Abstract
Next-generation sequencing technologies offer new approaches for global measurements of gene expression but are mostly limited to organisms for which a high-quality assembled reference genome sequence is available. We present a method for gene expression profiling called EDGE, or EcoP15I-tagged Digital Gene Expression, based on ultra-high-throughput sequencing of 27-bp cDNA fragments that uniquely tag the corresponding gene, thereby allowing direct quantification of transcript abundance. We show that EDGE is capable of assaying for expression in >99% of genes in the genome and achieves saturation after 6–8 million reads. EDGE exhibits very little technical noise, reveals a large (106) dynamic range of gene expression, and is particularly suited for quantification of transcript abundance in non-model organisms where a high-quality annotated genome is not available. In a direct comparison with RNA-seq, both methods provide similar assessments of relative transcript abundance, but EDGE does better at detecting gene expression differences for poorly expressed genes and does not exhibit transcript length bias. Applying EDGE to laboratory mice, we show that a loss-of-function mutation in the melanocortin 1 receptor (Mc1r), recognized as a Mendelian determinant of yellow hair color in many different mammals, also causes reduced expression of genes involved in the interferon response. To illustrate the application of EDGE to a non-model organism, we examine skin biopsy samples from a cheetah (Acinonyx jubatus) and identify genes likely to control differences in the color of spotted versus non-spotted regions.
Recent and ongoing advances in DNA sequencing technology have created new opportunities for measuring gene expression based on “counting,” in which a cDNA population is heavily oversampled by massively parallel sequencing, and transcript abundance is inferred from the relative frequencies with which different cDNAs are identified. The most widely used approach, RNA-seq, uses randomly sheared RNA or cDNA in which sequence reads generated by an Illumina or SOLiD instrument that align to a reference genome are analyzed with regard to transcript identity and read position within the transcript; these observations are then used to make inferences about transcript abundance (Cloonan et al. 2008; Mortazavi et al. 2008; Nagalakshmi et al. 2008; Wilhelm et al. 2008).
RNA-seq and related approaches (LQ-RNA-seq and digital transcriptome profiling with NSR primers) are best suited to organisms for which a high-quality assembled and annotated genome is available (Armour et al. 2009; Ozsolak et al. 2010). Mapping short reads to incomplete genome sequences entails both reduced power (reads that fail to align) and false-positive errors (reads that align uniquely to a partial genome sequence but arise from elsewhere). Furthermore, these approaches are especially challenging for natural populations with high levels of polymorphism. At the same time, sequencing-based approaches to assess gene expression are particularly appealing for non-model organisms with unique ecological, evolutionary, or developmental features. Cichlid fish, thirteen-lined ground squirrels, and songbirds are examples of animals for which there are significant biological questions that would benefit from transcriptome profiling but for which the respective research communities are insufficiently large to benefit from genomic resources associated with large economies of scale such as oligonucleotide microarrays (Renn et al. 2004; Replogle et al. 2008; Liu et al. 2010).
Here, we report molecular biologic and informatic development of a short-read sequence approach that is particularly suited for measuring gene expression in non-model organisms: EDGE, or EcoP15I-tagged Digital Gene Expression. Each expressed transcript in the genome is identified by a unique 27-bp tag; thus, the number of potential tags in an experiment corresponds to the number of genes in the genome, yielding a library of much less complexity than random shearing and that is less susceptible to amplification bias since every library molecule is exactly the same size. Consequently, the frequency at which a particular EDGE tag appears in a library serves as a proxy for quantifying and comparing transcript abundance. Importantly, the one-to-one correspondence between transcript and sequence tag allows gene expression differences to be measured by statistical analysis of relative tag frequencies, thus obviating the need to identify every sequence tag. Finally, tag-to-gene assignments can be accomplished effectively by leveraging a comparative genomics approach that relies on partially assembled transcriptomes.
We first describe the development of EDGE and its performance relative to RNA-seq in laboratory mice segregating a loss-of-function mutation for the melanocortin 1 receptor (Mc1r) gene, which underlies a fundamental aspect of pigmentary variation in many vertebrate species (Andersson 2003; Eizirik et al. 2003; Mundy et al. 2003; Rees 2003). Using a conventional approach in which individual tags are first mapped to a reference genome, we detect validated gene expression differences over a 106-fold dynamic range; we also identify a previously unappreciated component of MC1R signaling. We then apply the EDGE approach to a non-model organism, the cheetah, to investigate the molecular basis of black spotting. Our results illustrate various strategies for making tag-to-gene assignments and reveal gene expression signatures that provide new biologic insight into pigment patterning in a non-model organism.
Results
Overview of molecular biology and informatics
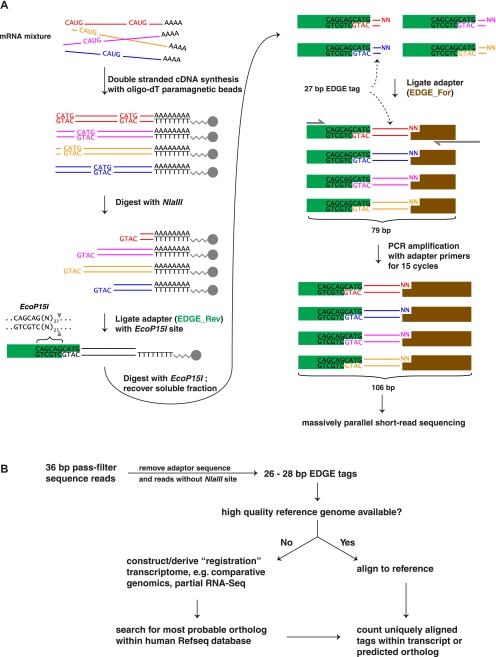
The EDGE approach starts with modest amounts of total RNA (1–2 μg) and uses paramagnetic oligo(dT) beads for mRNA enrichment and to facilitate subsequent biomolecular handling steps (Fig. 1A). Individual transcripts are directionally “tagged” according to a 27-bp sequence that begins with a 4-bp restriction site, NlaIII, and the 23 bp that lie immediately downstream, generated by the type III restriction endonuclease EcoP15I (Fig. 1A). Theoretically, each tag begins with the NlaIII site that lies closest to the poly(A) tail; in practice, we observe several-fold more tags than transcripts due to partial cleavage with NlaIII. We note that NlaIII sites are present in >99% of mouse or human cDNA sequences and that the application of EDGE to two types of mouse tissue captures ∼90% of the more than 20,000 genes represented in RefSeq (described below in Fig. 3C).
Figure 1.
Outline of EDGE methodology and informatic pipeline for tag identification. (A) Double-stranded cDNA synthesis is performed using paramagnetic oligo(dT) beads to capture polyadenylated RNA. Next, each cDNA molecule is “anchored” by NlaIII restriction cleavage that exposes the 3′-most “CATG” site within the transcript. Following this, the EDGE_Rev adaptor (green) carrying an EcoP15I recognition site (5′-CAGCAG-3′) is ligated, and the resulting molecule is “tagged” by EcoP15I restriction digest, generating a 27-bp sequence tag. The sticky end is ligated to the EDGE_For adaptor (blue). Finally, a 15-cycle PCR amplification using adaptor-specific primers (red half-arrows) is performed to add on the additional sequence required to complete the EDGE_For adaptor and to enrich the desired final product. (B) Thirty-six base-pair pass-filter reads from the Illumina Genome Analyzer were processed to obtain EDGE tags. If a high-quality reference transcriptome was available, e.g., mouse, EDGE tags were mapped to transcript sequence and uniquely aligned tags were counted for each gene. Otherwise, EDGE tags, e.g., cheetah, were mapped to a de novo assembled reference transcriptome, e.g., cat, which acts as a scaffold to identify the orthologous gene in the organism in which the EDGE tags were derived.
Figure 3.
