Abstract
Anopheles gambiae is a major mosquito vector responsible for malaria transmission, whose genome sequence was reported in 2002. Genome annotation is a continuing effort, and many of the approximately 13,000 genes listed in VectorBase for Anopheles gambiae are predictions that have still not been validated by any other method. To identify protein-coding genes of An. gambiae based on its genomic sequence, we carried out a deep proteomic analysis using high-resolution Fourier transform mass spectrometry for both precursor and fragment ions. Based on peptide evidence, we were able to support or correct more than 6000 gene annotations including 80 novel gene structures and about 500 translational start sites. An additional validation by RT-PCR and cDNA sequencing was successfully performed for 105 selected genes. Our proteogenomic analysis led to the identification of 2682 genome search–specific peptides. Numerous cases of encoded proteins were documented in regions annotated as intergenic, introns, or untranslated regions. Using a database created to contain potential splice sites, we also identified 35 novel splice junctions. This is a first report to annotate the An. gambiae genome using high-accuracy mass spectrometry data as a complementary technology for genome annotation.
Anopheles gambiae is a major vector for malaria, which is a main public health burden in many parts of the world. The first draft of the An. gambiae genome sequence was released in 2002 containing ∼278 Mb (Holt et al. 2002). Mongin et al. (2004) discussed the limitations associated with this genome assembly. A gene set annotated by VectorBase contains both manually annotated genes and predicted gene models from GeneWise (Birney et al. 2004), ClusterMerge (Eyras et al. 2004), and SNAP (Li et al. 2007) algorithms. The VectorBase bioinformatic resource provides several annotated and curated vector genomes in a Web-accessible integrated format including DNA and protein alignments (Lawson et al. 2009). Based on manual appraisal, the VectorBase (http://agambiae.VectorBase.org) updated the Anopheles gambiae genebuild (AgamP3.5) in September 2009, which contained 12,604 protein-coding genes. The updated gene sets include 765 novel genes, modification of 3726 gene models, and deletion of 456 genes. The latest genebuild, AgamP3.6, was released in December 2010, which contains 12,669 protein-coding genes. This release includes 227 new genes, changes to the structure of 443 gene models, and deletion of three genes as compared to the AgamP3.5 genebuild. In the VectorBase–Ensembl genome annotation pipeline, genes are annotated based on mRNA/cDNA sequences and comparative proteomic evidence, as well as manual appraisal. Manually annotated gene models are given the highest preference followed by comparative gene models, EST-based models, and ab initio gene models. GeneWise-based prediction uses alignment of dipterans and other protein sequences to the An. gambiae genome for building gene models. The ClusterMerge algorithm builds models based on EST evidence (Eyras et al. 2004). The SNAP and Genscan algorithms were used to predict ab initio models that are also included in the current genebuild (Korf 2004).
In the present study, we present many novel findings that were missed in spite of a robust annotation strategy and multiple revisions of An. gambiae genome annotations. The reverse process of genome annotation, i.e., from proteins to the genome, holds great promise for increasing the accuracy of the predicted gene structures. Annotation of genomes using mass spectrometry–based proteomics data is complementary to other gene prediction methods. Direct evidence for the protein-coding potential of the genome sequence can be obtained by searching tandem mass spectrometry data against nucleotide sequences like ESTs or genome sequence databases as against known protein databases (Pandey and Lewitter 1999; Pandey and Mann 2000; Choudhary et al. 2001; Mann and Pandey 2001; Xia et al. 2008). Certain features of peptides can provide definitive evidence pertaining to protein architecture that cannot be obtained from genome or transcript sequencing, e.g., acetylation of N termini of peptides, which indicates proximity to the translation start sites. An important outcome of such analyses is the identification of novel genes that have been entirely missed by other approaches. Protein-coding genes leading to splice variants, truncated proteins, and cSNPs can all also be directly studied by protein sequencing. Several studies have demonstrated the use of mass spectrometry–based proteomic approaches to validate or correct gene annotations in Homo sapiens (Molina et al. 2005; Suzuki and Sugano 2006; Sevinsky et al. 2008; Menon et al. 2009), Caenorhabditis elegans (Merrihew et al. 2008), Drosophila melanogaster (Brunner et al. 2007; Tress et al. 2008), An. gambiae (Pandey and Mann 2000; Kalume et al. 2005a,b; Okulate et al. 2007), Toxoplasma gondii (Xia et al. 2008), and Arabidopsis thaliana (Kuster et al. 2001; Baerenfaller et al. 2008).
Here, we present the results of an extensive qualitative proteomic analysis of An. gambiae to better understand gene structures and their functions. We report validation of existing genes, correction of existing gene models, identification of novel genes, identification of novel splice variants, confirmation of splice sites, and assignment of translational start sites based on high-resolution mass spectrometry–derived data. A total of 2682 peptides were identified that could not be mapped onto existing VectorBase annotations. We also used gene prediction models by SNAP, and in some cases by Fgenesh and GenMark, which supported the peptide evidence to identify novel genes or alternate gene models. Finally, we performed RT-PCR and sequencing to support the existence of a number of novel and modified coding regions identified in this study.
Results and Discussion
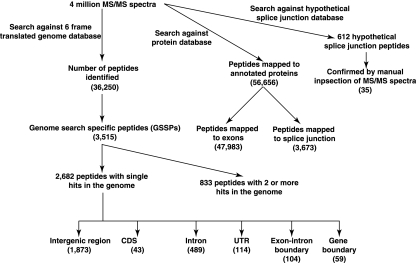
The goal of our study was to achieve deep coverage of the proteome of An. gambiae. To this end, proteins from nine different tissues such as larvae, pupae, salivary gland, midgut, malpighian tubules, ovaries, head, viscera, testis, and male accessory organ of An. gambiae were separated by SDS-PAGE, strong cation exchange chromatography, and by reversed-phase chromatography prior to trypsin digestion and LC-MS/MS analysis (Supplemental Fig. 1). In all, we performed 460 LC-MS/MS experiments using high-resolution Fourier transform mass spectrometry. The resulting mass spectral data were searched against a protein database, a database containing six-frame genome translation, and a database of exon–exon junction-spanning tryptic peptides of potential splice variants. The proteogenomic data analysis strategy and a summary of peptide identifications are shown in Figure 1. A correlation analysis of the number of identified peptides per protein and the number of proteins in each category showed a power-law distribution (r = 0.94) (Supplemental Fig. 2A). Supplemental Figure 2B shows the pattern of distribution of peptide coverage per gene from nine different tissues. Overall, 52% (SD ± 4%) of the identified proteins were represented by three or more peptides per protein, 14% (SD ± 1%) by two peptides, and the remaining 34% (SD ± 3%) by one peptide.
