Abstract
Dictyostelium discoideum (DD), an extensively studied model organism for cell and developmental biology, belongs to the most derived group 4 of social amoebas, a clade of altruistic multicellular organisms. To understand genome evolution over long time periods and the genetic basis of social evolution, we sequenced the genomes of Dictyostelium fasciculatum (DF) and Polysphondylium pallidum (PP), which represent the early diverging groups 1 and 2, respectively. In contrast to DD, PP and DF have conventional telomere organization and strongly reduced numbers of transposable elements. The number of protein-coding genes is similar between species, but only half of them comprise an identifiable set of orthologous genes. In general, genes involved in primary metabolism, cytoskeletal functions and signal transduction are conserved, while genes involved in secondary metabolism, export, and signal perception underwent large differential gene family expansions. This most likely signifies involvement of the conserved set in core cell and developmental mechanisms, and of the diverged set in niche- and species-specific adaptations for defense and food, mate, and kin selection. Phylogenetic dating using a concatenated data set and extensive loss of synteny indicate that DF, PP, and DD split from their last common ancestor at least 0.6 billion years ago.
The central and most fascinating problem in biology is how natural selection has acted on random changes in the genomes of individuals to generate the immense range and complexity of extinct and extant living forms. However, understanding the relationship between genotypic and phenotypic change on a genome-wide scale is complicated by the large number of loci involved and the range of phenotypic change. Comparative genomics is the tool of choice to define common gene sets and the first occurrence of genetic changes that may have caused phenotypic innovation. Genetic manipulation of altered genes can then reveal whether the genomic change was causal to the phenotypic alteration.
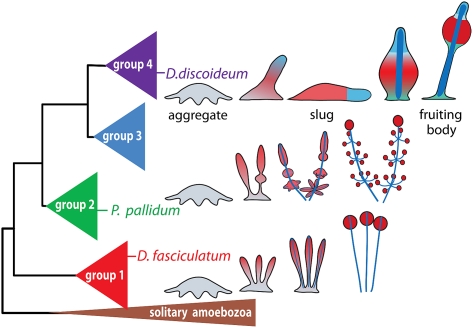
The social amoeba Dictyostelium discoideum (DD) is a widely used model system for studying a range of problems in cell and developmental biology and more recently the evolution of social behavior and multicellularity. Social amoebas are a single clade within the supergroup of Amoebozoa and are marked by their ability to sacrifice their life for the benefit of other members of the same species. They are common soil inhabitants that show the greatest species variety in the wet tropics, but can also live under arctic, alpine, and desert conditions (Swanson et al. 1999). A molecular phylogeny based on SSU rRNA and α-tubulin sequence data subdivides the social amoebas into four taxon groups, with DD residing in group 4 (Schaap et al. 2006). Groups 1–3 are characterized by forming clustered and branched fruiting structures that consist solely of stalk and spore cells (Fig. 1). Similar to their amoebozoan ancestors, many species in these groups can still encyst as individual cells. Group 4 species do not show encystation and mainly form large, solitary, and unbranched fruiting bodies, which support their stalk and spore mass with up to three additional cellular support structures. Furthermore, only group 4 uses cAMP as chemoattractant, while groups 1–3 use a range of other compounds (Fig. 1; Schaap et al. 1985, 2006).
Figure 1.
Phylogenetic position and life cycles of test species. Schematic representation of the SSU rRNA phylogeny of all social amoebas, with solitary amoebas as outgroup. DF and PP reside in groups 1 and 2, respectively. They differ from DD by generally forming small clustered and/or branched fruiting structures from a single aggregate, in which prespore cells differentiate first and the stalk is formed by dedifferentiation of prespore cells at the apex. In contrast, group 4 taxa form robust solitary unbranched fruiting structures and set aside appropiate proportions of prestalk cells after aggregation. They also differentiate into three additional cell types that support the stalk and spore head, and their sorogens can migrate extensively to seek optimal spots for fruiting body construction (Schaap et al. 2006; A Skiba and P Schaap, in prep.).
The DD genome was completely sequenced and comprises six chromosomes, an extrachromosomal element encoding rRNA genes, and a mitochondrial genome (Ogawa et al. 2000; Sucgang et al. 2003; Eichinger et al. 2005). Several exceptional features were described, among them a high A/T nucleotide content, surpassed only by Plasmodium falciparum (Gardner et al. 2002) and extreme richness in transposons and other repetitive elements (Glöckner et al. 2001). These elements not only had an impact on the genomic landscape in DD, but also contributed to basic genome organization by forming parts of telomeres and centromeres. Other interesting features were surprisingly large families of ABC transporters, polyketide synthases, and G-protein-coupled receptors. A draft genome sequence for another group 4 species D. purpureum (DP) was recently published. This genome shared most of the features described above with the DD genome (Sucgang et al. 2011).
To date, comparative genomics in the Amoebozoa was restricted to DD, DP, and the obligatory parasite Entamoeba histolytica (Eichinger and Noegel 2005; Loftus et al. 2005). EST data from a syncytial Amoebozoan, Physarum polycephalum, are available (Glöckner et al. 2008), but different life styles, incompleteness of EST data, and vast evolutionary distances between these taxa make a meaningful comparison difficult. Comparative genomics between more closely related species can be a powerful tool to retrace events that remodeled genome structure and to pinpoint positive selection events that may have contributed to phenotypic innovation and speciation, as is exemplified by the analyses of genomes from ∼30 yeast species and eight Drosophila species (Dujon et al. 2004; Kellis et al. 2004; Drosophila 12 Genomes Consortium 2007).
It is presently not known which genome features unite the social amoebas and which make them different. A previous study of the mitochondrial genomes (Ogawa et al. 2000; Heidel and Glöckner 2008) of several social amoebae showed conservation on the organizational level when compared with DD, but high sequence divergence. This study aims at the investigation of the extent of conservation and change in nuclear genome structures and gene content across the social amoebas using a relatively robustly developing taxon from group 1, Dictyostelium fasciculatum (DF), and Polysphondylium pallidum (PP), a taxon with full genetic tractability from group 2, for complete genome sequencing and assembly. Combined with the potential for gene replacement in both PP and DD, the availability of the comparative genome analysis will offer outstanding opportunities to test which altered gene features were instrumental in causing the appearance of novel phenotypes in the course of adaptive evolution.
Our results indicate that the three genomes show considerable changes in chromosome organization and striking examples of both extreme conservation and extreme change in specific gene families, with some DD developmental control genes missing in basal taxa. The data of the work presented here are accessible via a GenColors-based database (Romualdi et al. 2005) (http://sacgb.fli-leibniz.de).
Results
Genome structures
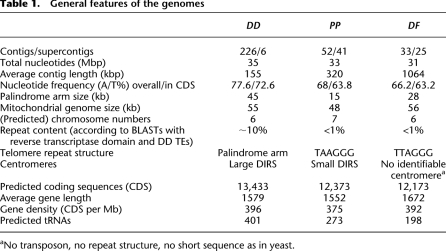
Definitive conclusions about genome structure and gains or losses of genes require complete or nearly complete genomic sequences. We achieved this by sequencing the PP and DF genomes to 15-fold coverage, construction of a dense physical map by end-sequencing of large insert clones, and primer walking to fill in gaps between assembled contigs (Supplemental Methods S6.2; Supplemental Table S1.1). Both genomes were finished to a very high standard, surpassing that of the model species DD with only a few gaps remaining, where no mapping data were available (Table 1).
