Carey and colleagues investigate the specificity of the chromatin remodeler CHD1 for H3K4me3-marked active genes. Mediator is found to be a key factor in CHD1 recruitment, with functional interactions between Mediator and CHD1 providing specificity for the latter's recruitment to active genes.
Keywords: Mediator, preinitiation complex, CHD1, H3K4me3
Abstract
Murine Chd1 (chromodomain helicase DNA-binding protein 1), a chromodomain-containing chromatin remodeling protein, is necessary for embryonic stem (ES) cell pluripotency. Chd1 binds to nucleosomes trimethylated at histone 3 Lys 4 (H3K4me3) near the beginning of active genes but not to bivalent domains also containing H3K27me3. To address the mechanism of this specificity, we reproduced H3K4me3- and CHD1-stimulated gene activation in HeLa extracts. Multidimensional protein identification technology (MuDPIT) and immunoblot analyses of purified preinitiation complexes (PICs) revealed the recruitment of CHD1 to naive chromatin but enhancement on H3K4me3 chromatin. Studies in depleted extracts showed that the Mediator coactivator complex, which controls PIC assembly, is also necessary for CHD1 recruitment. MuDPIT analyses of CHD1-associated proteins support the recruitment data and reveal numerous components of the PIC, including Mediator. In vivo, CHD1 and Mediator are recruited to an inducible gene, and genome-wide binding of the two proteins correlates well with active gene transcription in mouse ES cells. Finally, coimmunoprecipitation of CHD1 and Mediator from cell extracts can be ablated by shRNA knockdown of a specific Mediator subunit. Our data support a model in which the Mediator coordinates PIC assembly along with the recruitment of CHD1. The combined action of the PIC and H3K4me3 provides specificity in targeting CHD1 to active genes.
Histone H3 Lys 4 trimethylation (H3K4me3) near the beginning of genes correlates with active transcription. However, although most active genes contain this modification, not all H3K4me3-bearing genes are actively transcribed (Kim et al. 2005; Ruthenburg et al. 2007). H3K4me3 is also found in bivalent domains alongside H3K27me3, which correlates with gene silencing. Bivalent domains are found on promoters of important developmental regulator genes that are hypothesized to be “primed” but not yet active or fully silenced (Bernstein et al. 2006). The mechanisms by which the context-specific effects of H3K4me3 are achieved have not been fully explored.
The coordination of transcription with histone modifications allows RNA polymerase II (Pol II) to overcome nucleosomal barriers presented by chromatin (Li et al. 2007). Specific histone modifications recruit distinct effector proteins that alter the chromatin landscape of active genes, making them permissive for transcription. It is largely unknown how the binding and function of this chromatin machinery are coordinated with assembly of the preinitiation complex (PIC). The PIC comprises coactivators like the 30-subunit Mediator complex and the 14-subunit TFIID complex, along with Pol II and the general transcription factors (GTFs) TFIIA, TFIIB, TFIID, TFIIE, TFIIF, and TFIIH; TFIID is both a coactivator and a GTF (Kornberg 2005).
Mechanistic studies have established that activators contact and recruit TFIID and Mediator, while the GTFs and Pol II associate with these coactivators to complete PIC assembly (Roeder 1998; Johnson and Carey 2003). The GTFs bound at the promoter position Pol II at the start site, melt the DNA, and facilitate the catalytic steps of transcription initiation (Kornberg 2007). Cross-linking studies have shown that many of the GTFs contact promoter DNA (Lagrange et al. 1996). Moreover, Pol II, TFIIH, and the TBP-associated factor (TAFII) subunits of TFIID all bind both upstream of and downstream from the start site (Martinez 2002). Additionally, proteins such as the PAF1 complex and P-TEFb are recruited at or near the start site to facilitate Pol II elongation. Indeed, most genes contain a well-characterized NELF- and DSIF-mediated Pol II pause site located 30 base pairs (bp) downstream from the start site that must be overcome by the action of P-TEFb, possibly in the context of the super elongation complex (SEC) (Peterlin and Price 2006; Smith et al. 2011). Most human genes also contain nucleosomes positioned near the start site (Schones et al. 2008). The close proximity of the PIC and Pol II elongation machinery with nucleosomes emphasizes the need to understand how their functions are coordinated. Some chromatin factors—like the p300 histone acetyltransferase, which stimulates Pol II elongation, and the PAF1 complex, which controls H3K4me3—are known to be recruited to a functional PIC (Black et al. 2006; Wu et al. 2008; Kim et al. 2009). However, it is unknown whether the PIC or its associated factors control the recruitment of effector proteins that bind to H3K4me3.
