Abstract
Zinc finger nucleases (ZFNs) allow site-specific manipulation of the genome. So far, the use of ZFNs to create gene knockouts has been restricted to protein-coding genes. However, non-protein-encoding RNAs (ncRNA) play important roles in the cell, although the functions of most ncRNAs are unknown. Here, we describe a ZFN-based method suited for the silencing of protein-coding and noncoding genes. This method relies on the ZFN-mediated integration of RNA destabilizing elements into the human genome, e.g., poly(A) signals functioning as termination elements and destabilizing downstream sequences. The biallelic integration of poly(A) signals into the gene locus of the long ncRNA MALAT1 resulted in a 1000-fold decrease of RNA expression. Thus, this approach is more specific and 300 times more efficient than RNA interference techniques. The opportunity to create a variety of loss-of-function tumor model cell lines in different cancer backgrounds will promote future functional analyses of important long noncoding RNA transcripts.
Loss-of-function models are invaluable tools to assess the physiological function of any gene product. These analyses are greatly facilitated by the application of RNA interference (RNAi) using small interfering RNAs (siRNA) to knockdown a target gene of interest (Elbashir et al. 2001). Unfortunately, RNAi has multiple limitations: (1) siRNAs often do not only silence their specific target gene, but also influence the expression of other genes (off-target effects) (Svoboda 2007). (2) The efficiency of an siRNA is not predictable, so that finding a good—efficient and specific—siRNA can be time and cost intensive. (3) Some transcripts can be hard to target due to their strong secondary structure, incorporation into large protein complexes, or their intracellular localization. (4) The siRNA-mediated knockdown is not permanent, making it unsuitable for long-term studies.
Long non-protein-coding RNAs (ncRNA) are especially difficult to target by RNAi: Compared with protein-coding mRNAs, they often localize to the nucleus, have stronger secondary structures, and some are very abundantly expressed, so that a knockdown might not be sufficiently effective to evoke a phenotype and uncover their physiological function. This is of particular importance since ncRNAs are being recognized more and more as a large and important class of molecules with significant functions. Recent transcriptome analyses revealed that 70%–90% of the mammalian genome is transcribed, but only 1%–2% encode proteins (Carninci et al. 2005; Kapranov et al. 2007; Guttman et al. 2009). Thus, ncRNAs, also known as long intergenic ncRNA or lincRNA, greatly enlarge the human transcriptome. The size of these ncRNAs ranges from 200 nt to >100 kb. Functions have only been assigned to a few ncRNAs that participate in the regulation of gene expression at all stages, e.g., epigenetic and transcriptional regulation (Rinn et al. 2007; Mercer et al. 2009). However, for the vast majority of the newly discovered ncRNAs this functional analysis is lacking, but will be essential to fully understand the complex mechanisms underlying developmental, physiological, and pathological processes.
As a new and invaluable tool for gene silencing, synthetic zinc finger nucleases (ZFNs) allow the permanent manipulation of the genome (Bibikova et al. 2002). ZFNs are genetically engineered proteins comprised of a DNA-binding domain composed of at least three Cys2His2 zinc fingers and a nonspecific DNA cleavage domain derived from the endonuclease FokI (Kim et al. 1996). The zinc finger domains can be engineered to target a specific nucleotide sequence. The fused nuclease domain creates a DNA double-strand break (DSB) at this specific site after dimerization. The introduction of a DSB in a eukaryotic chromosome stimulates intracellular DNA repair by both homology-dependent and nonhomologous mechanisms (Jeggo 1998; van Gent et al. 2001). Non-homologous end joining (NHEJ) is an error-prone repair mechanism that produces short deletions at the break (Jeggo 1998).
When targeting the open reading frame (ORF) of a protein-coding gene, this deletion is a favorable event: The small deletion creates a frameshift in two-thirds of all cases disrupting the protein-coding potency of the mRNA, resulting in the desired knockout. Alternatively, the DSB can be repaired by Homologous Recombination (HR) that also allows the integration of ectopically provided sequences into the targeted locus. Consequently, the ZFN technique has broad applications and is used to create gene knockouts in mammalian cells (Maeder et al. 2008; Santiago et al. 2008) or integrate exogenous DNA sequences into plants (Shukla et al. 2009; Townsend et al. 2009) or human somatic, ES, or iPS cells (Urnov et al. 2005; Hockemeyer et al. 2009; Zou et al. 2009).
Despite their broad applicability to silencing of protein-coding genes, ZFNs have not been used for noncoding genes so far. As for RNAi, the ZFN technology also has to overcome major obstacles to silence ncRNAs: (1) Small deletions in ncRNAs are not effective since they do not harbor an open reading frame that could be interrupted. (2) It is not possible to predict functionally important domains in ncRNA molecules, so that a small deletion might hit a functionally irrelevant region of a large ncRNA. (3) Many ncRNAs are transcribed from multiple promoters, so that targeting a single promoter would not completely silence the respective ncRNA. Given the limitations of RNAi for the class of ncRNAs combined with the urgent need for loss-of-function models to uncover the physiological functions of ncRNAs, we provide a method to efficiently and permanently silence noncoding genes.
In our proof-of-principle study, the ncRNA gene MALAT1 (also known as MALAT-1) serves as a model for ZFN-mediated gene silencing. MALAT1 (Metastasis-associated lung adenocarcinoma transcript 1), also known as NEAT2, is an ∼8000-nt long, highly abundant and conserved transcript derived from multiple promoters. It was discovered as a predictive marker for metastasis in lung cancer (Ji et al. 2003). Overexpression of this ncRNA in other tumors such as breast (Guffanti et al. 2009) and liver cancer (Luo et al. 2006) points toward a general role of MALAT1 in carcinogenesis. MALAT1 is a nuclear transcript localizing to SC35 paraspeckles (Hutchinson et al. 2007), suggesting a function in alternative splicing. In fact, two recent studies uncovered that MALAT1 regulates alternative splicing of a subset of pre-mRNAs (Bernard et al. 2010; Tripathi et al. 2010).
To create MALAT1-deficient cells, we used a ZFN and integrated RNA destabilizing elements (RDE) into the human MALAT1 gene locus. The biallelic RDE integration resulted in a 1000-fold reduction of RNA expression and makes this synthetic biology approach far superior to the siRNA-mediated knockdown of MALAT1 in A549 cells. This method enables functional investigations on MALAT1 and other ncRNAs in ZFN-induced loss-of-function models. Moreover, we demonstrate the successful application of this method to silence the protein-coding gene IL2RG in K562 cells, which makes this method a universal tool that can be used to target any gene in the genome and can be combined with different RDEs.
