Abstract
SRC proteins are non-receptor tyrosine kinases that play key roles in regulating signal transduction by a diverse set of cell surface receptors. They contain N-terminal SH4 domains that are modified by fatty acylation and are functioning as membrane anchors. Acylated SH4 domains are both necessary and sufficient to mediate specific targeting of SRC kinases to the inner leaflet of plasma membranes. Intracellular transport of SRC kinases to the plasma membrane depends on microdomains into which SRC kinases partition upon palmitoylation. In the present study, we established a live-cell imaging screening system to identify gene products involved in plasma membrane targeting of SRC kinases. Based on siRNA arrays and a human model cell line expressing two kinds of SH4 reporter molecules, we conducted a genome-wide analysis of SH4-dependent protein targeting using an automated microscopy platform. We identified and validated 54 gene products whose down-regulation causes intracellular retention of SH4 reporter molecules. To detect and quantify this phenotype, we developed a software-based image analysis tool. Among the identified gene products, we found factors involved in lipid metabolism, intracellular transport, and cellular signaling processes. Furthermore, we identified proteins that are either associated with SRC kinases or are related to various known functions of SRC kinases such as other kinases and phosphatases potentially involved in SRC-mediated signal transduction. Finally, we identified gene products whose function is less defined or entirely unknown. Our findings provide a major resource for future studies unraveling the molecular mechanisms that underlie proper targeting of SRC kinases to the inner leaflet of plasma membranes.
The SRC protein family consists of non-receptor tyrosine kinases that play key roles in cellular signal transduction. They are involved in the regulation of fundamental cellular processes including cell growth and differentiation, as well as migration and survival (Parsons and Parsons 2004). The general SRC kinase architecture consists of four domains, three of which are directly linked to the signal transduction pathways in which they are involved. The SH3 domain mediates interactions with other signaling molecules, the SH2 domain is required for phosphotyrosine recognition, and the SH1 domain contains the kinase activity (Boggon and Eck 2004). In contrast, the fourth domain (SH4) is solely responsible for membrane attachment and targeting to plasma membranes.
The molecular mechanism by which SRC kinases and other peripheral membrane proteins such as Leishmania hydrophilic acylated surface protein B (HASPB) are anchored in the inner leaflet of plasma membranes is based on post-translational lipid modifications within their N-terminal SH4 domains. Following removal of the N-terminal methionine residue, SH4 proteins become myristoylated at the N terminus via an amide linkage (Resh 1999, 2004). The second acylation step involves the modification of at least one cysteine residue by palmitoylation, resulting in a thioester linkage (Linder and Deschenes 2007). Thus, most SRC kinases carry two binding sites for membrane anchoring. This feature has been generalized as the two-signal hypothesis that was originally developed for mammalian Ras proteins (Hancock et al. 1990). As a variation of this principle, instead of palmitoylation, a second membrane-binding site can be formed by a cluster of basic amino acids as is the case for the prototype member of the family, SRC (Smotrys and Linder 2004). While the enzymes involved in N-myristoylation of SH4 domains have been known for many years (Resh 1999), the enzymology of protein palmitoylation has been revealed only recently with so-called DHHC (Asp-His-His-Cys) proteins acting as S-acyltransferases (Mitchell et al. 2006; Linder and Deschenes 2007).
N-terminal SH4 domains are sufficient for intracellular targeting of, for example, GFP fusion proteins to the inner leaflet of plasma membranes (McCabe and Berthiaume 1999; Denny et al. 2000; McCabe and Berthiaume 2001; Stegmayer et al. 2005; Tournaviti et al. 2007, 2009). Myristoylation of SH4 domains occurs cotranslationally and results in a transient insertion into intracellular membranes in a perinuclear region where they colocalize with Golgi markers (Wolven et al. 1997; Denny et al. 2000; Stegmayer et al. 2005; Tournaviti et al. 2007). Recently, using semisynthetic substrates corresponding to various kinds of palmitoylated proteins, it has been suggested that palmitoylation of all cellular target proteins occurs specifically at the Golgi (Rocks et al. 2010). In this way, palmitoylation appears to be controlled spatially rather than being controlled by enzymatic specificity of acyl transferases, an observation that is consistent with the observed functional redundancy of DHHC acyl transferases that catalyze protein palmitoylation (Hou et al. 2009).
Interestingly, palmitoylation of SH4 domains is required for SRC kinases to properly partition into membrane microdomains (Brown and London 1998; Webb et al. 2000; Edidin 2003; Smotrys and Linder 2004; Linder and Deschenes 2007). This, in turn, causes retention of SRC kinases at the level of Golgi membranes; i.e., forward transport to the inner leaflet of the plasma membrane is blocked under these experimental conditions. These observations suggest that membrane microdomains serve as sorting and transport platforms required for proper targeting of SH4 proteins. Besides these insights, however, our knowledge remains poor with regard to mechanistic details of SH4-domain-dependent protein targeting to the inner leaflet of plasma membranes.
In the present study, we have conducted an unbiased RNAi screening approach analyzing the entire human genome for factors involved in proper targeting of SH4 proteins. We have established an experimental model system based on two different SH4 domains that are fused to GFP and mCherry, respectively, and are expressed simultaneously in HeLa cells in a doxycycline-dependent manner. Under control conditions both of these SH4 fusion proteins primarily localize to plasma membranes with only a minor fraction being detected intracellularly. For both primary screening and validation experiments, cells were grown on siRNA arrays (Erfle et al. 2004; Neumann et al. 2006, 2010; Simpson et al. 2007). A software tool was developed to detect and quantify perinuclear accumulation of one or both SH4 reporter molecules. Hits identified during primary screening were validated using independent siRNAs. All data on experimental conditions and results of the screening procedure were stored in a web-based database. The final list of validated gene products whose down-regulation causes intracellular retention of SH4 reporter molecules consists of 54 gene products. It contains proteins with well-defined functions in lipid metabolism, intracellular transport, and cellular signaling processes, as well as other functional classes. Additionally, several proteins were identified whose function is less defined or entirely unknown. Thus, this study provides a major resource for studying the mechanistic details of microdomain-dependent Golgi-to-plasma membrane trafficking of SRC kinases.
Results
An experimental model system to study SH4-domain-dependent protein targeting to plasma membranes
To study intracellular SH4-domain-dependent transport to the plasma membrane using a genome-wide RNAi screening approach, we engineered a human model cell line expressing two distinct SH4-domain-containing fusion proteins. These contained the N-terminal 18 amino acids of either Leishmania HASPB (Denny et al. 2000; Stegmayer et al. 2005) or of the mammalian SRC kinase YES1 (Tournaviti et al. 2009) fused to GFP (SH4-HASPB-GFP) and mCherry (SH4-YES1-mCherry), respectively. Both fusion proteins are substrates for N-terminal myristoylation and subsequent palmitoylation (cysteine 5 in HASPB; cysteine 3 in YES1) (Supplemental Fig. S1A). Simultaneous expression of both SH4-HASPB-GFP and SH4-YES1-mCherry in stably transducted HeLa cells was controlled by a doxycycline-sensitive transactivator and, therefore, could be induced by doxycycline as demonstrated by flow cytometry (Supplemental Fig. S1B). Upon expression, using confocal microscopy, both reporter proteins were found to localize at the plasma membrane establishing that N-terminal SH4 domains are sufficient for proper targeting (Supplemental Fig. S2C; McCabe and Berthiaume 1999; Denny et al. 2000; McCabe and Berthiaume 2001; Stegmayer et al. 2005).
Palmitoylation-dependent transport of SH4 reporter proteins to the plasma membrane
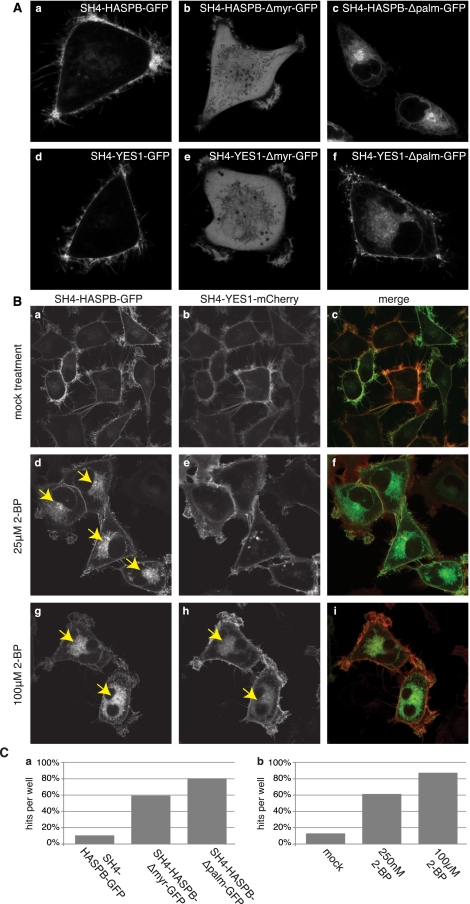
For both SH4-HASPB-GFP and SH4-YES1-mCherry, delivery to the plasma membrane depends on the acylation state of the respective SH4 domain. Substitution of the myristoylated glycine residue in SH4-HASPB not only prevents myristoylation, but also blocks palmitoylation (McCabe and Berthiaume 1999; Denny et al. 2000; Stegmayer et al. 2005; Tournaviti et al. 2009), i.e., N-terminal myristoylation is a prerequisite for palmitoylation to occur. A lack of acylation resulting from a deletion of the myristoylation site in turn renders both SH4-HASPB- and SH4-YES1-containing fluorescent reporter proteins incapable of interacting with cellular membranes, resulting in cytoplasmic localization (Fig. 1A, subpanels b and e). In contrast, disruption of the palmitoylation site in HASPB does not affect myristoylation (McCabe and Berthiaume 1999; Denny et al. 2000; Stegmayer et al. 2005) and results in intracellular retention at perinuclear membranes (Fig. 1A, subpanel c). This effect is more pronounced in case of SH4-HASPB-GFP but can also be observed for SH4-YES1-GFP (Fig. 1A, subpanel f). Alternatively, palmitoylation can also be blocked with a non-metabolizable palmitate analog, 2-bromo-palmitate (2-BP) (Webb et al. 2000; Resh 2006), that inhibits transport of SH4 fusion proteins to the plasma membrane (Fig. 1B). Again, pharmacological inhibition of palmitoylation caused a more pronounced transport block of SH4-HASPB-GFP when compared to SH4-YES1-mCherry (Fig. 1B). In the context of the present study, palmitoylation-deficient mutant forms of SH4 reporter proteins as well as pharmacological inhibition of SH4 palmitoylation were used as positive controls to implement the high-content screening system as it was designed to detect and quantify intracellular retention of SH4 fusion proteins.