The SET1 complex is responsible for the majority of H3K4me3 in yeast and mammalian cells (Ruthenburg et al. 2007; Wu et al. 2008). In budding yeast, recruitment of the SET1 complex (termed COMPASS) by PAF1, in combination with ubiquitinylation of H2B by Rad6-Bre1, leads to H3K4me3 (Wood et al. 2003; Kim et al. 2009). However, in mammals and other organisms, H3K4me3 is found at some gene promoters that have not yet generated productive transcript (Bernstein et al. 2006). Studies have shown that several DNA sequence-specific transcription factors associate directly with ASH2L, a core subunit of SET1 family complexes (Tan et al. 2008; Stoller et al. 2010). It is conceivable that SET1 might be recruited to a gene by such factors under conditions in which the gene is not actively transcribed.
The majority of H3K4me3-binding domains can be found in effector proteins associated with chromatin modification and remodeling (Ruthenburg et al. 2007). Chd1 (chromodomain helicase DNA-binding protein 1) is an H3K4me3-associated chromatin remodeler that is required for the expression of developmentally essential genes in mouse embryonic stem (ES) cells (Gaspar-Maia et al. 2009). However, although H3K4me3 is present across a large subset of genes, occupancy of mouse Chd1 correlates primarily with active gene promoters that display enrichment of Pol II. Human CHD1 has been reported to deposit the histone variant H3.3 in vivo (Konev et al. 2007). The CHD1 complex has been shown to associate with the PAF1 elongation complex in Saccharomyces cerevisiae (Warner et al. 2007). However, in S. cerevisiae, while CHD1 is capable of remodeling promoter nucleosomes, it does not exhibit preferential binding to H3K4me3 chromatin (Ehrensberger and Kornberg 2011). Thus, in higher eukaryotes, CHD1 is a model H3K4me3 effector used in developmental decisions and elsewhere; its enhancement of gene expression is linked to both transcription and H3K4me3. Since in vitro studies have demonstrated that the CHD1 double chromodomains bind H3K4me3 nucleosomes and tail peptides with seemingly high affinity (Sims et al. 2005; Bartke et al. 2010; Vermeulen et al. 2010), an important question is why Chd1 is found only at actively transcribed H3K4me3 genes and not at bivalent domains.
To investigate the biochemical mechanisms by which H3K4me3 effectors function in a transcription- and chromatin-specific context, we used multidimensional protein identification technology (MuDPIT) proteomics in conjunction with immunoblotting to identify factors found to be enriched in the context of a PIC assembled on chromatin in vitro. Proteomic techniques have been previously used to successfully analyze PICs on nonchromatin templates in yeast (Ranish et al. 2003). Surprisingly, we found that binding of CHD1 was stimulated by the activator on unmodified chromatin templates but was enhanced on methylated templates. CHD1 was observed to also participate in higher-order protein complex interactions with components of the transcription machinery located at or downstream from the start site and was a prime candidate for further mechanistic investigation. Using the immobilized chromatin template assay, we combined PIC-stimulated recruitment with functional transcription assays. Our studies show that the specific recruitment of CHD1 to an active gene is achieved by linking it to assembly of an active transcription complex via the action of Mediator.