Figure 1.
Flowchart illustrating the proteogenomics analysis steps.
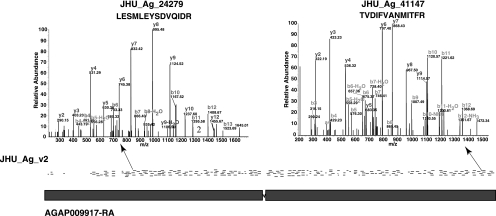
Figure 2 depicts the deep proteomic coverage that we obtained (>70%) for a high-molecular-weight (338 kDa) protein, salivary gland secreted protein 4 (AGAP009917-PA). This is an example where we found the highest number of peptides (>200) mapping to any single gene. The track named “JHU_Ag_v2” shows rectangle bars representing the peptides mapping to the genomic region and are designated as JHU_Ag_xxxx, where “JHU” and “Ag” stand for Johns Hopkins University and Anopheles gambiae, respectively, and “xxxx” indicates the serial number assigned to the peptide. This integrated graphical view allows researchers to rapidly access the transcriptomic and proteomic data for evaluation of the gene of interest along with all other annotations provided by Ensembl and VectorBase genome browsers (e.g., gene models, ESTs, transcripts). This feature can be used for confirming or altering the gene models of the existing annotations.
Figure 2.
Mapping of mass spectrometry-derived peptide data onto the VectorBase genome browser. The unique peptides identified by mass spectrometry (rectangle bars), which mapped to the known exons of the gene encoding salivary gland secreted protein 4 (SGS4) (AGAP009917-RA). The peptides identified in this study can be viewed as separate tracks on the VectorBase genome browser using the URL http://funcgen.vector base.org/gdav/das as DAS server and “JHU_Ag_v2” as the data source. The JHU_Ag_v2 track shows peptide data as JHU_Ag_xxxx, where JHU and Ag stand for Johns Hopkins University and An. gambiae, respectively; and “xxxx” denotes the serial number of the peptide. The MS/MS spectra of two representative peptides LESMLEYSDVQIDR (JHU_Ag_24279) and TVDIFVANMITFR (JHU_Ag_41147) are shown.
Overview of mass spectrometry data used for genome annotation
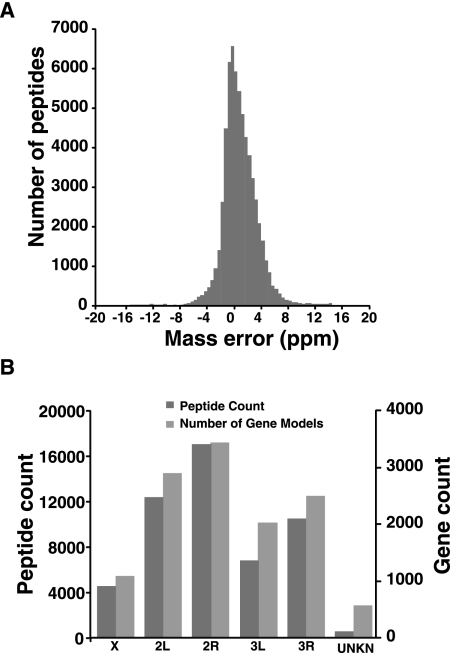
Approximately 4,000,000 MS/MS spectra acquired in this study resulted in approximately 529,287 peptide spectrum matches (PSMs), which yielded approximately 52,000 unique peptide sequences identified from genome and protein database searches. Figure 3A shows the distribution of mass error in parts per million for the entire peptide data set. Nearly 95% of the peptides were within ±5 ppm mass error, confirming the high accuracy of peptide data obtained from the mass spectrometer. It is important to note that we used high-resolution mass spectrometry for both MS and MS/MS experiments, which resulted in accurate measurement of both precursor as well as fragment ions. Figure 3B shows the chromosomal assignments of peptides identified by mass spectrometry. The number of peptides identified roughly parallels the number of gene models annotated for each chromosome. We have identified 5963 unique proteins from different An. gambiae organs excluding proteins that were entirely supported by redundant peptides. The number of proteins identified from nine different tissues include (1) head, 3460; (2) salivary glands, 1729; (3) Malpighian tubules, 2793; (4) male accessory glands and testis, 1716; (5) viscera, 1619; (6) ovaries, 2598; (7) midguts, 2448; (8) larvae, 1183; and (9) pupae, 1434. Supplemental Table 1 provides a complete list of peptides identified with their genomic coordinates and the organ(s) of origin. From our mass spectrometry data, 51,656 peptides (Supplemental Table 2) were mapped to protein-coding exons of >6000 genes. Although gene prediction programs, EST sequences, and sequence homology have already been used to build the existing annotation of gene structures by VectorBase, high-sequence-coverage peptide data can provide direct evidence for translated gene products. As an example, a novel protein (AGAP009323-PA) containing a prohibitin homolog domain was identified with 100% sequence coverage including an N-terminal acetylated peptide (Supplemental Fig. 3A). The gene encoding this protein has three exons, and the coding exon junction is confirmed by a splice junctional peptide. Another example showing extensive sequence coverage is shown in Supplemental Figure 3B, where the gene AGAP003153 (VATA_ANOGA), encoding a protein with three ATP synthase domains, was assigned 69 unique peptides that validated each of its five exons. In addition, all exon–exon junctions were confirmed by junctional peptides. AGAP007563-PA was the biggest protein (1.784 MDa, 15,844 amino acids) identified in our study from six tissues with sequence coverage up to 23% from mosquito head. A shorter splice variant of this protein, AGAP007563-PC, could also be identified based on one peptide unique to exon 13 of AGAP007563-RC. In light of the depth that can be achieved by mass spectrometry today, it should be possible to carry out such “peptide mapping” routinely to confirm predicted transcripts.
Figure 3.
Overview of mass spectrometry data used for genome annotation. (A) An estimation of the mass error of peptides in parts per million identified from mass spectrometric analysis of An. gambiae. (B) Chromosomal distribution of peptides identified by mass spectrometry. The number of peptides identified from each chromosome roughly parallels the estimated number of known and novel protein-coding genes in An. gambiae.