Table 1.
General features of the genomes
The DD, DF and PP genomes are around 30 Mb in size (Table 1), with the main size difference appearing to be related to varying amounts of transposable elements (TEs) (for description of TE search see Supplemental Material). Compared with DD, the PP and DF genomes have lower A/T biases, fewer tRNAs, and fewer TEs (Table 1). All analyzed Dictyostelia contain an amplified extrachromosomal palindromic sequence that codes for rRNA genes (Supplemental Fig. S2.1). Based on the relative abundance of the sequencing reads that match the palindromes compared with unique parts of the genome, the PP, DF, and DD rDNA palindromes represent 5%, 9%, and 25% of the genomic DNA content, respectively. This difference is mainly attributable to the shorter palindrome arms in PP (15 kb) and DF (26 kb) compared with 45 kb for DD.
Telomeres and centromeres
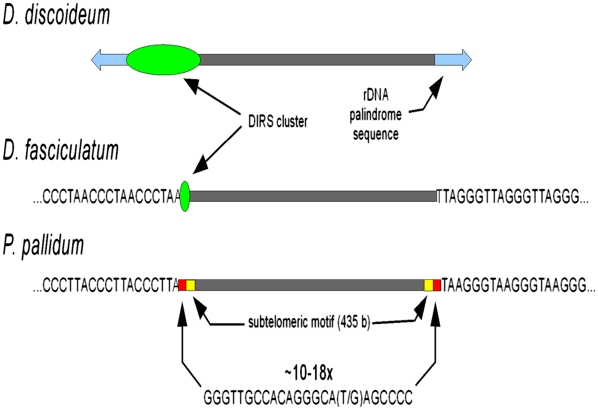
The telomere structures of the three genomes show striking differences (Table 1; Fig. 2). DD utilizes an unusual way of chromosome end maintenance by recruiting rDNA palindrome segments as “capping” entities to the chromosome ends (Eichinger et al. 2005). However, both the DF and PP chromosomes have normal eukaryote telomeres. The loss of telomere repeats in DD is correlated with a polyasparagine insertion in the telomerase reverse transcriptase protein (TERT) protein, which splits the functional domain of the protein into two halves (Supplemental Fig. S2.3). Polyasparagine stretches are common features of DD genes that are usually tolerated, but they normally do not disrupt functional domains. It is yet unclear whether the putative erosion of TERT function was a cause or a consequence of the lack of normal eukaryote telomeres in DD.
Figure 2.
Genome structures in social amoebae. Chromosomes are not drawn to scale. Telomere and centromere properties were deduced as described in Supplemental Material. Specific short sequence motifs are indicated as letters, identified common chromosomal regions as labeled areas. The polymorphism in the PP-specific sequence motif is indicated by the polymorphic nucleotides in brackets. Above this motif the number of observed repeat units is indicated.
Functional centromeres in DD consist of complex arrays of DIRS transposable elements at one end of each chromosome (Dubin et al. 2010). In DF we see such DIRS arrays, albeit much smaller than in DD (Fig. 2), since three of them are entirely spanned by fosmid clones. This indicates that the DIRS clusters here are <40 kb long, as compared with the DD clusters with >100 kb. In PP no DIRS elements or arrays of other TE types exist, and the nature of its centromeres, therefore, remains unclear. Based on nearly completed genomes and a karyotype for PP, we calculated the chromosome numbers for DF as six, and for PP as seven (Supplemental Methods; Supplemental Fig. S2.2).
Synteny
We used the orthology information to infer the number of syntenic regions between the genomes (Supplemental Table S3.4). Gene order conservation appears to be rare in the social amoebae, with only around 2000 CDS neighbors being conserved between DD and one of the other genomes. DF and PP show a slightly higher number of syntenic CDS, probably indicating a closer relationship between these taxa and suggesting that synteny breakup occurs in a time-dependent manner in social amoebae. Most synteny groups are small, the majority consisting of only two members. Only 1079 syntenic neighbors are conserved between all species. Thus, <10% of all protein-coding genes retained at least one neighboring gene in all species, indicating a frequent reshuffling of the genomes. In contrast, a recent comparison of the Dictyostelium purpureum and DD genomes showed that the majority of orthologous pairs seem to lie in syntenic regions (Sucgang et al. 2011), most likely highlighting that these two group 4 species diverged more recently from each other than any combination of DD, PP, and DF.
Protein-encoding potential
We detected a comparable number of protein-coding genes in the three genomes. Orthology relationships can be used to define the minimum conserved CDS set of the last common ancestor. We used the best bidirectional hit (BBH) approach to identify orthology relationships in all three genomes. Based on an identity threshold of 20%, over 50% of the protein length, we found that only half of the CDS (∼6500) are orthologous triplets (Supplemental Fig S3.5). Increasing the identity threshold yielded even less identifiable orthologs (Supplemental Fig. S3.3). Yet, even protein orthologs detected near the lowest threshold can be readily aligned and then yield slightly better identity scores than the BLAST based BBH analysis (Supplemental Fig. S3.4).
To complement the BBH approach, we additionally analyzed gene families using orthoMCL (Li et al. 2003). This analysis defined 8122 CDS groups, of which 5446 are common to the three species (Supplemental Fig. S3.2; Supplemental Table S3.3). Around 5000 gene families contain a single gene from each species and concomitantly have only one orthologous gene in each genome. The remaining genes are part of gene families with up to 210 members (Supplemental Table S3.2). Strikingly, some of the larger gene families are restricted to only one or two of the species (Supplemental Table S3.2). Thus, despite encoding approximately the same number of genes, all species underwent differential gene family expansions or gene losses (Supplemental Figs. S3.5, S3.6). Most of the proteins with functional domains can be clustered using the OrthoMCL analysis, and these proteins are also part of the common core set (Supplemental Fig. S3.6).
The OrthoMCL analysis suggests that species-specific duplications and deletions leave ∼3600 genes per species without one-to-one orthologs, while 5604 of the remaining 7054 genes have no assigned function and might represent either genes that have diverged too much for homology detection, genes in the making, or wrong gene predictions.
Homopolymer runs
Many DD proteins are characterized by long homopolymer runs of either Asparagine (N) or Glutamine (Q). In all species the A/T richer codons are preferred (AAT over AAC for N and CAA over CAG for Q), but this is most pronounced in DD, probably due to the overall higher A/T content in this genome (Table 1; Supplemental Table S2). Due to the high abundance of polyN tracts in DD, asparagine is by far the most abundant amino acid in this species (Eichinger et al. 2005), but not in PP and DF. PolyQ stretches are the second most abundant homopolymer runs in DD, and are similarly abundant in the PP and DF genomes (Supplemental Fig. S3.1; Supplemental Table S3.1). Thus, long homopolymer runs are a common feature of all social amoebae, but the polyN tracts are longer on average and more common in the DD genome. This could be due to species-specific amplifications in DD or deletions and shortenings in the other species.