Figure 1.
Plasma membrane targeting of SH4 fusion proteins depends on N-terminal acylation and can be measured using an image analysis tool. (A) Stable HeLa cells expressing GFP fusion proteins containing either wild-type or mutant forms of the SH4 domains of either HASPB or YES1 were induced for reporter protein expression and subjected to live cell confocal microscopy to determine the subcellular localization of the SH4 fusion proteins. (B) Stable HeLa cells expressing both SH4-HASPB-GFP and SH4-YES1-mCherry were treated with the palmitoylation inhibitor 2-bromopalmitate (2-BP). Subcellular localization of SH4 fusion proteins was analyzed by live-cell confocal microscopy. (C) Wide-field images from stable HeLa cells expressing wild-type or mutant SH4 domains of HASPB (a) or from HeLa cells expressing SH4-HASPB-GFP under treatment with 2-BP (b) were analyzed with the automated image analysis tool and evaluated for the detection of hit phenotypes.
A software-based image analysis tool to detect and quantify intracellular retention of SRC kinases
Microscopy-based high-content screening approaches result in extremely large sets of image-based data, requiring the development of tools to achieve an efficient, unbiased, and quantitative analysis of phenotypes of interest (Boland et al. 1998; Conrad et al. 2004; Carpenter et al. 2006; Neumann et al. 2006; Glory and Murphy 2007; Harder et al. 2008; Wang et al. 2008a; Walter et al. 2010). Based on general tools such as cell segmentation techniques (Elowitz et al. 2002; Megason and Fraser 2007; Wang et al. 2008b), we developed a software-based image analysis module specifically tailored to detect and quantify intracellular retention of SH4 fusion proteins. This software tool identifies individual cells, detects different cellular compartments, and classifies cells according to intensity distributions between such compartments (Supplemental Fig. S2). This classification can be conducted for different fluorescence channels simultaneously, i.e., the two different fusion proteins used in this study could be analyzed independently. Furthermore, the software module was designed in a way that a potential low quality of image-based screening data, such as low resolution and out-of-focus images, local contaminations, and areas of different cell densities as well as varying signal intensities, did not result in wrong classification of phenotypes.
A crucial step for automated image analysis was the correct identification and segmentation of individual cells since this step provides the basis for intensity feature extractions from different cellular areas and, therefore, has a direct impact on both quality and reliability of the results. Cell identification was based on the detection of stained cell nuclei by an adaptive thresholding approach. Due to the application of a stepwise convergence of intensities to the intensity of cell nuclei, this approach is invariant to different nuclear intensities between different images and therefore more reliable than fixed thresholding approaches. Using cell nuclei as a seed point, the shape of individual cells was identified by a classifier-enhanced region-growing approach. Cell boundaries were determined by two criteria: (1) a large GFP intensity difference between inner and outer pixels and (2) a lower GFP intensity of the outer pixels compared to a local brightness threshold for a given cell. These criteria were not only sufficient to determine the shape of cells independently of the distribution of reporter intensities inside the cell, but also provided reliable data when fluorescence intensities varied between cells. Following determination of cell shape, cells characterized by abnormal size and shape were classified as invalid and excluded from further analysis. The fluorescence distribution of the remaining cells was used to classify the localization of the reporter proteins within one of three possible intracellular compartments: (1) the nucleus, (2) the cytosol (including all membrane-compartments that are present in the cytoplasm such as the Golgi), and (3) the plasma membrane (Supplemental Fig. S2A). Intensity features from the complete cell area as well as the three cellular compartments were extracted and used for classification of individual cells (Supplemental Fig. S2B). Based on these criteria, perinuclear retention at, for example, Golgi membranes should result in an increased cytoplasmic intensity compared to nuclear and membrane-associated intensities. A detailed description of the software-based image analysis tool used in this study is given in Methods.
Based on the use of acylation-deficient variant forms of SH4 fusion proteins (Fig. 1A) and pharmacological inhibition of palmitoylation using 2-BP (Fig. 1B), intracellular accumulation of one or both reporter proteins and, therefore, redistribution of fluorescence intensity from the plasma membrane to intracellular compartments in individual cells was used to evaluate the image analysis software described above. Indeed, the phenotype of highest interest, i.e., perinuclear retention, could be detected in ∼80% of cells expressing the HASPB palmitoylation mutant (SH4-HASPB-Δpalm-GFP) (Fig. 1C, subpanel a). The software tool was also capable of identifying gradual effects evoked by treatment of cells expressing the wild-type form of SH4-HASPB-GFP with the palmitoylation inhibitor 2-BP. Here, a range between 60% and 85% of cells with significant perinuclear retention of SH4-HASPB-GFP was observed depending on the inhibitor concentration applied to the cells (Fig. 1C, subpanel b).
Automated high-content screening to identify gene products involved in SH4-domain-dependent targeting to plasma membranes
To identify proteins involved in SH4-domain-dependent transport to the plasma membrane, we used siRNA arrays covering all protein-coding genes known in the human genome. Each of the more than 21,000 genes was targeted with at least two independent siRNAs. As described previously (Erfle et al. 2004, 2007; Neumann et al. 2006, 2010; Simpson et al. 2007), siRNAs were immobilized on LabTek imaging chambers in 384-spot arrays and used for reverse transfection of a HeLa cell line stably transduced with SH4-HASPB-GFP and SH4-YES1-mCherry (Supplemental Fig. S1). Thirty-six hours after transfection, expression of SH4 fusion proteins was induced by doxycycline followed by live-cell imaging 48 h post-transfection (Supplemental Fig. S3). Various control siRNAs were tested such as a scrambled siRNA and an siRNA targeting GFP to down-regulate SH4-HASPB-GFP, as well as an siRNA to down-regulate INCENP, an inner centromere protein whose absence is known to cause mitotic defects and the formation of multinucleated cells (Neumann et al. 2010). Indeed, reverse transfection of a scrambled siRNA did not cause any pronounced effects on cellular morphology and SH4 fusion protein localization (Supplemental Fig. S4A–C). In contrast, an siRNA directed against GFP resulted in efficient silencing of the SH4-HASPB-GFP fusion protein without any detectable effect on SH4-YES1-mCherry (Supplemental Fig. S4D–F). Finally, as expected, siRNA-mediated down-regulation of INCENP resulted in the appearance of enlarged and multinucleated cells (Supplemental Fig. S4G–I). Thus, using the high-content screening platform described, the expected phenotypes of control siRNAs could be detected in a reliable manner.
Identification of candidate gene products during primary screening
For high-content screening purposes, acquisition of experimental data was followed by automated image analysis using a software-based image analysis tool. This analysis was used to determine scores as distance to the median of each 384-spot siRNA array expressed in standard deviations (Supplemental Fig. S3). To analyze the robustness of the system, the scores of 2286 negative controls (scrambled siRNA) contained in the siRNA library were analyzed separately for the two fusion proteins, SH4-HASPB-GFP and SH4-YES1-mCherry. In the case of SH4-HASPB-GFP (Fig. 2A,E), ∼97% of the negative controls did not return scores above a threshold of 2.5. In the case of SH4-YES1-Cherry (Fig. 2B,F), 98% of the negative controls were below a threshold of 3.5. Based on these data, scores of 2.5 for SH4-HASPB-GFP and 3.5 for YES1-mCherry in both replicates in any of the classes of phenotypes defined above were chosen as suitable thresholds for initial identification of potential phenotypes. This procedure was complemented by a manual inspection of images and resulted in the identification of 286 gene products in primary screening with a potential role in SH4-domain-dependent protein targeting (Supplemental Table S1).
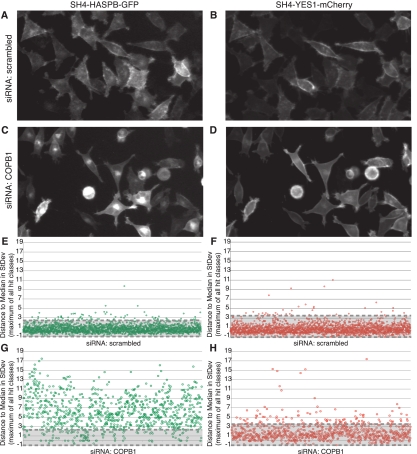
Figure 2.
Robustness and sensitivity of the experimental system at the level of primary screening. Stable HeLa cells were cultivated on 384-well siRNA arrays for 36 h, followed by the induction of expression of SH4-HASPB-GFP and SH4-YES1-mCherry. Nuclei were labeled and cells were subjected to live cell fluorescence microscopy in areas of immobilized siRNAs. Enlarged image details from primary screening data after transfection of a scrambled siRNA (A,B) and an siRNA targeted against the coatomer subunit COPB1 (C,D) are shown. Scores of all individual control experiments from primary screening were calculated using the image analysis tool described and are shown for SH4-HASPB-GFP (E,G) and SH4-YES1-mCherry (F,H), respectively. Negative controls (scrambled siRNA) are shown in panels E and F. Positive controls (siRNA targeted against COPB1) are shown in panels G and H. Scores below the defined hit threshold lie between the dashed lines within the gray shaded area.