Results
RNA interference against MALAT1 results in high remaining MALAT1 expression
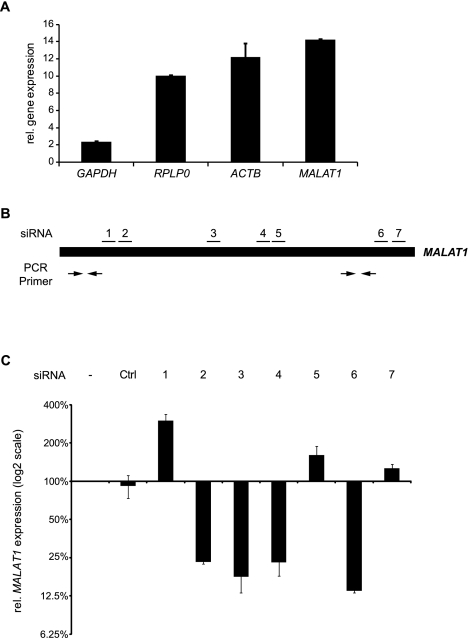
To characterize the abundance of MALAT1 RNA in A549 cells, we compared its expression with housekeeping genes known to be abundantly expressed (Fig. 1A). MALAT1 expression even exceeded the housekeeping genes GAPDH, RPLP0, or ACTB. Thus, MALAT1 is a highly abundant ncRNA expressed from a strong RNA Polymerase II promoter (Wilusz et al. 2008). For functional analysis, we aimed to reduce its expression level by RNAi and tested seven different siRNAs targeting MALAT1 in A549 cells. The binding sites were evenly distributed over the whole transcript with two siRNAs targeting the 5′-end, three siRNAs targeting the middle region, and two siRNAs targeting the 3′-end of MALAT1 (Fig. 1B). The siRNA sequences can be found in Supplemental Table 1. Notably, three siRNAs increased MALAT1 expression up to threefold, whereas the other four siRNAs were able to reduce the MALAT1 level to 13%–25% of its original level (Fig. 1C). However, given the high abundance of MALAT1, this approach still leaves large amounts of MALAT1 RNA in the cell comparable to the endogenous GAPDH mRNA level (Fig. 1A). Therefore, our results are in good concordance with other RNA interference or antisense approaches (siRNA/shRNA/antisense oligos) that also failed to reduce MALAT1 expression more effectively (Supplemental Table 2). The substantial remaining expression of MALAT1 could mask its loss-of-function phenotype and prevent the functional characterization of MALAT1. Therefore, we aimed to develop a new method to efficiently silence the expression of ncRNAs.
Figure 1.
Knockdown of the abundant ncRNA MALAT1. (A) The relative expression levels of GAPDH, RPLP0, ACTB, and MALAT1 in A549 cells were determined via qRT–PCR and analyzed using the 2(-ddCt) method (Livak and Schmittgen 2001). RN7SL1 was used as reference gene. Shown is the mean of measurements from two experiments (×10−7) and the standard deviation (SD). (B) Schematic overview of the siRNA and qPCR primer position in the MALAT1 transcript. (C) Targeting of MALAT1 with seven different siRNAs yielded a knockdown to, at maximum, 13% remaining expression. The transcript level was determined via qRT–PCR and analyzed using the 2(-ddCt) method (Livak and Schmittgen 2001) with RN7SL1 as the reference gene (mean + SD; n = 2).
A strategy to silence non-protein-coding genes using zinc finger nucleases
Recently, ZFNs became commercially available, but they can also be obtained from other sources (Kim et al. 2011), and methods have been described for in-house production (Maeder et al. 2008; Kim et al. 2009). To date, ZFNs are extensively used to target protein-coding genes (Urnov et al. 2005; Maeder et al. 2008; Santiago et al. 2008; Hockemeyer et al. 2009; Shukla et al. 2009; Townsend et al. 2009; Zou et al. 2009). The ZFN-induced DSB can be used to either create a knockout at the protein level by introducing a frameshift via NHEJ—or to integrate an exogenous DNA sequence via HR (Supplemental Fig. 1). The error-prone NHEJ pathway is of specific interest for knocking out protein-coding genes, but is not suitable for targeting long, non-protein-coding genes, which harbor no ORF that could be interrupted. In contrast, HR could be used to target and knockout any gene regardless of its protein-coding potential. We hypothesized that a stable site-specific integration of RNA destabilizing elements (RDEs) into the genome could yield fast and efficient permanent gene silencing. An overview about the potential RDEs that could be used for this gene-targeting approach is given in Figure 2A. These include RDEs that could be used in combination with genes transcribed by RNA Polymerase II as well as RNA Polymerase III.
Figure 2.
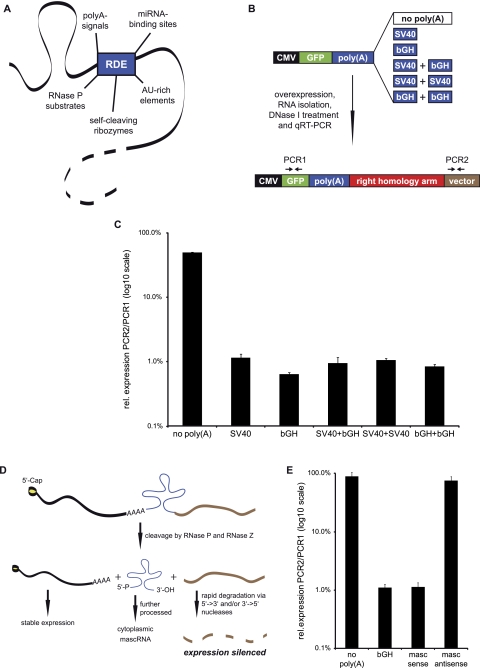
Use of RNA destabilizing elements for gene silencing. (A) RDEs that could be used for gene silencing show different silencing mechanisms. AU-rich elements and miRNA-binding sites influence the stability of the whole transcript, whereas poly(A) signals only silence downstream sequences. RNase P substrates and self-cleaving ribozymes can destabilize both upstream and/or downstream sequences, depending on the position and sequences used. (B) Different poly(A) signals were tested for their silencing potency in an in vitro combinatorial approach. A549 cells were transfected with plasmids containing combinations of poly(A) signals. Expression of GFP mRNA and vector-derived RNA was determined via qRT–PCR: Low vector encoded RNA expression indicated a strong silencing efficiency. (C) The poly(A) signals differ in silencing efficiency. The best inhibition of expression downstream from the RDE was observed with the bGH signal (mean of three experiments + SD). (D) The MALAT1-derived mascRNA sequence as RDE. Placing the mascRNA element immediately downstream from the ORF of a protein-coding gene leads to RNase P cleavage. The resulting mRNA upstream of the cleavage site is stabilized by a 5′-m7Cap and a short poly(A)-like moiety contributed by the fragment. The remaining 3′-sequence either gets degraded or is directly processed by RNase Z to yield a pre-mascRNA. The resulting 3′-end of the transcript is very unstable due to the lack of a 5′-m7Cap and a 3′-poly(A) tail, and is rapidly degraded. (E) The mascRNA element was tested for its silencing potency in the same in vitro approach used for the poly(A) signals. The 242-bp fragment was inserted immediately after the GFP ORF in sense or antisense orientation. The mascRNA element in sense orientation silenced downstream sequences as efficiently as a bGH signal (mean of three experiments + SD).