Genome annotation refinement using peptide sequence data
Gene prediction tools suffer from both false-negative and false-positive predictions, leading to incorrect exons or exon boundaries, wrong prediction of translational start/stop sites, and missed genes/exons. In this study, MS/MS data were searched against a six frame translated genome sequence to identify novel protein-coding regions. After excluding peptides that mapped to existing protein database entries (peptide sequences derived from Agam 3.6), the genome search-specific peptides, or GSSPs, were further analyzed to refine current gene predictions.
A total of 3515 peptides did not match any protein sequences of An. gambiae in VectorBase annotations, out of which 2682 peptides that mapped to only one location in the genome were considered for proteogenomic analysis. These peptides are listed in six groups as Supplemental Table 3A–3F in the following categories: (1) peptides mapping to intergenic regions; (2) peptides mapping within introns; (3) peptides mapping within annotated exons but not matching the frame of translation; (4) peptides overlapping exon–intron junctions; (5) peptides extending gene boundaries; and (6) peptides mapping to untranslated regions (UTRs). We used de novo gene predictions in combination with GSSPs to propose refinements/additions to the current genome annotation as described in the following sections.
Peptides mapping to intergenic regions
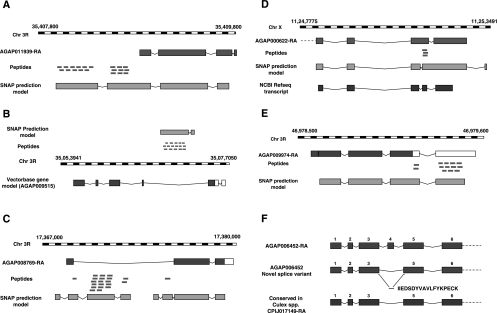
Peptides that map to intergenic regions could lead to identification of novel protein-coding genes or correction of existing gene models by extending them. One thousand eight hundred seventy-three peptides were found to map to intergenic regions (Supplemental Table 3A). Out of these, 454 peptides are supported by An. gambiae ESTs, whereas 52 peptides could be mapped to proteins from Culex quinquefasciatus and Aedes aegypti. Five hundred sixty-two peptides supported SNAP prediction models. Using the combination of intergenic peptide evidence and alternate gene prediction, we have confirmed the presence of 80 novel genes and more than 353 examples of gene model refinement. Figure 4A shows an example of N-terminal extension of a gene model AGAP011939-RA from 20 peptides that mapped upstream of the gene. SNAP predicts a longer gene model in the same region, which indicates an N-terminal extension of this gene. The extended part of this gene, which codes for alpha amylase, is conserved in Aedes aegypti and Culex quinquefasciatus. We have also validated it using RT PCR (GenBank accession no. GO935208). MS/MS spectra for identification of two representative genome search-specific peptides, EPGYEDYYVWHDGK and QQYYLHQFTVEQPDLNYR, are shown in the Supplemental Material. A similar example of gene structure modification of the AGAP010657 gene is shown in Supplemental Figure 4. SNAP predicts two gene structures in the genomic region where AGAP010657 is annotated. One of these SNAP models is supported by 15 intergenic peptides and one intronic peptide.
Figure 4.
(A) N-terminal extension of AGAP011939 using peptides mapping to an upstream intergenic region. Twenty peptides were mapped to an intergenic region upstream of the gene AGAP011939. SNAP predicts a longer gene model that is supported by novel peptides identified upstream of this gene. (B) Identification of a novel protein-coding gene using peptides mapping to an intergenic region. Sixteen peptides were mapped to an intergenic region on chromosome 3R, where the intron of a VectorBase gene model AGAP009515-RA was annotated on the opposite strand. The presence of a novel gene in this region is also indicated by the SNAP prediction program. (C) Correction of a gene structure using peptide mapping to an intron of an annotated gene. Fifteen peptides were identified in the intronic region of the gene AGAP008769. These peptides support two different gene models predicted by SNAP. (D) Identification of peptides translated in a different frame from the annotated protein sequence. Three GSSPs mapped within the coordinates of the sixth exon of the AGAP000622 gene that were not present in the predicted protein product of the gene. However, these peptides were present in the protein product of SNAP prediction and NCBI RefSeq annotation. (E) Identification of a novel protein-coding region using peptides mapping to the UTR of a gene. Five GSSPs mapped to the 3′-UTR region of the AGAP009974 gene. The SNAP prediction model for this genomic region supports a C-terminal extension of the protein encoded by the AGAP009974 gene. (F) Identification of a novel splice form. The peptide, IIEDSDYVAVLFYKPECK, was identified in the MS/MS ion search against the novel splice junction database of hypothetical splice isoforms. This novel splicing event, which occurred between exons 3 and 5 of the AGAP006452-RA gene, is also observed in Culex quinquefasciatus.
In another type of gene structure refinement, two adjacent gene models can be merged into a single longer gene. As shown in Supplemental Figure 5, 28 peptides were found to map to the intergenic region between AGAP011872 and AGAP011873. The identified peptides support an alternative gene prediction model by SNAP, which shows a single gene spanning both AGAP011872 and AGAP011873 plus two exons in the intergenic region. Comparative genomic analysis shows the presence of a protein orthologous to the longer gene model in both Ae. aegypti and Cx. quinquefasciatus. Finally, we have validated the presence of a transcript that connects the two genes (GenBank accession no. GO935137 and no. GO935138).
Identification of novel genes is an important finding of any proteogenomic analysis. One such example is shown in Figure 4B, where 16 unique intergenic peptides were found to map to a genomic region where the intron of transcript AGAP009515-RA is annotated on the opposite strand. SNAP predicts a model SNAP_ANOPHELES00000018835, which is supported by these intergenic peptides. No orthologous protein was found in the annotated proteomes of two other mosquito species, Ae. aegypti and Cx. quinquefasciatus, for this novel gene. However, a high protein sequence level conservation (>70%) is found in both Ae. aegypti and Cx. quinquefasciatus genomes if six frame translation is used to identify conserved regions. This example shows how this novel gene confirmed by proteomic analysis in this study is missed in the annotation of other mosquito species. MS/MS spectra for two representative peptides, NAFGQNVQELAEVLVR and LSGEYSTSVSTLVAAVR, supporting the novel gene annotation are shown in the Supplemental Material. A similar example of novel gene identification using evidence from 13 unique peptides is depicted in Supplemental Figure 6. In this example, the region is on the strand that is opposite to an annotated intron of a VectorBase transcript (AGAP007548-RB). All 13 peptides that were identified support a SNAP prediction model, SNAP_ANOPHELES 00000016290. In this example, the protein product of the novel gene does not have identifiable orthologous sequences in related species. In both of these cases, we have validated the existence of transcripts by sequencing the mRNA by RT-PCR. These examples emphasize the significance of proteogenomic analysis where protein-coding potential is proved unequivocally by proteomic evidence that may be further corroborated by transcriptomic or comparative genomic evidence.