Functional domains
The diversity and abundance of protein functional domains reflects the shared and specific potential for phenotypic complexity of species. We screened the three genomes for domain abundance using InterproScan (Supplemental Table S4.1; Mulder and Apweiler 2008). The majority of domains are present in roughly equal numbers in all genomes, but a subset is differentially amplified (Supplemental Table S4.2). Roughly 100 domains are absent from only one genome, whereas around the same number is present in one genome only. The five most amplified DD-specific domains are TE- or phage-integrase domains, or were defined using DD proteins as templates. Taking into account that Interpro domain accessions are partly redundant, with differing detection thresholds, there seems to be almost no differential domain usage in the three social amoebae species. An exception to this rule is, e.g., the Asp/Glu racemase domain (IPR018187), which is shared between PP and DF, but not present in DD. Furthermore, PP possesses five bacterial transcriptional regulators (IPR010499), of which three are located in the same chromosomal region (PPL_10904, PPL_10903, PPL_10898), indicating recent gene duplication events. Since these proteins are not found in other eukaryotes, they could be remnants of horizontal gene transfer.
Conservation and divergence in gene families
Loss and amplification of genes within specific gene families can be a major determinant for phenotypic innovation and speciation. We analyzed a number of gene families in each of the following functional categories: metabolism—cell shape and organization—gene transcription—export—cell adhesion—cell signaling.
Primary and secondary metabolism
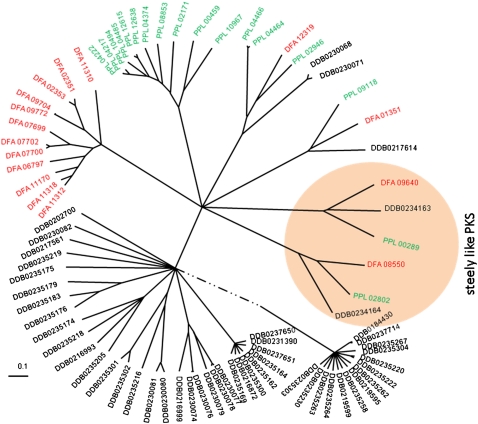
A comparison of the primary metabolism between the three species using the KEGG database (Kanehisa et al. 2008) revealed that this particular set of more than 400 proteins has remained remarkably unchanged since divergence from the last common ancestor (LCA). No gene family seems to have undergone a major expansion or shrinkage (Supplemental Tables S5.1, S5.2). This situation is entirely different for the polyketide synthases (PKS) that produce secondary metabolites, which have diverse functions as antibiotics and fungicides, and in defense against predation (Shen 2003). Type I iterative PKSs are large proteins, which consist of combinations of several distinct domains (Acyl transferase, AT; Acyl carier protein, ACP; Ketosynthase, KS; Ketoreductase, KR; Dehydratase, DH; Enoylreductase, ER; Methyltransferase, MT; Sulfhydrolase, SH; Thioesterase, TE). DD contains a wealth of such PKS genes (Supplemental Table S5.3), of which some were shown to produce compounds involved in cell differentiation (Austin et al. 2006). An alignment of the KS domains was used to calculate phylogenetic relationships between the PKS proteins (Fig. 3). Only steely-1 and steely-2 PKSs, which are involved in the synthesis of the spore maturation-inducing factor MPBP and the basal disc-inducing factor DIF (Austin et al. 2006; Narita et al. 2011) and a third uncharacterized gene form clear orthologous clusters. A second grouping contains members from all species, but with different copy numbers. Strikingly, three distinct expanded families are present exclusively in either DD, PP, or DF, indicating species or group-specific expansion from a single gene in the LCA (Fig. 3).
Figure 3.
Phylogenetic tree of PKS KS domains of social amoebae. The domains were extracted from the PKS proteins using hmmer models of DD-specific domains (Zucko et al. 2007). A ClustalW alignment was fed in the phylogeny software puzzle (Schmidt and von Haeseler 2007) and confirmed by neighbor-joining analysis using PHYLIP (Felsenstein 1989).
Export
The DD genome contains an unusually large number of 71 ATP-binding cassette (ABC) transporters, transmembrane proteins that are in eukaryotes used for export of natural products, toxic metabolites, and xenobiotics (Saurin et al. 1999). ABC transporters are either present as full transporters with two ATP binding cassettes (ABCs) and two sets of six transmembrane helices or half transporters with one of each (Anjard and Loomis 2002). PP and DF have in total 68 and 64 ABC proteins, respectively. Phylogenetic analysis of individual ABC domains shows that most of the half-transporters are conserved in 1:1:1 orthology relationships with occasional loss or gain of one or two genes. In contrast, the larger set of full transporters shows very extensive group or species-specific gene family expansions (Supplemental Fig. S5.9). It is remarkable that the divergent abilities of the DD, PP, and DF to synthesize unique repertoires of secondary metabolites (as shown above) match their divergent abilities to export toxic compounds. This suggests that diversification of chemical warfare strategies is an important aspect of adaptive evolution in the Dictyostelia. Interestingly, as is the case for the polyketide synthases, some of the ABC transporters came to play developmental roles. This is the case for the TagA-D subset of DD transporters with attached serine protease domains, which have roles in the processing of peptide signal molecules (Anjard and Loomis 2006). In PP and DF only, TagA and TagD appear to be conserved (Supplemental Fig. S5.9).
Cytoskeleton and movement
The microfilament system of DD has been extensively studied and much pioneering work on cytoskeleton and cell motility has been carried out in this model organism. In general terms, the repertoire of microfilament system components of DF and PP is almost a replica of that of DD, with only minor or no differences in many gene families (Supplemental Table S5.6). This is, for example, true for myosins, formins, and kinesins (Supplemental Figs. S5.1, S5.2; Supplemental Table S5.5).
The actin gene family has expanded in all three species. Since the sequences of all members of the expanded family are highly similar within one species, but differ between species, we conclude that the actin gene family frequently undergoes expansion and shrinkage. On the other hand, actin-related genes (ARP) are highly conserved between all species (Supplemental Fig. S5.3) and our results confirm and extend previous analyses (Joseph et al. 2008). Cofilin and comitin constitute examples of ARP families that have expanded differently. In DD, the cofilin subfamily of the actin depolymerizing factor family of proteins is represented by seven genes, compared with two in PP and one in DF (Supplemental Fig. S5.4; Supplemental Table S5.6), while comitin is represented by three genes in DD, six in PP, and five in DF.
Components involved in signaling to and from small GTPases of the Rho family are particularly important for cytoskeletal remodeling during chemotaxis and phagocytosis. Nearly every DD protein (Vlahou and Rivero 2006) has an ortholog in both DF and PP (Supplemental Table S5.10). Only the family of Rho GTPases seems to have diversified independently in the three species. This family comprises 20 rac genes and one pseudogene in DD, 14 genes in DF, and 21 genes in PP (Supplemental Fig. S5.8). Several rac genes have a direct ortholog in all three species, namely rac1a, racA–racE, racH, racL, and racP, indicating that this core of shared genes may play conserved roles in all social amoebae, while the others may mediate more species- or group-specific functions.
Gene transcription
The A/T content varies significantly between the three genomes (Table 1). Yet, the function of transcription factors (TFs) is tightly linked to their ability to detect their respective binding site in the genome. It is therefore of special interest to see whether a genome-wide nucleotide content difference affects the evolutionary trajectory of TFs. We identified TFs via their DNA-binding domains and found that each species contains roughly the same number of TFs in each TF class. Similar to DD, the basic helix–loop–helix TFs are missing in PP and DF (Eichinger et al. 2005; Supplemental Table S5.7). However, there is only limited conservation of specific TFs; only half of the identified TFs are part of the core set of proteins shared by all social amoebae. While in some classes, such as the STATs, an ortholog could be identified for each member, most members of other classes, such as the GATA-type zinc fingers, are not represented in the core orthologous set, indicating high divergence and adaptation to the genomic environment of individual TFs in the three species.
