Using the PAR-CLIP technique, Betel and colleagues identify a comprehensive target list for miR-302/367, the most highly expressed miRNAs in hESCs. They further find that miR-302/367 affects cell fate choice upon hESC differentiation via modulating TGF-β and BMP pathways.
Keywords: computational biology, stem cells, miR-302, miRNA, stem cell differentiation
Abstract
MicroRNAs are important regulators in many cellular processes, including stem cell self-renewal. Recent studies demonstrated their function as pluripotency factors with the capacity for somatic cell reprogramming. However, their role in human embryonic stem (ES) cells (hESCs) remains poorly understood, partially due to the lack of genome-wide strategies to identify their targets. Here, we performed comprehensive microRNA profiling in hESCs and in purified neural and mesenchymal derivatives. Using a combination of AGO cross-linking and microRNA perturbation experiments, together with computational prediction, we identified the targets of the miR-302/367 cluster, the most abundant microRNAs in hESCs. Functional studies identified novel roles of miR-302/367 in maintaining pluripotency and regulating hESC differentiation. We show that in addition to its role in TGF-β signaling, miR-302/367 promotes bone morphogenetic protein (BMP) signaling by targeting BMP inhibitors TOB2, DAZAP2, and SLAIN1. This study broadens our understanding of microRNA function in hESCs and is a valuable resource for future studies in this area.
The application of human embryonic stem (ES) cells (hESCs) and induced pluripotent stem cells (iPSCs) in cell therapy and human disease modeling requires detailed understanding of the complex signaling network responsible for self-renewal and differentiation. There is strong evidence that microRNAs play important roles in development, the establishment of cellular identity, and the regulation of stem cell behavior (Gangaraju and Lin 2009). Moreover, recent studies have demonstrated a remarkable ability of microRNAs in promoting reprogramming of somatic cells toward pluripotency during iPSC induction and, in some cases, in the absence of the classic reprogramming transcription factors (Anokye-Danso et al. 2011; Lin et al. 2011; Miyoshi et al. 2011; Subramanyam et al. 2011).
Initial profiling of microRNA in human and mouse ESCs identified the mir-302 cluster, which encodes for miR-302a/b/c/d and miR-367, as the most abundant microRNA transcript in hESCs and a related cluster, mir-290∼295, as the most abundant in mouse ESCs (Houbaviy et al. 2003; Suh et al. 2004). The function of ESC-specific microRNAs was initially studied in Dicer and DGCR8 mutants, which showed defects in differentiation and extended G1 phase (Bernstein et al. 2003; Kanellopoulou et al. 2005; Wang et al. 2007). Subsequent work implicated mir-302 in TGF-β regulation by targeting LEFTY1/2 (Rosa et al. 2009). Additional studies focused on the role of microRNAs in promoting rapid G1/S transition and the roles of let-7 and miR-145 in initiating differentiation (Xu et al. 2009; Melton et al. 2010). While these studies illustrate important aspects of microRNA regulation in pluripotent stem cells, a full description of microRNA function in ESCs requires the systematic identification of all microRNA targets in ESCs. A recent study in Dicer-null mouse ESCs pursued the global identification of microRNA targets (Leung et al. 2011); however, the resulting target list is missing many of the previously identified targets, such as Lefty2, Tgfbr2 (Subramanyam et al. 2011), and Rbl2 (Wang et al. 2008). Therefore, it remains critical to develop reliable strategies to identify the set of endogenous microRNA targets in ESCs that are readily applicable to the study of all pluripotent stem cells, including hESCs.
Here we report on the genome-wide identification of miR-302/367 endogenous targets in hESCs using a combination of computational and novel experimental approaches. We first performed an extensive microRNA profiling study in undifferentiated hESCs and their progeny at four distinct stages of differentiation. In agreement with previous studies, we found that the mir-302 cluster is the predominant microRNA transcript in hESCs and that its levels rapidly decline upon differentiation (Bar et al. 2008; Morin et al. 2008). We used a novel photoactivatable ribonucleoside-enhanced cross-linking and immunoprecipitation method (PAR-CLIP) (Hafner et al. 2010) that captures AGO2 binding to mRNAs, thereby identifying the full range of microRNA target sites in hESCs. Correlation of the CLIP data with miR-302/367 perturbation experiments identified a set of 146 high-confidence targets representing a wide range of functional categories, such as regulation of cell proliferation, chromatin organization, vesicle transport, actin cytoskeleton, and extracellular matrix constituents. We uncovered novel roles of miR-302/367 in hESCs that include modulation of pluripotency and self-renewal. Furthermore, we show that miR-302/367 inhibits neural differentiation and enhances trophectodermal fate by promoting bone morphogenetic protein (BMP) signaling. Modulation of BMP signaling is achieved, in part, by suppression of TOB2, DAZAP2, and SLAIN1, novel miR-302/367 targets with BMP inhibitory properties. This study presents a powerful approach for elucidating microRNA function in stem cells and is a valuable resource for the systematic exploration of miR-302/367 function in hESCs.
Results
MicroRNA profiling in undifferentiated hESCs and differentiated cell types derived from hESCs
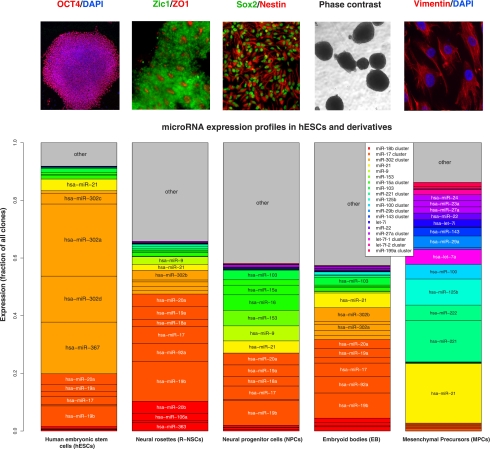
To characterize the role of microRNAs in hESCs, we first profiled microRNA expression in undifferentiated cells (hESCs) and in hESCs at four different differentiation stages—neural rosettes (R-NSCs) (Elkabetz et al. 2008), neural progenitors (NPCs), embryoid bodies (EBs), and mesenchymal precursors (MPCs)—by 454 sequencing of the small RNAs extracted from each cell type. To avoid biases from asynchronous differentiation and heterogeneous cell populations, the cells were sorted by stage-specific markers (hESCs, R-NSCs, and MPCs) or mechanically purified (NPCs) (Fig. 1; Elkabetz and Studer 2008). The microRNA profile in undifferentiated hESCs was dominated by the abundance of the mir-302 cluster, which accounted for >60% of all microRNAs expressed in hESCs (Fig. 1; Supplemental Table S1). Levels of mir-302 cluster drop sharply from the pluripotent to the rosette stage, suggesting that this cluster has a specific role at the hESC stage. Other abundant microRNA clusters highly expressed in hESCs included the mir-17∼92 cluster and its paralog, mir-106a∼363, which, unlike the mir-302 cluster, were also expressed in some of the hESC-derived cell populations. A number of microRNAs were persistently high in hESCs and most hESC-derived cell types, such as the oncomir mir-21 and mir-103. We further verified the expression profile in hESCs by Agilent microRNA arrays and Solexa sequencing. We found good concordance between array signal intensities and the sequence counts (Supplemental Fig. S1), as well as between the two deep-sequencing methods.
Figure 1.
MicroRNA sequencing profile in human stem cells. MicroRNA levels were measured by deep sequencing in five hESC-derived cell populations: hESCs, R-NSCs, NPCs, EBs, and MPCs. To obtain homogenous cell populations, the cells were sorted by stage-specific markers or mechanically purified. The top panel shows representative images of cell morphology and marker expression at each differentiation stage. In the bottom panel, color grouping indicates microRNAs that are members of the same primary transcript and therefore are cotranscribed. The mir-302 cluster that encodes miR-302a/b/c/d and miR-367 is the most abundant cluster in hESCs and its expression levels rapidly decline upon differentiation. Other abundant microRNAs include mir-17∼92 and its paralog, mir-106a; mir-21; and mir-103, which are persistently expressed in differentiated hESCs (see also Supplemental Table S1; Supplemental Fig. S1).