RDEs can be used to silence downstream sequences
To identify the most efficient silencers for the integration constructs, we tested the silencing potency of two widely used poly(A) sequences and different combinations of these: the poly(A) signals of the bovine growth hormone (bGH) and the simian virus 40 (SV40) (Fig. 2B). As a control, we used a construct that was lacking a poly(A) signal downstream from the GFP ORF. After transient transfection into A549 lung cancer cells, the silencing efficiency was determined by qRT–PCR. The expression of a vector-derived sequence downstream from the RDE was normalized to the GFP mRNA level upstream of the RDE. As expected, the transcript expression only displayed a minor difference between upstream and downstream sequences in the absence of a poly(A) signal. However, the integration of the different poly(A) sites drastically decreased the expression level of sequences downstream from the poly(A) signals, indicating a strong silencing effect (Fig. 2C). In this assay, the bGH signal was the most efficient signal, followed by the combination containing bGH. We also tested the poly(A) silencing potential in Huh7 cells, which yielded comparable results (Supplemental Fig. 2).
Additionally, we wanted to test alternative RDEs for this approach. We decided to target MALAT1 with a part of its own sequence: Wilusz and colleagues reported that MALAT1 is a substrate of RNase P and RNase Z (Wilusz et al. 2008). RNase P recognizes and cleaves a sequence element in the 3′-end of MALAT1. This defines a distinct 3′-end of MALAT1 and subsequent RNase Z-cleavage results in the formation of a cytoplasmic small RNA termed mascRNA. MALAT1 sequences downstream from this mascRNA element are rapidly degraded. Therefore, we speculated that this sequence element could function as an RDE to silence downstream sequences (Fig. 2D). We inserted the mascRNA with its surrounding sequence into the donor vector replacing the poly(A) signal and analyzed its silencing potency. In fact, the mascRNA region drastically inhibited the expression of downstream sequences, but only when inserted in sense orientation (Fig. 2E). The silencing efficiency was comparable to the bGH signal. When the same sequence element was inserted in an antisense orientation as control, no silencing effect was observed. This favors the idea of an RNase P/RNase Z-specific processing, which makes this sequence element an attractive tool also for the silencing of genes that are not RNA Pol II transcripts.
We decided to use poly(A) signals for integration into the MALAT1 gene locus, since these are universal cleavage signals that could be used for the overwhelming majority of genes.
Proof-of-principle
To achieve a site-specific integration of the RDEs, we used a pair of ZFNs that was designed to target the MALAT1 gene immediately downstream from the TATA box of the last MALAT1 promoter (Fig. 3A; Wilusz et al. 2008). The ZFN-mediated cleavage occurs right in front of the transcriptional start site, enabling us to target the gene at its beginning and avoiding the production of any truncated forms. The ZFNs themselves are composed of five zinc fingers in the case of “ZFN for” or six zinc fingers in the case of “ZFN rev.” The “ZFN for” binds to the negative strand with the specific 16-nt long sequence, including one skipped base (Fig. 3A, bold black), while “ZFN rev” binds the positive-strand sequence depicted in Figure 3A. The nucleotide sequences of both zinc fingers can be found in Supplemental Table 7. For the integration, we designed an integration construct that contains a CMV promoter or no exogenous promoter, the GFP gene, and the bGH or SV40+bGH poly(A) signals as RDE. The left homology arm was designed such that the endogenous TATA box was removed when a CMV promoter was present in the integration construct. Vice versa, the endogenous TATA box was preserved when no ectopic promoter was included. GFP expression was used for fast detection of integration-positive clones in subsequent FACS analysis. The cleavage of the pre-mRNA induced by the poly(A) signals has two effects: the upstream transcript is stabilized by the addition of a poly(A) tail and efficiently translated. In contrast, the sequences downstream are unstable due to a missing protective 5′-cap structure and are rapidly degraded.
Figure 3.
Silencing of endogenous genes by integration of RNA destabilizing elements. (A) Genomic MALAT1 region targeted by the ZFNs. The ZFNs cut between the MALAT1 TATA box (blue) and the transcriptional start site (arrow). The binding site for each ZFN is depicted in red. The site-specific integration via HR is mediated through the left and right homology arms surrounding the integration cassette containing GFP and the RDE. (B) Integration constructs used in our MALAT1 gene silencing approach. The constructs were either transfected with or without ZFNs. (C) Overview of the silencing approach. Cells were transfected with a pair of ZFNs and a repair template containing the RDE of choice, e.g., poly(A) signal. The successful gene silencing was validated via qRT–PCR and followed by genotyping. This protocol allowed the creation of single-cell clones with validated genotype and reduction in target gene expression within 6 to 8 wk. (D) Genotype–phenotype relationship of selected clones. Single cell clones (A549 wild-type, CMV-GFP, CMV-GFP-SV40+bGH) were genotyped via integration-sensitive PCR to discriminate between heterozygous and homozygous clones. The effective integration gave rise to a longer PCR product and was not present in wild-type cells. MALAT1 RNA expression was analyzed via RT–PCR using two independent primer pairs, detecting the 5′-end or the 3′-end of MALAT1. Only the homozygous, biallelic integration, including the RDE, yielded an efficient silencing of full-length MALAT1. The detection of RN7SL1 is shown as loading control.
To test our hypothesis that RNA destabilizing elements could be used for gene silencing, we treated A549 cells with a combination of different integration constructs and a pair of ZFNs (Fig. 3B). The complete experimental outline is presented in Figure 3C: Cells were transiently transfected and sorted for GFP-positive cells twice. In the second sort, individual cells were collected in a 96-well plate for clonal outgrowth. The expression of the target gene was then analyzed using the Cells-to-CT method in a 96-well format. This strategy allows the identification of single clones with significant knockdown within 5 wk. As optional steps, these clones can be cultivated in larger cell culture vessels and analyzed for their genotype and the MALAT1 expression using conventional RT–PCR (Fig. 3D).
In total, we analyzed 982 clones for their MALAT1 expression level using the Cells-to-CT method (Supplemental Fig. 3). Individual clones derived from single cells were directly analyzed in 96-well format to reduce handling time. The cells were lysed, cDNA was synthesized, and MALAT1 expression was analyzed by qRT–PCR according to the 2(-ddCt) method (Livak and Schmittgen 2001). RN7SL1 served as a reference gene and expression was normalized to the mean expression level in A549 wild-type clones (Supplemental Fig. 3). Seven weeks after transfection, we performed genotyping PCRs to test whether heterozygous, homozygous, or no integration had occurred (Fig. 3D, top) for one-third of all clones (n = 297) to validate the correlation between genotype and MALAT1 expression (Fig. 4). While this figure gives a comprehensive overview over the experiment, we also aimed to answer specific questions from this large data set. Thus, we calculated the average expression values and standard errors of mean (SEM) in the different groups and compared these groupwise with each other using statistical analyses (Fig. 5).