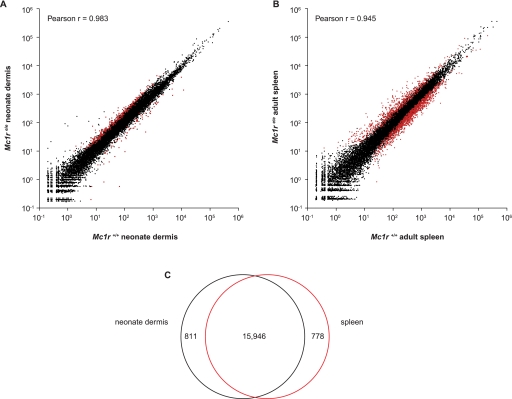
Application of EDGE to mouse tissues. (A) Using a Poisson log linear model, 327 and 945 genes were identified as differentially expressed between Mc1r+/+ and Mc1re/e in (A) neonate dermis (FDR < 5%) and (B) spleen (FDR < 0.1%), respectively. Average gene counts from wild-type (five libraries each for neonate dermis and spleen) and mutant (six for neonate dermis and five for spleen) EDGE libraries are plotted against each other on a log10 scale. Differentially expressed genes are plotted in red. (C) EDGE tags were detected for 17,535 unique mouse genes in at least one tissue, and 1589 genes were expressed in only a single tissue.
For organisms with high-quality assembled and annotated genomes, individual pass-filter EDGE tags from a massively parallel sequencing instrument that uniquely align to a reference transcriptome are “translated” to gene counts, and quantitative analysis of gene expression profiles is performed with a statistical model similar to SAM in which false discovery rates are estimated by permutation. For non-model organisms, tag-to-gene assignments are inferred using a comparative approach when there exists a closely related genome, and/or a stepwise approach using first-pass transcriptome data from 454 Life Sciences (Roche) or paired-end Illumina reads that serve as a scaffold to link EDGE tags to genes (Fig. 1B).
EDGE in a model organism: Technical characteristics
We first applied EDGE to laboratory mice carrying a loss-of-function alteration in the MC1R, a G-protein-coupled receptor mainly expressed in melanocytes. In this model, animals from the C57BL/6J strain exhibit a black coat color due to active MC1R signaling, whereas isogenic Mc1re/e mutants exhibit a yellow coat color (Robbins et al. 1993). Mc1r mutations are well recognized in a wide range of vertebrate species, including humans, where they cause red hair (Rees 2003) and have been proposed to underlie additional non-pigmentary phenotypes including increased susceptibility to skin cancer (Bastiaens et al. 2001; Kennedy et al. 2001) and altered sensitivity to general anesthesia (Liem et al. 2004; Mogil et al. 2005).
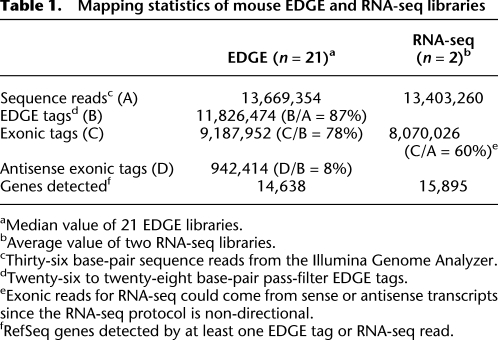
Summary alignment and mapping statistics from 21 mouse EDGE libraries (10 from Mc1r+/+ and 11 from Mc1re/e tissues) are presented in Table 1 and show that 87% of pass-filter sequence reads conform to expectation with a 26–28-bp read anchored at one end with the NlaIII recognition site. Of these, 86% could be aligned uniquely to the mouse transcriptome. This compares favorably with analogous results from two mouse skin RNA-seq libraries, in which 60% of the 36-bp pass-filter reads aligned uniquely to the mouse transcriptome (Table 1). In contrast, MmeI, a Type IIs restriction endonuclease commonly used in tag-based cDNA sequencing protocols (Asmann et al. 2009; Wu et al. 2010), generates a 21-bp tag that results in a smaller proportion of uniquely mapped tags—78% of a simulated MmeI-tagged data set mapped uniquely to the mouse transcriptome compared to 86% with EDGE—and that translates to 3% reduction in genes detected. Among the EDGE tags, 78% and 8% mapped uniquely to the sense and antisense strands of mouse transcripts, respectively (Table 1), and 6% mapped to multiple genomic locations or to introns and unannotated regions of the genome (Table 1).
Table 1.
Mapping statistics of mouse EDGE and RNA-seq libraries
The enzymology of the EDGE methodology ensures that each transcript is sampled by sequencing a single 26–28-bp tag that is anchored by NlaIII restriction digest. In theory, the one-to-one correspondence between transcript and EDGE tag would enable us to measure relative transcript levels by comparing tag frequencies between libraries. However, since there could be multiple transcript isoforms per gene and since NlaIII digestion is not 100% efficient, each transcript can, in theory, be represented by multiple tags. In practice, we found that, on average, 82% of tags for each transcript arise from a single site, indicating that the relative frequencies of most tags provide an accurate measure of gene expression. Furthermore, >99% of genes that showed considerable expression levels in an alternative method (>1.5 RPKM by RNA-seq) were also detected by EDGE, indicating that the efficiency of NlaIII cleavage does not limit the ability of EDGE to assay for transcript abundance.
To assess technical performance of the EDGE methodology, we examined correlations among libraries for both technical and biological replicates; we also compared both the general architecture of gene expression and specific biological findings obtained by EDGE to gene expression measurements obtained using alternative approaches. For this and subsequent work, we use the number of tags per million mapped exonic reads (TPM) as primary data for comparison of different libraries and for subsequent statistical analyses.
Pearson correlation coefficients of tag counts between libraries generated from the same pool of RNA or the same library sequenced at two different sites range from 0.927 to 0.992 with a mean of 0.975 (Supplemental Fig. S1). Correlations for biological replicates—tissues from age-matched isogenic animals—range from 0.869 to 0.992 with a mean of 0.955 (Supplemental Fig. S2). Thus, the EDGE protocol exhibits very little noise from library construction, amplification, and Illumina flow cell sequencing processes.
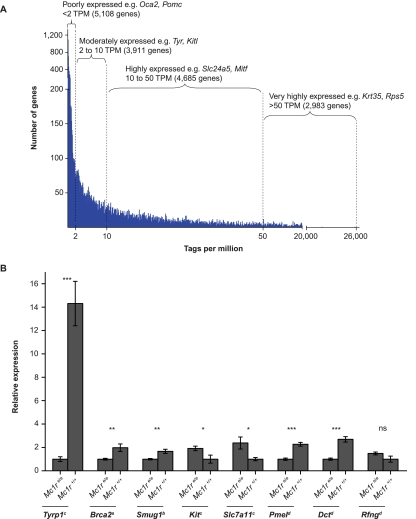
Like other sequence-based assays, EDGE reveals a wide spectrum of gene expression, with mean tag counts ranging from 0.09 to 25,846 TPM. Also like other sequence-based assays for gene expression, the distribution of tag counts is highly skewed toward a large number of genes expressed at low levels (Fig. 2A; Nagalakshmi et al. 2008; Asmann et al. 2009). In the skin, many of the genes expressed at low levels are melanocyte-specific including Tyrp1 (28.2 TPM), Tyr (5.7 TPM), Mc1r (9.8 TPM), and Oca2 (0.9 TPM), which indicates that EDGE is capable of detecting biologically relevant gene expression from a minor cell type in a heterogeneous tissue (we estimate that melanocytes represent 0.1%–1% of the cells in neonatal dermis).
Figure 2.