Cell adhesion and cell–cell communication
Adhesion
Soil amoebas generally require cell-adhesion proteins for attachment to the substratum and to prey during phagocytosis, but cell adhesion becomes particularly important during multicellular development. First, to provide general cohesion of the multicellular cell mass, but also to provide selective adhesion that allows the cells to sort and form tissues. Moreover, direct cell–cell interactions can have a signaling role. In DD, changes in the adhesive properties during development are mediated by a range of adhesion molecules that are expressed in a temporally and spatially regulated manner, correlating with the morphological changes (Siu et al. 2004).
We found that proteins involved in adhesion to particles, such as the Phg1A, Phg1B, and Phg1C, or to substrate, such as talin, vinculin, paxillin, and the integrin-like Sib proteins of DD, are also present in PP and DF (Supplemental Table S5.8). The calcium-dependent cell-adhesion proteins (cad; IPR015059) that mediate cell adhesion in pre-aggregative development are represented by three (DD), two (PP), and five (DF) members, showing 60%–70% identity amongst each other. The beta-catenin-like protein Aardvark and an interacting alpha-catenin-related protein are also present in PP and DF. Aardvark is part of adherens junction-like structures in the fruiting body tip and has additional signaling roles (Grimson et al. 2000; Dickinson et al. 2011). However, csA/gp80 and LagC/gp150, two members of the immunoglobulin superfamily of adhesion molecules, which in DD mediate cell adhesion during aggregation and contact-mediated signaling after aggregation, respectively, have no detectable orthologs in PP and DF. LagC and the related LagB have a role in kin discrimination, and both proteins are highly polymorphic in natural populations (Benabentos et al. 2009).
Cell–cell communication
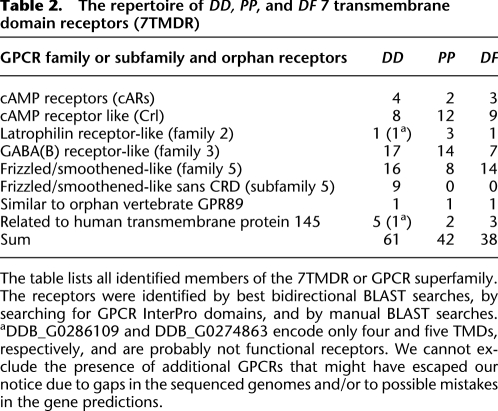
Cell-to-cell signaling is essential for the coordination of cell movement and cell differentiation that together allow multicellular development. The largest class of receptors for external stimuli in eukaryotes is the family of G-protein-coupled receptors (GPCRs) that generally activate intracellular responses by interacting with heterotrimeric G-proteins. They mediate diverse biological processes such as hormone action, odorant sensing, vision, cell migration, and development, and can be divided into six major families, of which four are present in DD (Eichinger et al. 2005; Xue et al. 2008), namely the secretin (family 2), metabotropic glutamate GABAB (family 3), the frizzled/smoothened (family 5), the cARs and Crls (cAMP receptor and cAMP receptor like) (family 6), and several orphan receptors. While all three dictyostelids have members in the same four families, the DD genome has the most expansive repertoire and harbors 61 members (Table 2). The total number of receptors rises from group 1 to group 2 by 10% and to group 4 by another 50%. Remarkably, within the cAR/Crl family, the Crl receptors show 1:1:1 orthology relationships, while the cARs show significant species-specific expansions. The largest differences were found for the GABAB and frizzled/smoothened receptors, which have strongly expanded in DD (Table 2; Supplemental Fig. S5.5). Interestingly, only the GPCRs are diverse, the subsequent links in the signal transduction chain, such as the components of the heterotrimeric G-proteins and downstream targets such as the adenylate and guanylate cyclases are nearly completely conserved in all species (Supplemental Figs. S5.6, S5.7, S5.8; Supplemental Table S5.9).
Table 2.
The repertoire of DD, PP, and DF 7 transmembrane domain receptors (7TMDR)
Cyclic nucleotide signaling
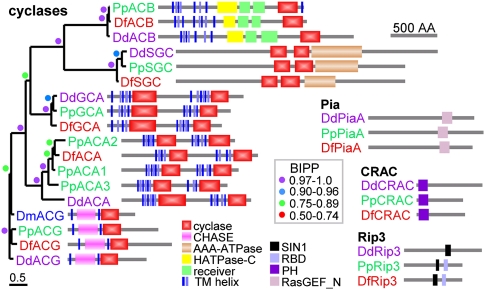
cAMP was initially characterized as the secreted chemoattractant that causes DD cells to aggregate, while cGMP is implicated as a second messenger in the chemotactic response to cAMP. Subsequent studies revealed that cAMP has a broad range of developmental roles. As a secreted signal acting on cAMP receptors (cARs), it up-regulates aggregation genes, triggers entry into the spore pathway, and inhibits stalk cell differentiation. As an intracellular messenger for other stimuli, it acts on cAMP-dependent protein kinases (PKA) to regulate initiation of development, maturation of spores and stalk cells, and germination of spores (Saran et al. 2002). DD uses the adenylate cyclases ACA, ACB, and ACG and the guanylate cyclases sGC and GCA for synthesis of cAMP and cGMP, respectively. All five cyclases are conserved in PP and DF, inclusive of their functional domain architecture (Fig. 4). PP carries two gene duplications of ACA, with the ACA1 flanking genes showing synteny with those of DD ACA, thus identifying PP ACA1 as the DD ACA ortholog. The transient activation of ACA by a complex signaling pathway generates the waves of cAMP that coordinate DD aggregation and fruiting body morphogenesis. Three essential components of this pathway: CRAC, PIA, and Rip3 (Lee et al. 2005) are also present in PP and DF, suggesting that the dynamic regulation of ACA is conserved throughout the Dictyostelia. In DD, seven phosphodiesterases (PDEs) are responsible for degradation of cyclic nucleotides (Bader et al. 2007). Except for the extracellular cAMP-PDEs, Pde4 and Pde7, these proteins are fully conserved in DF and PP, with the intracellular cAMP-PDE RegA being duplicated in both DF and PP, and the extracellular cAMP-PDE PdsA in DF (see Supplemental Fig. S5.6). Five intracellular cyclic nucleotide binding proteins, of which two also have PDE activity, are fully conserved between the three species, while the cell surface cAMP receptors show independent duplications in DD, PP, and DF (Supplemental Fig. S5.6A). In conclusion, the genes associated with cyclic nucleotide signaling appear to be well conserved, suggesting that the role of cGMP and most of the multiple roles of cAMP in DD are also present in DF and PP. This does not mean that these roles are invariant; additional functionality may have appeared through changes in gene expression. This is the case for cAR1, where addition of a distal “early” promoter to the existing “late” promoter caused cAR1 to be expressed before and during aggregation, and enabled the chemoattractant function of cAMP during DD aggregation (Alvarez-Curto et al. 2005).
Figure 4.