Results
H3K4me3 stimulates transcription in vitro
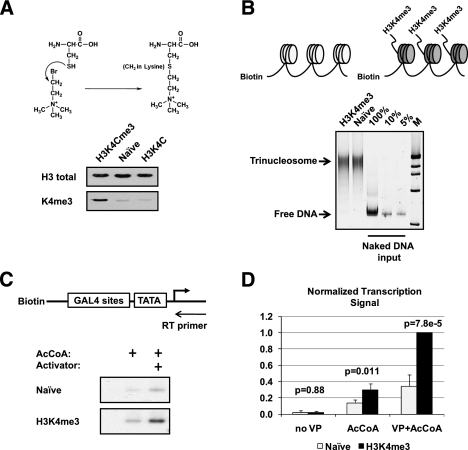
To study the mechanism by which H3K4me3 contributes to active gene transcription, we used the methods developed by Shokat and colleagues (Simon et al. 2007) to synthetically methylate chromatin templates for biochemical analysis. In this method, Lys 4 of histone H3 was mutated to a cysteine to generate H3K4C (Fig. 1A). Recombinant H3K4C was reacted with an ethylamine substrate [(2-bromoethyl) trimethylammonium bromide] to generate the methyl-lysine analog (MLA) of H3K4me3. The MLA was validated by immunoblotting with an antibody to H3K4me3 (Fig. 1A) and quantitated by electrospray ionization mass spectrometry, which revealed that 66% of H3 was modified (data not shown). As shown in the Figure 1B schematic, unmodified naive or synthetically methylated H3 was assembled into octamers and then into chromatin on biotinylated DNA templates containing a GAL4-responsive promoter (Black et al. 2006). The amounts of chromatin template were first normalized based on the extent of chromatin assembly as shown in Figure 1B. Subsequently, in vitro transcription was performed in HeLa nuclear extracts on equivalent amounts of chromatin. The data revealed stimulation by the activator GAL4-VP16 on the H3K4me3 versus the naive unmodified templates (Fig. 1C). Multiple repeats were quantitated, graphed, and subjected to statistical analysis to reveal that the stimulation was indeed reproducible and significant (Fig. 1D). This result demonstrated an ability to recreate, in a cell-free system, the transcription stimulation imparted by H3K4me3 and offered the possibility of generating mechanistic insights into its function. We addressed the molecular basis for this stimulation by first analyzing the composition of the PICs formed via immobilized template assays.
Figure 1.
H3K4me3 stimulates in vitro transcription from nuclear extract. (A) Schematic of the trimethyl-lysine analog synthesis at histone 3 Cys 4. Below the schematic is a Western blot showing the specific detection of H3K4me3 MLA by antibody (Abcam anti-K4me3). (B) Schematic of the immobilized chromatin template and chromatin normalization. The extent of chromatin assembly was monitored by EMSA; equivalent amounts of chromatin were used in all experiments. (C) Schematic and autoradiograph of in vitro transcription as detected by primer extension on unmodified (Naive) and synthetic H3K4me3 chromatin templates in the presence and absence of the activator GAL4-VP16 and acetyl-CoA (AcCoA) using HeLa nuclear extract. (AcCoA was necessary for optimal levels of transcription.) (D) Signal quantitation and statistical analysis of the stimulation by H3K4me3. Three repeats of the transcription experiment were quantitated using Imagequant TL software, graphed, and subjected to a Student's t-test as a measure of statistical significance between naive unmodified and H3K4me3 chromatin.
Activator-mediated recruitment of CHD1
The immobilized template assay is a powerful method for understanding the mechanism of gene activation in vitro because it permits a comparison of transcriptional activity with the composition of the PICs. In a typical experiment, biotinylated chromatin templates are attached to paramagnetic streptavidin-coated beads and incubated either in nuclear extract or with pure proteins. After washing the beads, the proteins captured by the template are eluted and analyzed by immunoblotting or MuDPIT.