As a positive control for intracellular transport processes, the library contained an siRNA directed against the coatomer subunit COPB1 (Rothman 1994; Rothman and Wieland 1996; Simpson et al. 2007), i.e., each 384-spot siRNA array contained up to four positive controls. Interestingly, down-regulation of COPB1 caused strong intracellular retention of SH4-HASPB-GFP (Fig. 2C,G); however, only a mild effect was observed for SH4-YES1-mCherry (Fig. 2D,H). Of the 898 individual experiments using an siRNA directed against COPB1, 86% showed a score above the threshold defined for SH4-HASPB-GFP. In contrast, only 14% caused a detectable intracellular retention of SH4-YES1-mCherry. Overall, these results indicate that the experimental system described in this study is characterized by a high robustness and sensitivity and thus generates well-reproducible data.
Validation and classification of gene products identified during primary screening
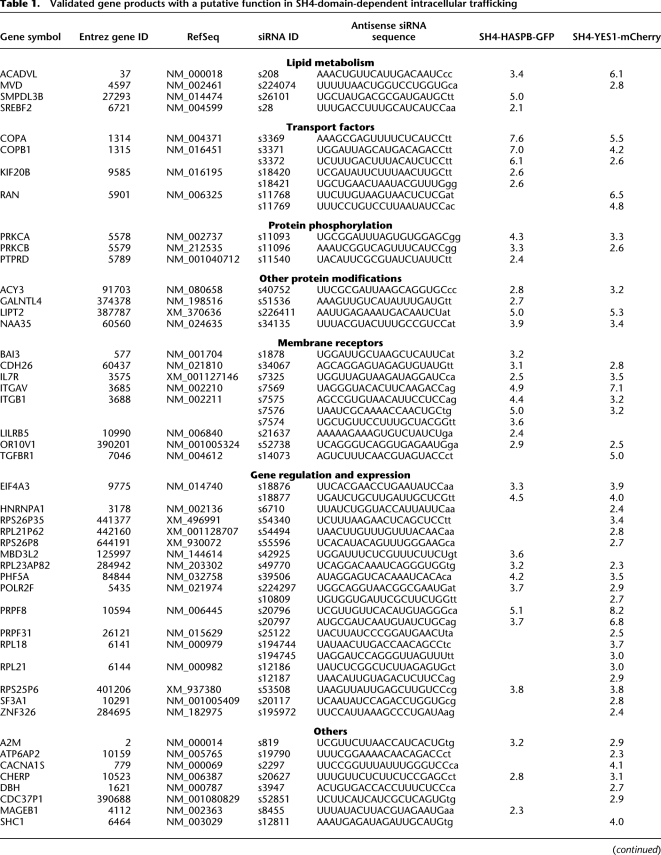
Candidate gene products identified during primary screening were validated using two additional siRNAs targeting different sequence areas of the respective mRNA. This analysis was conducted in triplicates using siRNA-coated 96-well plates (Erfle et al. 2008). Again, reverse transfection was achieved by cultivating cells on siRNA-coated wells for 36 h. Expression of SH4 fusion proteins was induced by doxycycline for 12 h. Forty-eight hours post-transfection, images were acquired and analyzed using the software-based image analysis tool. Scores were calculated based on 15 negative control experiments on each plate, and thresholds were adapted accordingly. In this set of experiments, down-regulation of 54 target genes resulted in a score above 2 in at least two out of three replicates (Table 1).
Table 1.
Validated gene products with a putative function in SH4-domain-dependent intracellular trafficking
Validated gene products were classified with regard to function as well as subcellular localization and membrane association (for detailed information, visit https://ichip.bioquant.uni-heidelberg.de, as indicated in Methods). The majority of gene products whose RNAi-mediated down-regulation was found to affect SH4-dependent targeting were membrane proteins. In addition, besides soluble cytoplasmic proteins, a small fraction of gene products were secretory proteins. As summarized in Table 1 and Supplemental Table S2, we found factors known to be involved in, for example, protein transport, lipid homeostasis, and protein modifications. Furthermore, several cell surface receptors and extracellular components were identified that may affect SH4-domain-dependent trafficking by various kinds of signaling processes.
A role of lipid homeostasis in SH4-dependent protein transport
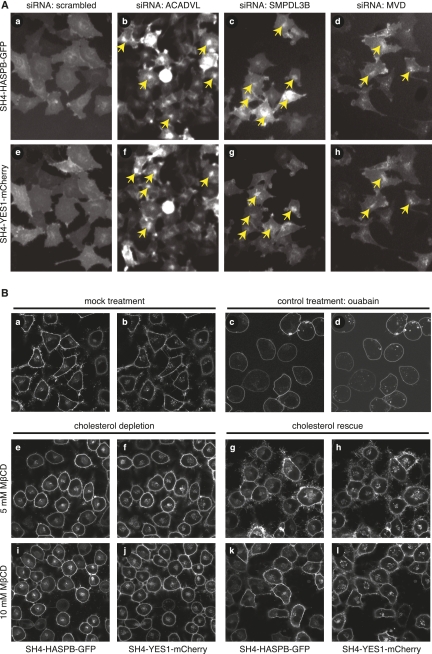
SH4-domain-dependent trafficking was shown to depend on cholesterol-dependent membrane microdomains (Brown and London 1998; Webb et al. 2000; Edidin 2003; Smotrys and Linder 2004; Linder and Deschenes 2007). Consistent with that, we retrieved genes whose products are involved in the biosynthesis of so-called raft lipids or in general lipid homeostasis. A prominent example turned out to be mevalonate-diphospho-dehydrogenase (MVD) (Toth and Huwyler 1996), an enzyme that decarboxylates mevalonate pyrophosphate and produces isopentenyl pyrophosphate (activated isoprene), a critical intermediate in the biosynthesis of cholesterol. As shown in Figure 3A (subpanels d and h), siRNA-mediated down-regulation of MVD caused intracellular retention of both SH4-HASPB-GFP and SH4-YES1-mCherry. Similarly, we identified sphingomyelin phosphodiesterase (acid-like 3B; SMPDL3B) (Marchesini and Hannun 2004), which catalyzes hydrolysis of sphingomyelin to phosphatidylcholine and ceramide. Again, its down-regulation inhibited transport of both SH4 fusion proteins (Fig. 3A, subpanels c and g). Finally, we discovered acyl-coenzyme-A dehydrogenase (ACADVL) as a gene product whose down-regulation affects SH4-dependent trafficking. ACADVL is a mitochondrial enzyme that catalyzes the first step in mitochondrial β-oxidation and is specific for very-long-chain fatty acids. As shown in Figure 3A (subpanels b and f), its down-regulation resulted in perinuclear accumulation of both SH4 fusion proteins. Thus, we have identified three enzymes involved in lipid metabolism, two of which directly participate in the biosynthesis of raft lipids such as cholesterol (Simons and Ikonen 1997; Simons and Toomre 2000; Simons and Vaz 2004) and ceramide (Bollinger et al. 2005). Additionally, depletion of ACADVL may affect SH4-dependent protein targeting by an indirect mechanism disturbing lipid homeostasis.
Figure 3.
Interference with cellular lipid metabolism by RNAi-mediated down-regulation of metabolic enzymes or pharmacological cholesterol depletion causes intracellular retention of SH4 proteins. (A) HeLa cells expressing SH4-HASPB-GFP and SH4-YES1-mCherry were reverse-transfected with a scrambled siRNA (a,e) and siRNAs targeted against acyl-coenzyme A-dehydrogenase, very long chain (ACADVL) (b,f), sphingomyelin phosphodiesterase, acid-like, 3B (SMPDL3B) (c,g), and mevalonate (diphospho) decarboxylase (MVD) (d,h) and subjected to live-cell fluorescence microscopy. Enlarged image details from original data from the validation screen are shown. (B) HeLa cells expressing SH4-HASPB-GFP and SH4-YES1-mCherry were treated with methyl-β-cyclodextrin (MβCD) to deplete cellular cholesterol and analyzed by live-cell confocal microscopy to determine the subcellular localization of SH4 fusion proteins. Subsequently, the same cells were supplemented with cholesterol and analyzed by live-cell confocal microscopy again. Control cells were treated with 50 mM ouabain and analyzed by live-cell confocal microscopy.
To further validate as to whether homeostasis of cholesterol as a raft lipid is, indeed, required for correct targeting of SH4-proteins to the plasma membrane, we tested whether down-regulation of MVD under the experimental conditions used causes cellular cholesterol levels to drop. Cholesterol levels were determined by nano-electrospray ionization mass spectrometry using phosphatidylcholine (PC) as a bulk lipid to normalize data. Relative to PC, upon treatment of cells with a siRNA targeted against MVD, we found a reduction of cellular cholesterol by 30% (Supplemental Fig. S5A). Under these conditions, MVD protein levels were reduced to ∼50% (Supplemental Fig. S5C), and MVD mRNA levels were reduced by >80% (Supplemental Fig. S5B). To further challenge the hypothesis that cholesterol homeostasis is essential for targeting of SH4-proteins to the plasma membrane, we treated cells with methyl-β-cyclodextrin (MβCD) to deplete cellular cholesterol. As indicated in Figure 3B, treatment with 5 to 10 mM MβCD resulted in pronounced intracellular retention of SH4-HASPB-GFP as well as SH4-YES1-mCherry (Fig. 3B, subpanels e,f,i,j), concomitant with a reduction of cellular cholesterol by 50% relative to PC (Supplemental Fig. S5A). These findings are consistent with other studies in which reduction of cholesterol levels by 30%–50% was found to have a significant impact on cholesterol-dependent processes such as shedding of the type XIII collagen ectodomain (Vaisanen et al. 2006). To test whether stress potentially exerted by MβCD causes a pleiotropic effect on SH4-dependent protein transport to the plasma membrane, we treated cells with ouabain, an unrelated inhibitor that blocks the activity of the Na+/K+-ATPase. Similar to MβCD, this treatment caused a mild effect on overall cell morphology; however, it did not result in perinuclear accumulation of SH4-HASPB-GFP or SH4-YES1-mCherry (Fig. 3B, subpanels c and d). To further analyze whether intracellular retention was, indeed, a direct result of cholesterol depletion, MβCD-treated cells were rescued by replenishment with exogenous cholesterol. This treatment resulted in a pronounced relocalization of both SH4-HASPB-GFP and SH4-YES1-mCherry to the plasma membrane (Fig. 3B, subpanels g,h and k,l).