Architectural conservation of adenylate and guanylate cyclases. Proteins were identified by TBLASTN search with DD cyclase sequences and query of SACGB with the IPR001054 A/G_cyclase domain. Protein phylogenies of nucleotidyl cyclases were constructed using Bayesian inference (Ronquist and Huelsenbeck 2003) from alignments of the shared functional domains as described in the legend to Supplemental Figure S5.6. For proteins with two cyclase domains, a tandem alignment of both domains was used, with the single domains of ACB and ACG used twice. The tree is presented unrooted and decorated with the domain architectures of the proteins as analyzed by SMART (Schultz et al. 1998). The posterior probabilities (BIPP) of nodes are indicated by colored circles. Locus and gene ids: (ACB) DD: DDB_G0267376, PP: PPL_10642, DF: DFA_10857; (SGC) DD: DDB_G0276269, PP: PPL_05930, DF: DFA_12195; (GCA) DD: DDB_G0275009, PP: PPL_11010, DF: DFA_10832; (ACA1) DD: DDB_G0281545, PP: PPL_01657, DF: DFA_04345; (ACA2) PP: PPL_12370; (ACA3) PP: PPL_10658; (ACG) DD: DDB_G0274569, PP: PPL_06767, DF: DFA_08118; (Pia) DD:DDB_G0277399, PP: PPL_02311, DF: DFA_00057; (CRAC) DD: DDB_G0285161, PP: PPL_07100; (Rip3) DD: DDB_G0284611, PP: PPL_11127, DF: DFA_06425.
At least five members of another set of 15 receptors, the sensor histidine kinases (HKs), regulate cAMP signaling by phosphorylating the response regulator domain of the cAMP PDE RegA, thereby activating the enzyme (Thomason et al. 1999). RegA in turn controls the activation state of PKA, which, as described above, regulates crucial steps in the developmental program. The set of histidine kinases is remarkably well conserved in PP and DF, except for DhkI and DhkA, which are not present in DF, and DhkH and DhkN, which are missing from PP and DD, respectively (Supplemental Fig. S5.7). Interestingly, the serine protease-coupled ABC transporter TagC is also missing in DF (Supplemental Fig. S5.10). TagC produces SDF-2, one of several spore maturation-inducing factors and the specific ligand of DhkA (Anjard and Loomis 2005). This whole pathway is therefore likely to be missing in DF. Apart from RegA, ACB (Supplemental Fig. S5.6), and the HKs, there are four other rather featureless proteins with response regulators in DD. Remarkably, this set is also present in DF and PP (Supplemental Fig. S5.7), suggesting that these proteins have core functions in HK initiated signal transduction.
Protein sequence divergence
Nucleotide exchanges in orthologous genes
Mutations accumulate in a more or less linear temporal manner if no selective constraint is imposed by the mutated nucleotide base. In the case of coding sequences, this is true for changes in the third bases of codons, which often do not affect the cognate amino acid identity (synonymous exchange), or in nonsynonymous exchanges that do not affect protein function. Over shorter evolutionary time scales, synonymous exchanges outweigh the nonsynonymous ones in most genes, due to the neutral accumulation of such mutations, while an increased or decreased ratio of synonymous to nonsynonymous changes in specific genes indicates whether those genes are under purifying or positive selection, respectively.
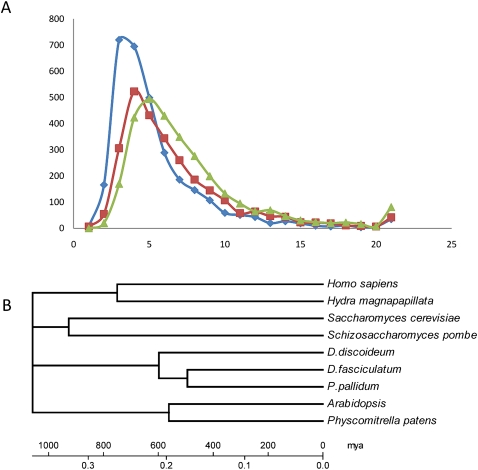
We analyzed the nucleotide exchanges in the set of orthologs with a minimum identity of 40% in each species pair. (dS/dN) ratios (Fig. 5A) show the protein variations between DD, PP, and DF. Peaks of all two-way comparisons are in the same range, but the most closely related pair PP/DF (green curve in Fig. 5A) seems to have accumulated slightly less nonsynonymous exchanges than the other species pairs (DD/PP and DD/DF) (see also Supplemental Fig. S6.1 for (Sd-Nd)/(Sd+Nd) values).
Figure 5.
(A) Ratio (dS/dN) of scaled synonymous (dS) to scaled nonsynynonymous (dN) base substitutions between species pairs. The summed number of proteins with a given value of dS/dN ratios is shown. Only true orthologs with a minimum sequence identity of 40% over at least 50% of the gene length were used for calculation. Protein pairs with a dS of 0 are omitted. (Blue) DD/DF species pair; (red) DD/PP species pair; (green) DF/PP species pair. (B) Calibrated phylogenetic tree of major eukaryote lineages. A concatenated data set of 33 orthologous genes (Supplemental Table S6.1) was aligned using ClustalW. A neighbor-joining tree was reconstructed with MEGA4 (Pairwise gap deletion, Poisson correction, bootstrap test with 1000 replicates). The scale was calibrated using an estimated divergence time of 750 mya for the human/hydra split. The sequence of early evolutionary events cannot be resolved with this data set. A similar scale was obtained by using the moss/vascular plant split at 550 mya.
Estimates for the timing of divergence from a LCA can be derived from phylogenetic analysis of protein sequences. To avoid errors associated with specific proteins being under selection in some lineages, we randomly selected a total of 33 genes that are well conserved between fungi (Saccharomyces cerevisiae, Schizosaccharomyces pombe), plants (Arabidopsis thaliana, Physcomitrella patens), animals (Hydra magnapapillata, Homo sapiens), and social amoebae (PP, DF, DD). The 33 genes were concatenated for each species (Supplemental Table S6.1), and alignments of all arrays were used to infer phylogenetic relationships. The root of the Dictyostelid clade is at a different position in the 33-gene tree (Fig. 5B) and the SSU rRNA-based tree (Fig. 1; Schaap et al. 2006), indicating that further phylogenetic analysis of all Dictyostelia with more genes is required.
The tree was linearized and calibrated on an estimated divergence time of 750 mya for the Hydra/Human split (Hedges 2002). We could then estimate a divergence time for the two plant lineages at 560 mya, for the fungal split at 900 mya, and for the social amoebae split at 600 mya, with DF/PP diverging at 494 mya (Fig. 5B). The divergence data for the plants, metazoa, and fungi are in good agreement with previous estimates (Hedges 2002). This provides a more recent estimate of 600 mya for the emergence of the four groups of Dictyostelia than provided by the comparison of (Sd-Nd)/(Sd+Nd) ratios (>750 mya), synteny breaks (1200 mya) (see Supplemental Section 3.3), or the genetic variation in SSU rRNA and α-tubulin sequences (1000 mya) as determined previously (Schaap et al. 2006). In the absence of a fossil record to calibrate molecular events, all of these estimates remain tenuous, but even the most conservative estimate of 600 mya indicates that the earliest social amobae speciation event is likely to have occurred before the divergence of plants (Rubinstein et al. 2010), shortly before or during the Cambrian explosion.
Discussion
The social amoebas represent an independent invention of multicellularity within the deeply branching supergroup of amoebozoa, and are themselves subdivided into four major groups. Combined with the published genome of the group 4 taxon DD, the finished genomes of group 1 and group 2 representative taxa now enabled the analysis of the entire breadth of genetic diversity in the social amoebas. Having the complete genomes in hand, we can date the split of groups 1–4 from their LCA at 600 to 1000 mya ago. At the time of this split no vascular plants existed and metazoans had not yet emerged from the oceans. Because fruiting body formation in the dictyostelids does not occur under water, it is not too farfetched to conclude that the social amoebas were among the first multicellular organisms on dry land. In the absence of a plant canopy, they would have been exposed to high levels of UV irradiation, possibly explaining their extremely high resistance to DNA damage by UV (Deering 1988).
