer, Berlin, Germany [Google Scholar]
- Krauss V 2008. Glimpses of evolution: heterochromatic histone H3K9 methyltransferases left its marks behind. Genetica 133: 93–106 [DOI] [PubMed] [Google Scholar]
- Oda M, Glass JL, Thompson RF, Mo Y, Olivier EN, Figueroa ME, Selzer RR, Richmond TA, Zhang X, Dannenberg L, et al. 2009. High-resolution genome-wide cytosine methylation profiling with simultaneous copy number analysis and optimization for limited cell numbers. Nucleic Acids Res 37: 3829–3839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng JC, Karpen GH 2007. H3K9 methylation and RNA interference regulate nucleolar organization and repeated DNA stability. Nat Cell Biol 9: 25–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plohl M, Luchetti A, Mestrovic N, Mantovani B 2008. Satellite DNAs between selfishness and functionality: structure, genomics and evolution of tandem repeats in centromeric (hetero)chromatin. Gene 409: 72–82 [DOI] [PubMed] [Google Scholar]
- Rein T, Zorbas H, DePamphilis ML 1997. Active mammalian replication origins are associated with a high-density cluster of mCpG dinucleotides. Mol Cell Biol 17: 416–426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rottach A, Frauer C, Pichler G, Bonapace IM, Spada F, Leonhardt H 2009. The multi-domain protein Np95 connects DNA methylation and histone modification. Nucleic Acids Res 38: 1796–1804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubeler D, Scalzo D, Kooperberg C, van Steensel B, Delrow J, Groudine M 2002. Genome-wide DNA replication profile for Drosophila melanogaster: a link between transcription and replication timing. Nat Genet 32: 438–442 [DOI] [PubMed] [Google Scholar]
- Selzer RR, Richmond TA, Pofahl NJ, Green RD, Eis PS, Nair P, Brothman AR, Stallings RL 2005. Analysis of chromosome breakpoints in neuroblastoma at sub-kilobase resolution using fine-tiling oligonucleotide array CGH. Genes Chromosomes Cancer 44: 305–319 [DOI] [PubMed] [Google Scholar]
- Sequeira-Mendes J, Diaz-Uriarte R, Apedaile A, Huntley D, Brockdorff N, Gomez M 2009. Transcription initiation activity sets replication origin efficiency in mammalian cells. PLoS Genet 5: e1000446 doi: 10.1371/journal.pgen.1000446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki M, Jing Q, Lia D, Pascual M, McLellan A, Greally JM 2010. Optimized design and data analysis of tag-based cytosine methylation assays. Genome Biol 11: R36 doi: 10.1186/gb-2010-11-4-r36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tasheva ES, Roufa DJ 1995. A densely methylated DNA island is associated with a chromosomal replication origin in the human RPS14 locus. Somat Cell Mol Genet 21: 369–383 [DOI] [PubMed] [Google Scholar]
- Thompson RF, Reimers M, Khulan B, Gissot M, Richmond TA, Chen Q, Zheng X, Kim K, Greally JM 2008. An analytical pipeline for genomic representations used for cytosine methylation studies. Bioinformatics 24: 1161–1167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viegas-Pequignot E, Dutrillaux B, Thomas G 1988. Inactive X chromosome has the highest concentration of unmethylated Hha I sites. Proc Natl Acad Sci 85: 7657–7660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel MJ, Guelen L, de Wit E, Peric-Hupkes D, Loden M, Talhout W, Feenstra M, Abbas B, Classen AK, van Steensel B 2006. Human heterochromatin proteins form large domains containing KRAB-ZNF genes. Genome Res 16: 1493–1504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeschnigk M, Martin M, Betzl G, Kalbe A, Sirsch C, Buiting K, Gross S, Fritzilas E, Frey B, Rahmann S, et al. 2009. Massive parallel bisulfite sequencing of CG-rich DNA fragments reveals that methylation of many X-chromosomal CpG islands in female blood DNA is incomplete. Hum Mol Genet 18: 1439–1448 [DOI] [PubMed] [Google Scholar]
- Zhang X, Yazaki J, Sundaresan A, Cokus S, Chan SW, Chen H, Henderson IR, Shinn P, Pellegrini M, Jacobsen SE, et al. 2006. Genome-wide high-resolution mapping and functional analysis of DNA methylation in Arabidopsis. Cell 126: 1189–1201 [DOI] [PubMed] [Google Scholar]
- Zilberman D, Gehring M, Tran RK, Ballinger T, Henikoff S 2007. Genome-wide analysis of Arabidopsis thaliana DNA methylation uncovers an interdependence between methylation and transcription. Nat Genet 39: 61–69 [DOI] [PubMed] [Google Scholar]