Our combined findings establish that interference with cellular cholesterol levels results in pronounced intracellular retention of SH4-proteins and further corroborate the role of cholesterol-dependent microdomains in SH4-domain-dependent protein transport to plasma membranes.
Analysis of protein kinase C alpha as a component of SH4-domain-dependent protein transport
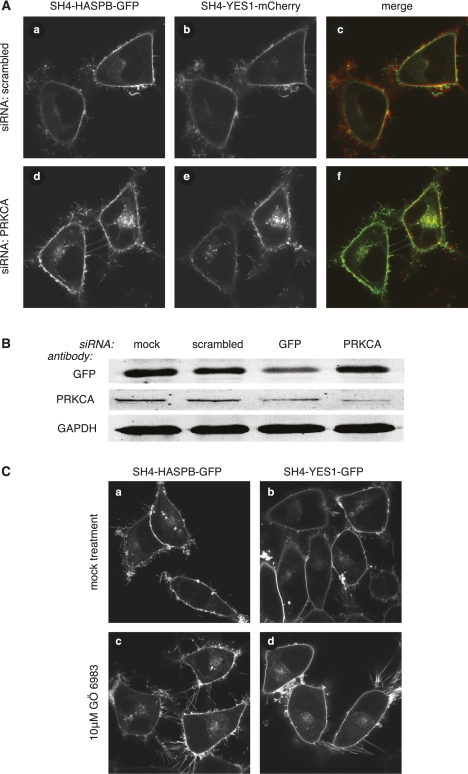
Since phosphorylation of SH4-domain-containing proteins such as SRC kinases has been implicated in regulation of endosomal recycling (Sandilands et al. 2004; Sandilands and Frame 2008; Tournaviti et al. 2009), another interesting result of this study was the identification of members of the protein kinase C family of serine/threonine kinases. As shown in Figure 4A, concomitant with reduction of PRKCA protein levels to ∼20% compared to a treatment with a scrambled siRNA (Fig. 4B; Supplemental Fig. S5D), siRNA-mediated down-regulation of PRKCA caused perinuclear accumulation of SH4 fusion proteins. This effect was not dependent on the fluorescent reporter protein fused to the respective SH4-domain, as knockdown of PRKCA in stable cell lines expressing either SH4-HASPB-mCherry (Supplemental Fig. S6A, subpanel c) or SH4-YES1-GFP (Supplemental Fig. S6A, subpanel f) also resulted in an intracellular retention. To test whether PKC activity is required for plasma-membrane targeting of SH4-proteins, stable HeLa cells were treated with the protein kinase C inhibitor GÖ 6983, resulting in pronounced intracellular retention of both SH4-HASPB-GFP (Fig. 4C, subpanel c) and SH4-YES1-GFP (Fig. 4C, subpanel d).
Figure 4.
Characterization and validation of PRKCA as a component in SH4-dependent protein transport. (A) HeLa cells expressing SH4-HASPB-GFP and SH4-YES1-mCherry were transfected with a scrambled siRNA (a–c), and a siRNA directed against PRKCA (d–f), and subjected to live-cell confocal microscopy to determine the localization of both SH4 fusion proteins. (B) Equal protein amounts of total cell lysates from transfected and induced cells were subjected to SDS-PAGE and Western blot analysis with antibodies directed against GFP, PRKCA, and GAPDH. (C) Stable HeLa cells expressing SH4-HASPB-GFP (a,c), or SH4-YES1-GFP (b,d), were cultured in the presence of 10 μM protein kinase C inhibitor GÖ 6983 for 16 h (c,d), and analyzed by live-cell confocal microscopy for subcellular localization of both SH4 fusion proteins.
These findings indicate that protein phosphorylation by PKC plays an important role in SH4-domain-dependent protein transport; however, it remains to be clarified whether PRKCA directly targets SH4 domains or whether other substrates of this kinase are affecting SH4-dependent trafficking.
Analysis of coatomer as a component of SH4-domain-dependent protein targeting
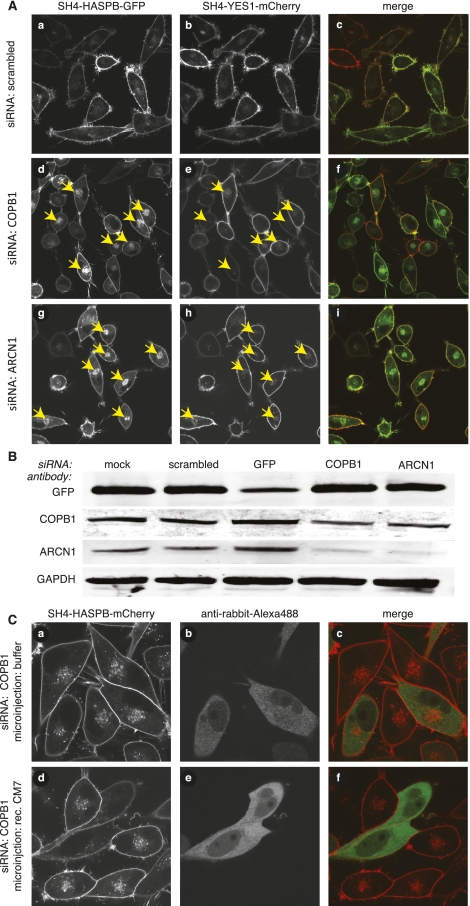
As mentioned above, COPB1 (β-COP) is a subunit of the coatomer complex that was identified and validated to be required for proper targeting of SH4 fusion proteins (Fig. 2; Table 1; Supplemental Table S2). Since the coatomer is a hetero-heptameric complex, we further evaluated the COPB1 phenotype based on the assumption that coatomer is required as a protein complex, i.e., coatomer subunits other than COPB1 should give a similar phenotype when down-regulated by RNA interference. In a first step, we extended our studies to ARCN1 (δ-COP) (Fig. 5A) and assessed down-regulation of both COPB1 and ARCN1 by quantitative Western blotting (Fig. 5B). These studies established efficient down-regulation at the protein level of each subunit by the respective siRNA to ∼40% or even below 20%, respectively (Supplemental Fig. S5E). As can be expected for a stoichiometric protein complex, down-regulation of COPB1 also caused a loss of ARCN1, and vice versa. As shown in Figure 5A, down-regulation of ARCN1 resulted in a pronounced intracellular retention of SH4-HASPB-GFP, while targeting of SH4-YES1-mCherry was only mildly affected. These differential effects were not caused by the different fluorescent reporter proteins fused to the SH4 domains of HASPB and YES1, since stable HeLa cells expressing SH4-HASPB-mCherry (Supplemental Fig. S6A, subpanel b) also showed strong intracellular retention of the SH4 fusion protein upon down-regulation of COPB1. Similarly, knockdown of COPB1 in stable HeLa cells expressing SH4-YES1-GFP resulted in a mild retention of the SH4 fusion protein (Supplemental Fig. S6A, subpanel e). These studies were further extended toward COPA (α-COP) and COPB2 (β′-COP), two additional subunits of coatomer. Again, following transfection of the corresponding siRNAs, strong intracellular retention was observed for SH4-HASPB-GFP (Supplemental Fig. S6B, subpanels a,d) but only a mild effect on targeting of SH4-YES1-mCherry (Supplemental Fig. S6B, subpanels b,e). To address whether intracellular retention evoked by knockdown of coatomer subunits was, indeed, a result of depletion of the coatomer protein complex, we microinjected stable HeLa cells expressing SH4-HASPB-mCherry with recombinant coatomer before transfection with siRNAs targeted against COPB1. Indeed, targeting of SH4 fusion proteins was unaffected in cells that were microinjected with coatomer, demonstrating a role for coatomer in proper targeting of SH4-HASPB-GFP; however, it does not appear to play an essential role in targeting of SH4-YES1-mCherry (Fig. 5C).
Figure 5.
Characterization and validation of coatomer as a component in SH4-dependent protein transport. (A) HeLa cells expressing SH4-HASPB-GFP and SH4-YES1-mCherry were transfected with a scrambled siRNA (a–c), siRNAs directed against COPB1 (d–f) and ARCN1 (g–i), and subjected to live-cell confocal microscopy to determine the localization of both SH4 fusion proteins. (B) Equal protein amounts of total cell lysates from transfected and induced cells were subjected to SDS-PAGE and Western blot analysis with antibodies directed against GFP, COPB1, ARCN1, and GAPDH. (C) HeLa cells expressing SH4-HASPB-mCherry were coinjected with recombinant coatomer and an Alexa-488-labeled secondary antibody before transfection with a siRNA directed against COPB1. Thirty hours post-transfection, cells were analyzed by live-cell confocal microscopy to determine the subcellular localization of the SH4 fusion protein in microinjected cells.