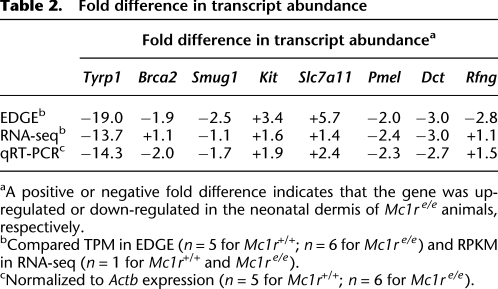
(A) Dynamic range of gene expression detected by EDGE. The TPM distribution (x-axis) for genes detected by EDGE is plotted against number of genes (y-axis) and identifies poorly expressed genes below 2 TPM, moderately expressed genes with 2 to 10 TPM, highly expressed genes with 10 to 50 TPM, and very highly expressed genes above 50 TPM. (B) Seven out of eight differentially expressed genes from EDGE showed significant differences when transcript abundance was measured by quantitative RT-PCR. Brca2: <2 TPM; Smug1: 2 to 10 TPM; Tyrp1, Kit, Slc7a11: 10 to 50 TPM; Pmel, Dct, Rfng: >50 TPM in EDGE libraries. (*) p < 0.05; (**) p < 0.001; (***) p < 0.0001; (ns) not significant.
EDGE achieves near saturation in genes detected after 6–8 million tags (Supplemental Fig. S3A). Furthermore, saturation of moderately to very highly expressed genes (>2 TPM) occurs with ∼3 million exonic EDGE tags (Supplemental Fig. S3B). Thus, barcoding strategies would allow multiple EDGE libraries to be sequenced efficiently and economically while still achieving robust measurements of the majority of the transcriptome.
EDGE in a model organism: A role for the MC1R in the interferon response
Using a Poisson log linear model to analyze gene counts from libraries of neonatal dermis—selected originally because Mc1r is expressed mainly on melanocytes and neonatal dermis is enriched for melanocytes relative to other skin compartments—we identified 72 genes that were down-regulated and 255 genes that were up-regulated in mutant (n = 6) compared with non-mutant (n = 5) tissue at an FDR of <5% (Fig. 3A; Witten et al. 2010). For eight differentially expressed genes chosen to represent a broad range of expression levels, quantitative RT-PCR confirmed the EDGE results for seven genes (Fig. 2B; Table 2); the eighth gene, Rfng, was down-regulated 2.8-fold as determined by EDGE (145.1 TPM in Mc1r+/+ vs. 51.3 TPM in Mc1re/e samples), but quantitative RT-PCR failed to detect a difference.
Table 2.
Fold difference in transcript abundance
Several genes down-regulated in mutant skin are expressed at very low to moderate levels, including Tyrp1 (56.2 TPM in Mc1r+/+ vs. 4.8 TPM in Mc1re/e skin), Brca2 (2.7 TPM in Mc1r+/+ vs. 1.4 TPM in Mc1re/e skin), and Smug1 (10.1 TPM in Mc1r+/+ vs. 4.1 TPM in Mc1re/e skin). Tyrp1, Dct, and Pmel encode melanogenic genes and are well-known targets of Mc1r based on studies of cultured melanocytes (Kobayashi et al. 1995; Lamoreux et al. 1995), but an effect of Mc1r on Brca2 and Smug1 has not been described previously and may contribute to differences in skin cancer susceptibility. We also note that Slc7a11, which encodes a melanocyte-specific cystine transporter that is essential for pheomelanin (yellow pigment) synthesis (and in which a loss-of-function is responsible for the subtle gray coat color mutation), is up-regulated (19.2 TPM in Mc1r+/+ vs. 44.5 TPM in Mc1re/e skin), which supports a hypothesis based on biochemical studies that cystine transport plays an instructive role in pigment-type switching (Chintala et al. 2005; Simon et al. 2009).
We carried out an unsupervised Gene Ontology analysis on the 327 differentially expressed genes and identified several unexpected biological processes affected by the Mc1r mutation (Supplemental Table S1). These functional categories are represented mostly by genes that are up-regulated in mutant skin except in one intriguing case, where genes down-regulated in mutant skin represent a functional classification category called “response to interferon-gamma” (Supplemental Table S1). Several of these genes, such as Oas2 and its family members Oasl1 and Oasl2, encode 2′–5′ oligoadenylate synthetases that play a direct role in anti-viral pathways (Baglioni et al. 1978; Hovanessian and Wood 1980; Perelygin et al. 2002). Others, such as Iigp1 and Gm12250, are involved in resistance to pathogens/viruses (Supplemental Table S2; Zerrahn et al. 2002; Uthaiah et al. 2003; Bernstein-Hanley et al. 2006; Miyairi et al. 2007). Notably, most of these genes are expressed at low levels in skin—the eight anti-viral genes are expressed 17.9 times lower (3.4 vs. 60.9 TPM) than the other 64 genes that were down-regulated in mutant skin—which probably explains why they were missed by previous studies that used microarrays (April and Barsh 2006; Le Pape et al. 2009).
To further investigate a possible role of MC1R signaling in interferon-mediated immunity, we constructed and analyzed EDGE libraries from spleen obtained from five Mc1r+/+ and five Mc1re/e adult animals. Surprisingly, a large number of genes were differentially expressed in adult spleen: 945 genes were differentially expressed at an FDR of <0.1% (Fig. 3B). Consistent with the functional signature from neonate dermis, genes involved in interferon-mediated immunity were also down-regulated in the adult mutant tissues (Supplemental Tables S1, S3).
Because EDGE detects transcripts expressed at extremely low levels, a large fraction of the transcriptome is sampled from a single tissue. For neonatal dermis and adult spleen, EDGE tags were detected in at least one tissue for 17,535 unique mouse genes; only 9% of the genes were limited to a single tissue (Fig. 3C).
Direct comparison with RNA-seq
We randomly selected an Mc1r+/+ and an Mc1re/e neonatal dermis RNA sample from which EDGE libraries had already been made, then constructed and sequenced conventional RNA-seq libraries from the same RNA samples, generating between 10 and 16 million reads per library. Summary statistics for the fraction of reads that aligned uniquely to the transcriptome and for the number of genes detected were all similar to that of EDGE (Table 1).
Estimates of transcript abundance from EDGE TPM values are correlated with those from RNA-seq values (based on reads per kilobase of exon model per million mapped reads, RKPM), as shown in Supplemental Figure S4. However, the extent of correlation, with Spearman coefficients of 0.82 and 0.81 for the Mc1r+/+ and Mc1re/e samples (Supplemental Fig. S4), respectively, is considerably less than observed for technical replicates by EDGE (mean 0.975) (Supplemental Fig. S1). Reduced correlation is most evident for genes expressed at lower levels and is symmetric; in other words, ∼20% of genes (about 1000 genes) that are poorly expressed (<1.5 RPKM by RNA-seq, or <2 TPM by EDGE) according to one platform are captured at moderate to high levels of expression by the reciprocal platform (Supplemental Fig. S4). In the case of the eight differentially expressed genes previously chosen for validation (and with the caveat that no biological replicates were generated in the case of RNA-seq), six displayed differences in RPKM values that were concordant with the EDGE and qRT-PCR results (Table 2).
Next, we explored the sensitivity and precision of EDGE and RNA-seq as a function of sequencing depth by random subsampling of sequence reads. Saturation of gene detection for moderately to very highly expressed genes (>2 TPM or >1.5 RPKM) occurs at ∼1 million exonic reads, whereas the detection of poorly expressed genes steadily increases up to ∼7 million exonic reads (Supplemental Fig. S3A). Furthermore, RPKM or TPM values for 80% of genes fall within 20% of the value in the total data set at ∼5 million and ∼6 million exonic reads for RNA-seq and EDGE, respectively (Supplemental Fig. S3B). Thus, both methods perform similarly across a broad range of expression levels.