We found interesting patterns of protein conservation across taxon groups. Only about half of the genes in each genome can be recognized as orthologs. However, the total content of protein functional domains shows little change between genomes, and proteins with well-defined functions in primary metabolism, cytoskeletal functions, and intracellular signal processing are generally conserved as 1:1:1 orthologs. A special case are the transcription factors; here the number of genes in each of ∼20 TF classes is conserved between genomes, but only half of these were recognized as orthologs. We therefore believe that while species-specific gene family expansion and loss explains part of the orthologless genes, a large proportion of orthologless genes in the three genomes is not due to extensive gene gain and loss. Instead, a likely cause is that the accumulation of mutations over 0.6 billion years eroded sequence similarities to such an extent that orthologs are no longer recognizable as such. A corollary of this conclusion is that the coding potential of the three genomes is actually well conserved.
The interwoven networks of interacting proteins that participate in processes such as primary metabolism, cytokinesis, cell motility, vesicle trafficking, and intracellular transduction of external stimuli are extremely well conserved between DD, DF, and PP. Some gene families like polyketide synthases (PKS), ABC transporters, and the G-protein-coupled receptors (GPCR) show large differential amplifications. A few members of these large families have roles in producing, processing, and detection of developmental signals, but the functions for most family members are unknown. It was shown in another system that an albino mutant is caused by the mutation of a PKS gene (Tanguay et al. 2006). Thus, it is possible that some colors of the fruiting bodies are caused by different sets of PKS genes in the Dictyostelidae. Furthermore, the species-specific expansions of PKSs, which synthesize noxious compounds, and the ABC transporters that export them suggest habitat-specific development of attack and defense strategies against predation.
Large families of GPCRs are involved in odorant and chemoattractant sensing in vertebrates and worms, and the latter functions may also be their predominant role in social amoebas. We speculate that these gene family expansions possibly mark adaptation to available bacterial food sources in different ecological niches and may thus be the drivers of speciation.
Despite their ancient origins, the social amoebae do not seem to have achieved a great degree of complexity in their body plans when compared with animals. Nevertheless, there are trends in the morphological evolution of Dictyostelia. Group 4 species uniquely have sophisticated cellular support structures that require differentiation into three more cell types. They also set aside stalk cell precursors before fruiting body formation, instead of forming stalks by dedifferentiation of prespore cells (Schaap et al. 2006).
It is not yet possible to associate the morphological differences between group 4 and groups 1–3 with alterations in specific genes. However, the genome of the group 4 species DD has many unique features. It has lost conventional telomeres and gained large amounts of transposable elements, repeats of tRNA and rRNA genes, and polyasparagine repeats in coding sequences. Furthermore, protein families showing extensive gene gain and loss between species tend to have more members in DD than in PP and DF. Transposable elements and repeats are known agents for generating genome plasticity and diversification in prokaryotes and vertebrates (Bohne et al. 2008; Kersulyte et al. 2009), so they may have also contributed to increased morphological complexity in DD.
The sequenced genomes offer tremendous opportunities to identify correlations between changes in genotype and phenotype during Dictyostelid evolution at all levels of their cellular and developmental processes. As a prerequisite for such analyses, methodologies have been established for gene disruption and allelic replacement in two species, DD and PP. Future challenges are to map both genotypic and phenotypic change across the Dictyostelium phylogeny in much greater detail and to establish genetic tractability across all four taxon groups.
Methods
Sequencing and assembly
To avoid contamination with bacterial DNA, which would be preferentially cloned (Glöckner 2000), PP PN500 was grown in axenic medium, and DF SH3, which cannot grow axenically, was harvested from bacterial growth plates well after the plates had cleared, and starved for another 6 h, while shaking in phosphate buffer. Plasmid (pUC)-based sequencing libraries were constructed as previously described (Glöckner et al. 2002). Fosmid libraries with average insert sizes of 33 kb in vector pCC2FOS were prepared according to the manufacturer's instructions (Epicentre Biotechnologies). DNA sequencing was done with the BigDye kit from ABI using standard forward and reverse primers. Pre-assembly trimming of sequences was performed with a modified version of phred. The sequencing libraries for the 454 Life Sciences (Roche) FLX system were prepared according to the manufacturer's protocols. The 454 data were assembled using the newbler software. All contigs larger than 500 bases were entered in the Staden package database, including the newbler-derived quality values. The Sanger-based sequencing reads were then added using the gap assembler.
Chromosome structure analysis
The consensus sequences of the telomere repeats were used to search for contigs and single reads containing these repeats using BLASTn in the whole data set. Consensus sequences of the DD repetitive elements were used as query to find similar sequences in the genomes of the other social amoebae. Regions adjacent to telomeres were searched for other repeated sequence motifs. Detected repeat units were then used as query to find additional identical sequences throughout the genome.
Gene prediction, BLAST, and synteny analysis
tRNA genes were predicted using trnascan (Lowe and Eddy 1997). For protein-coding gene prediction we used geneid (Parra et al. 2000; Glöckner et al. 2002) after training with transcript data obtained by 454 sequencing. Specifically, single-transcript reads were aligned to the reference genome sequence using Exalign (http://159.149.109.9/exalign/). Intron-Exon boundaries were extracted from the resulting alignments and used as a training set for geneid. Furthermore, coding sequences from defined orthologs covered by transcript reads were used as a training set to define coding sequence features. For the analysis of specific gene families the predicted gene models were thoroughly examined and corrected where needed and possible. We examined more than 700 genes for this study. Nearly 1/3 of those are large multidomain genes, and therefore more prone to error. We found that 233 of the examined gene models required correction, since they were split or fused genes or contained incorrectly predicted intron/exon boundaries. Thus, we would estimate 1/3 of gene models to be incorrect, but since most of the predictions were small, we are confident that our global analysis of gene families and domains is only slightly compromised by these errors.
To define gene families irrespective of direct alignability of the proteins, we used orthoMCL with default values. This gave us an overview on the general structure of gene family expansions, but this method yields only a rough clustering of similar sequences. For the definition of orthologs, we used BLAST analysis of all CDS of one genome against the other, yielding best bidirectional hits (BBH). We used a filter threshold for significant hits of 20% identity between amino acid sequences over at least 50% of the protein to define the core proteome of social amoebae. A synteny group was defined as being a chromosomal segment encoding the same CDS in both species, irrespective of the order. Within a synteny group orthologous pairs can be separated by as much as five unrelated genes, including tRNAs. Clear orthology relationships are confounded by differential gene family expansions. This is why we compared each protein set with each of the others and obtained slightly differing numbers for shared genes.
Gene family detection using domain analysis
We took into account that not all members of a specific family would be detectable using simple BLAST algorithms, due to the occlusion of similarities, by accumulating mutations over such long time periods as are spanned by the evolution of the species examined. Thus, in all cases, the prospective members of the analyzed gene families were retrieved by identifying domain structures using IPRscan (Mulder and Apweiler 2008). In this way, we also were able to detect family members, obscured by erroneous predictions. Similarities detected by our BBH approach were not counted if a required functional domain could not be detected. PKS domains were detected using available software (SearchPKS) (Yadav et al. 2003).