Coatomer and PRKCA as components in plasma membrane transport of endogenous YES1 and other SH4 proteins
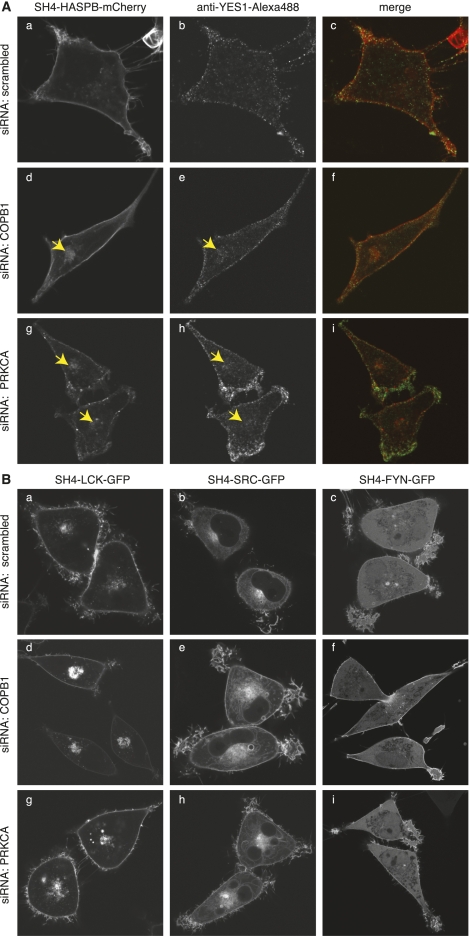
To address whether our experimental system is capable of identifying components required for transport of endogenous SH4 proteins, we analyzed the localization of endogenous YES1 after knockdown of the factors identified and validated in this study. Transfection of siRNAs directed against COPB1 (Fig. 6A, subpanels d–f) and PRKCA (Fig. 6A, subpanels g–i) not only resulted in an intracellular retention of SH4-HASPB-mCherry, but also affected plasma membrane targeting of endogenous YES1 visualized by immunolabeling with specific antibodies. We further analyzed whether the identified components required for plasma membrane targeting of HASPB and YES1 also affected transport of other SH4-domain-containing proteins. Among those were other members of the SRC family kinases (SFKs) such as SRC (non-palmitoylated), LCK and FYN (both dually palmitoylated) (Shenoy-Scaria et al. 1993; Koegl et al. 1994; Resh 1994), whose SH4 domains were expressed as GFP fusion proteins in stable HeLa cells (Tournaviti et al. 2007, 2009). Upon down-regulation of COPB1 and PRKCA, we observed differential effects on targeting of the different SH4 fusion proteins. RNAi-mediated down-regulation of COPB1 (Fig. 6B, subpanels d–f) resulted in an enhanced intracellular retention of SH4-LCK-GFP and SH4-SRC-GFP; however, it did not influence plasma membrane targeting of SH4-FYN-GFP. Knockdown of PRKCA (Fig. 6B, subpanels g–i) caused strong intracellular accumulation of SH4-LCK-GFP and also affected the localization of SH4-SRC-GFP. Again, transport of SH4-FYN-GFP was not affected. These findings support previous data showing that different SH4-domain-containing proteins localize to different subcellular membranes and traffic along different routes, often dependent on the acylation pattern present in the respective SH4 domain (Shenoy-Scaria et al. 1994; Sandilands et al. 2004; Kasahara et al. 2007). The finding that both coatomer and PRKCA exert effects on SH4-LCK-GFP and SH4-SRC-GFP but do not affect transport of SH4-FYN-GFP, which is directly targeted to the plasma membrane without passing through the Golgi system, may indicate that both factors play a role in SH4-dependent transport at the level of the Golgi or the endosomal system.
Figure 6.
PRKCA and coatomer play differential roles in transport of endogenous YES1 and other SH4 proteins. (A) Stable HeLa cells expressing SH4-HASPB-mCherry were transfected with a scrambled siRNA (a–c), and siRNAs directed against COPB1 (d–f) and PRKCA (g–i), and induced for reporter protein expression. Forty-eight hours post-transfection, cells were fixed, subjected to immunostaining against endogenous YES1, and subsequently analyzed by confocal microscopy. (B) Stable HeLa cells expressing fusion proteins of the SH4 domains of the acylated SRC kinases LCK, SRC, and FYN were transfected with a scrambled siRNA (a–c), and siRNAs directed against COPB1 (d–f) and PRKCA (g–i). Forty-eight hours post-transfection, cells were analyzed by live-cell confocal microscopy to determine the subcellular localization of the respective SH4-protein.
Besides the examples given above and as documented in Table 1 and Supplemental Table S2, we were also able to identify gene products with so far unknown functions. Additionally, gene products were identified that are likely to have a rather indirect impact on SH4-domain-dependent targeting such as factors regulating gene expression. This observation is also consistent with a relatively large number of factors that localize to the nucleus.
Discussion
SRC kinases and other peripheral membrane proteins contain acylated SH4 domains that mediate both association with membranes and proper targeting to specific subcellular membranes. While the physical association of SH4 domains with membranes is understood in great detail, their role in intracellular sorting and plasma membrane targeting as well as the function of membrane microdomains in this process is poorly understood. The aim of the present study was to conduct a global analysis of the human genome to reveal gene products involved in SH4-domain-dependent plasma membrane targeting of acylated proteins, such as SRC kinases. Stable cell lines were engineered that express two different kinds of SH4 fusion proteins in a doxycycline-dependent manner. An experimental system was developed to detect and quantify intracellular retention of SH4 proteins, which was combined with siRNA arrays to individually down-regulate every single gene known in the human genome. Live-cell imaging was conducted using an automated wide-field microscopy platform. Using a software-based image analysis tool, intracellular retention of SH4 fusion proteins could be detected and quantified for each individual experimental condition of down-regulation of a specific gene product. The two different SH4 fusion proteins were analyzed independently in order to identify potential differences in SH4-dependent trafficking depending on the individual properties of a given SH4 domain.
To characterize the experimental system described, we reasoned that general components of intracellular transport might be required for targeting of SH4 fusion proteins. More specifically, since all palmitoylated proteins have been proposed to contact the Golgi (Rocks et al. 2010), we hypothesized that the heterooligomeric coatomer complex may play a role in SH4-dependent trafficking. Indeed, siRNA-mediated down-regulation of the COPB1 gene consistently resulted in perinuclear retention of SH4 fusion proteins. We further concluded that down-regulation of coatomer subunits that exist in just one isoform (e.g., COPA, COPB1, COPB2, and ARCN1 [δ-COP]) (Wegmann et al. 2004) should result in a similar phenotype, which was indeed the case. In addition, we could show that down-regulation of single coatomer subunits affected protein levels of other subunits as well and, hence, most likely results in a destabilization of the complete coatomer complex. Moreover, RNAi-mediated down-regulation of endogenous coatomer subunits did not affect SH4-dependent trafficking when cells were microinjected with recombinant coatomer. Therefore, using our experimental system, we were able to identify COPB1 as a component of a multi-protein complex, which plays an important role in SH4-dependent protein transport.
Another protein whose down-regulation caused a pronounced effect on SH4-dependent protein transport was a protein kinase, PRKCA. Moreover, using a pharmacological inhibitor of protein kinase C, we could demonstrate a requirement for protein kinase C activity in correct targeting of SH4 proteins to the plasma membrane. Whether SH4 domains are direct targets for phosphorylation by PRKCA or whether other target proteins play direct roles in SH4-domain-dependent protein transport remains a subject for further investigation.
Regarding further requirements of SH4-dependent trafficking, a major result of this study was the identification of enzymes that play key roles in lipid biosynthesis and homeostasis. Among those was a key factor of cholesterol biosynthesis, MVD, the enzyme that generates activated isoprene. Down-regulation of MVD, indeed, caused reduced cellular cholesterol levels that were correlated with intracellular accumulation of SH4 proteins. Furthermore, pharmacological depletion of cellular cholesterol also inhibited transport of SH4 proteins, an effect that could be reversed upon replenishment with exogenous cholesterol. Taken together, our findings corroborate the role of cholesterol-dependent membrane microdomains in SH4 trafficking (McCabe and Berthiaume 1999, 2001; Webb et al. 2000; Tournaviti et al. 2009). They are further consistent with other large-scale RNAi approaches, suggesting that RNAi-mediated down-regulation of MVD affects cholesterol-dependent processes such as Dengue virus replication (Rothwell et al. 2009).
Another interesting observation of this study was differences between the two SH4 domains used as targeting motifs. Compared to the YES1 SH4 domain, down-regulation of coatomer subunits had a much more pronounced inhibitory effect on plasma membrane targeting of a fusion protein containing the SH4 domain of HASPB, indicating differential requirements depending on different kinds of SH4 domains. Consistently, factors involved in transport of those two SH4 domains were shown to have differential effects on fusion proteins containing SH4 domains of other SRC kinases, such as LCK, SRC, and FYN, as well as on endogenous YES1. Differential transport requirements are also evident from the complete list of validated gene products shown in Table 1 and Supplemental Table S2. Of the 54 gene products shown to play a role in SH4-dependent trafficking, 13 were found to function primarily in HASPB targeting, 18 appeared to be specific for YES1 targeting, and 23 were found to be required for both transport processes. These observations are generally consistent with earlier findings from other groups suggesting the existence of multiple pathways of SH4 trafficking (Sato et al. 2009). Furthermore, our data show that the experimental model system is not only suited to identify general components involved in SH4-dependent transport processes but also reveals differential requirements when different SH4 domains are directly compared. These findings also directly exclude potential pleiotropic effects and emphasize the capability of the experimental system to reveal differences between various kinds of SH4-dependent trafficking.
The 54 gene products validated in this study to play a role in SH4-domain-dependent protein targeting were classified into eight groups according to experimentally shown or annotated functions (Table 1; Supplemental Table S2). As expected, we found several membrane receptors as well as kinases and phosphatases whose down-regulation affects SH4-dependent trafficking. Besides gene products involved in the formation or the removal of other post-translational modifications as well as a relatively large group of gene products involved in the regulation of gene expression, we also revealed four gene products whose functions are entirely unknown.