Identification of AGO-binding sites by PAR-CLIP
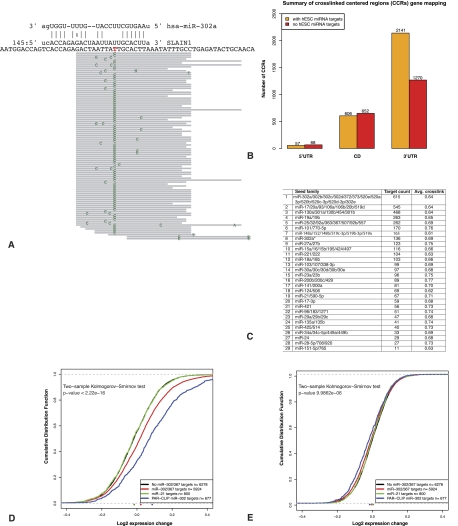
To directly identify microRNA targets in hESCs, we performed a genome-wide identification of RISC-binding sites using the PAR-CLIP approach (Hafner et al. 2010). Briefly, the binding sites of AGO2, the core component of RISC, in cultured cells were captured by UV cross-linking of mRNAs labeled with 4-thiouridine to interacting RNA-binding proteins. Following immunoprecipitation of the native AGO2 protein, the cross-linked RNA segments were recovered, transformed into a cDNA library, and sequenced. Binding sites were determined as clusters of overlapping reads where at least 20% of the reads contained a characteristic T-to-C mutation that marked the cross-linked site (Fig. 2A). The cross-linked centered regions (CCRs) were defined by a window of 60 nucleotides (nt) around the most frequent cross-linked site, which were then used to search for microRNA targets. In total, 7527 CCRs were identified, of which 3411, 1258, and 125 were mapped to known 3′ untranslated regions (UTRs), CDSs, and 5′ UTRs, respectively (Fig. 2B), and the remaining 2733 were intergenic (data not shown). The majority of the intergenic CCRs are likely fortuitous binding events to unannotated transcripts. However, we cannot exclude the possibility that some of these represent functional microRNA-mediated regulation of noncoding transcripts. While there is an appreciable number of CCRs in coding regions, we found that their regulatory effect is significantly lower than CCRs in 3′ UTRs (Supplemental Fig. S2). Of the CCRs in 3′ UTRs, 2141 (63%) had at least one predicted microRNA target site (Betel et al. 2010) to the top 29 expressed seed classes in hESCs (see the Materials and Methods; Supplemental File S1), whereas the remaining 1270 CCRs did not have predicted target sites for these seed sequences. Consistent with the microRNA expression levels in hESCs, the constituents of the mir-302 cluster—miR-302a/b/c/d and miR-367—had the largest number of binding sites in CCRs, followed by members of the mir-17∼92 cluster (Fig. 2C). However, many of the CCRs are predicted to be regulated by more than one of the top 29 endogenously expressed seed groups in hESCs. Therefore, the true identity of the regulating microRNA is ambiguous, and some CCRs are likely to be targeted by multiple microRNAs. Due to sequence similarities in the seed regions, ∼70% of the 3′ UTR CCRs predicted to be regulated by miR-302a/b/c/d are also predicted to be regulated by miR-17/20a/106a, which are members of the highly expressed mir-17∼92 and mir-106a clusters in hESCs (Supplemental Fig. S3; Supplemental File S1). We found a significant number of CCRs that mapped to coding sequences. However, in contrast to the sites in 3′ UTRs, the number of CCRs in coding regions that contained target sites for any of the hESC-expressed microRNAs is similar to the number of CCRs with no such target sites (Fig. 2B). This is consistent with previous CLIP results that found no enrichment of target sites in CCRs from coding regions (Chi et al. 2009; Hafner et al. 2010). Contrary to a previous report of microRNA-mediated regulation in the coding regions of Oct4, Nanog, and Sox2 in mouse ESCs (Tay et al. 2008), we found no evidence of AGO2-binding events in these genes, suggesting that such regulation may be specific to mice or not detected by our approach due to technical reasons. Furthermore, it was previously reported that miR-145, which is expressed at low levels in H9 cells and up-regulated upon EB differentiation, targets the 3′ UTRs of OCT4, SOX2, and KLF4 (Xu et al. 2009). In this study, we detected only low levels of miR-145 in the EB and MPC data and did not find evidence for AGO2 binding of pluripotency factors in undifferentiated hESCs.
Figure 2.
Experimental and computational identification of miR-302/367 targets in hESCs. (A) PAR-CLIP data provide precise location of the AGO2-binding site with hallmark T-to-C mutation at the cross-linked site. This position is typically upstream of the complement microRNA seed sequence shown in the novel SLAIN1 target site identified in this study. The top panel represents an miR-302a predicted target site (at position 145 in the 3′ UTR), and the bottom panel depicts the mapped sequence reads and mismatched positions in green. (B) Summary of the genomic mapping of the CCRs identified by PAR-CLIP. Of the total 7527 CCRs, 4794 (>63%) were mapped to annotated genes. Orange bars indicate the number of CCRs with predicted target sites for one or more of the endogenously expressed (top 29) seed sequences in hESCs, whereas red bars indicate the number of CCRs with no predicted sites for these microRNAs. (C) A summary of the number of CCRs targeted by each of the top 29 hESC-expressed microRNA seed families. Average cross-link is the average fraction of reads in the CCRs with T-to-C mutations (see also Supplemental File S1; Supplemental Table S2). (D) Inhibition of miR-302/367 results in up-regulation of their predicted targets. The cumulative distribution of the log2 expression change of miR-302/367 targets is significantly increased after miR-302/367 inhibition relative to the background set that lacks miR-302/367 predicted targets. Similarly, the miR-302/367 targeted genes identified in the PAR-CLIP experiments are also significantly up-regulated following miR-302/367 inhibition. In contrast, targets of miR-21, another highly expressed microRNA in hESCs, do not undergo a significant change in expression, indicating specific inhibition of miR-302/367. (E) In contrast to inhibition, ectopic expression of miR-302/367 results in only a minor decrease in the expression of their targets.
Genome-wide identification of miR-302/367 targets in undifferentiated hESCs
The PAR-CLIP data provide a genome-wide map of the AGO2-binding sites; however, the data do not indicate which of those sites leads to microRNA-mediated down-regulation of the target genes. To identify the targets that are under direct mir-302 cluster regulation in hESCs, we performed miR-302/367 perturbation experiments by overexpression or inhibition of miR-302/367 to identify the genes whose expression levels are regulated by this cluster.
Transfection of hESCs with miR-302/367 mimics or cholesterol-modified antagomirs (Krützfeldt et al. 2007) led to the increase or decrease of miR-302/367 function, respectively, as measured by a dual-luciferase reporter containing artificial tandem miR-302a target sites (Supplemental Fig. S4). Gene expression analysis and computational target prediction following the inhibition of miR-302/367 showed a strong up-regulation of the miR-302/367 putative targets (P-value ≤ 2.22 × 10−16 KS test) (Fig. 2D; Supplemental Table S2). We also confirmed by additional analysis that the antagomir treatment inhibited both miR-302 and miR-367 activity independently and did not affect microRNAs with similar seed sequences (Supplemental Fig. S5). In contrast, there was only marginal, although significant, down-regulation of miR-302/367 targets following the overexpression of this cluster (Fig. 2E). Similarly, in the dual-luciferase system, miR-302/367 overexpression had only a modest effect on luciferase activity compared with miR-302/367 inhibition. We reasoned that the existing high levels of endogenous miR-302/367 saturated the repression of their targets, and therefore any additional increase in miR-302/367 resulted in only minor additional down-regulation. However, the lack of significant target down-regulation may also be attributed to poor incorporation of the transfected miR-302/367 into the RISC complex.
Identification of high-confidence miR-302/367 target genes
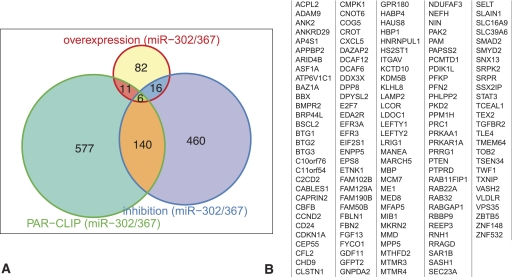
The correlation of the PAR-CLIP experiment with the miR-302/367 perturbation experiments identified a set of 146 high-confidence targets that (1) had 3′ UTR AGO2-binding sites corresponding to miR-302/367 target sites and (2) were significantly up-regulated after inhibition of miR-302/367 (Fig. 3A). These 146 genes were significantly more up-regulated by miR-302/367 inhibition than the other 476 up-regulated targets that did not contain miR-302/367 CCRs (P-value = 3.7 × 10−7, KS test) (Supplemental Fig. S6a). Similarly, there was an increase in the number of cross-linked reads in this gene set relative to the set of the other 588 genes with miR-302/367 CCRs that were not differentially up-regulated following miR-302/367 inhibition (P-value = 1.6 × 10−5) (Supplemental Fig. S6B). Thus, the experimental results and the enrichment analysis indicate that these 146 genes represent a high-confidence set of bona fide miR-302/367 targets in hESC.
Figure 3.
High-confidence microRNA-302/367 targets. (A) The intersection between PAR-CLIP data and the miR-302/367 perturbation experiments identifies the set of most regulated targets. The set of 146 high-confidence miR-302/367 targets is defined as those genes with favorable mirSVR scores (Betel et al. 2010) that are significantly up-regulated after miR-302/367 inhibition and have identified PAR-CLIP sites in their 3′ UTRs. (B) The full list of high-confidence miR-302/367 targets in undifferentiated hESCs. Included in this list are a number of previously identified miR-302 targets (e.g., LEFTY1/2 [Rosa et al. 2009], CDKN1A [Wang et al. 2008], and TGFBR2 [Subramanyam et al. 2011]), many that are implicated in cell cycle regulation (e.g., BTG1,2,3), and negative regulators of BMP and TGF-β signaling.