Phylogeny and species split dating
We established the orthology relationships between proteins of social amoebaes (PP, DF, DD), plants (Arabidopsis thaliana, Physcomitrella patens), Metazoa (Hydra magnapapillata, Homo sapiens), and fungi (Saccharomyces cerevisiae, Schizosaccharomyces pombe) by using best reciprocal BLAST hits. Out of the orthologous proteins, we randomly chose 33 for further phylogenetic analysis. The 33 protein sequences were concatenated and then aligned using ClustalW.
Phylogenetic analyses were conducted in MEGA4 (Tamura et al. 2007). The evolutionary history was inferred using the Neighbor-Joining method, with a total of 34,111 positions in the final data set. All branches had 100% support from 1000 bootstrap replicates. The evolutionary distances were computed using the Poisson correction method, and a phylogenetic tree was linearized assuming equal evolutionary rates in all lineages. The clock calibration to convert distance to time was 2852.5146482082 (time/node height). All positions containing alignment gaps and missing data were eliminated only in pairwise sequence comparisons.
Data access
Whole Genome Shotgun projects have been deposited at GenBank (http://www.ncbi.nlm.nih.gov/genbank/) under accession numbers ADHC00000000 and ADBJ00000000. The versions described in this study are the first versions, ADHC01000000 and ADBJ01000000.
Acknowledgments
The DFG supported A.A.N., L.E., M.P., and G.G. with a grant (Gl235/1; NO113/18). The BBSRC supported P.S., H.M.L., C.S., and N.R.H. with grant BB/E016308/. G.G. and P.S. devised the study; M.F., H.M.L., N.R.H. were involved in assembly, quality control, and annotation; and M.F. set up the database. A.A.N., A.J.H., B.T., F.R, U.J., C.S., M.S., L.E., and M.P. contributed analyses and comments; A.A.N., P.S., and G.G. wrote the manuscript.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.121137.111.
References
- Alvarez-Curto E, Rozen DE, Ritchie AV, Fouquet C, Baldauf SL, Schaap P 2005. Evolutionary origin of cAMP-based chemoattraction in the social amoebae. Proc Natl Acad Sci 102: 6385–6390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anjard C, Loomis WF 2002. Evolutionary analyses of ABC transporters of Dictyostelium discoideum. Eukaryot Cell 1: 643–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anjard C, Loomis WF 2005. Peptide signaling during terminal differentiation of Dictyostelium. Proc Natl Acad Sci 102: 7607–7611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anjard C, Loomis WF 2006. GABA induces terminal differentiation of Dictyostelium through a GABAB receptor. Development 133: 2253–2261 [DOI] [PubMed] [Google Scholar]
- Austin MB, Saito T, Bowman ME, Haydock S, Kato A, Moore BS, Kay RR, Noel JP 2006. Biosynthesis of Dictyostelium discoideum differentiation-inducing factor by a hybrid type I fatty acid-type III polyketide synthase. Nat Chem Biol 2: 494–502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bader S, Kortholt A, Van Haastert PJ 2007. Seven Dictyostelium discoideum phosphodiesterases degrade three pools of cAMP and cGMP. Biochem J 402: 153–161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benabentos R, Hirose S, Sucgang R, Curk T, Katoh M, Ostrowski EA, Strassmann JE, Queller DC, Zupan B, Shaulsky G, et al. 2009. Polymorphic members of the lag gene family mediate kin discrimination in Dictyostelium. Curr Biol 19: 567–572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohne A, Brunet F, Galiana-Arnoux D, Schultheis C, Volff JN 2008. Transposable elements as drivers of genomic and biological diversity in vertebrates. Chromosome Res 16: 203–215 [DOI] [PubMed] [Google Scholar]
- Drosophila 12 Genomes Consortium 2007. Evolution of genes and genomes on the Drosophila phylogeny. Nature 450: 203–218 [DOI] [PubMed] [Google Scholar]
- Deering RA 1988. DNA repair in Dictyostelium. Dev Genet 9: 483–493 [DOI] [PubMed] [Google Scholar]
- Dickinson DJ, Nelson WJ, Weis WI 2011. A polarized epithelium organized by β- and α-catenin predates cadherin and metazoan origins. Science 331: 1336–1339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubin M, Fuchs J, Graf R, Schubert I, Nellen W 2010. Dynamics of a novel centromeric histone variant CenH3 reveals the evolutionary ancestral timing of centromere biogenesis. Nucleic Acids Res 38: 7526–7537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dujon B, Sherman D, Fischer G, Durrens P, Casaregola S, Lafontaine I, De Montigny J, Marck C, Neuveglise C, Talla E, et al. 2004. Genome evolution in yeasts. Nature 430: 35–44 [DOI] [PubMed] [Google Scholar]
- Eichinger L, Noegel AA 2005. Comparative genomics of Dictyostelium discoideum and Entamoeba histolytica. Curr Opin Microbiol 8: 606–611 [DOI] [PubMed] [Google Scholar]
- Eichinger L, Pachebat JA, Glöckner G, Rajandream MA, Sucgang R, Berriman M, Song J, Olsen R, Szafranski K, Xu Q, et al. 2005. The genome of the social amoeba Dictyostelium discoideum. Nature 435: 43–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J 1989. PHYLIP - Phylogeny Inference Package. Cladistics 5: 164–166 [Google Scholar]
- Gardner MJ, Hall N, Fung E, White O, Berriman M, Hyman RW, Carlton JM, Pain A, Nelson KE, Bowman S, et al. 2002. Genome sequence of the human malaria parasite Plasmodium falciparum. Nature 419: 498–511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glöckner G 2000. Large scale sequencing and analysis of AT rich eukaryote genomes. Curr Genomics 1: 289–299 [Google Scholar]
- Glöckner G, Szafranski K, Winckler T, Dingermann T, Quail MA, Cox E, Eichinger L, Noegel AA, Rosenthal A 2001. The complex repeats of Dictyostelium discoideum. Genome Res 11: 585–594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glöckner G, Eichinger L, Szafranski K, Pachebat JA, Bankier AT, Dear PH, Lehmann R, Baumgart C, Parra G, Abril JF, et al. 2002. Sequence and analysis of chromosome 2 of Dictyostelium discoideum. Nature 418: 79–85 [DOI] [PubMed] [Google Scholar]
- Glöckner G, Golderer G, Werner-Felmayer G, Meyer S, Marwan W 2008. A first glimpse at the transcriptome of Physarum polycephalum. BMC Genomics 9: 6 doi: 10.1186/1471-2164-9-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimson MJ, Coates JC, Reynolds JP, Shipman M, Blanton RL, Harwood AJ 2000. Adherens junctions and β-catenin-mediated cell signalling in a non-metazoan organism. Nature 408: 727–731 [DOI] [PubMed] [Google Scholar]
- Hedges SB 2002. The origin and evolution of model organisms. Nat Rev Genet 3: 838–849 [DOI] [PubMed] [Google Scholar]
- Heidel AJ, Glöckner G 2008. Mitochondrial genome evolution in the social amoebae. Mol Biol Evol 25: 1440–1450 [DOI] [PubMed] [Google Scholar]