Figure 4.
Genotype–phenotype correlation in single-cell clones. For 297 clones, the genotype was determined. In a pristine correlation, the lowest MALAT1 levels were only found in homozygous clones harboring biallelic RDE integration (red). The homozygous integration of “CMV-GFP-SV40+bGH” (last column) yielded clones with <0.1% MALAT1 expression (median = 0.14%). Displayed are the MALAT1 expression levels of individual clones, as well as the median expression levels in the individual groups (Cells-to-CT; reference gene: RN7SL1).
Figure 5.
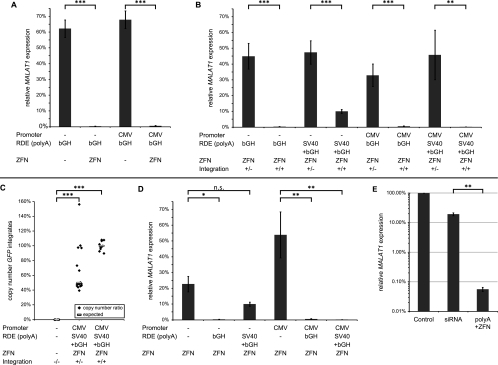
Gene silencing requires ZFN and homozygous poly(A) signal integration and is more effective than siRNA-mediated knockdown. MALAT1 expression levels were determined by Cells-to-CT qRT–PCR of individual clones with RN7SL1 as standardization control. For each group, average expression levels (±SEM) normalized to MALAT1 expression levels in untreated A549 cells (=100%) are shown. The statistical significance of the detected differences was calculated in t-tests; (*) P < 0.05; (**) P < 0.01; (***) P < 0.001. (A) Comparison of the same targeting constructs in the presence or absence of the ZFN. Integration of the poly(A) signal was only observed in the presence of the ZFN, and thus, the MALAT1 expression in clones with homozygous integration was more than 100-fold lower than in clones lacking the ZFN. (B) Comparison of heterozygous and homozygous integration of the poly(A) signal. Homozygous integration of the RDE gave rise to significantly more effective gene silencing. (C) Copy number determination of GFP integration sites in 30 clones. Quantitative RT–PCR of genomic DNA from wild-type, heterozygous, and homozygous clones unraveled the number of GFP integration sites relative to sequences upstream of the MALAT1 locus. The number of GFP integration sites matched the copy number of the control sequence in homozygous clones corroborating the specificity of the integration reaction. (D) Comparison of targeting constructs with and without the silencing element, the poly(A) signal(s). While the integration of GFP without an RDE had only a minor effect on MALAT1 expression, homozygous integration of a poly(A) signal significantly reduced MALAT1 expression with one exception. (E) Comparison between RNA interference and the ZFN-mediated RDE integration. Transfection of the four most effective siRNAs was compared with the four clones with strongest poly(A)-induced gene silencing. MALAT1 knockdown was significantly more than 300-fold stronger in the clones with homozygous poly(A) signal integration.
To determine the efficiency and validity of our approach, we designed the experiment to answer multiple questions:
(1) Is the GFP expression level strong enough for subsequent FACS analysis? We either left the endogenous MALAT1 promoter intact to drive GFP expression or disrupted it by removing the TATA box and included the CMV promoter to drive GFP expression (Fig. 3B). In fact, both promoters were strong enough to yield sufficient GFP expression for subsequent FACS analysis. Another rationale for the use of two different promoters was that promoters with different strengths and origins could result in different sensitivities to the poly(A) signals used. Indeed, we found that the CMV promoter seemed to enhance the silencing in combination with the bGH+SV40 poly(A) signal combination, while the bGH poly(A) signal alone was effective independent of the promoter (Fig. 4).
(2) Is the ZFN necessary to mediate site-specific integration? We compared the expression of the target gene, MALAT1, after transfection of the targeting construct with or without the ZFN. In the absence of the ZFN, we did not detect a strong decrease in MALAT1 expression and no integration (Fig. 5A). Thus, the ZFN is necessary to induce the DSB as a prerequisite for integration of the target construct.
(3) Most importantly, is the integration of an RDE, the poly(A) signal, effective for downstream gene silencing? We tested two poly(A) signals that both were effective in our combinatorial approach (Fig. 2C), the bGH signal and the SV40+bGH signal combination, and compared these with untreated cells for their silencing potency. Using conventional RT–PCR to determine the MALAT1 expression (Fig. 3D, second and third panels) with two independent primer pairs detecting the 5′- or the 3′-region of MALAT1 (Fig. 1B), the silencing effect was already obvious: The integration of the CMV-GFP construct lacking a poly(A) signal (hetero- or homozygous) had no impact on the expression level. However, MALAT1 expression was efficiently reduced in clones with a homozygous integration of the CMV-GFP-SV40+bGH construct, whereas heterozygous integration still left substantial amounts of MALAT1 in the cell.
After quantification by qRT–PCR (Cells-to-CT), lowest MALAT1 levels were detected in multiple clones with homozygous integration of CMV-GFP-SV40+bGH, which displayed a more than 1000-fold reduction in MALAT1 expression down to 0.03% (Fig. 4). Comparing the average expression levels revealed statistically significant (t-test) differences. The integration of GFP alone did not drastically alter MALAT1 expression levels. Only homozygous integration of the poly(A) signals induced strong and lasting gene silencing, whereas heterozygous integration reduced MALAT1 expression by around 50% as expected (Fig. 5B). Notably, the efficiency of transfection, integration, and sorting was sufficient to generate clones with homozygous integration in each group, and in each out of four experiments. Thus, the homozygous (biallelic) integration of poly(A) signals was required and sufficient to shut down expression of the long noncoding RNA MALAT1.
(4) How efficient does our ZFN mediate targeted integration into the genome? For MALAT1, we analyzed the phenotype–genotype relationship of 311 single A549 clones after ZFN transfection. We found 232 clones (74.6%) with a heterozygous integration, while 48 clones (15.4%) showed homozygous integration events. A total of 31 clones (10.0%) did not show any site-specific integration. For IL2RG, we analyzed 270 K562 clones identifying 174 (64.4%) heterozygous and 42 (15.6%) homozygous clones, whereas 54 (20.0%) clones lacked any site-specific integration. Thus, our approach, including two sorting steps, generates homozygous functional knockout clones with high efficiency, displaying some variability between cell lines and integration constructs (Supplemental Table 8).