Identification of peptides in introns
Supplemental Table 3B shows 459 GSSPs that were found to map to intronic regions of 247 genes. Peptides that are identified in intronic regions on annotated genes can lead to either correction of gene structure or identification of novel splice isoforms. For example, we identified 15 peptides in the intronic region of the AGAP008769-RA gene. Alternative gene prediction programs predict multiple gene structures in this intronic region of the AGAP008769-RA gene. Out of these prediction models, two are supported by the intronic peptides identified by us as shown in Figure 4C. Comparative genomic analysis shows that the alternative gene models show higher sequence conservation with Ae. aegypti and Cx. quinquefasciatus protein orthologs than the existing gene model. MS/MS spectra for two representative peptides, LFVALTGIQYAGSHLK and SIGYGGTDLSAFVADPLK, supporting the correction of annotated gene structure are shown in the Supplemental Material. In another instance, we found about 200 unique peptides within several exons of the AGAP010021-RA gene. Interestingly, we identified one peptide in the intron between exons 6 and 7, 11 peptides in the intron between exons 9 and 10, and three peptides in the intron between exons 10 and 11 (Supplemental Fig. 7). We also found one peptide in the upstream flanking region, 10 peptides in the downstream flanking region, and two peptides that partially mapped to exons 7 and 10. In all, we identified 27 GSSPs that did not agree with the VectorBase model for AGAP010021-RA. The alternative gene model by SNAP is supported by 23 out of 27 GSSPs in this region, whereas the Fgenesh model is supported by 26 GSSPs. In 22 cases in which the peptide was found in the intron of a gene, we also found a peptide spanning the exon–exon junction of exons flanking that intron, indicating the presence of alternatively spliced variants.
Identification of peptides translated in a different frame from existing annotations
Most of the GSSPs map to regions of genomes where no gene structure is present, hence enabling us to find new genomic regions with protein-coding potential. However, some peptides mapped to regions with existing annotations of coding regions but in a different frame of translation. More than 40 peptides were found to map to coding exons of 37 genes, but in a different frame of translation (Supplemental Table 3C). Figure 4D shows an example where three GSSPs were found to map within the fifth exon of the AGAP000622-RB transcript. An MS/MS spectrum for identification of genome search-specific peptide DEDTDVESFR is shown in the Supplemental Material. Not only is the protein product of the SNAP model consistent with these peptides, but also a transcript reported in the NCBI RefSeq database for this gene. Interestingly, the exon structures of the SNAP model and RefSeq transcript vary considerably, indicating that additional molecular biology experiments will be necessary to establish the gene structure accurately.
Identification of peptides partially overlapping with exon boundaries
Peptides that map partially to annotated exons clearly suggest change in exon structure by extending them. We categorized such peptides into two categories: (1) peptides that span exon–intron boundaries; and (2) peptides that extend gene boundaries. One hundred four unique peptides were found to span exon–intron boundaries, which indicates changes in the structures of 77 genes (Supplemental Table 3D). We also checked whether the portion of the peptides that maps onto exons is translated in the same frame as the exon itself. In 13 cases, the peptides were found to be translated in a frame other than that of the annotated exon, suggesting a possible correction in the reading frame along with a change in the exon coordinates. In the second category, we identified 59 peptides that extended gene boundaries of 45 genes (Supplemental Table 3E). Of these, in 20 genes, peptides mapped to the N-terminal boundary of genes, indicating an N-terminal extension. It should be pointed out that in 13 out of these 20 cases, the N terminus of the protein was annotated to begin with an amino acid other than methionine, again suggesting an error in the start site annotation. In 25 genes, the peptides mapped beyond the most 5′ exon, suggesting a C-terminal extension. In 23 out of these 25 genes, the encoded protein sequences were annotated to terminate without any stop codon, indicating erroneous annotation of the stop codon. In the remaining two cases, the peptides that we identified were translated in a different frame from the existing annotation.
Peptides mapping to untranslated regions (UTRs)
We found a total of 88 peptides that mapped to regions annotated as untranslated regions (UTRs) of transcripts in VectorBase. In all, UTRs of 45 different transcripts were shown to be translated based on peptide evidence—18 of these transcripts had multiple peptides mapping to them. The 5′ UTR was shown to be translated in 20 transcripts, while the 3′ UTR was found to have coding potential in the remaining 25 transcripts. Interestingly, eight of the encoded protein sequences were annotated to terminate without any stop codon, indicating erroneous annotation of the stop codon. One such example is illustrated in Figure 4E. The AGAP009974-RA gene had 13 peptides mapping to its 3′ UTR, providing strong evidence of translation in the predicted UTR. An alternative gene prediction model by SNAP also indicates that this region is translated. The current annotation of protein encoded by the AGAP009974-PA gene is erroneous as it does not end in a stop codon. A list of all of the peptides identified in UTRs is provided in Supplemental Table 3F. MS/MS spectra for identification of two representative genome search-specific peptides, ELDDGLIER and EQELSDCIVDK, are shown in the Supplemental Material.
Confirmation of splice sites with peptides spanning exon–exon junctions
Identification of peptide sequences spanning exon–exon junctions provides evidence for confirmation of splice sites of predicted transcripts and novel splice variants. Peptides identified in the protein database search were mapped onto the transcript sequences from An. gambiae, which also included splice isoforms to identify splice junction–spanning peptides. We found a total of 3673 unique peptides that spanned exon–exon junctions of VectorBase transcripts which supported 2996 splice junctions from 1918 genes (Supplemental Table 4). As shown in Supplemental Figure 3B, all four splice junction peptides for VATA_ANOGA proteins were identified in our study. Additional examples of confirmation of splice sites include the following: (1) the AGAP011026 gene encoding 5′-nucleotidase, which has seven exons—a total of 31 peptides mapped entirely within exons and an additional four peptides derived from exon–exon junctions confirmed splice sites for exon1–exon2, exon4–exon5, exon5–exon6, and exon6–exon7 junctions (Supplemental Fig. 8); (2) the AGAP009833 gene encoding porin, which has four exons—we identified 25 peptides mapping completely within exons along with three junctional peptides confirming all three splice sites of the predicted transcript; (3) the AGAP002350-RA gene—30 peptides mapped completely to the exons, and three peptides were mapped to splice sites confirming three splice sites predicted for this gene. Unique splice junction peptides along with other peptides also confirmed other isoforms (AGAP002350-PB and AGAP002350-PE) encoded by the gene AGAP002350 as shown in Supplemental Figure 9.