Included in our set of high-confidence targets are previously characterized miR-302 targets such as LEFTY1, LEFTY2 (Rosa et al. 2009), p21 (CDKN1A) (Wang et al. 2008), TGFBR2 (Subramanyam et al. 2011), and Cyclin D2 (CCND2) (Lee et al. 2008) (Fig. 3B). Our screen also identified a novel miR-302 target site in PTEN's 3′ UTR in addition to the previously described site (Poliseno et al. 2010). Functional analysis of this gene set revealed a significant overrepresentation of genes regulating cell cycle and proliferation as well as additional biological pathways, suggesting that the mir-302 cluster modulates a wide range of functionalities in undifferentiated hESCs in a fashion similar to the programming factors (Cerami et al. 2010; Warde-Farley et al. 2010).
miR-302/367 is a positive regulator of pluripotency and self-renewal in hESCs
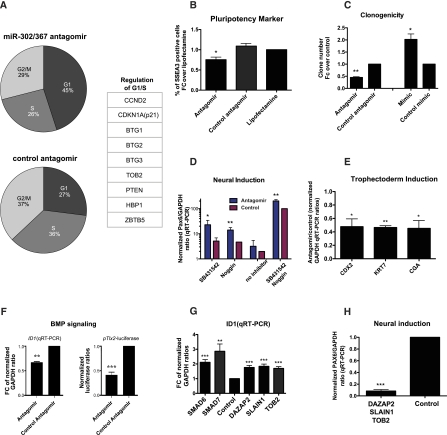
To further investigate the role of miR-302/367 in regulating pluripotency, we performed gain-of-function and loss-of-function studies for miR-302/367 in undifferentiated hESCs using microRNA antagomirs and mimics followed by quantitative analyses of pluripotent behavior and differentiation. Previous studies have demonstrated a role for ESC-specific microRNAs in suppressing regulators of the G1/S transition (Wang et al. 2008). We observed that inhibiting miR-302/367 function in hESCs increased the percentage of cells in the G1 phase of the cell cycle (Fig. 4A). In addition to the previously described miR-302 target, p21, which is a known negative regulator of G1/S transition, other high-confidence targets were previously shown to regulate cell cycle in other cell types, including BTG1/2/3, TOB2 (Winkler 2010), PTEN (Ramaswamy et al. 1999; Mamillapalli et al. 2001), and HBP1 (Berasi et al. 2004). Cyclin D2 (CCND2), which was identified in our screen and previously reported as an miR-302 target (Lee et al. 2008), is a positive regulator of the G1/S transition in differentiated cells. The role of D-type cyclins in hESCs is less clear, and it is thought that hESCs are less dependent on D-type cyclins for rapid cell cycles, presumably due to the constitutively hyperphosphorylated state of RB (Fluckiger et al. 2006).
Figure 4.
The miR-302/367 cluster promotes pluripotency and modulates BMP signaling during differentiation. (A) FACS-based cell cycle analysis on propidium iodide (PI)-stained cells was used to determine the percentage of cells in a given cell cycle phase. Inhibition of miR-302/367 by antagomirs increased the fraction of cells in G1 phase relative to control (n = 3 independent experiments: antagomir vs. control: P < 0.05 for G1, S, and G2/M comparisons). (B) Inhibition of miR-302/367 by antagomirs results in reduction of pluripotency as measured by FACS analysis of the pluripotency marker SSEA-3. (C) Quantification of hESC clonogenicity as determined by the number of OCT4+ colonies 7 d after replating at clonal density. (D) qRT–PCR for early neural marker PAX6 after 7 d of differentiation in knockout serum replacement (KSR)-based medium containing inhibitors of BMP (Noggin) and TGF-β/Activin/Nodal (SB-431542) signaling pathways. (E) qRT–PCR for trophectoderm markers CDX2, KRT7, and CGA after 3 d of differentiation in RPMI medium with BMP4. (F) qRT–PCR analysis for BMP target gene ID1 (left panel) and luciferase assay of BMP reporter Tlx2-lux (right panel) 36 h after miR-302/367 inhibition by antagomir. (G) qRT–PCR of ID1 48 h after siRNA-mediated knockdown of targeted genes. (H) qRT–PCR of PAX6 after 7 d of differentiation in KSR-based medium containing inhibitor of TGF-β/Activin/Nodal signaling and simultaneous siRNA knockdown of DAZAP2, SLAIN1, and TOB2. (*) P < 0.05; (**) P < 0.01; (***) P < 0.001 compared with control antagomir mimic or siRNA (see also Supplemental Figs. S7, S8).
We next tested whether perturbation of miR-302/367 impacts other aspects of hESC maintenance, such as the expression of pluripotency markers and hESC self-renewal. Stage-specific embryonic antigen-3 (SSEA-3) is a well-characterized marker in pluripotent stem cells and preimplantation embryos, and is rapidly down-regulated upon differentiation (Henderson et al. 2002; International Stem Cell Initiative 2007). Inhibition of miR-302/367 led to a significant decrease in the relative percentage of cells expressing SSEA-3 (Fig. 4B). In contrast to other pluripotency markers such as SSEA-4, expression of SSEA-3 is limited to the most deeply pluripotent fraction of hESCs. The SSEA-3-negative hESC subfractions have been shown to differentiate more readily and exhibit decreased clonogenic potential (Stewart et al. 2006). We therefore next tested the clonogenic behavior of undifferentiated hESCs following miR-302/367 perturbation experiments. Upon transfection with miR-302/367 and control antagomirs or mimics, cells were replated at clonal density (Franken et al. 2006). After 7 d of culture, the total number of OCT4-positive colonies was determined. Inhibition of miR-302/367 led to a significant decrease in the number of OCT4-positive clones, while overexpression of miR-302/367 led to an increase in OCT4-positive clones as compared with their respective control conditions (Fig. 4C). This suggests that miR-302/367 acts as a positive regulator for the expression of pluripotency markers and self-renewal behavior in undifferentiated hESCs, promoting robustness of the pluripotent stage and preventing premature differentiation.
miR-302/367 modulates TGF-β and BMP signaling during neural induction
Previous studies have suggested a role for miR-302/367 in neural differentiation mediated by LEFTY1/2 during hESC-derived EB culture and in early frog development (Rosa et al. 2009). Here, we took advantage of our recently established neural induction protocol based on dual SMAD inhibition upon exposure of hESCs to recombinant Noggin (BMP inhibition) and SB-431542 (inhibition of TGF-β/Activin/Nodal) (Chambers et al. 2009). The defined nature of the protocol allows for selective interrogation of the BMP versus TGF-β/Activin/Nodal signaling branches during neural induction. Cells were transfected with miR-302/367 or control antagomirs and treated for 7 d (1) with both inhibitors (Noggin and SB), (2) with TGF-β/Activin/Nodal inhibitor only (SB), (3) with BMP inhibitor only (Noggin), or (4) in the absence of either inhibitor. At day 7 of differentiation, cells were collected for expression analysis of the early neural marker PAX6 by quantitative RT–PCR (qRT–PCR) (Zhang et al. 2010). In all four conditions, PAX6 expression was higher in antagomir-treated cells compared with the control. However, the most dramatic differences were observed in the SB condition. Similar results were obtained using qRT–PCR for another early neural marker, SOX1 (Pevny et al. 1998), and using quantitative analysis of PAX6 induction by intracellular flow cytometry (Supplemental Fig. S7). These data suggest that even in the presence of TGF-β/Activin/Nodal pathway blockade, inhibition of miR-302/367 exerts a strong positive effect on neural induction. The most obvious explanation for this result might be regulation of the BMP pathway by miR-302/367, presumably by targeting an endogenous BMP inhibitor. Treatment with miR-302/367 antagomirs alone, in the absence of Noggin and SB-431542, did not significantly promote neural induction (Fig. 4D), suggesting that the regulation of TGF-β and BMP pathways by miR-302/367 inhibition alone did not reach levels sufficient to trigger neural induction.
We then proceeded to test the effect of miR-302/367 inhibition on induction of other lineages. Endoderm is induced by Activin/Nodal signaling (D'Amour et al. 2005), and mesendoderm is induced by both Activin/Nodal and BMP signaling (Winnier et al. 1995; Kimelman 2006; Zhang et al. 2008). After transfection of miR-302/367 antagomirs and controls, we carried out directed differentiation toward mesoderm or endoderm. Consistent with previous reports (Rosa et al. 2009), miR-302/367 inhibition led to a decrease in the level of expression of markers specific for mesoderm (BRACHYURY) and endoderm (SOX17) lineages (Supplemental Fig. S8), likely due to the derepression of TGF-β/Activin/Nodal inhibitors such as LEFTY1/2. On the other hand, induction of trophectoderm requires BMP signaling but is inhibited by TGF-β/Activin/Nodal signaling (Xu et al. 2002; Chen et al. 2008; Wu et al. 2008). Upon miR-302/367 inhibition, we observed a decrease in the efficiency of trophectoderm induction, as measured by the level of expression of trophectoderm markers CDX2, KRT7, and CGA (Fig. 4E; Schulz et al. 2008). The decreased trophectoderm induction cannot be explained by a decrease in TGF-β/Activin/Nodal signaling due to derepression of LEFTY1/2, which would be expected to increase trophectodermal yield (Wu et al. 2008). Our data rather suggest that miR-302/367 inhibition mediates derepression of a BMP-inhibitor responsible for decreased trophectodermal yield. These findings were further corroborated by measuring modulation of BMP signaling levels in reporter assays for Tlx2 and ID1. Both of these genes are regulated selectively by BMP but not by TGF-β/Activin/Nodal signaling (Tang et al. 1998; Korchynskyi and ten Dijke 2002). We observed decreased Tlx2-luciferase activity and ID1 transcript upon miR-302/367 inhibition, consistent with a positive regulation of BMP-induced transcriptional activity by miR-302/367 (Fig. 4F).