- Joseph JM, Fey P, Ramalingam N, Liu XI, Rohlfs M, Noegel AA, Muller-Taubenberger A, Glockner G, Schleicher M 2008. The actinome of Dictyostelium discoideum in comparison to actins and actin-related proteins from other organisms. PLoS ONE 3: e2654 doi: 10.1371/journal.pone.0002654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Araki M, Goto S, Hattori M, Hirakawa M, Itoh M, Katayama T, Kawashima S, Okuda S, Tokimatsu T, et al. 2008. KEGG for linking genomes to life and the environment. Nucleic Acids Res 36: D480–D484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellis M, Birren BW, Lander ES 2004. Proof and evolutionary analysis of ancient genome duplication in the yeast Saccharomyces cerevisiae. Nature 428: 617–624 [DOI] [PubMed] [Google Scholar]
- Kersulyte D, Lee W, Subramaniam D, Anant S, Herrera P, Cabrera L, Balqui J, Barabas O, Kalia A, Gilman RH, et al. 2009. Helicobacter Pylori's plasticity zones are novel transposable elements. PLoS ONE 4: e6859 doi: 10.1371/journal.pone.0006859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Comer FI, Sasaki A, McLeod IX, Duong Y, Okumura K, Yates JR III, Parent CA, Firtel RA 2005. TOR complex 2 integrates cell movement during chemotaxis and signal relay in Dictyostelium. Mol Biol Cell 16: 4572–4583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Stoeckert CJ, Roos DS 2003. OrthoMCL: Identification of ortholog groups for eukaryotic genomes. Genome Res 13: 2178–2189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loftus B, Anderson I, Davies R, Alsmark UC, Samuelson J, Amedeo P, Roncaglia P, Berriman M, Hirt RP, Mann BJ, et al. 2005. The genome of the protist parasite Entamoeba histolytica. Nature 433: 865–868 [DOI] [PubMed] [Google Scholar]
- Lowe TM, Eddy SR 1997. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic. Nucleic Acids Res 25: 955–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulder NJ, Apweiler R 2008. The InterPro database and tools for protein domain analysis. Curr Protoc Bioinformatics 21: 2.7.1–2.7.18 [DOI] [PubMed] [Google Scholar]
- Narita TB, Koide K, Morita N, Saito T 2011. Dictyostelium hybrid polyketide synthase, SteelyA, produces 4-methyl-5-pentylbenzene-1.3-diol and induces spore maturation. FEMS Microbiol Lett 319: 82–87 [DOI] [PubMed] [Google Scholar]
- Ogawa S, Yoshino R, Angata K, Iwamoto M, Pi M, Kuroe K, Matsuo K, Morio T, Urushihara H, Yanagisawa K, et al. 2000. The mitochondrial DNA of Dictyostelium discoideum: complete sequence, gene content and genome organization. Mol Gen Genet 263: 514–519 [DOI] [PubMed] [Google Scholar]
- Parra G, Blanco E, Guigo R 2000. GeneID in Drosophila. Genome Res 10: 511–515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romualdi A, Siddiqui R, Glöckner G, Lehmann R, Suhnel J 2005. GenColors: accelerated comparative analysis and annotation of prokaryotic genomes at various stages of completeness. Bioinformatics 21: 3669–3671 [DOI] [PubMed] [Google Scholar]
- Ronquist F, Huelsenbeck JP 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19: 1572–1574 [DOI] [PubMed] [Google Scholar]
- Rubinstein CV, Gerrienne P, de la Puente GS, Astini RA, Steemans P 2010. Early Middle Ordovician evidence for land plants in Argentina (eastern Gondwana). New Phytol 188: 365–369 [DOI] [PubMed] [Google Scholar]
- Saran S, Meima ME, Alvarez-Curto E, Weening KE, Rozen DE, Schaap P 2002. cAMP signaling in Dictyostelium. Complexity of cAMP synthesis, degradation and detection. J Muscle Res Cell Motil 23: 793–802 [DOI] [PubMed] [Google Scholar]
- Saurin W, Hofnung M, Dassa E 1999. Getting in or out: early segregation between importers and exporters in the evolution of ATP-binding cassette (ABC) transporters. J Mol Evol 48: 22–41 [DOI] [PubMed] [Google Scholar]
- Schaap P, Pinas JE, Wang M 1985. Patterns of cell differentiation in several cellular slime mold species. Dev Biol 111: 51–61 [Google Scholar]
- Schaap P, Winckler T, Nelson M, Alvarez-Curto E, Elgie B, Hagiwara H, Cavender J, Milano-Curto A, Rozen DE, Dingermann T, et al. 2006. Molecular phylogeny and evolution of morphology in the social amoebas. Science 314: 661–663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt HA, von Haeseler A 2007. Maximum-likelihood analysis using TREE-PUZZLE. Curr Protoc Bioinformatics 17: 6.6.1–6.6.23 [DOI] [PubMed] [Google Scholar]
- Schultz J, Milpetz F, Bork P, Ponting CP 1998. SMART, a simple modular architecture research tool: identification of signaling domains. Proc Natl Acad Sci 95: 5857–5864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen B 2003. Polyketide biosynthesis beyond the type I, II and III polyketide synthase paradigms. Curr Opin Chem Biol 7: 285–295 [DOI] [PubMed] [Google Scholar]
- Siu CH, Harris TJ, Wang J, Wong E 2004. Regulation of cell-cell adhesion during Dictyostelium development. Semin Cell Dev Biol 15: 633–641 [DOI] [PubMed] [Google Scholar]
- Sucgang R, Chen G, Liu W, Lindsay R, Lu J, Muzny D, Shaulsky G, Loomis W, Gibbs R, Kuspa A 2003. Sequence and structure of the extrachromosomal palindrome encoding the ribosomal RNA genes in Dictyostelium. Nucleic Acids Res 31: 2361–2368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sucgang R, Kuo A, Tian X, Salerno W, Parikh A, Feasley CL, Dalin E, Tu H, Huang E, Barry K, et al. 2011. Comparative genomics of the social amoebae Dictyostelium discoideum and Dictyostelium purpureum. Genome Biol 12: R20 doi: 10.1186/gb-2011-12-2-r20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson AR, Vadell EM, Cavender JC 1999. Global distribution of forest soil dictyostelids. J Biogeogr 26: 133–148 [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol 24: 1596–1599 [DOI] [PubMed] [Google Scholar]
- Tanguay P, Loppnau P, Morin C, Bernier L, Breuil C 2006. A spontaneous albino mutant of Ceratocystis resinifera results from a point mutation in the polyketide synthase gene, PKS1. Can J Microbiol 52: 501–507 [DOI] [PubMed] [Google Scholar]
- Thomason PA, Traynor D, Stock JB, Kay RR 1999. The RdeA-RegA system, a eukaryotic phospho-relay controlling cAMP breakdown. J Biol Chem 274: 27379–27384 [DOI] [PubMed] [Google Scholar]
- Vlahou G, Rivero F 2006. Rho GTPase signaling in Dictyostelium discoideum: insights from the genome. Eur J Cell Biol 85: 947–959 [DOI] [PubMed] [Google Scholar]
- Xue C, Hsueh YP, Heitman J 2008. Magnificent seven: roles of G protein-coupled receptors in extracellular sensing in fungi. FEMS Microbiol Rev 32: 1010–1032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadav G, Gokhale RS, Mohanty D 2003. SEARCHPKS: A program for detection and analysis of polyketide synthase domains. Nucleic Acids Res 31: 3654–3658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zucko J, Skunca N, Curk T, Zupan B, Long PF, Cullum J, Kessin RH, Hranueli D 2007. Polyketide synthase genes and the natural products potential of Dictyostelium discoideum. Bioinformatics 23: 2543–2549 [DOI] [PubMed] [Google Scholar]