Identification of novel splice variants
To identify novel splice variants, MS/MS data were searched against the database of exon–exon junction spanning tryptic peptides of hypothetical splice variants generated using all possible forward combinations of exons in a gene. The spectral assignments of all identified peptides (that would correspond to novel splice junctions) were further confirmed by manual inspection of the MS/MS spectra. Thirty-five peptides qualified manual inspection with 13 peptides being identified from multiple spectra. Using these data we were able to identify novel splice events in 32 genes (Supplemental Table 5). Out of these, the splice variant described in Figure 4F is also seen in other mosquito species. The peptide sequence, IIEDSDYVAVLFYKPECK, resulting from splicing together of exons 3 and 5 of the AGAP006452 gene that was detected in the head and male reproductive organs, has been previously described for the Cx. quinquefasciatus protein CPIJ017149-PA. An annotated MS/MS spectrum of the peptide, IIEDSDYVAVLFYKPECK, that led to identification of this novel exon–exon junction in the AGAP006452-RA gene is shown in the Supplemental Material. The peptide sequence, DCSDGEDEICEAQR, was identified as a result of splicing of exon 3 to exon 6 of the AGAP003656-RA gene. The MS/MS spectrum along with the exon–exon junction identified using the peptide DCSDGEDEICEAQR are shown in Supplemental Figure 10. Taken together, these data demonstrate that high-resolution mass spectrometry is a unique tool in annotating novel splice variants even in the absence of transcript evidence.
Translational start site assignments using N-terminally acetylated peptides
Because of the unavailability of simple experimental methods to assign a translational start site in predicted transcripts or cDNA sequences, most translational start sites are annotated by predictions that are generally based on the longest open reading frame. Translation initiation can deviate significantly from the predicted start sites, necessitating the validation by other methods such as homology-based sequence alignments (Peri and Pandey 2001) and mass spectrometry–based determination of protein N-terminal acetylation sites (Gevaert et al. 2003; Molina et al. 2005; Oyama et al. 2007; Goetze et al. 2009). The acetylation of N termini of proteins is a common modification carried out by N-acetyl transferases mostly following the cleavage of the initiator methionine residue. Thus, global mass spectrometric analyses of N-terminally modified sites can provide confirmatory evidence for determination of translational start sites. Such modified peptides can be identified by choosing N-terminal acetylation as a modification during database searching of mass spectrometry data. Nearly 74% of protein N-terminal peptides identified in our analysis were found to be acetylated. Among the 616 protein N-terminally acetylated peptides that we identified, the majority had alanine (31%) or serine (29%) residues modified, which is in agreement with previous observations (Driessen et al. 1985), while 28% peptides were found to have an acetylated methionine residue itself. Supplemental Table 6 lists the N-terminally acetylated peptides and their corresponding protein entries that were identified in this study.
Validation of mass spectrometry–derived data using RT-PCR and cDNA sequencing
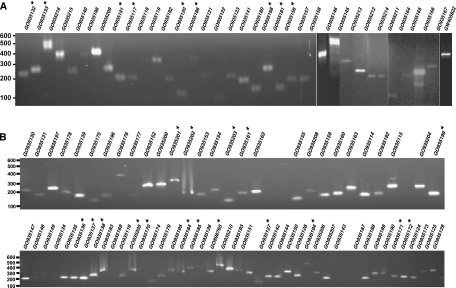
We performed an additional level of validation by performing RT-PCR on selected examples of novel genes and changes in gene structures. We carried out RT-PCR for 105 instances in which at least one of the alternative gene prediction programs predicted a coding exon. Figure 5 shows the RT-PCR amplification products along with the corresponding GenBank accession numbers from submitted sequences. The details of sequences submitted to GenBank including sequences, primers, and organs are provided as Supplemental Data 1. We would like to note that 105 RT-PCR products that corresponded to novel protein-coding regions discovered were absent in genebuild Agamp3.4. These data can be found in VectorBase as JHU_Ag_v1 entries. However, in the subsequent builds, AgamP3.5 and AgamP3.6, many of these examples were included in the standard annotations. Instances in which RT-PCR products corresponding to annotations that are still not included in AgamP3.6 are marked with an asterisk in Figure 5. Supplemental Table 7 provides a list of selected novel genes validated by RT-PCR and cDNA sequencing along with their corresponding peptides. Supplemental Figure 5 illustrates a novel gene, which was validated by RT-PCR and sequencing of products Anogamb_JHU77 and Anogamb_JHU78 (GenBank accession no. GO935137 and no. GO935138). More than 30 peptides mapped to the “intergenic” region between VectorBase genes AGAP011872 and AGAP011873 (Supplemental Table 7). Gene prediction programs predicted a six-exon gene in this region merging the two VectorBase-annotated genes into a longer gene structure. Using the exons predicted by SNAP, we designed two sets of primer pairs, one within the first exon of the AGAP011872 gene and the other located within a novel exon (flanking an intron of 2 kb). The other set of primers was designed within the second exon of the AGAP011873 gene and a novel exon (flanking an intron of 2.6 kb). The sequence analysis of RT-PCR products from these two regions confirmed the model for joining of two adjacent genes.
Figure 5.
Validation of mass spectrometry-derived data using RT-PCR and cDNA sequencing. RT-PCR products were sequenced on both strands, and the resulting sequences were submitted to GenBank. GenBank accession numbers are indicated above each lane. (A) RT-PCR validation of 35 novel genes. (B) RT-PCR validation of 70 gene models that led to the correction of existing VectorBase gene annotations in genebuild AgambP3.4. The genes that belong to novel categories with respect to genebuild AgambP3.6 are marked with an asterisk (*).
Conclusions
Our study illustrates the application of high-resolution mass spectrometry data for analysis and annotation of the An. gambiae genome. The peptide data served as corroboratory evidence in the validation of known and novel protein-coding genes as well as in correction of some of the existing gene models. Overall, our study demonstrates the power of high mass accuracy mass spectrometry–derived data to complement other approaches for genome annotation. Rapid advancement in high-throughput DNA sequencing has led to a great increase in the number of sequenced genomes, which, in turn, provides an excellent opportunity for carrying out proteogenomic analyses (Gupta et al. 2008). As of March 2011, 1652 completely sequenced genomes have been published, and an additional 8053 genome projects are under way (Genomes OnLine; http://www.genomesonline.org). The present study and several other previously published data support the proteogenomic approach as a valuable tool to complement genome sequencing. With the growing number of genome sequencing projects, we anticipate that proteogenomic approaches will become a popular method for confirmation of predicted gene structures and to accelerate the process of genome curation.