miR-302/367 up-regulates BMP signaling by inhibition of DAZAP2, SLAIN1, and TOB2
To determine which of the 146 high-confidence targets function as BMP inhibitors, we performed siRNA knockdown of 11 candidate genes selected based on their robust regulation by miR-302/367 or their known involvement in BMP signaling in other cell types. hESCs were transfected with siRNAs targeting each of the 11 genes. In addition, we used a negative control siRNA and siRNAs targeting SMAD6 or SMAD7 (inhibitory, I-SMADs known to repress BMP signaling) (Hata et al. 1998; Souchelnytskyi et al. 1998). After 48 h, cells were assayed for ID1 levels by qRT–PCR. Among the 11 genes, knockdown of DAZAP2, SLAIN1, or TOB2 resulted in the most robust up-regulation of ID1, to levels comparable with knockdown of I-SMADs (Fig. 4G; Supplemental Fig. S9A), suggesting that each of these genes is a negative regulator of BMP signaling. Knockdown levels for each of the three genes and I-SMAD were comparable (Supplemental Fig. S9b), suggesting that our results were not skewed by differences in siRNA efficiencies. We also confirmed by luciferase assay that the three genes are regulated by miR-302 (Supplemental Fig. S9C–E). We next tested whether knockdown of these genes can impact neural differentiation propensity. While down-regulation of each gene did not have a significant effect on differentiation (data not shown), simultaneous knockdown of all three genes dramatically decreased neural induction efficiency in the presence of the TGF-β/Activin/Nodal signaling inhibitor (Fig. 4H). Our data demonstrate that miR-302/367 promotes BMP signaling by targeting DAZAP2, SLAIN1, and TOB2, and that inhibition of these three genes can repress neural differentiation of hESCs.
Discussion
In this study, we presented the first unbiased, genome-wide survey of miR-302/367 targets and function in hESCs. Our novel strategy for target identification was based on the combination of (1) global identification of genes regulated by miR-302/367 in perturbation studies, (2) global identification of genes cross-linked to AGO2 using PAR-CLIP technology, and (3) computational prediction of miR-302/367 targets. This approach enabled the identification of a comprehensive list of high-confidence targets, including most known targets and many novel genes regulated by miR-302/367. Gain-of-function and loss-of-function studies of miR-302/367 in undifferentiated hESCs performed in parallel confirmed a functional role in promoting G1/S transition and identified novel roles in stabilizing pluripotency and in regulating both the TGF-β and BMP branches of SMAD signaling during neural differentiation. Finally, we linked the proposed role of miR-302/367 in regulating BMP signaling to a set of novel targets—including TOB2, DAZAP2, and SLAIN1—that show BMP inhibitory activity in undifferentiated hESCs.
A key component of our study was the combination of complementary experimental and computational approaches that resulted in the identification of a set of high-confidence miR-302/367 targets. On their own, microRNA perturbation, PAR-CLIP, and computational target prediction were not sufficient for accurate identification of functional target sites. Inhibition of miR-302/367 resulted in relatively modest changes in expression levels for most of the predicted miR-302/367 targets, which makes it difficult to identify the true targets strictly by significance of log expression changes. In contrast, the PAR-CLIP method provides a comprehensive mapping of AGO2-binding sites in the genome but does not provide a direct measure of the extent of microRNA-mediated regulation, and the identity of the regulating microRNAs is often ambiguous. Finally, accurate prediction of microRNA target sites is a difficult computational challenge and, despite algorithmic advancements, the number of false predictions remains high. The combined target identification approach allows for accurate discovery of the endogenously regulated microRNA targets without the need for individual verification of each site.
This study is also the first application of the PAR-CLIP method using antibodies toward the native AGO2 proteins, rather than a Flag/HA-tagged form that requires engineering of cells and is prone to artifacts caused by nonphysiological AGO2 levels. Accordingly, our method is broadly applicable to identify microRNA regulation in most cell types. A primary feature of the PAR-CLIP method is the introduction of T-to-C mutation at the site of cross-linking, which experimentally identifies, almost at a nucleotide-level resolution, the site of AGO2 binding. However, due to seed sequence similarities among the highly expressed microRNAs in hESCs, 52% of the CCRs in 3′ UTRs are potentially targeted by more than one seed family. In addition, 29% of the 1432 genes with multiple CCRs in their 3′ UTRs are regulated by more than one microRNA seed sequence (Supplemental File S1). These results underscore the multiplicative nature of microRNA regulation, where a single UTR, or even a single AGO-binding site, can be regulated by multiple distinct microRNAs.
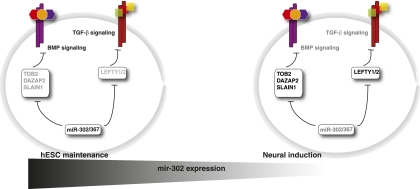
Our functional studies revealed a novel role for miR-302/367 in promoting BMP signaling in undifferentiated hESCs in addition to the well-known function of regulating TGF-β signaling. Loss of function of DAZAP2, SLAIN1, and TOB2 leads to up-regulation of the BMP-responsive ID1 with concomitant decrease of neural induction efficiency (Fig. 5). This is consistent with previous findings that TOB2, a member of BTG/TOB family, acts on SMAD6/7 and inhibits BMP signaling (Yoshida et al. 2003) as well as BTG3, which inhibits BMP in osteoblasts (Miyai et al. 2009). There is evidence that suggests that SLAIN1 and DAZAP2 have important developmental functions, but their role in BMP inhibition has not been previously demonstrated. SLAIN1 is a poorly characterized gene expressed in mouse and human ESCs and in some tissues of the developing mouse embryo, predominantly in the nervous system (Hirst et al. 2010; Smith et al. 2010). In Xenopus, DAZAP2 is required for FGF2-mediated posterior patterning (Roche et al. 2009) and FGFs are known to negatively regulate BMP signaling (Sapkota et al. 2007), suggesting that DAZAP2 may act with additional factors to modulate BMP indirectly.
Figure 5.
miR-302/367 targets modulate TGF-β and BMP signaling. In hESCs, neural induction is initiated by inhibition of TGF-β and BMP signaling. The rapid decline in mir-302 levels upon neural induction suggests that miR-302/367 promotes self-renewal and pluripotency and represses neural differentiation, possibly by suppressing inhibitors of TGF-β and BMP pathways. In this study, we identified three novel miR-302/367 targets that inhibit BMP signaling. Thus, by promoting TGF-β and BMP signaling at the ESC stage through down-regulation of pathway inhibitors, miR-302/367 promotes the pluripotency stage and attenuates neural differentiation.
TGF-β/Activin/Nodal signaling in hESCs promotes pluripotency (Xu et al. 2008), while BMP signaling promotes differentiation toward mesoderm and trophectoderm (Xu et al. 2002; Zhang et al. 2008); therefore, it is striking that a microRNA cluster that positively regulates pluripotency and self-renewal in hESCs would also promote BMP signaling by targeting its inhibitors. One possible explanation for this result is the default model of neural specification, which states that in the absence of cell–cell signaling, hESCs are fated to differentiate along the neural lineage rather than to mesendodermal or trophectodermal fates (Muñoz-Sanjuán and Brivanlou 2002). Therefore, by targeting inhibitors of BMP and TGF-β/Activin/Nodal signaling pathways, which in turn activates those pathways, miR-302/367 both suppresses neural induction and promotes pluripotency. Based on our set of validated miR-302/367 targets, several additional mechanisms may contribute to promoting pluripotency and self-renewal. For example, targeting of PTEN by miR-302 suggests regulation of AKT/PI3K signaling, a pathway that was previously linked to expression of pluripotency markers in hESCs (Armstrong et al. 2006) and Nanog regulation in spermatagonial cells (Kuijk et al. 2010). Alternatively, prolonging the G1 phase of the cell cycle following miR-302/367 inhibition may predispose hESCs to spontaneous differentiation, as inhibition of G1/S progression in hESCs has been shown to promote loss of pluripotency marker SSEA-4 and hESC morphology (Neganova et al. 2009). Interestingly, some targets identified in this study, such as members of the BTG/TOB family, regulate concomitantly cell cycle progression as well as BMP signaling. BTG/TOB proteins have also been shown to regulate nuclear receptor activity, transcription factor binding, and arginine methylation during embryonic development and are typically associated with differentiation (Prévôt et al. 2000; Berthet et al. 2002; Busson et al. 2005; Kawate et al. 2005; Passeri et al. 2006).