(5) Beyond effectiveness, is the integration of the targeting construct specific? To assess the number of integration sites in the MALAT1-deficient clones, we determined the copy number of the inserted GFP gene relative to an unaffected control locus upstream of MALAT1 by qPCR of genomic DNA (Fig. 5C). In all 10 homozygous clones analyzed, we found the same copy number for GFP and the control locus, indicating that the ZFN mediated specifically the integration only into both copies of the MALAT1 gene but nowhere else in the genome, demonstrating extraordinary specificity. In a few heterozygous clones, however, we found additional GFP copies pointing toward the need to establish the copy number for individual clones before experimental analysis.
(6) Does the integration event itself—in the absence of an RDE—affect gene expression, or is the RDE required for silencing? To exclude the possibility that the integration negatively influenced the expression of MALAT1, we included constructs without any poly(A) signal. In the presence of the ZFN, these sequences were efficiently integrated into the genome, giving rise to heterozygous as well as homozygous clones. However, they were only weakly effective in silencing the downstream MALAT1 gene (Fig. 5D). Hence, the integration itself does not have a major impact on gene expression, whereas the combination of the ZFN and the RDE is essential and sufficient for efficient and effective gene silencing.
In summary, poly(A) signals can be used as RNA destabilizing elements for efficient and highly effective gene silencing of downstream sequences. The ZFN-induced integration of RDEs was ∼300-fold more effective in gene silencing than the siRNA-mediated knockdown (Fig. 5E). A more than 1000-fold reduction of the target gene was achieved in individual clones, rendering them MALAT1 deficient and most likely functional MALAT1 knockouts. Hence, this methodology enables the use of the ZFN technology in the field of long noncoding RNAs to uncover their physiological roles in genetic loss-of-function models.
Silencing IL2RG, a protein-coding gene with RDEs
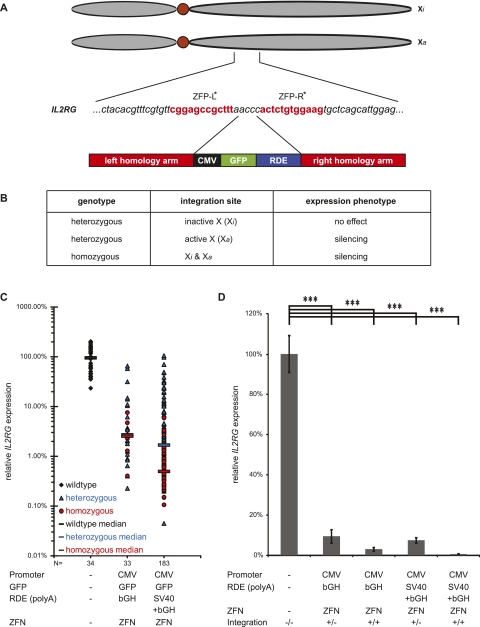
As shown for the ncRNA MALAT1, RNA destabilizing elements are a powerful tool to achieve gene silencing. To test their applicability in silencing protein-coding genes and to analyze the versatility of our approach in a different cell line, we integrated poly(A) signals into the IL2RG locus on chromosome Xq13.1 in K562 leukemia cells (Fig. 6). Dr. Toni Cathomen (Hannover Medical School) provided the plasmids encoding the ZFN targeting exon 5 (Fig. 6A). This ZFN was shown to enable IL2RG gene repair (Urnov et al. 2005) and exogenous gene integration (Moehle et al. 2007). We cotransfected the ZFN with a donor vector encoding the “CMV-GFP-bGH” or “CMV-GFP-SV40+bGH” integration cassettes. The IL2RG mRNA level was determined via qRT–PCR using PCR primers positioned downstream from the integration site in exon 8. The experiment was performed in K562 cells, a female chronic myelogenous leukemia cell line that contains an active (Xa) and an inactivated (Xi) X-chromosome (Fig. 6A). Therefore, different outcomes were expected when IL2RG was targeted (Fig. 6B). For this X-chromosomal locus, heterozygous integration could be sufficient to yield efficient gene silencing if the active X chromosome was targeted. In turn, heterozygous clones could also show IL2RG expression levels similar to wild-type clones if the integration took place on the inactivated X chromosome (Fig. 6B). Indeed, we observed a broad range of IL2RG mRNA expression levels in heterozygous clones reaching from wild-type expression level to more than 1000-fold IL2RG silencing in other heterozygous clones (Fig. 6C). On average, integration of a single bGH poly(A) signal or the combination of the SV40 and the bGH poly(A) signals for RNA destabilization yielded 174 heterozygous and 42 homozygous clones with statistically significant decreased expression of IL2RG (Fig. 6D). As for MALAT1, individual clones achieved a 200-fold to 1900-fold silencing of IL2RG (Fig. 6C).
Figure 6.
Silencing of the protein-coding gene IL2RG with RDE. (A) Genomic IL2RG region on chromosome Xq13.1 targeted by the ZFNs in K562 cells, a female chronic myelogenous leukemia cell line containing an active (Xa) and an inactivated (Xi) X-chromosome. The ZFNs cut in exon 5. The binding site for each zinc finger protein (ZFP) is depicted in red. The site-specific integration via HR is mediated through the left and right homology arms surrounding the integration cassette containing GFP and the RDE (here: bGH or SV40+bGH poly(A) signals). (B) Overview about possible experimental outcomes. Even heterozygous integrations of the RDE can lead to a knockout phenotype if the active X chromosome harbors the integration site. (C) IL2RG expression in individual clones after ZFN-mediated RDE integration. The IL2RG mRNA expression levels were determined by qRT–PCR for 34 wild-type clones of untreated K562 cells as well as for 216 clones with a heterozygous or homozygous RDE integration mediated by a ZFN. Individual clones displayed expression levels around or below 0.1% expression compared with the average IL2RG expression level in untreated K562 clones. (D) Average IL2RG expression after ZFN-mediated RDE integration. On average, the IL2RG mRNA expression was significantly decreased in heterozygous as well as homozygous clones compared with the expression level in wild-type K562 cells. (***) P < 0.001, t-tests.
These results for a non-protein-coding as well as a protein-coding gene in two independent cell lines prove the universality and broad applicability of the RDE-mediated silencing for efficient and effective gene silencing in cell lines.
Discussion
Loss-of-function models are an important and informative way to characterize the physiological and pathological function of mammalian genes. RNAi-mediated gene knockdown is a common strategy, but this widely used approach has multiple limitations (Jackson and Linsley 2010). For our model ncRNA MALAT1, its size, nuclear localization, and its high expression level could be hindrances for an efficient siRNA-mediated knockdown.