Methods
Collection of An. gambiae organs, pupae, and larvae
An. gambiae mosquitoes (G-3 strain from the Laboratory of Parasitic Diseases, National Institutes of Health) were grown in an insectary under ambient conditions (humidity 80% ± 5% and temperature 27°C ± 1°C). Adult mosquitoes were grown using 10% Karo dark corn syrup at least 12 h before dissection. The different body parts of the female mosquito such as head, salivary gland, malpighian tubules, ovary, midgut, viscera, and reproductive organs from male adult mosquitoes were dissected by an expert using an Olympus SZX12 stereomicroscope and stored at −80°C until use. In addition to dissected organs, third and fourth instar larvae and late pupae were also collected and preserved at −80°C until used.
Protein separation and trypsin digestion
Protein level fractionation was carried out for larvae, pupae, midguts, salivary glands, malpighian tubules, ovaries, heads, and viscera from female mosquitoes and reproductive organs from male mosquitoes. The tissues were lysed in 0.1% SDS and sonicated using a probe sonicator (Branson Sonifier) for 3 min on ice. Nearly 100 μg of protein lysate was loaded on an SDS-PAGE gel (4%–12% gel, NuPAGE gel; Invitrogen) and stained using Colloidal Coomassie stain. After removing excess stain, the lanes were cut into 20 to 25 gel pieces depending on the complexity of sample and subjected to in-gel tryptic digestion. In-gel reduction was performed using 5 mM DTT followed by alkylation using 20 mM iodoacetamide. In-gel digestion was carried out using trypsin (Promega, 1:50) for 12 h at 37°C. Peptides were extracted from the gel and dried using the vacuum drying process as explained earlier (Harsha et al. 2008). Peptide level fractionation was performed for head, midgut, ovary, and Malpighian tubule proteins by strong cation exchange chromatography as follows. In-solution trypsin digestion was carried out separately for each sample for ∼250 μg of protein. After reduction and alkylation, trypsin (Sequencing grade, Promega) digestion (ratio 1:20) was performed for 12 h at 37°C. The desalted peptides were fractionated on a polysulfoethyl A strong cation exchange column (PolyLC, 200 mm × 2.1 mm, 5 μm, 200 Å). The reversed-phase protein fractionation of midgut proteins was carried out on an mRP-C18 High-Recovery Protein Column (Agilent) as described earlier (Molina et al. 2007). The fractions were digested using the standard in-solution digestion protocol described above.
LC-MS/MS analysis
We performed a total of 460 LC-MS/MS analyses. One hundred forty-three runs were performed on an LTQ-Orbitrap XL ETD mass spectrometer interfaced with an Eksigent 2D nano scale pump, while the remaining 319 runs were performed on an LTQ-Orbitrap Velos ETD mass spectrometer interfaced with an Agilent 1200 series HPLC system. In both, the reversed-phase-LC system consisted of a desalting column (75 μm × 3 cm, Magic AQ C18 material, 5–10 μm, 100 Å) and an analytical column (75 μm × 10 cm, Magic AQ C18 material, 5 μm, 100 Å) with an electrospray (i.d. 8 μm) emitter tip (New Objective) that was maintained at 2.0 KV ion spray voltage. The mass spectrometry analysis was carried out in data-dependent analysis mode with survey scans (MS) acquired at a resolution of 30,000 at m/z 400 and fragment ion scan (MS/MS) acquired at a resolution of 15,000 at m/z 400. Both MS and MS/MS scans were acquired in Orbitrap mass analyzer after fragmentation by collision induced dissociation (CID; normalized collision energy value of 35%). The parameter settings for LTQ-Orbitrap XL analysis were (a) up to 5 MS/MS scans per duty cycle. (b) Precursor ions were dynamically excluded for a period of 30 sec. (c) For MS/MS analysis, monoisotopic precursor mass selection and rejection of singly charged ion criteria were enabled. (d) Capillary temperature was set at 175°C. The parameter settings used for analysis on LTQ-Orbitrap Velos were (a) acquisition of up to 20 MS/MS scan per duty cycle. (b) Precursors ions were dynamically excluded for a period of 30 sec. For MS/MS analysis, monoisotopic precursor mass selection was enabled. Capillary temperature was set at 220°C. Up to 2000 topmost abundant precursor ions were excluded during the consecutive analysis of peptide samples from same tissues.
Database searches for peptide identification
The protein database (agambiae.PEPTIDES-AgamP3.6.fa) used for MS/MS ion searches was downloaded from VectorBase. The genome sequence of An. gambiae was downloaded from the Ensembl ftp site (ftp://ftp.ensembl.org), and a six-frame translated database was created. Additionally, a database of novel splice variants was created as described in the following section—MS/MS ion searches submitted through Proteome Discoverer software (Thermo Scientific) to the Mascot search engine (version 2.2). The search parameters used were as follows: (1) trypsin as a proteolytic enzyme (with up to one missed cleavage); (2) peptide mass error tolerance of 15 ppm; (3) fragment mass error tolerance of 0.05 Da. (4) The carbamidomethylation of cysteine as fixed modification and oxidation of methionine, acetylation of protein N termini, and deamidation of glutamine and asparagine were included as variable modifications. Peptide identifications using 1% false discovery rate were considered for further analysis. The false discovery rate was estimated using a decoy database.
Peptide database for identification of novel alternative splice isoforms
The transcript database and coding sequence information were downloaded from VectorBase FTP for genebuild AgamP3.6. All possible forward combinations of two exons at a time from a given gene were generated and translated in the frame in which the 5′ exon of the combination was translated in the known protein isoform. Only tryptic peptides that spanned exon–exon junctions and that were within the range of 8 to 25 amino acid length were selected. The reason for selecting this size range for peptides is that confident peptide identification can be more stringently obtained in a database search for peptides within this range. From these exon junction spanning peptides, those that were also present in the protein database (i.e., from already known isoforms) were filtered out. Further peptide sequences that mapped to the database of a six-frame translation of genome sequence were removed. Finally, 22,777 tryptic peptide sequences that qualified all above criteria were used as a database for the MS/MS ion search.