Recent studies have demonstrated that expression of miR-302/367 in fibroblasts is sufficient to induce pluripotency or enhance iPSC induction in combination with pluripotency transcription factors (Anokye-Danso et al. 2011; Lin et al. 2011; Miyoshi et al. 2011; Subramanyam et al. 2011). It has been suggested that miR-302/367 may act by promoting mesenchymal-to-epithelial transition (MET) through suppression of TGF-β signaling-mediated epithelial-to-mesenchymal transition (EMT) by targeting TGFBR2 (Subramanyam et al. 2011). Consistent with this role, our target list includes, in addition to TGFBR2, several other genes associated with EMT, such as SMAD2, the zinc transporter gene SLC39A6 (Yamashita et al. 2004), and DDX3X. Furthermore, positive regulation of BMP signaling by miR-302/367 could also promote BMP-dependent MET during reprogramming (Samavarchi-Tehrani et al. 2010). By computational analysis (Cerami et al. 2010; Warde-Farley et al. 2010) of the 146 genes regulated by miR-302/367, we identified a number of additional pathways that may be involved in modulating pluripotent cell behavior, including vesicular transfer (SEC23A, RAB22A, SAR1B, and AP4S1), chromatin regulators (ASF1A, SMYD2, KDM5B, and ARID4B), and cytoskeletal modulators (PFN2, TWF1, CFL2, and FBLN1).
Our list of 146 validated targets is a valuable resource for future functional studies to comprehensively define the role of miR-302/367 in all aspects of human pluripotent cell behavior as well as in the reprogramming process. Our strategy for the global identification of microRNA targets is a powerful approach for elucidating microRNA-mediated regulatory networks in many cell types and biological systems beyond pluripotent hESCs.
Materials and methods
Cell culture, differentiation, and characterization
hESCs (WA-09), passages 33–50, were maintained on irradiated mouse embryonic fibroblast (MEF) feeder layers as described previously (Perrier et al. 2004), and, for some of the experiments, under feeder-free conditions on Matrigel according to Xu et al. (2001), with a daily change of medium—consisting of DMEM/F12 medium, 20% knockout serum replacement (KSR) (Invitrogen), nonessential amino acids, 1 mM L-glutamine, 10 μM β-mercaptoethanol, and 8 ng/mL FGF2—and passaged weekly using 4 U/mL dispase in hESC medium. For microRNA profiling, R-NSCs were generated as described previously (Elkabetz et al. 2008). Briefly, dissociated hESCs were plated on irradiated (50 Gy) stromal cells (MS5s) and maintained in knockout DMEM medium (Invitrogen); 15% KSR medium (Invitrogen) with 2 mM L-glutamine, 10 μM β-mercaptoethanol, and nonessential amino acids and supplemented with Noggin (250 ng/mL) for 7 d; and with SHH (200 ng/mL), FGF8 (100 ng/mL), ascorbic acid (200 μg/mL), and BDNF (20 ng/mL) for an additional 4 d. Subsequently R-NSCs were mechanically passaged onto culture dishes precoated with 15 μg/mL polyornithine and 1 μg/mL laminin (Po/Lam) in N2 medium supplemented with SHH (200 ng/mL), FGF8 (100 ng/mL), ascorbic acid (200 μg/mL), and BDNF (20 ng/mL) and harvested after 1 wk in culture. NPCs were derived from R-NSCs upon extended culture in FGF2/EGF (day 95 of differentiation) as described previously (Elkabetz et al. 2008). MPCs were generated as in Barberi et al. (2007). In brief, hESCs were dissociated with 0.05% Trypsin and plated at 500 cells per square centimeter on irradiated MEFs and maintained in hES conditions until day 4. Cells were further differentiated in ITS medium (days 5–20), followed by exposure to α-MEM medium with 10% fetal bovine serum (days 21–35). For EB formation, hESC colonies were detached by dispase, washed, and plated into ultralow adhesion plates in knockout DMEM supplemented with glutamine and 20% defined FBS (Hyclone). The resulting EB cultures were maintained for 21 d.
Each cell type was characterized as described previously (Barberi et al. 2007; Elkabetz et al. 2008), including immunocytochemistry for the pluripotency marker Oct4 (Santa Cruz Biotechnologies), the neuroepithelial marker Zic1 (Novus Biologicals), the polarity marker ZO1 (BD Pharmingen), the neural stem cell markers Sox2 (Abcam) and Nestin (Neuromics), and the MPC marker Vimentin (Sigma).
MicroRNA profiling
Small RNA libraries from the five differentiation states (undifferentiated hESCs, N-RNCs, NPCs, EBs, and MPCs) were generated from 5 μg of total RNA as described in Hafner et al. (2008) and sequenced by 454 sequencing. The reads were processed similarly to Rajasethupathy et al. (2009). Briefly, the sequence reads were mapped to a database of known RNAs, and expression levels were determined by the number of sequence reads mapped to known microRNAs and normalized by the total number of microRNA reads in each library. A second profiling experiment was performed using Agilent microRNA arrays (human microRNA version 1.0), and signal intensities were processed by vsn and limma Bioconductor R packages. Additional microRNA sequencing for undifferentiated hESCs was performed using Solexa sequencing.
MicroRNA target prediction
MicroRNA target predictions were performed using a variant of the miRanda algorithm (John et al. 2004) using a score cutoff of 120 and gap opening and gap extension of −9 and −4, respectively. Target sites were scored by mirSVR, which ranks predicted target sites using a combination of local and global features of the target site (Betel et al. 2010). The top expressed microRNAs in hESCs (54 microRNAs with sequence counts of more than two representing 29 seed classes) were used to predict target sites in the PAR-CLIP CCRs. Predictions were filtered for perfect 6-mer seed complementarity or targets with a mirSVR score −0.05 or lower, which allowed prudent inclusion of noncanonical targets with favorable features (e.g., conservation). Similar prediction was performed for miR-302/367 targets in the overexpression and inhibition experiments.
PAR-CLIP
PAR-CLIP was performed as described in Hafner et al. (2010) with small modifications. hESCs were passaged onto Matrigel (BD)-coated plates and maintained with MEF-conditioned medium and 10 ng/mL FGF2. After 3 d, the cells were treated with 100 μM 4SU for 6 h and 100 × 106 cells were irradiated with 0.15 mJ/cm2 365-nm UV light to cross-link RNA to RNA-binding proteins. Cells were harvested and lysed in NP40 lysis buffer. The cleared cell lysates were treated with RNase T1. AGO2 was immunoprecipitated with monoclonal anti-AGO2 antibodies (C1.9E8.2) bound to Protein G Dynabeads (0.25 mg of antibody per milliliter of beads). RNase T1 (100 U/mL) was added to the immunoprecipitate. Beads were washed and resuspended in dephosphorylation buffer. Calf intestinal alkaline phosphatase was added to dephosphorylate the RNA. Beads were washed and incubated with polynucleotide kinase and γ32P-ATP to radioactively label the cross-linked RNA. The protein–RNA complexes were separated by SDS-PAGE, the radioactive band migrating at ∼100 kDa was excised, and the protein–RNA complex was electroeluted from the gel. The protein was removed by digestion with 0.12 mg/mL proteinase K. The RNA was recovered by acidic phenol/chloroform extraction and subsequent ethanol precipitation. The recovered RNA was converted into a cDNA library as described (Hafner et al. 2008) and Solexa-sequenced. The extracted sequence reads were mapped to the human genome (hg18), human mRNAs, and microRNA precursor regions. Clusters were defined as genomic regions with more than five sequence reads, of which 20% cross-linked (i.e., with T-to-C mutations). CCRs were generated from the clusters by extracting 30 nt upstream of and downstream from the most frequently cross-linked nucleotide. The genomic locations of the CCRs were matched to annotated genomic regions (5′ UTRs, CDSs, and 3′ UTRs) downloaded from the University of California at Santa Cruz Genome Browser. For comparison with array expression values, genes with CCR sites were matched to Illumina probe annotation by RefSeq IDs or gene names.
Transfections with miR-302/367 antagomirs and mimics
MicroRNA perturbation studies for global gene expression profiles
For miR-302/367 inhibition, antagomir design was used as described previously (Krützfeldt et al. 2007). Briefly, RNA antisense oligonucleotides (ASO) harbored complete 2-OMe sugar modifications, partial phosphothiroate linkage modifications, and a cholesterol moiety at the 3′ end. Random sequence was used for control antagomir. For miR-302/367 overexpression, mimics (Dharmacon) were used. Mixes included antagomir or mimics to each individual microRNA in the cluster (miR-302a/b/c/d/367). For transfections, hESCs were dissociated with dispase and subsequently with accutase, and replated as single cells on Matrigel (BD)-coated 24-well plates at a density of 40,000 cells per square centimeter in MEF-conditioned medium spiked with 10 ng/mL FGF2 and ROCK inhibitor Y-27632 (Sigma-Aldrich). After 24–48 h, cells were transfected with either 50–100 nM antagomirs, 60–120 nM mimics, or the corresponding controls using Lipofectamine 2000 (Invitrogen) in 30% Opti-MEM (Invitrogen) and 70% CM with 10 ng/mL FGF2. After transfection, cells were washed and fed with MEF-conditioned medium spiked with 10 ng/mL FGF2. For global mRNA analysis, 24 h after transfection, cells were harvested in Trizol and processed for Illumina BeadArrays according to the specifications of the manufacturer, and the data were processed using the lumi Bioconductor package. Log expression changes were computed using the limma package, which implements a variant of the t-test. For genes with multiple probes, the probe with the lowest adjusted P-value was selected (false discovery rate [FDR] < 0.05). Genes with a log2 expression change of >0.2 for the antagomir experiment and less than −0.2 for the mimic experiment were considered for subsequent analysis.