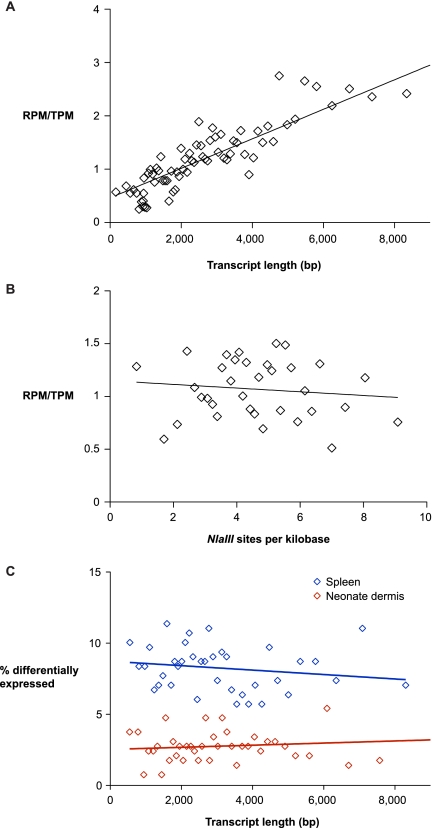
For genes that were differentially detected by either method, several observations suggest that the underlying explanation appears to be transcript length bias and the frequency of NlaIII sites. Because RNA-seq reads are randomly distributed and EDGE relies on the availability of NlaIII sites to generate tags, we expect the sensitivity of RPKM-based and TPM-based estimates to be inversely correlated with transcript length and the frequency of NlaIII sites, respectively. Indeed, in our direct comparison data sets—with the caveat that RNA-seq does not discriminate between sense or antisense reads—the mean length of the 436 genes detected only by EDGE is 548 bp shorter than the 1295 genes detected only by RNA-seq (p < 1×10−8) (Supplemental Fig. S5A). On the other hand, genes that were only detected by EDGE and genes that were only detected by RNA-seq have 5.4 and 4.6 NlaIII sites per kilobase of transcript, respectively (p < 1×10−6) (Supplemental Fig. S5B).
To further explore potential bias in the entire data set, we examined the relationship between the relative number of RNA-seq reads or EDGE tags per gene as a function of transcript length and the frequency of NlaIII sites. Not surprisingly, compared with EDGE, RNA-seq exhibits a strong bias toward detecting reads from longer transcripts (p < 1×10−4) (Fig. 4A). In contrast, the relative rate of RNA-seq reads and EDGE tags does not depend on the frequency of NlaIII sites within transcripts (p = 0.51), implying that EDGE is capable of providing robust measurements for transcript abundance using tags from one or a few NlaIII sites in each transcript (Fig. 4B). As a consequence of transcript length bias caused by random sampling, statistical power for detecting differentially expressed genes by RNA-seq has been found to depend on transcript length (Oshlack and Wakefield 2009). Conversely, among the 21 EDGE libraries from mouse tissue, our ability to detect differentially expressed genes is independent of transcript length (Fig. 4C).
Figure 4.
Systematic biases in RNA-seq and EDGE. The relative frequency of RNA-seq reads and EDGE tags is dependent on transcript length (A) and independent of NlaIII site frequency within transcripts (B). RefSeq genes were sorted by transcript length (A) and frequency of NlaIII sites (B) and placed into bins of 300 genes and 500 genes, respectively. The relative ratio of reads per million exonic RNA-seq reads (RPM) and tags per million exonic EDGE tags (TPM) within each bin is plotted (diamonds). Linear regression lines are plotted for each graph and show a significant correlation in RPM/TPM ratio with transcript length (p < 1×10−4) and an insignificant relationship in RPM/TPM ratio with NlaIII site frequency (p = 0.51). (C) The ability of EDGE to detect differential gene expression is not dependent on transcript length. Genes that were detected by EDGE were sorted by transcript length and placed into bins of 300 genes. The percentage of differentially expressed genes within each bin is plotted (diamonds). Linear regression lines are plotted for neonate dermis (p = 0.68) and spleen (p = 0.29).
To assess the performance of EDGE and RNA-seq in situations in which a complete reference transcriptome is unavailable, we simulated an incomplete reference that represented a subsample of the existing mouse reference in which each transcript contained 30% of contiguous sequence selected randomly from mouse RefSeq genes. Using this simulated reference, we assigned genes to fastq reads from an EDGE and a RNA-seq library generated from mouse neonatal dermis and calculated the rate of tag-to-gene assignment relative to the complete reference. Consistent with the results described above, EDGE and RNA-seq performed equally well—34% of EDGE tags and 32% of RNA-seq reads were correctly assigned to mouse genes in the incomplete reference, while 3.0% and 2.8%, respectively, were incorrectly assigned due to multiple locations in the full transcriptome (Supplemental Table S4).
In summary, both EDGE and RNA-seq provide similar estimates of transcript abundance for most genes, but the two approaches have different strengths and weaknesses, and EDGE is likely to perform better for short genes.
Analyzing transcript abundance with a tag-based approach
A principal advantage of EDGE over RNA-seq or related methods is the opportunity to study gene expression without a high-quality reference genome, by first identifying differentially expressed tags and then inferring tag-to-gene assignments with partial and/or comparative information (Fig. 1B, see below). We used the existing mouse data to compare the previous “by-gene” approach to what would have been obtained with a “by-tag” approach (had a reference genome not been available).
We applied the Poisson log-linear model to tag counts from EDGE library reads of mutant (n = 6) and non-mutant (n = 5) neonatal dermis, ranked all unique tags by increasing FDR (or decreasing statistical significance for differential expression), and compared the results with the by-gene approach.
Overall, there was good agreement between the by-gene and the by-tag approaches. Among the genes that were previously identified as differentially expressed (<5% FDR) in mouse neonatal dermis, 52% were detected as differentially expressed tags at <5% FDR, and 90% were detected as differentially expressed tags at <10% FDR (data not shown). Thus, in the absence of a high-quality reference genome, an approach that relies on statistical analysis of EDGE tag frequencies is adequate for profiling differences in transcript abundance.
Applying EDGE to a non-model organism: Color variation in the cheetah
As a direct test of EDGE profiling in a non-model organism, we carried out a pilot study to compare gene expression in areas of differently colored skin regions of a cheetah (Acinonyx jubatus). Periodic color patterns of black versus yellow hair, such as spots on a cheetah or stripes on a tiger—for which a suitable model organism does not exist—represent a subject of long-standing interest to developmental and evolutionary biologists.
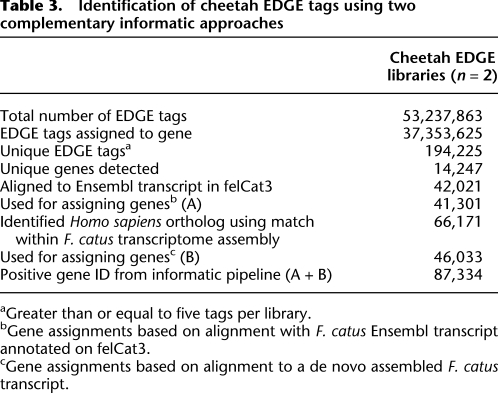
Two EDGE libraries were generated from cheetah skin, one from a black-pigmented region (hereafter referred to as “black spot”) and the other from an adjacent yellow-pigmented region (hereafter referred to as “yellow background”). Each library was sequenced on one lane of the Illumina Genome Analyzer IIx, generating an average of ∼27 million EDGE tags per library. After removing poorly expressed tags, 194,225 unique tag sequences were used for tag-to-gene assignments (Table 3).
Table 3.
Identification of cheetah EDGE tags using two complementary informatic approaches
We used two different approaches for tag-to-gene assignments, both of which are based on existing genome resources in the domestic cat (Felis catus), which diverged from the cheetah ∼4–6 million years ago, and therefore predict >98% sequence identity between the two species for most regions of the genome, including non-protein-coding transcribed regions where the majority of EDGE tags are located. The two genomic resources include a 2×-coverage cat genome that has been partially annotated by comparison to other mammalian genomes and a partial cat transcriptome generated by 454 sequencing of 10 different cat tissues, but that has not yet been integrated with the genome assembly.