It is clear that the present screening approach cannot be considered as comprehensive since obviously a number of gene products that are known to play a role in SH4-dependent targeting were not returned as hits. For example, we could not identify a single DHHC protein, the enzymes that mediate palmitoylation of SH4 proteins (Smotrys et al. 2005; Mitchell et al. 2006). In the light of recent findings, it is clear that this phenomenon is due to functional redundancy within the family of DHHC proteins (Hou et al. 2009; Rocks et al. 2010), making it impossible to evoke a phenotype by down-regulation of individual members of the DHHC family. In this sense, the siRNA screening effort presented in this study cannot be considered as an attempt to identify all gene products that play a role in SH4-dependent protein targeting but rather provides a major resource of a subgroup of proteins with non-redundant functions in this process.
Methods
Generation of stable HeLa cell lines expressing acylated SH4 fusion proteins
Constructs encoding acylated fluorescent reporter proteins (SH4-YES1-mCherry, SH4-HASPB-GFP [see Supplemental Material 1], SH4-YES1-GFP, SH4-HASPB-mCherry, SH4-LCK-GFP, SH4-SRC-GFP, SH4-FYN-GFP) were cloned into the retroviral expression vector pRevTRE2. Following retroviral transduction of HeLa cells, constructs were stably inserted into the target genome followed by the isolation of cell lines using fluorescence-activated cell sorting as described previously (Engling et al. 2002). Doxycycline-dependent expression of the respective SH4 fusion proteins was confirmed by analytical flow cytometry.
Analysis of the subcellular localization of SH4 fusion proteins using live-cell confocal and wide-field fluorescence microscopy
HeLa cells were cultivated on eight-well LabTek imaging chambers (Nunc) and induced to express fluorescent SH4 fusion proteins by cultivation in the presence of 1 μg/mL doxycycline for 12 to 24 h. To inhibit protein palmitoylation, cells were cultivated in the presence of 2-bromopalmitate (2-bromohexadecanoic acid) (Sigma-Aldrich). Cells were cultivated in the presence of the protein kinase C inhibitor GÖ 6983 (Sigma-Aldrich) for 16 h. Thirty minutes prior to imaging, the medium was replaced by prewarmed CO2-independent medium (Invitrogen), and cells were analyzed on an LSM 510 confocal microscope (Zeiss), an Axiovert fluorescence wide-field microscope (Zeiss), or a Scan^R screening microscope (Olympus).
Genome-wide primary screening for factors involved in subcellular localization of acylated SH4 fusion proteins
Genome-wide primary screening was conducted by reverse transfection of cells with an siRNA library (Ambion) consisting of about 52,000 siRNAs targeting all genes known in the human genome (Ensembl version 27) with at least two siRNAs per gene. This siRNA library was arranged in 384-spot siRNA arrays (Erfle et al. 2004, 2007; Neumann et al. 2006, 2010; Simpson et al. 2007). Per array, 120,000 cells were seeded and, 36 h post-transfection, expression of SH4 fusion protein was induced by incubation of cells for 12 h in the presence of 1 μg/mL doxycycline. Growth medium was replaced by prewarmed CO2-independent medium (without phenol-red) (Invitrogen) containing 0.2 μg/mL Hoechst 33342 (Sigma-Aldrich) for labeling of cell nuclei. Forty-eight hours post-transfection, siRNA arrays were loaded onto an Olympus Scan^R screening microscope with a 37°C incubation chamber, and images from the GFP, Cherry, and Hoechst channel were acquired by automated live-cell fluorescence microscopy at 10× magnification. Images were analyzed using the image analysis tool described below.
Validation of gene products identified during primary screening
Validation screening was conducted by reverse transfection of a newly designed siRNA library consisting of 776 silencer select siRNAs (Ambion) targeting 286 human genes. Validation was conducted on siRNA-coated 96-well plates (Erfle et al. 2008) in four replicates. Four thousand cells were seeded per well using a FlexDrop system (Perkin Elmer). Thirty-six hours post-transfection, SH4 fusion protein expression was induced by cultivation in the presence of 1 μg/mL doxycycline for 12 h. Growth medium was replaced by prewarmed CO2-independent medium (without phenol-red) (Invitrogen) containing 0.2 μg/mL Hoechst 33342 (Sigma-Aldrich) to label cell nuclei. Forty-eight hours post-transfection, siRNA-coated 96-well plates were loaded onto an Olympus Scan^R screening microscope with a 37°C incubation chamber, and two images per well were acquired for each channel by automated live-cell fluorescence microscopy at 10× magnification. Images were analyzed using the image analysis tool described below.
A software-based image analysis tool to detect and quantify intracellular retention of SH4 fusion proteins
Based on general techniques to analyze microscopy-based imaging data such as cell segmentation (Elowitz et al. 2002; Megason and Fraser 2007; Wang et al. 2008b), an image analysis tool was developed to automatically detect and quantify intracellular retention of SH4 fusion proteins as a desired phenotype. To achieve optimal cell detection, cell nuclei were segmented by Hoechst staining in a first step as a prerequisite for cell segmentation (Rosenfeld et al. 2005; Gordon et al. 2007). Since intensity values of nuclei in any given Hoechst image were within a certain range, thresholding techniques could be applied (Wu et al. 2000). However, due to varying nuclear intensities within the complete screening data set, an adaptive thresholding technique was required for reliable determination of cell nuclei. Starting with a high value, the threshold was gradually decreased until the size of segmented nuclei was consistent with the assumed size. In an additional step, segmented nuclei were post-processed, removing local contaminations and other artifacts in the region of the nucleus.
Segmented cell nuclei were the basis of a classifier-enhanced region growing method used to segment complete cells. This method requires a seed point provided by cell nuclei and a criterion to detect cell boundaries, which was achieved by a rule-based classifier. The classifier uses both local cell brightness in the area of the nucleus and a gradient of fluorescence intensities across the whole area of the cell. The cell boundary is recognized when the brightness of an inspected pixel is below the minimal brightness in the area of the nucleus and the gradient magnitude is above a certain threshold. This approach is suitable to segment the whole cell area and allows for the detection of changes of gradient magnitudes within the cytoplasm, which is the case when the fluorescent reporter protein accumulates in a perinuclear region, a distribution indicative for localization in Golgi membranes or recycling endosomes. An important feature of this method is its ability to segment cells independently of local brightness. Due to the choice of nuclei as seed points and the methodology to detect cell boundaries, it was possible to detect individual cells with similar fluorescence intensities even when multiple cells formed direct plasma membrane contacts. Additionally, cell clusters with overlapping cell boundaries were detected by the proximity of their nuclei and were excluded from further analysis.
Cell segmentation was finally used to define three cellular regions, the nucleus, the cytoplasm and the cell boundary. To detect the cell boundary, the difference between the original and an eroded cell segmentation result was determined. Following the determination of the nucleus and the cell boundary, the remaining cellular area was defined as the interior of the cell along with all organelles such as the Golgi that are embedded in the cytoplasm. Ratios of mean values and standard deviations of each cell and its constituent parts were calculated for brightness-invariant classification of phenotypes. This was achieved using a decision tree based on the analysis of the extracted features for all phenotypes.
Calculation of scores from classification results derived from the automated image analysis tool
Results of cell classification were subjected to further analysis to calculate scores. The percentage of cells displaying a classified phenotype was determined for each image. In the validation screening procedure, cells from both images of the same well were combined for further analysis. Both the median and the standard deviation of a complete array were calculated and used to determine a score for each experimental condition. This score was expressed as the distance in standard deviations to the median of the respective 384-spot siRNA array. In validation screening, the median and standard deviation from all negative controls analyzed on the same plate were used for the determination of scores. In primary screening, the hit threshold for SH4-HASPB-GFP was set to a score of 2.5 for both replicates and to 2 (3/4 or 2/3 replicates) in validation screening. For potential hits from the so-called druggable genome, SH4-HASPB-GFP hits were included in the validation screening procedure when the hit threshold was above 5 on only one replicate. The hit threshold for SH4-YES1-Cherry was adjusted to a score of 3.5 in primary screening (both replicates) and 2 in validation screening (3/4 or 2/3 replicates). All potential hits derived from primary screening that were identified with the automated image analysis tool were further subjected to manual inspection.
Liquid-phase transfection of stable HeLa cells with siRNAs directed against MVD, coatomer subunits, and PRKCA
Stably transduced HeLa cells were transfected with siRNAs directed against MVD (Ambion, s9094), the coatomer subunits COPA (Ambion, 146404), COPB1 (Ambion, 24546), COPB2 (Ambion, 137699), ARCN1 (Ambion, 10234), and a siRNA directed against PRKCA (Ambion, s11093) as well as a scrambled siRNA (Ambion, 103860 or neg. control #1) in 24-well plates using oligofectamine (Invitrogen). For each well, 50 pmol of siRNA was preincubated with 12 μL of diluted (1:4) oligofectamine for 20 min at room temperature before addition to cells. Thirty-two to 36 h post-transfection, cells were passaged and cultivated in eight-well LabTek imaging chambers (Nunc). Expression of SH4 fusion proteins was induced by cultivating cells in the presence of 1 μg/mL doxycycline for 12–18 h. For MVD knockdowns, cells were cultivated in DMEM containing 10% delipidated FCS (PAN Biotech).