Directed differentiation assays
For miR-302/367 inhibition functional assays, differentiations were started 12–24 h after antagomir transfection. For trophectoderm differentiation, cells were grown in RPMI medium supplemented with 0.5% HyClone FBS, 1 mM L-Glu, and 10 ng/mL BMP4 and harvested on day 3. For mesoderm differentiation, cells were grown in RPMI medium supplemented with 0.5% HyClone FBS and 1 mM L-Glu, spiked with 50 ng/mL Activin on day 1 and 10 ng/mL BMP4 on day 2, and harvested on day 3. For endoderm differentiation, cells were grown in DMEM/F12 medium supplemented with 1% FBS and 50 ng/mL Activin and harvested on day 10. For neural induction experiments, hESCs were seeded and transfected at reduced FGF2 and CM levels (5 ng/mL FGF2; MEF-conditioned medium was 50% diluted by regular FGF2-free hESC medium). Neural induction was carried out in KSR-based medium: knockout DMEM medium (Invitrogen), 15% KSR (Invitrogen) with 2 mM L-glutamine, 10 μM β-mercaptoethanol, and nonessential amino acids. Cells were induced in (1) KSR medium supplemented with SB-431542 (10 mM), (2) KSR medium supplemented with Noggin (250 ng/mL), (3) KSR medium supplemented with both SB-431542 and Noggin, or (4) KSR medium alone. Cells were harvested at day 7 of differentiation.
Clonogenic assay
Twenty-four hours post-transfection, hESCs were dissociated with accutase and replated on MEF-coated six-well plates at a density of 50,000 cells per well. After 7 d in culture in hES medium, cells were fixed in 4% paraformaldehyde and stained with anti-OCT4 antibody (Santa Cruz Biotechnologies), and the number of OCT4-positive colonies was counted.
Flow cytometry
For the microRNA profiling study, hESCs were dissociated into single cells using accutase followed by labeling with anti-SSEA-4 antibody (Developmental Studies Hybridoma Bank). MPCs were dissociated with 0.05% Trypsin and labeled with anti-CD73 antibody (BD Pharmingen). R-NSCs were dissociated with HBSS (Invitrogen) and labeled with anti-N-cadherin antibody (Santa Cruz Biotechnologies). All cell sorting studies were performed on a Moflo flow cytometer (Dako). For FACS analysis in miR-302/367 perturbation experiments, 36–48 h post-transfection cells were dissociated with accutase and stained with Alexa Fluor 488-conjugated anti-SSEA-3 antibody (BD Pharmingen). 7-AAD was used for cell death detection. For cell cycle analysis, dissociated cells were incubated with RNase A and propidium iodide (PI). For intracellular PAX6 analysis, cells were dissociated with accutase on day 11 of differentiation, fixed with paraformaldehyde, permeabilized, and stained with Alexa Fluor 488-conjugated anti-PAX6 antibody (BD Pharmingen) using Cytofix/Cytoperm Fixation/Permeabilization kit (BD Pharmingen). Analysis was performed using FACSCalibur (Becton Dickinson) and Flow Jo software (Tree Star, Inc.).
qRT–PCR analysis and luciferase reporter assays
RNA was extracted using Trizol (Invitrogen), treated for DNA contamination, and reverse-transcribed using Quantitect RT kit (Qiagen). The mRNA levels were assayed using the Quantitect primer assays and SYBR Green PCR kit (Qiagen) on a MasterCycler RealPlex2 (Eppendorf). All results were normalized to a GAPDH control.
For initial miR-302 perturbation assessment, hESCs were first transfected with psi-check2 plasmid (Promega) containing four tandem miR-302a target sites in the 3′ UTR. For BMP reporter assays, Tlx2-lux (Addgene 17567) firefly and pRL-TK Renilla control plasmid (Promega) were transfected at a 10:1 ratio. After 2–3 h, DNA plasmid transfections were followed by transfection with 50–100 nM antagomirs, 60–120 nM mimics, or corresponding controls. Lysates were harvested 24 h (for psi-check2) or 36 h (for Tlx2-lux) after transfection, and reporter activity was measured using the Dual-Luciferase assay (Promega).
3′ UTR luciferase reporter assays
The 3′ UTRs of DAZAP2, SLAIN1, and TOB2 were PCR-amplified from H9 hESC cDNA, cloned into pMiR-Report (Ambion) downstream from the firefly luciferase gene, and verified by sequencing. For DAZAP2 and SLAIN1, full-length 3′ UTRs were cloned, and for TOB2, the first 1.4 kb (which contained the target site) out of a total 2.8 kb was cloned. Site-directed mutagenesis was carried out using QuikChange Lightning Site-Directed Mutagenesis kit (Stratagene). miR-302 target sites in DAZAP2 were mutated both individually and together. pRL-TK plasmid was used as a control. HEK293T cells were transfected with 120 nM mimics and 1 ng of pMIR-Report and pRL-TK plasmids per 8 × 104 cells. hESC were transfected with 170 nM antagomirs and 6 ng of pMIR-Report and pRL-TK plasmids per 8 × 104 cells. Cell lysates were harvested 24 h after transfection, and reporter activity was measured using the dual-luciferase assay.
siRNA assays
Matrigel-plated hESCs were transfected with a cocktail of three Silencer Select siRNAs (Ambion) or siRNA-negative controls at 200 nM total concentration using Lipofectamine RNAiMAX (Invitrogen). Alternatively, siRNA cocktails for three genes were transfected simultaneously at a total concentration of 600 nM matched by negative control at 600 nM. For ID1 expression analysis and knockdown efficiency determination, cells were harvested 48 h after transfection. For neural induction experiments, differentiation was carried out in the same manner as for antagomir transfection assays.
Computational pathway and functional analysis
Pathway analysis was performed using the Netbox algorithm, which performs a module analysis on a human interaction network (Cerami et al. 2010). Using the set of 146 high-confidence miR-302/367 targets, the algorithm identified regions in the network that are enriched for those genes. Additional analysis was performed using GeneMania (Warde-Farley et al. 2010). For miR-302/367 perturbation experiments, statistical analysis was performed using GraphPad Prism version 5.0b (GraphPad Software). Significance of differences was examined using two-tailed Students t-test. Data are presented as mean ± SEM against control antagomir or control mimic.
Acknowledgments
We thank A. Viale (MSKCC Genomics Core Laboratory), J. Hendrikx (MSKCC Flow Cytometry Core Laboratory), and S. Dewell (Genomics Resource Center, Rockefeller University). M.H. is supported by a fellowship of the Charles Revson, Jr., Foundation. T.T. is an HHMI investigator, and work in his laboratory was supported by NIH grants GM073047 and MH08442. This work was supported by grants from the Starr Foundation (Tri-Institutional Stem Cell Initiative).
Footnotes
Supplemental material is available for this article.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.17221311.