Approximately 21% of the unique cheetah tags could be assigned to genes by alignment to the cat genome, and an additional 24% could be assigned to genes by alignment to the cat transcriptome (Table 3). As with the mouse data, the distribution of unique cheetah EDGE tags is highly skewed toward those that are expressed at low levels (Supplemental Fig. S6); thus, of ∼53 million tags from the two cheetah libraries, ∼37 million could be assigned to genes. Overall, this provided information for 14,247 different genes and illustrates how EDGE can capture the majority of variation in gene expression in the absence of a high-quality genome sequence.
The Pearson correlation coefficient of gene counts between the two EDGE libraries was 0.945; thus, patterned control of color variation in cheetahs is not accompanied by significant differences in gene expression at a genome-wide level (Supplemental Fig. S6). Because the lack of biological replicates does not allow statistical evaluation of genome-wide expression differences, we instead examined tag counts for sets of known pigmentation genes based on whether they lie upstream of or downstream from MC1R signaling.
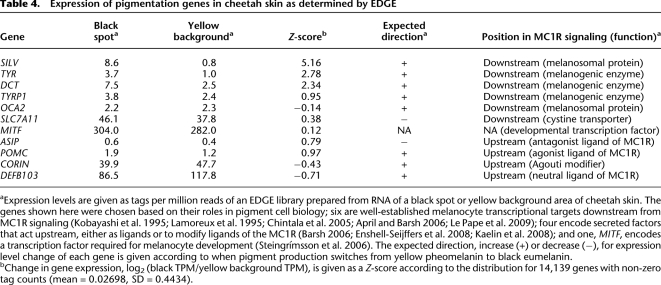
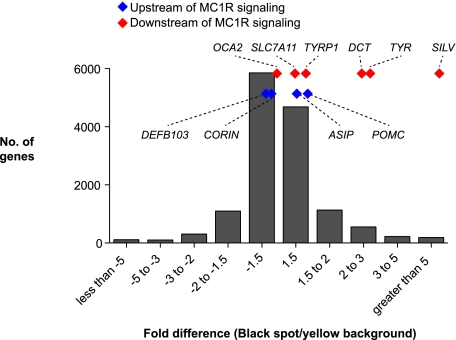
As described above, an Mc1r loss-of-function mutation in laboratory mice and many other mammals converts black hair to yellow hair in the entire animal by altering the expression of genes involved in the synthesis of eumelanin versus pheomelanin, so-called pigment-type switching. Comparing black spot to yellow background RNA for cheetah skin (Table 4), we observed substantially higher tag counts in several genes that lie downstream from MC1R signaling and that promote switching from pheomelanin to eumelanin: SILV (+11-fold), TYR (+3.7-fold), DCT (+3-fold), and TYRP1 (+1.6-fold). One gene that lies downstream from MC1R signaling exhibited small changes in expression whose direction was opposite to that predicted from laboratory mouse studies, SLC7A11 (+1.2-fold). In contrast, genes that encode upstream regulators of MC1R signaling exhibited relatively small changes in tag count, including ASIP (+1.5-fold), POMC (+1.6-fold), CORIN (−1.2-fold), and DEFB103 (−1.4-fold).
Table 4.
Expression of pigmentation genes in cheetah skin as determined by EDGE
The significance of the changes described is difficult to evaluate without replicate samples; however, we note that the direction of change for three of the upstream genes (ASIP, CORIN, and DEFB103) occurs in a direction opposite to that expected for an instructive role in pigment-type switching. Furthermore, considered as a group (Fig. 5), the distribution of Z-scores for the downstream genes is significantly different from the entire data set (p = 2.7×10−6); in contrast, neither the range nor the values of individual Z-scores for upstream genes stand out from the entire data set (Fig. 5; Table 4). Taken together, these results suggest that black spots in cheetahs are brought about by localized alterations downstream from MC1R signaling that engage known components of the pigment type-switching apparatus.
Figure 5.
Expression of pigment-type switching genes in cheetah skin. Fold difference in gene expression between black spot and background was determined by EDGE. The relative fold difference for genes that encode components of pigment-type switching that lie upstream (blue) or downstream (red) of MC1R signaling is shown, as in Table 4.
Discussion
Established and emerging technologies for ultra-high-throughput sequencing are being increasingly applied to measure gene expression in a variety of basic science and translational settings. Like other so-called digital gene expression approaches (pioneered with serial analysis of gene expression, or SAGE), EDGE is based on a molecular biologic strategy in which the relative frequencies of unique cDNA tags are used to infer transcript abundance. However, unlike classical SAGE methods that use Sanger sequencing, EDGE relies on ultra-high-throughput sequencing technology to generate millions of cDNA tags per RNA sample with increased time and cost savings. Compared with classical SAGE, EDGE provides substantially improved sensitivity for detecting rare transcripts and more robust measurements of transcript abundance across a broad range of expression levels, resulting in stronger statistical power to detect differentially expressed transcripts. In addition, the EDGE method is facilitated by the high cleavage efficiency of EcoP15I, resulting in improved transcriptome coverage compared to other tag-sequencing approaches that rely on shorter tags generated by MmeI. Like RNA-seq, EDGE is extraordinarily sensitive, able to detect transcripts present at low levels or in a minority of cells in a heterogeneous tissue. Unlike RNA-seq, EDGE is not subject to transcript length bias; however, EDGE provides little or no information about transcript structure. An important application of EDGE as shown here is the ability to evaluate transcriptomic changes in non-model organisms where a high-quality reference genome is not available.
Applied to the skin of laboratory mice carrying a classical coat color mutation, EDGE detects expression from approximately 17,500 genes. Most of these are represented at very low levels, including components of the interferon response that are differentially expressed between Mc1r+/+ and Mc1re/e animals and that were not detected in previous microarray analyses. Additional studies will be required to investigate the potential mechanisms and consequences of differences in innate immunity between Mc1r+/+ and Mc1re/e animals, but we speculate that differences in the chemistry of eumelanin and pheomelanin may have secondary effects on the ability of the innate immune system to respond to environmental pathogens or stress. For example, pheomelanin is associated with very different antioxidant levels from eumelanin (Chedekel et al. 1978; Samokhvalov et al. 2005), and it is interesting to note that melanin plays an important and established role in innate immunity in insects (Eleftherianos and Revenis 2011).
Compared with RNA-seq, EDGE provides little information about transcript structure; however, the ability of EDGE to detect differential gene expression is not influenced by transcript length or potential size amplification bias during PCR amplification. Hence, EDGE is particularly attractive for experiments that require sensitive and robust measurements of relative transcript levels across the genome. Furthermore, EDGE achieves near saturation in gene detection with 6–8 million sequence reads, making it possible to assay for gene expression differences in multiple biological replicates by using a molecular barcoding strategy, thus substantially decreasing the cost of using EDGE while still providing significant advantages over microarrays.
In a pilot study to investigate the effectiveness of EDGE in a non-model organism, we compared tag counts in skin of the cheetah taken from adjacent areas of different color. By taking advantage of the reduced complexity of sequence tags in EDGE relative to RNA-seq (and using a partially annotated, low-coverage genome and an independently generated transcriptome assembly from the domestic cat), we assigned ∼70% of cheetah EDGE tags to about 14,000 unique genes, which is comparable to a 78% tag-to-gene assignment rate in a parallel comparison to mouse EDGE libraries. Our results suggest that black spotting in cheetahs arises via patterned control of the same melanocyte-based pathways used in other mammals but that the mechanism of patterning does not involve known components of pigment type-switching that lie upstream of the MC1R. Studies of additional cheetah samples will be required to confirm this suggestion and can easily be extended to analogous questions in other patterned mammals such as tigers, leopards, and zebras.