Supplemental Figure 1A shows a typical time-course experiment on naive chromatin in which we determined the optimal conditions for chromatin binding of various GTFs, coactivators (Mediator and TAFs), and complexes associated with H3K4me3, including PAF1 and SET1. Immunoblotting for each subunit of every transcription factor is nearly impossible given the large number of polypeptides constituting the PIC. We therefore chose specific subunits to represent various factors within the PIC. The activator recruited the Mediator with the fastest kinetics as previously reported (Black et al. 2006). Importantly, we observed activator-stimulated recruitment of the PAF1 and SET1 complexes, although they bound more slowly than Mediator. The coordinated recruitment of PAF1, SET1, and the PIC was expected based on Shilatifard's previous studies in yeast (Wu et al. 2008; Kim et al. 2009), with the caveat that PAF1 and SET1 bound in the absence of transcription, whereas cotranscriptional phosphorylation of Ser 5 in the Pol II C-terminal domain (CTD) is necessary for PAF1 binding in yeast. Immunoblotting revealed that no detectable Ser 5 phosphorylated Pol II was observed bound to the immobilized template in the absence of ATP (Supplemental Fig. 1B; data not shown). This result was consistent with numerous studies showing that phosphorylated Pol II does not join the PIC (Drapkin et al. 1994).
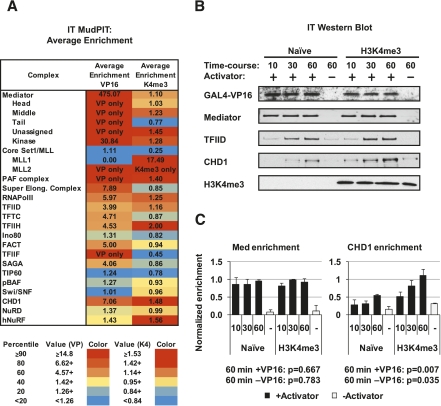
To more thoroughly interrogate and compare the components of PICs formed on naive and H3K4me3 chromatin, we scaled up the immobilized template reactions and performed MuDPIT analysis. We compared two primary criteria in our analysis—the proteins whose binding appeared to be stimulated by activator, and those that were further enriched by H3K4 methylation. Our goal was to identify H3K4me3-enriched factors that interact with the PIC and participate in transcription initiation on chromatin. PIC-related complexes detected in the MudPIT data are summarized as a table in Figure 2A. (The annotated data set with accompanying graphs representing peptide spectra focused on transcription/chromatin proteins are shown in Supplemental Table 1 and Supplemental Fig. 2A–D.) Relative enrichment of specific PIC components was calculated by average NSAF (normalized spectral abundance factor) for all subunits of a complex and ranked by color. A gradient of red to yellow to light blue represents proteins ranked in the 90th to 20th percentiles, while values shown in solid blue represent proteins ranking below the 20th percentile. The most enriched factor was Mediator. The Mediator is a direct target of VP16 (Uhlmann et al. 2007), and its recruitment by activator represented a validation of the overall approach (Fig. 2A). Indeed, for most of the Mediator subunits, the MuDPIT data indicated that no binding occurred in the absence of activator. Among other transcription factors, the number of peptides decreased substantially, but Pol II, TFIID, and TFIIH were readily detected. Surprisingly, among the known or suspected H3K4me3 effector proteins, CHD1 binding was strongly stimulated by activator on naive templates. CHD1 was further enriched on methylated templates in the absence of activator. In addition, we observed activator-stimulated enrichment of the PAF complex. The PAF1 complex is necessary for recruitment of the SET1 complex COMPASS in yeast (Krogan et al. 2003; Wood et al. 2003). CHD1 has been reported to associate with PAF1 in HeLa extracts (Sims et al. 2007; Warner et al. 2007).
Figure 2.
H3K4me3 effectors are recruited to the PIC. (A) MuDPIT analyses were performed on PICs assembled on chromatin arrays: a chart of PIC components, H3K4me3 effector proteins, and other factors recruited. Average enrichment in the presence of GAL4-VP16 and further enrichment on H3K4me3 chromatin are shown as a heat map from average NSAFe5 values for the factors in the complex. Average NSAFe5 values for each complex are ranked by percentile in a three-color gradient from high (red) to medium (yellow) to low (blue) as shown. (B) Time courses of PIC recruitment in HeLa extracts were compared between Naive and H3K4me3 templates by immunoblotting to validate enrichment of select factors as determined by the MuDPIT screen. (C) Quantitation and statistical analysis of Western blot signals for CHD1 and Mediator enriched on H3K4me3 versus Naive chromatin. Signals were normalized to VP16. Student's t-test was used to calculate the statistical significance of the differences at the 60-min time point. Assays were performed in triplicate.