References
- Anokye-Danso F, Trivedi CM, Juhr D, Gupta M, Cui Z, Tian Y, Zhang Y, Yang W, Gruber PJ, Epstein JA, et al. 2011. Highly efficient miRNA-mediated reprogramming of mouse and human somatic cells to pluripotency. Cell Stem Cell 8: 376–388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong L, Hughes O, Yung S, Hyslop L, Stewart R, Wappler I, Peters H, Walter T, Stojkovic P, Evans J, et al. 2006. The role of PI3K/AKT, MAPK/ERK and NFκβ signalling in the maintenance of human embryonic stem cell pluripotency and viability highlighted by transcriptional profiling and functional analysis. Hum Mol Genet 15: 1894–1913 [DOI] [PubMed] [Google Scholar]
- Bar M, Wyman SK, Fritz BR, Qi J, Garg KS, Parkin RK, Kroh EM, Bendoraite A, Mitchell PS, Nelson AM, et al. 2008. MicroRNA discovery and profiling in human embryonic stem cells by deep sequencing of small RNA libraries. Stem Cells 26: 2496–2505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barberi T, Bradbury M, Dincer Z, Panagiotakos G, Socci ND, Studer L 2007. Derivation of engraftable skeletal myoblasts from human embryonic stem cells. Nat Med 13: 642–648 [DOI] [PubMed] [Google Scholar]
- Berasi SP, Xiu M, Yee AS, Paulson KE 2004. HBP1 repression of the p47phox gene: cell cycle regulation via the NADPH oxidase. Mol Cell Biol 24: 3011–3024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein E, Kim SY, Carmell MA, Murchison EP, Alcorn H, Li MZ, Mills AA, Elledge SJ, Anderson KV, Hannon GJ, et al. 2003. Dicer is essential for mouse development. Nat Genet 35: 215–217 [DOI] [PubMed] [Google Scholar]
- Berthet C, Guéhenneux F, Revol V, Samarut C, Lukaszewicz A, Dehay C, Dumontet C, Magaud J-P, Rouault J-P 2002. Interaction of PRMT1 with BTG/TOB proteins in cell signalling: molecular analysis and functional aspects. Genes Cells 7: 29–39 [DOI] [PubMed] [Google Scholar]
- Betel D, Koppal A, Agius P, Sander C, Leslie C 2010. Comprehensive modeling of microRNA targets predicts functional non-conserved and non-canonical sites. Genome Biol 11: R90 doi: 10.1186/gb-2010-11-8-r90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busson M, Carazo A, Seyer P, Grandemange S, Casas F, Pessemesse L, Rouault J-P, Wrutniak-Cabello C, Cabello G 2005. Coactivation of nuclear receptors and myogenic factors induces the major BTG1 influence on muscle differentiation. Oncogene 24: 1698–1710 [DOI] [PubMed] [Google Scholar]
- Cerami E, Demir E, Schultz N, Taylor BS, Sander C 2010. Automated network analysis identifies core pathways in glioblastoma. PLoS ONE 5: e8918 doi: 10.1371/journal.pone.0008918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers SM, Fasano CA, Papapetrou EP, Tomishima M, Sadelain M, Studer L 2009. Highly efficient neural conversion of human ES and IPS cells by dual inhibition of SMAD signaling. Nat Biotechnol 27: 275–280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G, Ye Z, Yu X, Zou J, Mali P, Brodsky RA, Cheng L 2008. Trophoblast differentiation defect in human embryonic stem cells lacking PIG-A and GPI-anchored cell-surface proteins. Cell Stem Cell 2: 345–355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi SW, Zang JB, Mele A, Darnell RB 2009. Argonaute hits-clip decodes microRNA-mRNA interaction maps. Nature 460: 479–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Amour KA, Agulnick AD, Eliazer S, Kelly OG, Kroon E, Baetge EE 2005. Efficient differentiation of human embryonic stem cells to definitive endoderm. Nat Biotechnol 23: 1534–1541 [DOI] [PubMed] [Google Scholar]
- Elkabetz Y, Studer L 2008. Human ESC-derived neural rosettes and neural stem cell progression. Cold Spring Harb Symp Quant Biol 73: 377–387 [DOI] [PubMed] [Google Scholar]
- Elkabetz Y, Panagiotakos G, Al Shamy G, Socci ND, Tabar V, Studer L 2008. Human ES cell-derived neural rosettes reveal a functionally distinct early neural stem cell stage. Genes Dev 22: 152–165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fluckiger A-C, Marcy G, Marchand M, Négre D, Cosset F-L, Mitalipov S, Wolf D, Savatier P, Dehay C 2006. Cell cycle features of primate embryonic stem cells. Stem Cells 24: 547–556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franken NAP, Rodermond HM, Stap J, Haveman J, van Bree C 2006. Clonogenic assay of cells in vitro. Nat Protoc 1: 2315–2319 [DOI] [PubMed] [Google Scholar]
- Gangaraju VK, Lin H 2009. MicroRNAs: key regulators of stem cells. Nat Rev Mol Cell Biol 10: 116–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafner M, Landgraf P, Ludwig J, Rice A, Ojo T, Lin C, Holoch D, Lim C, Tuschl T 2008. Identification of microRNAs and other small regulatory RNAs using CDNA library sequencing. Methods 44: 3–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafner M, Landthaler M, Burger L, Khorshid M, Hausser J, Berninger P, Rothballer A, Ascano M Jr, Jungkamp A-C, Munschauer M, et al. 2010. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell 141: 129–141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hata A, Lagna G, Massagué J, Hemmati-Brivanlou A 1998. Smad6 inhibits BMP/smad1 signaling by specifically competing with the smad4 tumor suppressor. Genes Dev 12: 186–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson JK, Draper JS, Baillie HS, Fishel S, Thomson JA, Moore H, Andrews PW 2002. Preimplantation human embryos and embryonic stem cells show comparable expression of stage-specific embryonic antigens. Stem Cells 20: 329–337 [DOI] [PubMed] [Google Scholar]
- Hirst CE, Lim S-M, Pereira LA, Mayberry RA, Stanley EG, Elefanty AG 2010. Expression from a βgeo gene trap in the Slain1 gene locus is predominantly associated with the developing nervous system. Int J Dev Biol 54: 1383–1388 [DOI] [PubMed] [Google Scholar]
- Houbaviy HB, Murray MF, Sharp PA 2003. Embryonic stem cell-specific microRNAs. Dev Cell 5: 351–358 [DOI] [PubMed] [Google Scholar]
- International Stem Cell Initiative 2007. Characterization of human embryonic stem cell lines by the International Stem Cell Initiative. Nat Biotechnol 25: 803–816 [DOI] [PubMed] [Google Scholar]
- John B, Enright AJ, Aravin A, Tuschl T, Sander C, Marks DS 2004. Human microRNA targets. PLoS Biol 2: e363 doi: 10.1371/journal.pbio.0020363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanellopoulou C, Muljo SA, Kung AL, Ganesan S, Drapkin R, Jenuwein T, Livingston DM, Rajewsky K 2005. Dicer-deficient mouse embryonic stem cells are defective in differentiation and centromeric silencing. Genes Dev 19: 489–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawate H, Wu Y, Ohnaka K, Nawata H, Takayanagi R 2005. Tob proteins suppress steroid hormone receptor-mediated transcriptional activation. Mol Cell Endocrinol 230: 77–86 [DOI] [PubMed] [Google Scholar]
- Kimelman D 2006. Mesoderm induction: from caps to chips. Nat Rev Genet 7: 360–372 [DOI] [PubMed] [Google Scholar]
- Korchynskyi O, ten Dijke P 2002. Identification and functional characterization of distinct critically important bone morphogenetic protein-specific response elements in the id1 promoter. J Biol Chem 277: 4883–4891 [DOI] [PubMed] [Google Scholar]
- Krützfeldt J, Kuwajima S, Braich R, Rajeev KG, Pena J, Tuschl T, Manoharan M, Stoffel M 2007. Specificity, duplex degradation and subcellular localization of antagomirs. Nucleic Acids Res 35: 2885–2892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuijk EW, van Mil A, Brinkhof B, Penning LC, Colenbrander B, Roelen BAJ 2010. PTEN and TRP53 independently suppress nanog expression in spermatogonial stem cells. Stem Cells Dev 19: 979–988 [DOI] [PubMed] [Google Scholar]
- Lee NS, Kim JS, Cho WJ, Lee MR, Steiner R, Gompers A, Ling D, Zhang J, Strom P, Behlke M, et al. 2008. mir-302b maintains ‘stemness’ of human embryonal carcinoma cells by post-transcriptional regulation of cyclin D2 expression. Biochem Biophys Res Commun 377: 434–440 [DOI] [PubMed] [Google Scholar]
- Leung AKL, Young AG, Bhutkar A, Zheng GX, Bosson AD, Nielsen CB, Sharp PA 2011. Genome-wide identification of Ago2 binding sites from mouse embryonic stem cells with and without mature microRNAs. Nat Struct Mol Biol 18: 237–244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin S-L, Chang DC, Lin C-H, Ying S-Y, Leu D, Wu DTS 2011. Regulation of somatic cell reprogramming through inducible mir-302 expression. Nucleic Acids Res 39: 1054–1065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mamillapalli R, Gavrilova N, Mihaylova VT, Tsvetkov LM, Wu H, Zhang H, Sun H 2001. PTEN regulates the ubiquitin-dependent degradation of the CDK inhibitor p27(KIP1) through the ubiquitin E3 ligase SCF(SKP2). Curr Biol 11: 263–267 [DOI] [PubMed] [Google Scholar]
- Melton C, Judson RL, Blelloch R 2010. Opposing microRNA families regulate self-renewal in mouse embryonic stem cells. Nature 463: 621–626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyai K, Yoneda M, Hasegawa U, Toita S, Izu Y, Hemmi H, Hayata T, Ezura Y, Mizutani S, Miyazono K, et al. 2009. ANA deficiency enhances bone morphogenetic protein-induced ectopic bone formation via transcriptional events. J Biol Chem 284: 10593–10600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyoshi N, Ishii H, Nagano H, Haraguchi N, Dewi DL, Kano Y, Nishikawa S, Tanemura M, Mimori K, Tanaka F, et al. 2011. Reprogramming of mouse and human cells to pluripotency using mature microRNAs. Cell Stem Cell 8: 633–638 [DOI] [PubMed] [Google Scholar]