Here, we introduce a new method that overcomes these limitations for a stable gene knockdown and is 300 times more efficient than siRNA-mediated knockdown for a model ncRNA. Due to the methods' concept, it is also more specific, resulting in fewer off-target effects, and therefore prospectively producing more reliable results. The ZFN technology has been proven to specifically target the desired genomic locus with great specificity (Miller et al. 2007). In our experiments, none of the 10 homozygous clones analyzed showed any additional nontarget integration corroborating the specificity of the reaction. Thus, by definition of the copy number of inserted constructs, the specificity of the silencing can be tested for each individual clone, whereas the off-target effects of individual siRNAs are largely unknown and cannot be elucidated. In comparison to classical knockout strategies, our approach is fast, simple, and very versatile to create gene-deficient mammalian cells and requires only one specific pair of ZFNs to target virtually any gene in a cell. This makes it a valuable tool, especially for studying molecular and cellular functions of non-protein-coding genes. However, this approach has some limitations, as well. The cell line of interest needs to have at least a minimum transfection efficiency and HR activity to deliver the ZFN and RDE-containing template into the cell and integrate the construct via HR repair of the ZFN-induced DSB. Additionally, cell lines of interest should be tested in advance for their cloning efficiency, and sorting protocols should be optimized to reduce mechanical and environmental stress. Alternatively, the GFP marker can be replaced by a gene conferring antibiotic resistance or any other marker gene to select cells with successful integration. Taken together, only previously optimized protocols should be used for the ZFN and donor plasmid delivery, sorting, and single-cell cloning to increase the success rate. Lastly—as for every genetic manipulation—researchers should carefully evaluate the target locus and determine whether other transcripts, e.g., antisense transcripts, microRNA precursors, or regulatory sites, might be affected by the integration. Rescue experiments reintroducing the silenced gene can serve as controls for the specificity of the phenotype.
Beyond the use of poly(A) signals, other RDE options make the methodology flexible and versatile. Here, we tested the use of an RNase P substrate that efficiently silenced downstream sequences as well (Fig. 3D). In addition, other destabilizing elements and/or their combinations could be used for gene silencing, such as AU-rich elements, microRNA binding sites, or ribozymes, so that it appears reasonable to test different RDE combinations for targeting of individual genes. In our experiments, the bGH poly(A) signal gave the most consistent phenotype independent of the construct and promoter in use. Thus, we recommend the use of the bGH poly(A) signal as a universal and strong silencing element.
Currently, the limited availability of customized ZFNs for individual genes restricts the broad application of ZFN-mediated genetic approaches. However, fast and promising progress is made in the field of ZFN engineering (Sander et al. 2010) and application (Perez et al. 2008). This will enable broad adoption of ZFN technology and allow self-made ZFN design and production.
Importantly, when targeting non-protein-coding genes with the approach introduced here, the ZFN target site should be immediately downstream from the (last) promoter to avoid the formation of potentially functional truncated transcripts.
Long, non-protein-coding RNAs are a growing class of naturally occurring transcripts, whose cellular functions are largely unknown. Their expression is frequently linked to human cancer and other diseases (Panzitt et al. 2007; Taft et al. 2009). Some ncRNAs have critical functions in development (Caley et al. 2010) and in the derivation of pluripotent stem cells (Loewer et al. 2010). With this gene silencing approach suitable also for the generation of ncRNA-deficient cells, we provide a method that will help to unravel the cellular and molecular functions of these noncoding transcripts.
Methods
Cell culture
A549 lung adenocarcinoma cells were purchased from ATCC (CCL-185). Huh7 cells were a kind gift of Dr. Kai Breuhahn (University Hospital Heidelberg). Cells were cultivated at 37°C, 5% CO2 in DMEM + 10% FBS; 0.2 mM Glutamine and antibiotics. K562 chronic myelogenous leukemia cells were a kind gift of Dr. Christoph Plass (DKFZ Heidelberg). Cells were cultivated at 37°C, 5% CO2 in RPMI + 10%FBS; 0.2 mM Glutamine; 0.1 mM Sodium Pyruvate and antibiotics. For transfection experiments, cells were grown in antibiotic-free medium.
ZFN design
ZFNs for MALAT1 were purchased from Sigma-Aldrich. They were custom designed for targeting the TATA box present at nucleotides 1254–1259 of the human MALAT1 gene on chromosome 11 (Gene ID: 378938). The ZFN activity was tested by Sigma-Aldrich in K562 cells using a Surveyor Mutation Detection Assay. The cutting efficiency was 9.1% when ZFNs were transfected as plasmids, or 18.5% when ZFNs were transfected as RNA. The ZFN binding sites (uppercase) and cutting site (lowercase) are as follows: TACGCCTCGCCCGAGCtgtgcgGTAGGCATTGAGGCAGCC. The skipped base is marked in italics (position 7). Detailed sequence information about the zinc finger modules can be found in Supplemental Table 7.
ZFNs for IL2RG were a kind gift of Dr. Toni Cathomen (Hannover Medical School) and had been published by Urnov et al. (2005).
Cloning of integration constructs
The pCRII-TOPO vector (Invitrogen) was used as backbone. The left homology arm was inserted into the HindIII/BamHI site and the right homology arm was inserted into the NotI/XhoI site. Homology sequences were cloned from A549 genomic DNA. The CMV promoter and the bGH poly(A) signal were subcloned via PCR from the pcDNA3.1D V5-His-TOPO vector (Invitrogen). The SV40 poly(A) site was PCR amplified from pFLAG/HA-DGCR8 (a kind gift from Dr. Thomas Tuschl [Rockefeller University, New York]) and the cGFP ORF was PCR amplified from pRNAT-H1.1/Neo (GenScript). For cloning of the mascRNA sequence element, nucleotides 6581–6822 of mMALAT1 (GenBank: NR_002847.2) were PCR amplified from genomic DNA of murine embryonic fibroblasts. A list of all cloning primers can be found in Supplemental Table 3. The sequences contain a random overhang for restriction enzyme binding and the restriction site used for cloning. An overview about the integration constructs and their sequences can be found in Supplemental Table 5.
For IL2RG integration construct cloning the left and right MALAT1 homology arms were exchanged by the IL2RG homology arms. Therefore, vectors were digested with BamHI/HindIII (left arm) and EcoRI/XhoI (right arm). Homology arms were cloned from K562 genomic DNA. A list of all cloning primers can be found in Supplemental Table 3.
Transfection
Cells were seeded into 6-well plates the day before transfection (∼300.000 cells A549 or ∼500.000 K562). For site-specific integration, cells were transfected with a mixture of 3 μg of donor plasmid + 1 μg of ZFN-Mix (0.5 μg per ZFN construct; A549) or 2μg of ZFN-Mix (1μg per ZFN construct; K562). For random integration, cells were transfected with 3 μg of donor plasmid only. Turbofect transfection reagent (Fermentas) was used at a 2:1 ratio (reagent:DNA). Transfection medium was changed after 4–6 h and cells were grown for an additional 42 h in 6-well plates before being transferred into larger dishes. For poly(A) testing, cells were transfected with 2 μg of donor plasmid and 4 μL of Turbofect transfection reagent. Cells were grown for 48 h in 6-well plates before lysis, RNA isolation and qRT–PCR. In the case of K562, cells were grown for 24h at 37°C and moved to 30°C for an additional 48 h to induce a transient cold shock. After this, cells were further cultivated at 37°C.