Workflow for genome annotation
Peptides obtained after application of the 1% FDR cutoff were selected for further analysis using a computational pipeline developed for identification of novel genes, correction of gene models, and validation of known genes of An. gambiae. From the genome database search results, peptides mapping to known proteins from the protein database (AgamP3.6) were excluded to obtain novel peptides that we are calling as genome search-specific peptides (GSSPs). TBLASTN was performed to obtain the genome coordinates of GSSPs. Peptides that mapped to more than one place in the genome were not included for further analysis. GSSPs were programmatically categorized in six categories as (1) peptides mapping to intergenic regions; (2) peptides mapping to intronic regions; (3) peptides overlapping exon–intron junctions; (4) peptides mapping onto gene boundaries; (5) peptides mapping to UTRs; and (6) peptides mapping within exons but translated in a different frame. Intergenic peptides that mapped within the 2-kb region of the genome were grouped and prioritized for analysis. The peptides of interest were analyzed as follows. Alternative gene models by SNAP, Fgenesh, and Genmark were checked in regions where GSSPs were mapping. Peptide sequences were aligned to An. gambiae EST sequences (downloaded from the NCBI EST database and VectorBase) using the TBLASTN algorithm to find additional EST evidence for the GSSPs. Similarly, the protein BLAST algorithm was used to align GSSPs to protein sequences from Ae. aegypti and Cx. quinquefasciatus to find ortholog evidence. In-house scripts were used to fetch the peptides that spanned splice junctions from the protein database search results.
Primer design strategy
Gene-specific primers were designed using the Primer 3 (v.0.4.0) software (Rozen and Skaletsky 2000). Some of the primers were designed manually. The primers were designed based on the annotation of the exons for the gene models predicted by the alternate gene prediction programs described in the text. The gene model selected was one that either gave a good alignment with other related species using NCBI BLASTX and TBLASTX or that was consistent among all the alternate gene prediction models used. The primer pairs spanned an intron wherever possible except in cases of single exon gene models and where the amplicon size was exceeding 2 kb.
RT-PCR validation
Total RNA was isolated from 346 midguts, 226 Malpighian tubules, 164 ovaries, and 102 salivary glands dissected from female adult An. gambiae mosquitoes. Total RNA was isolated from pooled mosquitoes using the QIAGEN miRNeasy mini kit. All RNA was treated with RNase-free DNase to remove any contaminating genomic DNA. cDNA synthesis was performed using Quantiscript Reverse Transcriptase provided using QIAGEN's QuantiTect Reverse Transcription kit. The resulting cDNA was used as a template for the PCR reactions. The PCR was performed using Platinum Taq polymerase (Invitrogen), 30 to 50 ng of cDNA, and 200 nM each primer pair. PCR products were analyzed by agarose gel electrophoresis to determine the presence or absence of the product fragments. The size was determined using a 100-bp DNA ladder (Invitrogen). Actin was used as a positive control, and the isolated RNA as a template was used as negative control to ensure that there is no genomic DNA contamination. Once bands were observed at the expected sizes, they were excised and purified using QIAGEN's QIAquick Gel Extraction kit. The purified PCR products were subjected to automated DNA sequencing using the Applied Biosystems 3730xl DNA Analyzer at the Johns Hopkins sequencing facility.
Data access
The complete set of raw mass spectrometry data (.raw files) generated from this study has been made available through the Tranche server (http://proteomecommons.org/tranche). The raw data files used for genome annotation can be retrieved using the stable URL https://proteomecommons.org/tranche/data-downloader.jsp?i=75536. Additionally, all the peptide data are made available as a DAS track that will allow users to visualize the identified peptides on VectorBase genome browsers. This will allow users to get an integrated view of the genomic, transcriptomic, and proteomic information in a single location. The cDNA sequence of novel and corrected genes can be retrieved from the NCBI EST database (http://www.ncbi.nlm.nih.gov/nucest) and Genbank (http://www.ncbi.nlm.nih.gov/genbank/) using the accession numbers listed in the Supplemental Material.
Competing interest statement
B.D. is an employee of Thermo Fisher Scientific. All other authors have expressed no conflict of interest.
Acknowledgments
We thank the Department of Biotechnology (DBT) of the Government of India for research support to the Institute of Bioinformatics, Bangalore. D.S.K. and S.R. are recipients of a Senior Research Fellowship from the University Grants Commission (UGC), India. N.A.S., S.B.D., and H.P. are recipients of the Senior Research Fellowship from the Council for Scientific and Industrial Research (CSIR), India. T.S.K.P. is a recipient of a Young Investigator award from the DBT, Government of India. R.S. is a research associate supported by DBT. This study was supported in part by a pilot grant from Johns Hopkins Malaria Research Institute (JHMRI), the Bloomberg Family Foundation, and grant S10RR023025 from the High End Instrumentation Program of the National Institutes of Health.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.127951.111.