Continuing advances in the cost and scale of sequencing technology and increased sophistication of de novo assembly algorithms are likely to provide reference genome sequences for thousands of mammalian species in the not too distant future (Zerbino and Birney 2008; Metzker 2009; Li et al. 2010; Robertson et al. 2010; Grabherr et al. 2011). Like EDGE, this will further blur the distinction between model and non-model organisms and provide opportunities to investigate many aspects of phenotypic variation that occur only in natural populations.
Methods
Mouse biological samples
C57BL/6J-Mc1r+/+ and Mc1re/e animals were obtained from The Jackson Laboratory (Bar Harbor, MA). Analysis of differential gene expression was based on RNA samples prepared from neonatal dermis (P3.5) and adult spleen (8 wk old). The neonatal dermis samples were obtained by first removing whole dorsal skin and then separating the epidermal and dermal layers using fine forceps after a 12-h incubation with 0.25% trypsin (GIBCO) at 4°C.
For technical replicates, we created two pools of skin RNA from three Mc1r+/+ and three Mc1re/e animals and prepared two EDGE libraries from each pool. For analysis of differential gene expression, tissue samples from individual animals were used to build independent EDGE libraries (21 libraries). Two neonate dermis samples (one Mc1r+/+ and one Mc1re/e) were also used to prepare RNA-seq libraries.
Total RNA from adult skin was prepared using the RNeasy Fibrous Tissue Midi Kit (QIAGEN). Total RNA from neonate dermis and spleen was prepared using TRIzol reagent (Invitrogen) followed by an additional purification using the RNeasy Mini Kit (QIAGEN). Both RNA isolation methods include an on-column DNase I treatment.
Cheetah biological samples
Skin biopsies from cheetahs were obtained using 4-mm biopsy punches at the Cheetah Conservation Fund (Namibia) when animals were placed under general anesthesia during regular veterinary sessions. From a single individual, a pair of skin biopsies was obtained from a black-haired region and an adjacent yellow-haired region and preserved in RNAlater (Ambion). Following the isolation of total RNA using the RNeasy Fibrous Tissue Mini Kit (QIAGEN), EDGE libraries were constructed, and each library was sequenced on one lane of an Illumina Genome Analyzer IIx.
EDGE library preparation
Between 2 and 10 μg of total RNA was used for EDGE library preparation. Briefly, each RNA sample was used for double-stranded cDNA synthesis using paramagnetic oligo(dT) beads to capture polyadenylated RNA. Next, each cDNA molecule was “anchored” by NlaIII restriction digest that cleaves up to the 3′-most restriction site. cDNA fragments carrying the 4-bp overhang (5′-CATG-3′) that remain attached to the paramagnetic beads were ligated to an Illumina adaptor carrying an EcoP15I recognition site (5′-CAGCAG-3′). (EcoP15I is a Type III restriction endonuclease that cleaves 27 bp away from the 3′ end of its recognition site and requires two inversely oriented recognition sites for efficient cleavage [Meisel et al. 1992]. However, we determined optimal reaction conditions that allow for efficient EcoP15I cleavage on linear DNA carrying a single recognition site, obtaining an ∼6.4-fold improvement in cleavage efficiency compared with standard NEB reaction conditions [Supplemental Fig. S7].) Next, cDNA fragments were “tagged” by EcoP15I restriction digest, generating a 27-bp sequence tag with a 2-bp overhang. After restriction digest, the supernatant was saved for the subsequent step, and the paramagnetic beads were removed. Another Illumina adaptor carrying the sequencing primer was ligated to the sticky end, and the 79-bp ligation product was obtained by gel purification. Finally, a 15-cycle PCR enrichment step was performed to enrich for the desired library molecule, and the PCR product was purified using the AMPure XP Kit (Beckman Coulter). A detailed protocol is available in the Supplemental Methods. Cluster generation and sequencing was performed on an Illumina Genome Analyzer II at Stanford University (Stanford, CA) or on an Illumina Genome Analyzer IIx at the HudsonAlpha Institute for Biotechnology (Huntsville, AL).
RNA-seq library preparation
RNA-seq libraries were prepared according to the method described by Mortazavi et al. (2008). Briefly, we started with 2 μg of total RNA and performed a double selection of polyadenylated RNA using oligo(dT) magnetic beads. Next, the RNA was fragmented with RNA fragmentation buffer (200 mM Tris acetate at pH 8.1, 500 mM potassium acetate, 150 mM magnesium acetate) and free ions were removed with a G-50 Sepharose spin column (USA Scientific). Fragmented mRNA was used as a template to synthesize single-stranded cDNA with SuperScript II reverse transcriptase and random hexamer primers in the presence of RNaseOUT (Invitrogen). Double-stranded cDNA was synthesized in a modified buffer of 500 mM Tris-HCl (pH 7.8), 50 mM MgCl2, and 10 mM DTT (Illumina). To prepare cDNA for sequencing, we performed end repair using T4 DNA polymerase and Klenow DNA polymerase (NEB), addition of an “A” base to the 3′ ends of the cDNA using Klenow fragment (NEB), followed by ligation of adaptors designed for the Illumina sequencing platform. The ligation product was purified by gel electrophoresis and purification of the 175–225-bp region on a 1.5% NuSieve GTG agarose gel (Lonza) using the QIAquick Gel Extraction Kit (QIAGEN). Finally, we enriched the library with 15 cycles of PCR amplification using Illumina sequencing primers. Cluster generation and 36-bp single-end sequencing were performed on an Illumina Genome Analyzer IIx at the HudsonAlpha Institute for Biotechnology (Huntsville, AL).
Data processing and analysis: mouse
For each EDGE library, EDGE tags were obtained by selecting sequence reads that passed the quality filter defined by the default Illumina pipeline and trimming off the adaptor sequence at the end of each read. Sequence reads that were not anchored by an NlaIII site, i.e., “CATG,” were also removed, resulting in EDGE tags that were 26, 27, or 28 bp in length (26%, 67%, and 7%, respectively).
For the EDGE libraries, EDGE tags were uploaded onto DNAnexus (http://www.dnanexus.com) and aligned to the mm9 reference genome (NCBI Build 37) using default parameters. Next, the RNA-seq analysis tool was used to count sequence reads that aligned to the sense strand of mouse RefSeq transcripts. An EDGE tag is counted when its posterior probability of mapping to its match is 0.9 or greater, and the posterior probabilities contribute to the sum of the reads. For the RNA-seq libraries, the fastq file from each sequencing run was uploaded onto DNAnexus and analyzed in a similar fashion to the EDGE data. Since our RNA-seq protocol is non-directional, reads that mapped to either orientation of the transcript were counted.
Data processing and analysis: cheetah
Cheetah EDGE libraries were processed (as described above) to obtain EDGE tags. We removed poorly expressed tags, i.e., less than five tags in both libraries, and assigned cheetah EDGE tags to genes using two complementary strategies. The first strategy involved aligning EDGE tags, using ELAND (Illumina), and allowing up to two mismatches, to an Ensembl-annotated, 2×-coverage domestic cat genome assembly (felCat3, UCSC). EDGE tags were assigned to genes if they aligned uniquely to an Ensembl transcript. However, a substantial proportion of EDGE tags aligned to the region immediately downstream from many cat Ensembl genes because the majority of cat Ensembl genes are poorly annotated beyond its coding sequence. To increase our ability to align tags, we created “virtual 3′ UTRs” by extending each Ensembl transcript in the 3′ direction by 1.8 kb (Supplemental Fig. S8). This “virtual 3′ UTR” region contained an ∼34-fold over-representation of EDGE tags compared with the background tag frequency observed in unannotated regions of the genome and corresponds to a 1% false discovery rate. The second strategy relied on a de novo assembled transcriptome from domestic cat that was generated by the Genome Center at Washington University (WC Warren, RK Wilson, unpubl.). In brief, oligo(dT) primed cDNA libraries were obtained from 10 different cat tissues—cerebrum, hypothalamus, thalamus, retina, kidney, ovary, cochlea, vallate tongue, fetal body, and fetal head—and each library was sequenced on a full single-end run on the GS FLX system (Roche). Raw sequence reads from each tissue were then assembled into contigs using Newbler (Roche), resulting in 10 partially assembled cat transcriptomes. EDGE tags were aligned to the cat transcriptome, using ELAND and allowing up to two mismatches, and partial transcripts within the best stratum, i.e., least number of mismatches, were used as a query to identify the most probable human ortholog within RefSeq (release 41) using discontiguous megablast. The hits returned by BLAST were filtered for matches with significant e-values smaller than 10×10−20. Using this conservative threshold, EDGE tags were assigned to a RefSeq gene associated with the best BLAST match (i.e., lowest e-value) to a partial cat transcript.