RNA interference
SiRNAs were designed with the BLOCK-iT RNAi Designer (Invitrogen). Control-siRNA was obtained from Qiagen (AllStars Negative Control siRNA). For reverse transfection, 180,000 cells were seeded into 6-well plates and transfected with 5 μL of RNAiMax (Invitrogen) and 100 pmol of siRNA (40 nM f.c.). Knockdown was analyzed 48 h after transfection. For a complete list of siRNAs tested, see Supplemental Table 1.
RNA isolation and DNase I digest
RNA was isolated with TRIzol reagent (Invitrogen) according to the manufacturer's recommendations. Samples were treated with DNase I (Roche) for 30 min at 37°C followed by phenol:chloroform extraction and ethanol precipitation at −80°C.
Reverse transcription and PCR
RNA (1 μg) was reverse transcribed with RevertAid H Minus Reverse Transcriptase (Fermentas) according to the manufacturer's recommendations. Complete removal of genomic DNA or plasmid DNA was controlled in minus-RT samples in which the Reverse Transcriptase was replaced by water. For qRT–PCR, the PowerSYBR Green PCR Master Mix (Applied Biosystems) was used. For RT–PCR, the DreamTaq Green DNA Polymerase (Fermentas) was used according to the manufacturer's recommendations. Primer sequences can be found in Supplemental Table 4.
Cells-to-CT analysis
The PowerSYBR Green Cells-to-CT Kit (Applied Biosystems) was used for rapid screening of single cell clones in 96-well plates according to the manufacturer's recommendations. MALAT1 (5′-region), IL2RG, and RN7SL1 gene expression was analyzed via qRT–PCR. See Supplemental Table 4 for primer sequences.
Isolation of genomic DNA, genotyping PCR, and copy number determination
Genomic DNA of individual A549 clones was isolated with the GenElute mammalian genomic DNA MiniPrep Kit (Sigma) according to the manufacturer's recommendations. A total of 200 ng of genomic DNA was used for subsequent Integration-PCR with DreamTaq Green DNA Polymerase (Fermentas) according to the manufacturer's recommendations. The following primer pair was used for the MALAT1 gene locus: 5′-TTGCAGCTCAAATCTTTCCA (forward); 5′-CGTTAAAAACTTAACGCTAAGCAA (reverse). A list of expected product sizes can be found in Supplemental Table 5. For GFP copy number determination, 10 ng of genomic DNA of 30 randomly chosen single cell clones was analyzed in qPCR with primers detecting GFP. For copy number reference, a genomic sequence upstream of the MALAT1 gene unaffected by the ZFN treatment was used (Supplemental Table 4).
Genotyping of K562 single cell clones was done with the DirectPCR lysis reagent (Peqlab) according to the manufacturer's recommendations. Cells were lysed in 70 μL overnight at 55°C. After heat inactivation, 5 μL of the reaction mixture were directly used for genotyping with DreamTaq Green DNA Polymerase (Fermentas). The following primer pair was used for the IL2RG gene locus: 5′-GGTGGGTGTTCAGGAGTATGTT (forward); 5′-AAGTGGAGCAAAAGACAGTGGT (reverse).
FACS analysis
GFP-positive cells were sorted on a FACS Aria II (BD Biosciences, DKFZ Core Facility). Cells were either bulk sorted or single-cell sorted into 96-well plates.
Statistical analysis
Statistical analyses were performed using Excel and SPSS 17.0. Significance was assessed using t-tests after determination of the variance equality using an f-test.
Acknowledgments
We thank G. Stoecklin, D. Ostareck, E. Fiskin, and H. Uckelmann for helpful discussions and critical reading of the manuscript. We thank Dr. Cathomen for providing the IL2RG ZFN. We thank S. Schmitt and the DKFZ Flow Cytometry Core Facility for excellent technical assistance. Our research is supported by the German Research Foundation (DFG Transregio TRR77, TP B03), the Marie Curie Program of the European Commission, the Helmholtz Society (VH-NG-504), the German Cancer Research Center (DKFZ), and the Institute of Pathology, University of Heidelberg. T.G. is supported by a DKFZ PhD Fellowship.
Authors’ contributions: T.G. and S.D. conceived and planned the experiments and analyzed and interpreted the results. T.G. and M.B. performed the experiments. T.G. and S.D. wrote the manuscript.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.122358.111.
References
- Bernard D, Prasanth KV, Tripathi V, Colasse S, Nakamura T, Xuan Z, Zhang MQ, Sedel F, Jourdren L, Coulpier F, et al. 2010. A long nuclear-retained non-coding RNA regulates synaptogenesis by modulating gene expression. EMBO J 29: 3082–3093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibikova M, Golic M, Golic KG, Carroll D 2002. Targeted chromosomal cleavage and mutagenesis in Drosophila using zinc-finger nucleases. Genetics 161: 1169–1175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caley DP, Pink RC, Trujillano D, Carter DR 2010. Long noncoding RNAs, chromatin, and development. TheScientificWorldJOURNAL 10: 90–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carninci P, Kasukawa T, Katayama S, Gough J, Frith MC, Maeda N, Oyama R, Ravasi T, Lenhard B, Wells C, et al. 2005. The transcriptional landscape of the mammalian genome. Science 309: 1559–1563 [DOI] [PubMed] [Google Scholar]
- Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T 2001. Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 411: 494–498 [DOI] [PubMed] [Google Scholar]
- Guffanti A, Iacono M, Pelucchi P, Kim N, Solda G, Croft LJ, Taft RJ, Rizzi E, Askarian-Amiri M, Bonnal RJ, et al. 2009. A transcriptional sketch of a primary human breast cancer by 454 deep sequencing. BMC Genomics 10: 163 doi: 10.1186/1471-2164-10-163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, Huarte M, Zuk O, Carey BW, Cassady JP, et al. 2009. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature 458: 223–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hockemeyer D, Soldner F, Beard C, Gao Q, Mitalipova M, DeKelver RC, Katibah GE, Amora R, Boydston EA, Zeitler B, et al. 2009. Efficient targeting of expressed and silent genes in human ESCs and iPSCs using zinc-finger nucleases. Nat Biotechnol 27: 851–857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutchinson JN, Ensminger AW, Clemson CM, Lynch CR, Lawrence JB, Chess A 2007. A screen for nuclear transcripts identifies two linked noncoding RNAs associated with SC35 splicing domains. BMC Genomics 8: 39 doi: 10.1186/1471-2164-8-39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson AL, Linsley PS 2010. Recognizing and avoiding siRNA off-target effects for target identification and therapeutic application. Nat Rev Drug Discov 9: 57–67 [DOI] [PubMed] [Google Scholar]