Quantification of RNAi-mediated knockdown efficiencies of MVD, coatomer subunits, and PRKCA using Western blot analysis and quantitative real-time PCR
Stable HeLa cells expressing SH4-HASPB-GFP and SH4-YES1-mCherry were transfected with a scrambled siRNA (Ambion, 103860) and siRNAs directed against GFP (Ambion, 4626), MVD (Ambion, s9094), COPB1 (Ambion, 24546), and ARCN1 (Ambion, 10234) or PRKCA (Ambion, s11093) on six-well plates using 300 pmol of siRNA. For MVD knockdowns, cells were cultivated in DMEM containing 10% delipidated FCS (PAN Biotech). Twenty-four hours post-transfection, reporter protein expression was induced by cultivation in the presence of 1 μg/mL doxycycline for a further 24 h. Forty-eight hours post-transfection, cells were harvested. For Western blot analysis, 60 μg of total protein from the post-nuclear supernatant was subjected to SDS-PAGE on NuPAGE 4%–12% gradient gels (Invitrogen). Gels were blotted on PVDF membranes, and specific proteins were detected using polyclonal antibodies raised against GFP, MVD (Santa Cruz, sc-160550), COPB1, and ARCN1 and mouse monoclonal antibodies against PRKCA (Abcam, ab31) and GAPDH (Ambion, 4300). Signals were detected and quantified using a LICOR infrared imaging system. For real-time PCR, RNA was extracted using the RNeasy Mini Kit (QIAGEN) and used for cDNA synthesis applying the ImProm-II Reverse Transcription System (Promega). Using the TaqMan Gene Expression assay (Ambion, Hs00159403_m1), mRNA levels were determined using a StepOne Real-Time PCR System (Applied Biosystems).
Depletion of cellular cholesterol and cholesterol rescue
Stable HeLa cells were incubated for 20 h in the presence of 1 μg/mL doxycycline to induce expression of SH4-HASPB-GFP and SH4-YES1-mCherry in DMEM containing 10% delipidated FCS (PAN Biotech). For cholesterol depletion, cells were incubated for 1 h in the presence of 5–10 mM methyl-β-cyclodextrin (MβCD) (Sigma-Aldrich) before analysis by live-cell confocal microscopy. Subsequently, for cholesterol replenishment, water-soluble cholesterol (Sigma-Aldrich) was added to the MβCD-treated cells in a 1:10 ratio (0.5 mM cholesterol to 5 mM MβCD-treated cells and 1 mM cholesterol to 10 mM MβCD-treated cells) and incubated for 1.5 h before analysis by live-cell confocal microscopy. Control cells were incubated for 20 h with 50 nM ouabain in DMEM containing 10% delipidated FCS and analyzed by live-cell confocal microscopy.
Immunostaining of stable HeLa cells
Stable HeLa cells grown on coverslips were fixed in 5% PFA for 5 min at room temperature, quenched in 50 mM NH4Cl for 5 min, and permeabilized in 0.1% TX-100 for 5 min before blocking in 1% BSA for 1 h. Endogenous YES1 was labeled by incubation with a specific antibody (BD, 610375) diluted 1:250 in 1% BSA for 1 h. Samples were incubated with a Alexa488-labeled secondary antibody (Invitrogen) diluted 1:1000 in 1% BSA for 1 h and mounted in LinMount (Linaris) mounting medium.
Microinjection of recombinant coatomer
Recombinant coatomer (∼1.3 mg/mL) (Sahlmuller et al. 2011) or the respective buffer was co-injected with 1:10 diluted Alexa488-labeled secondary antibodies (Invitrogen) into stable HeLa cells using a FemtoJet microinjection device (Eppendorf) mounted on an Axiovert 35 microscope (Zeiss). Three to 6 h after microinjection, cells were transfected with a siRNA against COPB1 (Ambion, 24546) and induced for reporter protein expression 4 h post-transfection. Cells were analyzed by live-cell confocal microscopy 30 h after transfection.
Determination of cellular cholesterol levels using nano-electrospray ionization mass spectrometry
Stable HeLa cells were cultivated in DMEM containing 10% delipidated FCS (PAN Biotech GmbH) and transfected with a siRNA directed against MVD (Ambion, s9094) for 48 h or treated with 10 mM methyl-β-cyclodextrin (Sigma-Aldrich) for 30 min. Control cells were cultivated in normal DMEM (untreated) or in DMEM containing 10% delipidated FCS (mock). Either whole cells or membranes isolated by ultracentrifugation of postnuclear supernatants from homogenized cells were subjected to lipid extraction and mass spectrometric quantification of cholesterol and PC as described previously (Merz et al. 2011).
Data access
All original data along with analysis results derived from this study have been stored in two web-based databases. They can be accessed at https://ichip.bioquant.uniheidelberg.de (User: “ichipguest”; Password: “Dec5ex2h”) and http://www.mitocheck.org/cgi-bin/mtc.
Acknowledgments
We thank Timo Sachsenheimer and Iris Leibrecht for technical support. This work was supported by the Peter and Traudl Engelhorn Foundation (J.R.), the DFG Collaborative Research Center TRR83 (W.N., B.B., F.T.W.), as well as the DFG Cluster of Excellence CellNetworks (W.N., R.P., J.E., B.B., F.T.W.).
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.116087.110.
References
- Boggon TJ, Eck MJ 2004. Structure and regulation of Src family kinases. Oncogene 23: 7918–7927 [DOI] [PubMed] [Google Scholar]
- Boland MV, Markey MK, Murphy RF 1998. Automated recognition of patterns characteristic of subcellular structures in fluorescence microscopy images. Cytometry 33: 366–375 [PubMed] [Google Scholar]
- Bollinger CR, Teichgraber V, Gulbins E 2005. Ceramide-enriched membrane domains. Biochim Biophys Acta 1746: 284–294 [DOI] [PubMed] [Google Scholar]
- Brown DA, London E 1998. Functions of lipid rafts in biological membranes. Annu Rev Cell Dev Biol 14: 111–136 [DOI] [PubMed] [Google Scholar]
- Carpenter AE, Jones TR, Lamprecht MR, Clarke C, Kang IH, Friman O, Guertin DA, Chang JH, Lindquist RA, Moffat J, et al. 2006. CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biol 7: R100 doi: 10.1186/gb-2006-7-10-r100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conrad C, Erfle H, Warnat P, Daigle N, Lorch T, Ellenberg J, Pepperkok R, Eils R 2004. Automatic identification of subcellular phenotypes on human cell arrays. Genome Res 14: 1130–1136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denny PW, Gokool S, Russell DG, Field MC, Smith DF 2000. Acylation-dependent protein export in Leishmania. J Biol Chem 275: 11017–11025 [DOI] [PubMed] [Google Scholar]
- Edidin M 2003. The state of lipid rafts: From model membranes to cells. Annu Rev Biophys Biomol Struct 32: 257–283 [DOI] [PubMed] [Google Scholar]
- Elowitz MB, Levine AJ, Siggia ED, Swain PS 2002. Stochastic gene expression in a single cell. Science 297: 1183–1186 [DOI] [PubMed] [Google Scholar]
- Engling A, Backhaus R, Stegmayer C, Zehe C, Seelenmeyer C, Kehlenbach A, Schwappach B, Wegehingel S, Nickel W 2002. Biosynthetic FGF-2 is targeted to non-lipid raft microdomains following translocation to the extracellular surface of CHO cells. J Cell Sci 115: 3619–3631 [DOI] [PubMed] [Google Scholar]
- Erfle H, Simpson JC, Bastiaens PI, Pepperkok R 2004. siRNA cell arrays for high-content screening microscopy. Biotechniques 37: 454–462 [DOI] [PubMed] [Google Scholar]
- Erfle H, Neumann B, Liebel U, Rogers P, Held M, Walter T, Ellenberg J, Pepperkok R 2007. Reverse transfection on cell arrays for high content screening microscopy. Nat Protoc 2: 392–399 [DOI] [PubMed] [Google Scholar]
- Erfle H, Neumann B, Rogers P, Bulkescher J, Ellenberg J, Pepperkok R 2008. Work flow for multiplexing siRNA assays by solid-phase reverse transfection in multiwell plates. J Biomol Screen 13: 575–580 [DOI] [PubMed] [Google Scholar]
- Glory E, Murphy RF 2007. Automated subcellular location determination and high-throughput microscopy. Dev Cell 12: 7–16 [DOI] [PubMed] [Google Scholar]
- Gordon A, Colman-Lerner A, Chin TE, Benjamin KR, Yu RC, Brent R 2007. Single-cell quantification of molecules and rates using open-source microscope-based cytometry. Nat Methods 4: 175–181 [DOI] [PubMed] [Google Scholar]
- Hancock JF, Paterson H, Marshall CJ 1990. A polybasic domain or palmitoylation is required in addition to the CAAX motif to localize p21ras to the plasma membrane. Cell 63: 133–139 [DOI] [PubMed] [Google Scholar]
- Harder N, Eils R, Rohr K 2008. Automated classification of mitotic phenotypes of human cells using fluorescent proteins. Methods Cell Biol 85: 539–554 [DOI] [PubMed] [Google Scholar]
- Hou H, John Peter AT, Meiringer C, Subramanian K, Ungermann C 2009. Analysis of DHHC acyltransferases implies overlapping substrate specificity and a two-step reaction mechanism. Traffic 10: 1061–1073 [DOI] [PubMed] [Google Scholar]
- Kasahara K, Nakayama Y, Kihara A, Matsuda D, Ikeda K, Kuga T, Fukumoto Y, Igarashi Y, Yamaguchi N 2007. Rapid trafficking of c-Src, a non-palmitoylated Src-family kinase, between the plasma membrane and late endosomes/lysosomes. Exp Cell Res 313: 2651–2666 [DOI] [PubMed] [Google Scholar]