To integrate the tag to gene assignments from the two informatic approaches, we selected gene assignments based on the number of mismatches for each EDGE tag when it was aligned to the cat genome or transcriptome. Therefore, if a tag can be assigned with either approach, we selected the assignment with the lower number of mismatches. Also, if the number of mismatches was equal, the assignment to an Ensembl gene was chosen as the default.
Identification of differentially expressed genes
To analyze the gene expression profile in mouse tissues, we converted raw gene counts from each EDGE library to TPM and removed genes within each tissue type where the most highly expressed library did not exceed 2 TPM. We applied a Poisson log-linear model described in Witten et al. (2010) to identify genes that were differentially expressed between mutant and wild-type mouse samples.
Quantitative RT-PCR
Quantitative RT-PCR was performed on the same mouse neonate dermis RNA samples used to prepare EDGE libraries. Two micrograms of total RNA was first treated with DNase I (Invitrogen) before reverse transcription with Superscript III (Invitrogen). cDNA samples were diluted fivefold and used for real-time PCR using the Lightcycler Faststart DNA Master Plus SYBR Green I Kit (Roche). Primer sequences used for quantitative PCR were designed to span exon–intron boundaries and are available upon request. The P-values for differences in transcript levels were calculated using the Student's t-test.
Data access
The data from this study have been submitted to the NCBI Sequence Read Archive (http://www.ncbi.nlm.nih.gov/Traces/sra/sra.cgi) under accession number SRA027301.
Acknowledgments
We are extremely grateful to Chris Kaelin for advice on the technical development of EDGE, and Laurie Marker for providing cheetah samples. We also thank Stephen Clark, Ziming Weng, Phil Lacroute, Flo Pauli, Jason Dilocker, Mike Muratet, and Barbara Pusey for their work on generating sequencing data; and Stephen O'Brien, Marilyn Menotti-Raymond, Victor David, and Melody Roelke for providing the source of domestic cat RNA used by the Washington University Genome Center. This work was supported by funds from the HudsonAlpha Institute, the Department of Genetics at Stanford University, a grant from the NIH (to G.S.B.), and by a Genentech Foundation Fellowship and a Stanford Graduate Fellowship (to L.Z.H).
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.122135.111.
References
- Andersson L 2003. Melanocortin receptor variants with phenotypic effects in horse, pig, and chicken. Ann N Y Acad Sci 994: 313–318 [DOI] [PubMed] [Google Scholar]
- April CS, Barsh GS 2006. Skin layer-specific transcriptional profiles in normal and recessive yellow (Mc1re/Mc1re) mice. Pigment Cell Res 19: 194–205 [DOI] [PubMed] [Google Scholar]
- Armour CD, Castle JC, Chen R, Babak T, Loerch P, Jackson S, Shah JK, Dey J, Rohl CA, Johnson JM, et al. 2009. Digital transcriptome profiling using selective hexamer priming for cDNA synthesis. Nat Methods 6: 647–649 [DOI] [PubMed] [Google Scholar]
- Asmann YW, Klee EW, Thompson EA, Perez EA, Middha S, Oberg AL, Therneau TM, Smith DI, Poland GA, Wieben ED, et al. 2009. 3′ tag digital gene expression profiling of human brain and universal reference RNA using Illumina Genome Analyzer. BMC Genomics 10: 531 doi: 10.1186/1471-2164-10-531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baglioni C, Minks MA, Maroney PA 1978. Interferon action may be mediated by activation of a nuclease by pppA2′p5′A2′p5′A. Nature 273: 684–687 [DOI] [PubMed] [Google Scholar]
- Barsh GS 2006. Regulation of pigment type switching by Agouti, melanocortin signaling, attractin, and mahoganoid. In The pigmentary system: Physiology and pathophysiology (ed. Nordlund JJ et al. ), pp. 395–409 Blackwell, Oxford [Google Scholar]
- Bastiaens MT, ter Huurne JA, Kielich C, Gruis NA, Westendorp RG, Vermeer BJ, Bavinck JN, Team LSCS 2001. Melanocortin-1 receptor gene variants determine the risk of nonmelanoma skin cancer independently of fair skin and red hair. Am J Hum Genet 68: 884–894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein-Hanley I, Coers J, Balsara ZR, Taylor GA, Starnbach MN, Dietrich WF 2006. The p47 GTPases Igtp and Irgb10 map to the Chlamydia trachomatis susceptibility locus Ctrq-3 and mediate cellular resistance in mice. Proc Natl Acad Sci 103: 14092–14097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chedekel MR, Smith SK, Post PW, Pokora A, Vessell DL 1978. Photodestruction of pheomelanin: Role of oxygen. Proc Natl Acad Sci 75: 5395–5399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chintala S, Li W, Lamoreux ML, Ito S, Wakamatsu K, Sviderskaya EV, Bennett DC, Park YM, Gahl WA, Huizing M, et al. 2005. Slc7a11 gene controls production of pheomelanin pigment and proliferation of cultured cells. Proc Natl Acad Sci 102: 10964–10969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cloonan N, Forrest AR, Kolle G, Gardiner BB, Faulkner GJ, Brown MK, Taylor DF, Steptoe AL, Wani S, Bethel G, et al. 2008. Stem cell transcriptome profiling via massive-scale mRNA sequencing. Nat Methods 5: 613–619 [DOI] [PubMed] [Google Scholar]
- Eizirik E, Yuhki N, Johnson WE, Menotti-Raymond M, Hannah SS, O'Brien SJ 2003. Molecular genetics and evolution of melanism in the cat family. Curr Biol 13: 448–453 [DOI] [PubMed] [Google Scholar]
- Eleftherianos I, Revenis C 2011. Role and importance of phenoloxidase in insect hemostasis. J Innate Immun 3: 28–33 [DOI] [PubMed] [Google Scholar]
- Enshell-Seijffers D, Lindon C, Morgan BA 2008. The serine protease Corin is a novel modifier of the Agouti pathway. Development 135: 217–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabherr M, Haas B, Yassour M, Levin J, Thompson D, Amit I, Adiconis X, Fan L, Raychowdhury R, Zeng Q, et al. 2011. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol 29: 1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hovanessian AG, Wood JN 1980. Anticellular and antiviral effects of pppA(2′p5′A)n. Virology 101: 81–90 [DOI] [PubMed] [Google Scholar]
- Kaelin CB, Candille SI, Yu B, Jackson P, Thompson DA, Nix MA, Binkley J, Millhauser GL, Barsh GS 2008. New ligands for melanocortin receptors. Int J Obes (Lond) (Suppl 7) 32: S19–S27 [DOI] [PubMed] [Google Scholar]
- Kennedy C, ter Huurne J, Berkhout M, Gruis N, Bastiaens M, Bergman W, Willemze R, Bavinck JN 2001. Melanocortin 1 receptor (MC1R) gene variants are associated with an increased risk for cutaneous melanoma which is largely independent of skin type and hair color. J Invest Dermatol 117: 294–300 [DOI] [PubMed] [Google Scholar]