References
- Baerenfaller K, Grossmann J, Grobei MA, Hull R, Hirsch-Hoffmann M, Yalovsky S, Zimmermann P, Grossniklaus U, Gruissem W, Baginsky S 2008. Genome-scale proteomics reveals Arabidopsis thaliana gene models and proteome dynamics. Science 320: 938–941 [DOI] [PubMed] [Google Scholar]
- Birney E, Clamp M, Durbin R 2004. GeneWise and Genomewise. Genome Res 14: 988–995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunner E, Ahrens CH, Mohanty S, Baetschmann H, Loevenich S, Potthast F, Deutsch EW, Panse C, de Lichtenberg U, Rinner O, et al. 2007. A high-quality catalog of the Drosophila melanogaster proteome. Nat Biotechnol 25: 576–583 [DOI] [PubMed] [Google Scholar]
- Choudhary JS, Blackstock WP, Creasy DM, Cottrell JS 2001. Matching peptide mass spectra to EST and genomic DNA databases. Trends Biotechnol 19: S17–S22 [DOI] [PubMed] [Google Scholar]
- Driessen HP, de Jong WW, Tesser GI, Bloemendal H 1985. The mechanism of N-terminal acetylation of proteins. CRC Crit Rev Biochem 18: 281–325 [DOI] [PubMed] [Google Scholar]
- Eyras E, Caccamo M, Curwen V, Clamp M 2004. ESTGenes: Alternative splicing from ESTs in Ensembl. Genome Res 14: 976–987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gevaert K, Goethals M, Martens L, Van Damme J, Staes A, Thomas GR, Vandekerckhove J 2003. Exploring proteomes and analyzing protein processing by mass spectrometric identification of sorted N-terminal peptides. Nat Biotechnol 21: 566–569 [DOI] [PubMed] [Google Scholar]
- Goetze S, Qeli E, Mosimann C, Staes A, Gerrits B, Roschitzki B, Mohanty S, Niederer EM, Laczko E, Timmerman E, et al. 2009. Identification and functional characterization of N-terminally acetylated proteins in Drosophila melanogaster. PLoS Biol 7: e1000236 doi: 10.1371/journal.pbio.1000236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta N, Benhamida J, Bhargava V, Goodman D, Kain E, Kerman I, Nguyen N, Ollikainen N, Rodriguez J, Wang J, et al. 2008. Comparative proteogenomics: Combining mass spectrometry and comparative genomics to analyze multiple genomes. Genome Res 18: 1133–1142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harsha HC, Molina H, Pandey A 2008. Quantitative proteomics using stable isotope labeling with amino acids in cell culture. Nat Protoc 3: 505–516 [DOI] [PubMed] [Google Scholar]
- Holt RA, Subramanian GM, Halpern A, Sutton GG, Charlab R, Nusskern DR, Wincker P, Clark AG, Ribeiro JM, Wides R, et al. 2002. The genome sequence of the malaria mosquito Anopheles gambiae. Science 298: 129–149 [DOI] [PubMed] [Google Scholar]
- Kalume DE, Okulate M, Zhong J, Reddy R, Suresh S, Deshpande N, Kumar N, Pandey A 2005a. A proteomic analysis of salivary glands of female Anopheles gambiae mosquito. Proteomics 5: 3765–3777 [DOI] [PubMed] [Google Scholar]
- Kalume DE, Peri S, Reddy R, Zhong J, Okulate M, Kumar N, Pandey A 2005b. Genome annotation of Anopheles gambiae using mass spectrometry-derived data. BMC Genomics 6: 128 doi: 10.1186/1471-2164-6-128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korf I 2004. Gene finding in novel genomes. BMC Bioinformatics 5: 59 doi: 10.1186/1471-2105-5-59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuster B, Mortensen P, Andersen JS, Mann M 2001. Mass spectrometry allows direct identification of proteins in large genomes. Proteomics 1: 641–650 [DOI] [PubMed] [Google Scholar]
- Lawson D, Arensburger P, Atkinson P, Besansky NJ, Bruggner RV, Butler R, Campbell KS, Christophides GK, Christley S, Dialynas E, et al. 2009. VectorBase: a data resource for invertebrate vector genomics. Nucleic Acids Res 37: D583–D587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Ma L, Li H, Vang S, Hu Y, Bolund L, Wang J 2007. Snap: an integrated SNP annotation platform. Nucleic Acids Res 35: D707–D710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann M, Pandey A 2001. Use of mass spectrometry-derived data to annotate nucleotide and protein sequence databases. Trends Biochem Sci 26: 54–61 [DOI] [PubMed] [Google Scholar]
- Menon R, Zhang Q, Zhang Y, Fermin D, Bardeesy N, DePinho RA, Lu C, Hanash SM, Omenn GS, States DJ 2009. Identification of novel alternative splice isoforms of circulating proteins in a mouse model of human pancreatic cancer. Cancer Res 69: 300–309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrihew GE, Davis C, Ewing B, Williams G, Kall L, Frewen BE, Noble WS, Green P, Thomas JH, MacCoss MJ 2008. Use of shotgun proteomics for the identification, confirmation, and correction of C. elegans gene annotations. Genome Res 18: 1660–1669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molina H, Bunkenborg J, Reddy GH, Muthusamy B, Scheel PJ, Pandey A 2005. A proteomic analysis of human hemodialysis fluid. Mol Cell Proteomics 4: 637–650 [DOI] [PubMed] [Google Scholar]
- Molina H, Horn DM, Tang N, Mathivanan S, Pandey A 2007. Global proteomic profiling of phosphopeptides using electron transfer dissociation tandem mass spectrometry. Proc Natl Acad Sci 104: 2199–2204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mongin E, Louis C, Holt RA, Birney E, Collins FH 2004. The Anopheles gambiae genome: an update. Trends Parasitol 20: 49–52 [DOI] [PubMed] [Google Scholar]
- Okulate MA, Kalume DE, Reddy R, Kristiansen T, Bhattacharyya M, Chaerkady R, Pandey A, Kumar N 2007. Identification and molecular characterization of a novel protein Saglin as a target of monoclonal antibodies affecting salivary gland infectivity of Plasmodium sporozoites. Insect Mol Biol 16: 711–722 [DOI] [PubMed] [Google Scholar]
- Oyama M, Kozuka-Hata H, Suzuki Y, Semba K, Yamamoto T, Sugano S 2007. Diversity of translation start sites may define increased complexity of the human short ORFeome. Mol Cell Proteomics 6: 1000–1006 [DOI] [PubMed] [Google Scholar]
- Pandey A, Lewitter F 1999. Nucleotide sequence databases: a gold mine for biologists. Trends Biochem Sci 24: 276–280 [DOI] [PubMed] [Google Scholar]
- Pandey A, Mann M 2000. Proteomics to study genes and genomes. Nature 405: 837–846 [DOI] [PubMed] [Google Scholar]
- Peri S, Pandey A 2001. A reassessment of the translation initiation codon in vertebrates. Trends Genet 17: 685–687 [DOI] [PubMed] [Google Scholar]
- Rozen S, Skaletsky H 2000. Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol 132: 365–386 [DOI] [PubMed] [Google Scholar]
- Sevinsky JR, Cargile BJ, Bunger MK, Meng F, Yates NA, Hendrickson RC, Stephenson JL Jr 2008. Whole genome searching with shotgun proteomic data: Applications for genome annotation. J Proteome Res 7: 80–88 [DOI] [PubMed] [Google Scholar]
- Suzuki Y, Sugano S 2006. Transcriptome analyses of human genes and applications for proteome analyses. Curr Protein Pept Sci 7: 147–163 [DOI] [PubMed] [Google Scholar]
- Tress ML, Wesselink JJ, Frankish A, Lopez G, Goldman N, Loytynoja A, Massingham T, Pardi F, Whelan S, Harrow J, et al. 2008. Determination and validation of principal gene products. Bioinformatics 24: 11–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia D, Sanderson SJ, Jones AR, Prieto JH, Yates JR, Bromley E, Tomley FM, Lal K, Sinden RE, Brunk BP, et al. 2008. The proteome of Toxoplasma gondii: integration with the genome provides novel insights into gene expression and annotation. Genome Biol 9: R116 doi: 10.1186/gb-2008-9-7-r116 [DOI] [PMC free article] [PubMed] [Google Scholar]