Despite the fact that our approach detected activator-stimulated recruitment of proteins like Mediator, the spectral counting-based quantitation in MuDPIT is not ideal for detecting small differences in abundance as compared with other mass spectrometry techniques like SILAC (Bartke et al. 2010). We therefore used immunoblotting to validate candidates chosen for further mechanistic analysis. Figure 2B shows a recruitment time course of CHD1 versus the Mediator and TFIID on both naive and H3K4me3 templates. Quantitation of the blots from multiple experiments revealed that the amount of Mediator remained roughly constant between naive versus H3K4me3 chromatin (Fig. 2C). In contrast, the activator stimulated a significant twofold to threefold increase with CHD1 on the H3K4me3 chromatin, with the effect being most evident at the 60-min time point. Moreover, CHD1 bound with a slightly higher affinity to the H3K4me3 versus naive templates in the absence of activator, although not to the extent observed in the MuDPIT analysis (Fig. 2B,C). The data suggest that the activator and H3K4me3 both contribute to the recruitment of CHD1. To distinguish between the effect of chromatin and the effect of activator on the recruitment of CHD1, we compared its binding on chromatin and naked DNA templates. The data in Supplemental 2E show a significant increase in CHD1 binding in response to activator even on naked DNA templates. These data imply that the recruitment of CHD1 might be linked directly to PIC assembly.
The role of Mediator in recruitment of CHD1
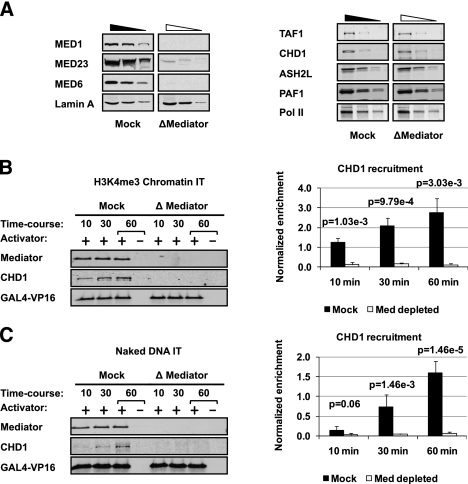
Biochemical analysis of PICs assembled in vitro revealed a dual requirement for the coactivators TFIID and Mediator to achieve efficient binding of the GTFs and Pol II (Johnson and Carey 2003). To assess the role of the coactivators in recruitment of CHD1, we prepared nuclear extracts depleted of either TFIID or Mediator. Our immunodepletion protocol removed ∼90% or more of the Mediator and TFIID but did not significantly affect the levels of CHD1, other H3K4me3-related proteins, or Pol II (Fig. 3A; Supplemental Fig. 3A). We found that activator-dependent CHD1 recruitment was significantly reduced in immobilized template assays performed in the Mediator-depleted extracts on both chromatin and naked DNA (Fig. 3B,C). Unlike Mediator, immunodepletion of TFIID had little effect on recruitment of CHD1 (Supplemental Fig. 3B). Our data suggest that the Mediator is required for recruitment of CHD1. Importantly, the Mediator dependence of CHD1 recruitment observed on naked DNA reinforces the idea that chromatin need not be present for CHD1 recruitment to PICs. The results suggest that Mediator coordinates the recruitment of a key chromatin remodeling enzyme with PIC assembly. These data reinforce the findings in Figure 2 and provide a basis for how Chd1 is preferentially localized to active genes in mouse ES cells.
Figure 3.