- Koegl M, Zlatkine P, Ley SC, Courtneidge SA, Magee AI 1994. Palmitoylation of multiple Src-family kinases at a homologous N-terminal motif. Biochem J 303: 749–753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linder ME, Deschenes RJ 2007. Palmitoylation: policing protein stability and traffic. Nat Rev Mol Cell Biol 8: 74–84 [DOI] [PubMed] [Google Scholar]
- Marchesini N, Hannun YA 2004. Acid and neutral sphingomyelinases: roles and mechanisms of regulation. Biochem Cell Biol 82: 27–44 [DOI] [PubMed] [Google Scholar]
- McCabe JB, Berthiaume LG 1999. Functional roles for fatty acylated amino-terminal domains in subcellular localization. Mol Biol Cell 10: 3771–3786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCabe JB, Berthiaume LG 2001. N-terminal protein acylation confers localization to cholesterol, sphingolipid-enriched membranes but not to lipid rafts/caveolae. Mol Biol Cell 12: 3601–3617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Megason SG, Fraser SE 2007. Imaging in systems biology. Cell 130: 784–795 [DOI] [PubMed] [Google Scholar]
- Merz A, Long G, Hiet MS, Brugger B, Chlanda P, Andre P, Wieland F, Krijnse-Locker J, Bartenschlager R 2011. Biochemical and morphological properties of hepatitis C virus particles and determination of their lipidome. J Biol Chem 286: 3018–3032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell DA, Vasudevan A, Linder ME, Deschenes RJ 2006. Protein palmitoylation by a family of DHHC protein S-acyltransferases. J Lipid Res 47: 1118–1127 [DOI] [PubMed] [Google Scholar]
- Neumann B, Held M, Liebel U, Erfle H, Rogers P, Pepperkok R, Ellenberg J 2006. High-throughput RNAi screening by time-lapse imaging of live human cells. Nat Methods 3: 385–390 [DOI] [PubMed] [Google Scholar]
- Neumann B, Walter T, Heriche JK, Bulkescher J, Erfle H, Conrad C, Rogers P, Poser I, Held M, Liebel U, et al. 2010. Phenotypic profiling of the human genome by time-lapse microscopy reveals cell division genes. Nature 464: 721–727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons SJ, Parsons JT 2004. Src family kinases, key regulators of signal transduction. Oncogene 23: 7906–7909 [DOI] [PubMed] [Google Scholar]
- Resh MD 1994. Myristylation and palmitylation of Src family members: the fats of the matter. Cell 76: 411–413 [DOI] [PubMed] [Google Scholar]
- Resh MD 1999. Fatty acylation of proteins: new insights into membrane targeting of myristoylated and palmitoylated proteins. Biochim Biophys Acta 1451: 1–16 [DOI] [PubMed] [Google Scholar]
- Resh MD 2004. Membrane targeting of lipid modified signal transduction proteins. Subcell Biochem 37: 217–232 [DOI] [PubMed] [Google Scholar]
- Resh MD 2006. Use of analogs and inhibitors to study the functional significance of protein palmitoylation. Methods 40: 191–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocks O, Gerauer M, Vartak N, Koch S, Huang ZP, Pechlivanis M, Kuhlmann J, Brunsveld L, Chandra A, Ellinger B, et al. 2010. The palmitoylation machinery is a spatially organizing system for peripheral membrane proteins. Cell 141: 458–471 [DOI] [PubMed] [Google Scholar]
- Rosenfeld N, Young JW, Alon U, Swain PS, Elowitz MB 2005. Gene regulation at the single-cell level. Science 307: 1962–1965 [DOI] [PubMed] [Google Scholar]
- Rothman JE 1994. Mechanisms of intracellular protein transport. Nature 372: 55–63 [DOI] [PubMed] [Google Scholar]
- Rothman JE, Wieland FT 1996. Protein sorting by transport vesicles. Science 272: 227–234 [DOI] [PubMed] [Google Scholar]
- Rothwell C, Lebreton A, Young Ng C, Lim JY, Liu W, Vasudevan S, Labow M, Gu F, Gaither LA 2009. Cholesterol biosynthesis modulation regulates dengue viral replication. Virology 389: 8–19 [DOI] [PubMed] [Google Scholar]
- Sahlmuller MC, Strating JR, Beck R, Eckert P, Popoff V, Haag M, Hellwig A, Berger I, Brugger B, Wieland FT 2011. Recombinant heptameric coatomer complexes: novel tools to study isoform-specific functions. Traffic 12: 682–692 [DOI] [PubMed] [Google Scholar]
- Sandilands E, Frame MC 2008. Endosomal trafficking of Src tyrosine kinase. Trends Cell Biol 18: 322–329 [DOI] [PubMed] [Google Scholar]
- Sandilands E, Cans C, Fincham VJ, Brunton VG, Mellor H, Prendergast GC, Norman JC, Superti-Furga G, Frame MC 2004. RhoB and actin polymerization coordinate Src activation with endosome-mediated delivery to the membrane. Dev Cell 7: 855–869 [DOI] [PubMed] [Google Scholar]
- Sato I, Obata Y, Kasahara K, Nakayama Y, Fukumoto Y, Yamasaki T, Yokoyama KK, Saito T, Yamaguchi N 2009. Differential trafficking of Src, Lyn, Yes and Fyn is specified by the state of palmitoylation in the SH4 domain. J Cell Sci 122: 965–975 [DOI] [PubMed] [Google Scholar]
- Shenoy-Scaria AM, Gauen LK, Kwong J, Shaw AS, Lublin DM 1993. Palmitylation of an amino-terminal cysteine motif of protein tyrosine kinases p56lck and p59fyn mediates interaction with glycosyl-phosphatidylinositol-anchored proteins. Mol Cell Biol 13: 6385–6392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shenoy-Scaria AM, Dietzen DJ, Kwong J, Link DC, Lublin DM 1994. Cysteine3 of Src family protein tyrosine kinase determines palmitoylation and localization in caveolae. J Cell Biol 126: 353–363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons K, Ikonen E 1997. Functional rafts in cell membranes. Nature 387: 569–572 [DOI] [PubMed] [Google Scholar]
- Simons K, Toomre D 2000. Lipid rafts and signal transduction. Nat Rev Mol Cell Biol 1: 31–39 [DOI] [PubMed] [Google Scholar]
- Simons K, Vaz WL 2004. Model systems, lipid rafts, and cell membranes. Annu Rev Biophys Biomol Struct 33: 269–295 [DOI] [PubMed] [Google Scholar]
- Simpson JC, Cetin C, Erfle H, Joggerst B, Liebel U, Ellenberg J, Pepperkok R 2007. An RNAi screening platform to identify secretion machinery in mammalian cells. J Biotechnol 129: 352–365 [DOI] [PubMed] [Google Scholar]
- Smotrys JE, Linder ME 2004. Palmitoylation of intracellular signaling proteins: Regulation and function. Annu Rev Biochem 73: 559–587 [DOI] [PubMed] [Google Scholar]
- Smotrys JE, Schoenfish MJ, Stutz MA, Linder ME 2005. The vacuolar DHHC-CRD protein Pfa3p is a protein acyltransferase for Vac8p. J Cell Biol 170: 1091–1099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stegmayer C, Kehlenbach A, Tournaviti S, Wegehingel S, Zehe C, Denny P, Smith DF, Schwappach B, Nickel W 2005. Direct transport across the plasma membrane of mammalian cells of Leishmania HASPB as revealed by a CHO export mutant. J Cell Sci 118: 517–527 [DOI] [PubMed] [Google Scholar]
- Toth MJ, Huwyler L 1996. Molecular cloning and expression of the cDNAs encoding human and yeast mevalonate pyrophosphate decarboxylase. J Biol Chem 271: 7895–7898 [DOI] [PubMed] [Google Scholar]
- Tournaviti S, Hannemann S, Terjung S, Kitzing TM, Stegmayer C, Ritzerfeld J, Walther P, Grosse R, Nickel W, Fackler OT 2007. SH4-domain-induced plasma membrane dynamization promotes bleb-associated cell motility. J Cell Sci 120: 3820–3829 [DOI] [PubMed] [Google Scholar]
- Tournaviti S, Pietro ES, Terjung S, Schafmeier T, Wegehingel S, Ritzerfeld J, Schulz J, Smith DF, Pepperkok R, Nickel W 2009. Reversible phosphorylation as a molecular switch to regulate plasma membrane targeting of acylated SH4 domain proteins. Traffic 10: 1047–1060 [DOI] [PubMed] [Google Scholar]
- Vaisanen T, Vaisanen MR, Pihlajaniemi T 2006. Modulation of the cellular cholesterol level affects shedding of the type XIII collagen ectodomain. J Biol Chem 281: 33352–33362 [DOI] [PubMed] [Google Scholar]
- Walter T, Held M, Neumann B, Heriche JK, Conrad C, Pepperkok R, Ellenberg J 2010. Automatic identification and clustering of chromosome phenotypes in a genome wide RNAi screen by time-lapse imaging. J Struct Biol 170: 1–9 [DOI] [PubMed] [Google Scholar]
- Wang J, Zhou X, Bradley PL, Chang SF, Perrimon N, Wong ST 2008a. Cellular phenotype recognition for high-content RNA interference genome-wide screening. J Biomol Screen 13: 29–39 [DOI] [PubMed] [Google Scholar]
- Wang M, Zhou X, Li F, Huckins J, King RW, Wong ST 2008b. Novel cell segmentation and online SVM for cell cycle phase identification in automated microscopy. Bioinformatics 24: 94–101 [DOI] [PubMed] [Google Scholar]
- Webb Y, Hermida-Matsumoto L, Resh MD 2000. Inhibition of protein palmitoylation, raft localization, and T cell signaling by 2-bromopalmitate and polyunsaturated fatty acids. J Biol Chem 275: 261–270 [DOI] [PubMed] [Google Scholar]
- Wegmann D, Hess P, Baier C, Wieland FT, Reinhard C 2004. Novel isotypic γ/ζ subunits reveal three coatomer complexes in mammals. Mol Cell Biol 24: 1070–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolven A, Okamura H, Rosenblatt Y, Resh MD 1997. Palmitoylation of p59fyn is reversible and sufficient for plasma membrane association. Mol Biol Cell 8: 1159–1173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu HS, Barba J, Gil J 2000. Iterative thresholding for segmentation of cells from noisy images. J Microsc 197: 296–304 [DOI] [PubMed] [Google Scholar]