- Morin RD, O'Connor MD, Griffith M, Kuchenbauer F, Delaney A, Prabhu A-L, Zhao Y, McDonald H, Zeng T, Hirst M, et al. 2008. Application of massively parallel sequencing to microRNA profiling and discovery in human embryonic stem cells. Genome Res 18: 610–621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muñoz-Sanjuán I, Brivanlou AH 2002. Neural induction, the default model and embryonic stem cells. Nat Rev Neurosci 3: 271–280 [DOI] [PubMed] [Google Scholar]
- Neganova I, Zhang X, Atkinson S, Lako M 2009. Expression and functional analysis of G1 to S regulatory components reveals an important role for CDK2 in cell cycle regulation in human embryonic stem cells. Oncogene 28: 20–30 [DOI] [PubMed] [Google Scholar]
- Passeri D, Marcucci A, Rizzo G, Billi M, Panigada M, Leonardi L, Tirone F, Grignani F 2006. Btg2 enhances retinoic acid-induced differentiation by modulating histone H4 methylation and acetylation. Mol Cell Biol 26: 5023–5032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perrier AL, Tabar V, Barberi T, Rubio ME, Bruses J, Topf N, Harrison NL, Studer L 2004. Derivation of midbrain dopamine neurons from human embryonic stem cells. Proc Natl Acad Sci 101: 12543–12548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pevny LH, Sockanathan S, Placzek M, Lovell-Badge R 1998. A role for SOX1 in neural determination. Development 125: 1967–1978 [DOI] [PubMed] [Google Scholar]
- Poliseno L, Salmena L, Riccardi L, Fornari A, Song MS, Hobbs RM, Sportoletti P, Varmeh S, Egia A, Fedele G, et al. 2010. Identification of the mir-106b 25 microRNA cluster as a proto-oncogenic PTEN-targeting intron that cooperates with its host gene MCM7 in transformation. Sci Signal 3: ra29 doi: 10.1126/scisignal.2000594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prévôt D, Voeltzel T, Birot AM, Morel AP, Rostan MC, Magaud JP, Corbo L 2000. The leukemia-associated protein Btg1 and the p53-regulated protein Btg2 interact with the homeoprotein Hoxb9 and enhance its transcriptional activation. J Biol Chem 275: 147–153 [DOI] [PubMed] [Google Scholar]
- Rajasethupathy P, Fiumara F, Sheridan R, Betel D, Puthanveettil SV, Russo JJ, Sander C, Tuschl T, Kandel E 2009. Characterization of small RNAs in Aplysia reveals a role for miR-124 in constraining synaptic plasticity through CREB. Neuron 63: 803–817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramaswamy S, Nakamura N, Vazquez F, Batt DB, Perera S, Roberts TM, Sellers WR 1999. Regulation of G1 progression by the PTEN tumor suppressor protein is linked to inhibition of the phosphatidylinositol 3-kinase/AKT pathway. Proc Natl Acad Sci 96: 2110–2115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roche DD, Liu KJ, Harland RM, Monsoro-Burq AH 2009. Dazap2 is required for FGF-mediated posterior neural patterning, independent of wnt and cdx function. Dev Biol 333: 26–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosa A, Spagnoli FM, Brivanlou AH 2009. The mir-430/427/302 family controls mesendodermal fate specification via species-specific target selection. Dev Cell 16: 517–527 [DOI] [PubMed] [Google Scholar]
- Samavarchi-Tehrani P, Golipour A, David L, Sung H-K, Beyer TA, Datti A, Woltjen K, Nagy A, Wrana JL 2010. Functional genomics reveals a BMP-driven mesenchymal-to-epithelial transition in the initiation of somatic cell reprogramming. Cell Stem Cell 7: 64–77 [DOI] [PubMed] [Google Scholar]
- Sapkota G, Alarcón C, Spagnoli FM, Brivanlou AH, Massagué J 2007. Balancing BMP signaling through integrated inputs into the Smad1 linker. Mol Cell 25: 441–454 [DOI] [PubMed] [Google Scholar]
- Schulz LC, Ezashi T, Das P, Westfall SD, Livingston KA, Roberts RM 2008. Human embryonic stem cells as models for trophoblast differentiation. Placenta 29: S10–S16 doi: 10.1016/j.placenta.2007.10.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith B, Treadwell J, Zhang D, Ly D, McKinnell I, Walker PR, Sikorska M 2010. Large-scale expression analysis reveals distinct microRNA profiles at different stages of human neurodevelopment. PLoS ONE 5: e11109 doi: 10.1371/journal.pone.0011109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souchelnytskyi S, Nakayama T, Nakao A, Morén A, Heldin CH, Christian JL, ten Dijke P 1998. Physical and functional interaction of murine and Xenopus Smad7 with bone morphogenetic protein receptors and transforming growth factor-β receptors. J Biol Chem 273: 25364–25370 [DOI] [PubMed] [Google Scholar]
- Stewart MH, Bossé M, Chadwick K, Menendez P, Bendall SC, Bhatia M 2006. Clonal isolation of hESCs reveals heterogeneity within the pluripotent stem cell compartment. Nat Methods 3: 807–815 [DOI] [PubMed] [Google Scholar]
- Subramanyam D, Lamouille S, Judson RL, Liu JY, Bucay N, Derynck R, Blelloch R 2011. Multiple targets of mir-302 and mir-372 promote reprogramming of human fibroblasts to induced pluripotent stem cells. Nat Biotechnol 29: 443–448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh M-R, Lee Y, Kim JY, Kim S-K, Moon S-H, Lee JY, Cha K-Y, Chung HM, Yoon HS, Moon SY, et al. 2004. Human embryonic stem cells express a unique set of microRNAs. Dev Biol 270: 488–498 [DOI] [PubMed] [Google Scholar]
- Tang SJ, Hoodless PA, Lu Z, Breitman ML, McInnes RR, Wrana JL, Buchwald M 1998. The Tlx-2 homeobox gene is a downstream target of BMP signalling and is required for mouse mesoderm development. Development 125: 1877–1887 [DOI] [PubMed] [Google Scholar]
- Tay Y, Zhang J, Thomson AM, Lim B, Rigoutsos I 2008. MicroRNAs to Nanog, Oct4 and Sox2 coding regions modulate embryonic stem cell differentiation. Nature 455: 1124–1128 [DOI] [PubMed] [Google Scholar]
- Wang Y, Medvid R, Melton C, Jaenisch R, Blelloch R 2007. DGCR8 is essential for microRNA biogenesis and silencing of embryonic stem cell self-renewal. Nat Genet 39: 380–385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Baskerville S, Shenoy A, Babiarz JE, Baehner L, Blelloch R 2008. Embryonic stem cell-specific microRNAs regulate the G1–S transition and promote rapid proliferation. Nat Genet 40: 1478–1483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warde-Farley D, Donaldson SL, Comes O, Zuberi K, Badrawi R, Chao P, Franz M, Grouios C, Kazi F, Lopes CT, et al. 2010. The genemania prediction server: biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res 38: W214–W220 doi: 10.1093/nar/gkq537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler GS 2010. The mammalian anti-proliferative BTG/Tob protein family. J Cell Physiol 222: 66–72 [DOI] [PubMed] [Google Scholar]
- Winnier G, Blessing M, Labosky PA, Hogan BL 1995. Bone morphogenetic protein-4 is required for mesoderm formation and patterning in the mouse. Genes Dev 9: 2105–2116 [DOI] [PubMed] [Google Scholar]
- Wu Z, Zhang W, Chen G, Cheng L, Liao J, Jia N, Gao Y, Dai H, Yuan J, Cheng L, et al. 2008. Combinatorial signals of activin/nodal and bone morphogenic protein regulate the early lineage segregation of human embryonic stem cells. J Biol Chem 283: 24991–25002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C, Inokuma MS, Denham J, Golds K, Kundu P, Gold JD, Carpenter MK 2001. Feeder-free growth of undifferentiated human embryonic stem cells. Nat Biotechnol 19: 971–974 [DOI] [PubMed] [Google Scholar]
- Xu R-H, Chen X, Li DS, Li R, Addicks GC, Glennon C, Zwaka TP, Thomson JA 2002. BMP4 initiates human embryonic stem cell differentiation to trophoblast. Nat Biotechnol 20: 1261–1264 [DOI] [PubMed] [Google Scholar]
- Xu R-H, Sampsell-Barron TL, Gu F, Root S, Peck RM, Pan G, Yu J, Antosiewicz-Bourget J, Tian S, Stewart R, et al. 2008. NANOG is a direct target of TGFβ/activin-mediated SMAD signaling in human ESCs. Cell Stem Cell 3: 196–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu N, Papagiannakopoulos T, Pan G, Thomson JA, Kosik KS 2009. MicroRNA-145 regulates Oct4, Sox2, and KLF4 and represses pluripotency in human embryonic stem cells. Cell 137: 647–658 [DOI] [PubMed] [Google Scholar]
- Yamashita S, Miyagi C, Fukada T, Kagara N, Che Y-S, Hirano T 2004. Zinc transporter LIVI controls epithelial–mesenchymal transition in zebrafish gastrula organizer. Nature 429: 298–302 [DOI] [PubMed] [Google Scholar]
- Yoshida Y, von Bubnoff A, Ikematsu N, Blitz IL, Tsuzuku JK, Yoshida EH, Umemori H, Miyazono K, Yamamoto T, Cho KWY, et al. 2003. Tob proteins enhance inhibitory SMAD-receptor interactions to repress BMP signaling. Mech Dev 120: 629–637 [DOI] [PubMed] [Google Scholar]
- Zhang P, Li J, Tan Z, Wang C, Liu T, Chen L, Yong J, Jiang W, Sun X, Du L, et al. 2008. Short-term BMP-4 treatment initiates mesoderm induction in human embryonic stem cells. Blood 111: 1933–1941 [DOI] [PubMed] [Google Scholar]
- Zhang X, Huang CT, Chen J, Pankratz MT, Xi J, Li J, Yang Y, Lavaute TM, Li X-J, Ayala M, et al. 2010. Pax6 is a human neuroectoderm cell fate determinant. Cell Stem Cell 7: 90–100 [DOI] [PMC free article] [PubMed] [Google Scholar]