- Jeggo PA 1998. DNA breakage and repair. Adv Genet 38: 185–218 [DOI] [PubMed] [Google Scholar]
- Ji P, Diederichs S, Wang W, Boing S, Metzger R, Schneider PM, Tidow N, Brandt B, Buerger H, Bulk E, et al. 2003. MALAT-1, a novel noncoding RNA, and thymosin β4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene 22: 8031–8041 [DOI] [PubMed] [Google Scholar]
- Kapranov P, Cheng J, Dike S, Nix DA, Duttagupta R, Willingham AT, Stadler PF, Hertel J, Hackermuller J, Hofacker IL, et al. 2007. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science 316: 1484–1488 [DOI] [PubMed] [Google Scholar]
- Kim YG, Cha J, Chandrasegaran S 1996. Hybrid restriction enzymes: zinc finger fusions to Fok I cleavage domain. Proc Natl Acad Sci 93: 1156–1160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HJ, Lee HJ, Kim H, Cho SW, Kim JS 2009. Targeted genome editing in human cells with zinc finger nucleases constructed via modular assembly. Genome Res 19: 1279–1288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S, Lee MJ, Kim H, Kang M, Kim JS 2011. Preassembled zinc-finger arrays for rapid construction of ZFNs. Nat Methods 8: 7 doi: 10.1038/nmeth0111-7a [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Loewer S, Cabili MN, Guttman M, Loh YH, Thomas K, Park IH, Garber M, Curran M, Onder T, Agarwal S, et al. 2010. Large intergenic non-coding RNA-RoR modulates reprogramming of human induced pluripotent stem cells. Nat Genet 42: 1113–1117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo JH, Ren B, Keryanov S, Tseng GC, Rao UN, Monga SP, Strom S, Demetris AJ, Nalesnik M, Yu YP, et al. 2006. Transcriptomic and genomic analysis of human hepatocellular carcinomas and hepatoblastomas. Hepatology 44: 1012–1024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeder ML, Thibodeau-Beganny S, Osiak A, Wright DA, Anthony RM, Eichtinger M, Jiang T, Foley JE, Winfrey RJ, Townsend JA, et al. 2008. Rapid “open-source” engineering of customized zinc-finger nucleases for highly efficient gene modification. Mol Cell 31: 294–301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer TR, Dinger ME, Mattick JS 2009. Long non-coding RNAs: insights into functions. Nat Rev Genet 10: 155–159 [DOI] [PubMed] [Google Scholar]
- Miller JC, Holmes MC, Wang J, Guschin DY, Lee YL, Rupniewski I, Beausejour CM, Waite AJ, Wang NS, Kim KA, et al. 2007. An improved zinc-finger nuclease architecture for highly specific genome editing. Nat Biotechnol 25: 778–785 [DOI] [PubMed] [Google Scholar]
- Moehle EA, Rock JM, Lee YL, Jouvenot Y, DeKelver RC, Gregory PD, Urnov FD, Holmes MC 2007. Targeted gene addition into a specified location in the human genome using designed zinc finger nucleases. Proc Natl Acad Sci 104: 3055–3060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panzitt K, Tschernatsch MM, Guelly C, Moustafa T, Stradner M, Strohmaier HM, Buck CR, Denk H, Schroeder R, Trauner M, et al. 2007. Characterization of HULC, a novel gene with striking up-regulation in hepatocellular carcinoma, as noncoding RNA. Gastroenterology 132: 330–342 [DOI] [PubMed] [Google Scholar]
- Perez EE, Wang J, Miller JC, Jouvenot Y, Kim KA, Liu O, Wang N, Lee G, Bartsevich VV, Lee YL, et al. 2008. Establishment of HIV-1 resistance in CD4+ T cells by genome editing using zinc-finger nucleases. Nat Biotechnol 26: 808–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E, et al. 2007. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell 129: 1311–1323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sander JD, Dahlborg EJ, Goodwin MJ, Cade L, Zhang F, Cifuentes D, Curtin SJ, Blackburn JS, Thibodeau-Beganny S, Qi Y, et al. 2010. Selection-free zinc-finger-nuclease engineering by context-dependent assembly (CoDA). Nat Methods 8: 67–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santiago Y, Chan E, Liu PQ, Orlando S, Zhang L, Urnov FD, Holmes MC, Guschin D, Waite A, Miller JC, et al. 2008. Targeted gene knockout in mammalian cells by using engineered zinc-finger nucleases. Proc Natl Acad Sci 105: 5809–5814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla VK, Doyon Y, Miller JC, DeKelver RC, Moehle EA, Worden SE, Mitchell JC, Arnold NL, Gopalan S, Meng X, et al. 2009. Precise genome modification in the crop species Zea mays using zinc-finger nucleases. Nature 459: 437–441 [DOI] [PubMed] [Google Scholar]
- Svoboda P 2007. Off-targeting and other non-specific effects of RNAi experiments in mammalian cells. Curr Opin Mol Ther 9: 248–257 [PubMed] [Google Scholar]
- Taft RJ, Pang KC, Mercer TR, Dinger M, Mattick JS 2009. Non-coding RNAs: regulators of disease. J Pathol 220: 126–139 [DOI] [PubMed] [Google Scholar]
- Townsend JA, Wright DA, Winfrey RJ, Fu F, Maeder ML, Joung JK, Voytas DF 2009. High-frequency modification of plant genes using engineered zinc-finger nucleases. Nature 459: 442–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripathi V, Ellis JD, Shen Z, Song DY, Pan Q, Watt AT, Freier SM, Bennett CF, Sharma A, Bubulya PA, et al. 2010. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol Cell 39: 925–938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urnov FD, Miller JC, Lee YL, Beausejour CM, Rock JM, Augustus S, Jamieson AC, Porteus MH, Gregory PD, Holmes MC 2005. Highly efficient endogenous human gene correction using designed zinc-finger nucleases. Nature 435: 646–651 [DOI] [PubMed] [Google Scholar]
- van Gent DC, Hoeijmakers JH, Kanaar R 2001. Chromosomal stability and the DNA double-stranded break connection. Nat Rev Genet 2: 196–206 [DOI] [PubMed] [Google Scholar]
- Wilusz JE, Freier SM, Spector DL 2008. 3′ end processing of a long nuclear-retained noncoding RNA yields a tRNA-like cytoplasmic RNA. Cell 135: 919–932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou J, Maeder ML, Mali P, Pruett-Miller SM, Thibodeau-Beganny S, Chou BK, Chen G, Ye Z, Park IH, Daley GQ, et al. 2009. Gene targeting of a disease-related gene in human induced pluripotent stem and embryonic stem cells. Cell Stem Cell 5: 97–110 [DOI] [PMC free article] [PubMed] [Google Scholar]