- Kobayashi T, Vieira WD, Potterf B, Sakai C, Imokawa G, Hearing VJ 1995. Modulation of melanogenic protein expression during the switch from eu- to pheomelanogenesis. J Cell Sci 108: 2301–2309 [DOI] [PubMed] [Google Scholar]
- Lamoreux ML, Zhou BK, Rosemblat S, Orlow SJ 1995. The pinkeyed-dilution protein and the eumelanin/pheomelanin switch: In support of a unifying hypothesis. Pigment Cell Res 8: 263–270 [DOI] [PubMed] [Google Scholar]
- Le Pape E, Passeron T, Giubellino A, Valencia JC, Wolber R, Hearing V 2009. Microarray analysis sheds light on the dedifferentiating role of agouti signal protein in murine melanocytes via the Mc1r. Proc Natl Acad Sci 106: 1802–1807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R, Zhu H, Ruan J, Qian W, Fang X, Shi Z, Li Y, Li S, Shan G, Kristiansen K, et al. 2010. De novo assembly of human genomes with massively parallel short read sequencing. Genome Res 20: 265–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liem EB, Lin CM, Suleman MI, Doufas AG, Gregg RG, Veauthier JM, Loyd G, Sessler DI 2004. Anesthetic requirement is increased in redheads. Anesthesiology 101: 279–283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Hu W, Wang H, Lu M, Shao C, Menzel C, Yan Z, Li Y, Zhao S, Khaitovich P, et al. 2010. Genomic analysis of miRNAs in an extreme mammalian hibernator, the Arctic ground squirrel. Physiol Genomics 42A: 39–51 [DOI] [PubMed] [Google Scholar]
- Meisel A, Bickle TA, Krüger DH, Schroeder C 1992. Type III restriction enzymes need two inversely oriented recognition sites for DNA cleavage. Nature 355: 467–469 [DOI] [PubMed] [Google Scholar]
- Metzker ML 2009. Sequencing technologies—the next generation. Nat Rev Genet 11: 31–46 [DOI] [PubMed] [Google Scholar]
- Miyairi I, Tatireddigari VR, Mahdi OS, Rose LA, Belland RJ, Lu L, Williams RW, Byrne GI 2007. The p47 GTPases Iigp2 and Irgb10 regulate innate immunity and inflammation to murine Chlamydia psittaci infection. J Immunol 179: 1814–1824 [DOI] [PubMed] [Google Scholar]
- Mogil JS, Ritchie J, Smith SB, Strasburg K, Kaplan L, Wallace MR, Romberg RR, Bijl H, Sarton EY, Fillingim RB, et al. 2005. Melanocortin-1 receptor gene variants affect pain and μ-opioid analgesia in mice and humans. J Med Genet 42: 583–587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B 2008. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods 5: 621–628 [DOI] [PubMed] [Google Scholar]
- Mundy NI, Kelly J, Theron E, Hawkins K 2003. Evolutionary genetics of the melanocortin-1 receptor in vertebrates. Ann N Y Acad Sci 994: 307–312 [DOI] [PubMed] [Google Scholar]
- Nagalakshmi U, Wang Z, Waern K, Shou C, Raha D, Gerstein M, Snyder M 2008. The transcriptional landscape of the yeast genome defined by RNA sequencing. Science 320: 1344–1349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oshlack A, Wakefield MJ 2009. Transcript length bias in RNA-seq data confounds systems biology. Biol Direct 4: 14 doi: 10.1186/1745-6150-4-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozsolak F, Goren A, Gymrek M, Guttman M, Regev A, Bernstein BE, Milos PM 2010. Digital transcriptome profiling from attomole-level RNA samples. Genome Res 20: 519–525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perelygin AA, Scherbik SV, Zhulin IB, Stockman BM, Li Y, Brinton MA 2002. Positional cloning of the murine flavivirus resistance gene. Proc Natl Acad Sci 99: 9322–9327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rees JL 2003. Genetics of hair and skin color. Annu Rev Genet 37: 67–90 [DOI] [PubMed] [Google Scholar]
- Renn SC, Aubin-Horth N, Hofmann HA 2004. Biologically meaningful expression profiling across species using heterologous hybridization to a cDNA microarray. BMC Genomics 5: 42 doi: 10.1186/1471-2164-5-42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Replogle K, Arnold AP, Ball GF, Band M, Bensch S, Brenowitz EA, Dong S, Drnevich J, Ferris M, George JM, et al. 2008. The Songbird Neurogenomics (SoNG) Initiative: Community-based tools and strategies for study of brain gene function and evolution. BMC Genomics 9: 131 doi: 10.1186/1471-2164-9-131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbins LS, Nadeau JH, Johnson KR, Kelly MA, Roselli-Rehfuss L, Baack E, Mountjoy KG, Cone RD 1993. Pigmentation phenotypes of variant extension locus alleles result from point mutations that alter MSH receptor function. Cell 72: 827–834 [DOI] [PubMed] [Google Scholar]
- Robertson G, Schein J, Chiu R, Corbett R, Field M, Jackman S, Mungall K, Lee S, Okada H, Qian J, et al. 2010. De novo assembly and analysis of RNA-seq data. Nat Methods 7: 909–912 [DOI] [PubMed] [Google Scholar]
- Samokhvalov A, Hong L, Liu Y, Garguilo J, Nemanich RJ, Edwards GS, Simon JD 2005. Oxidation potentials of human eumelanosomes and pheomelanosomes. Photochem Photobiol 81: 145–148 [DOI] [PubMed] [Google Scholar]
- Simon JD, Peles D, Wakamatsu K, Ito S 2009. Current challenges in understanding melanogenesis: bridging chemistry, biological control, morphology and function. Pigment Cell Melanoma Res 22: 563–579 [DOI] [PubMed] [Google Scholar]
- Steingrímsson E, Copeland NG, Jenkins NA 2006. Mouse coat color mutations: From fancy mice to functional genomics. Dev Dyn 235: 2401–2411 [DOI] [PubMed] [Google Scholar]
- Uthaiah RC, Praefcke GJ, Howard JC, Herrmann C 2003. IIGP1, an interferon-γ-inducible 47-kDa GTPase of the mouse, showing cooperative enzymatic activity and GTP-dependent multimerization. J Biol Chem 278: 29336–29343 [DOI] [PubMed] [Google Scholar]
- Wilhelm BT, Marguerat S, Watt S, Schubert F, Wood V, Goodhead I, Penkett CJ, Rogers J, Bähler J 2008. Dynamic repertoire of a eukaryotic transcriptome surveyed at single-nucleotide resolution. Nature 453: 1239–1243 [DOI] [PubMed] [Google Scholar]
- Witten D, Tibshirani R, Gu SG, Fire A, Lui WO 2010. Ultra-high throughput sequencing-based small RNA discovery and discrete statistical biomarker analysis in a collection of cervical tumours and matched controls. BMC Biol 8: 58 doi: 10.1186/1741-7007-8-58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z, Meyer C, Choudhury S, Shipitsin M, Maruyama R, Bessarabova M, Nikolskaya T, Sukumar S, Schwartzman A, Liu J, et al. 2010. Gene expression profiling of human breast tissue samples using SAGE-Seq. Genome Res 20: 1730–1739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerbino D, Birney E 2008. Velvet: Algorithms for de novo short read assembly using de Bruijn graphs. Genome Res 18: 821–829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerrahn J, Schaible UE, Brinkmann V, Guhlich U, Kaufmann SH 2002. The IFN-inducible Golgi- and endoplasmic reticulum- associated 47-kDa GTPase IIGP is transiently expressed during listeriosis. J Immunol 168: 3428–3436 [DOI] [PubMed] [Google Scholar]