Mediator plays a key role in CHD1 recruitment to the PIC. (A) Immunoblot of Mediator subunits and control proteins in Mediator-depleted (ΔMediator) and mock-depleted (Mock) nuclear extract loaded in threefold steps. (B) Immunoblot (left panel) and quantitation (right panel) comparing CHD1 recruitment time courses in an immobilized template assay (IT) on H3K4me3 chromatin in Mock versus ΔMediator extracts. Western blot signal was quantified using LiCOR imaging software, and the statistical significance between Mock- and Mediator-depleted extracts was calculated using Student's t-test. Signals were normalized to VP16. (C) CHD1 recruitment in Mock- versus Mediator-depleted extracts repeated using naked DNA templates.
Mediator stimulates CHD1 function
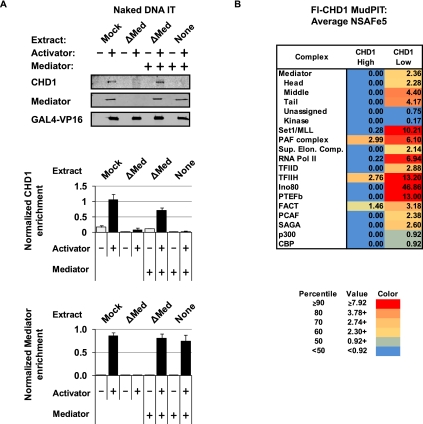
We took two approaches to determine the role of Mediator in recruiting CHD1. First, Mediator was immunoaffinity-purified from HeLa cell lines expressing Flag-tagged Med 29 and used to supplement our Mediator-depleted extract in add-back experiments to rescue CHD1 recruitment. Second, we expressed and purified Flag-CHD1 from the baculovirus system and used MuDPIT analysis to identify other factors that were consistently associated with CHD1, albeit in low abundance, in solution in HeLa extracts. This is a standard proteomic approach for identifying candidate proteins that might interact in the context of a PIC. The caveats in such analyses are that associated factors are usually present in substoichiometric amounts in solution, and the technology does not distinguish between direct and indirect associations.
We found that adding back Mediator, purified under high-stringency conditions, to Mediator-depleted extract was able to rescue CHD1 recruitment to the immobilized template on naked DNA (Fig. 4A). Quantitation of triplicate repeats, normalized to VP16 activator binding, revealed that restoration of CHD1 recruitment in Mediator-depleted extract is reproducible and significant. One prediction of the hypothesis that CHD1 somehow attaches to the PIC is that CHD1 should interact with PIC components. To test this hypothesis, recombinant CHD1 was incubated with HeLa nuclear extracts in 100 mM KCl using lower and higher DNase/heparin conditions (see the Materials and Methods) and then repurified at high salt (300 mM NaCl) to examine interacting proteins. MuDPIT analysis of proteins bound to CHD1 revealed subunits from the Mediator and other PIC components present in the lower heparin conditions (Fig. 4B; Supplemental Table 2A). These components included the SEC, TFIID, TFIIH, INO80, and the SET1 and PAF1 complexes. TFIIH, PAF1, and FACT complexes were still detected at the higher heparin conditions (Fig. 4B; Supplemental Table 2B) and may possibly help bridge the CHD1–Mediator interaction. Nonetheless, the observation that CHD1 pull-down experiments mainly detect components of the PIC and associated chromatin remodeling factors adds additional strength to the idea that CHD1 is indeed a component of the PIC bound at a promoter.
Figure 4.
Characterization of CHD1 recruitment using purified complexes. (A) Immunoblot analysis of CHD1 recruitment to naked DNA templates in a Mediator-depleted extract supplemented with highly purified Mediator ± activator. Mediator was normalized to levels found in the HeLa nuclear extract used in Figures 2 and 3. The top panel is a representative immunoblot of the Mediator complementation experiment, while the bottom panels are graphs analyzing triplicate experiments. Quantitation was performed using LiCOR imaging software. Signals were normalized to VP16. (B) Chart of PIC-related complexes found associated with recombinant Flag-tagged human CHD1 incubated with HeLa nuclear extract at higher and lower heparin conditions. Average NSAFe5 values for each complex are ranked by percentile in a three-color gradient from high (red) to mid (yellow) to low (blue) as shown. Note that the cutoff values differ from the chart in Figure 2.
CHD1 is necessary for efficient transcription in vitro
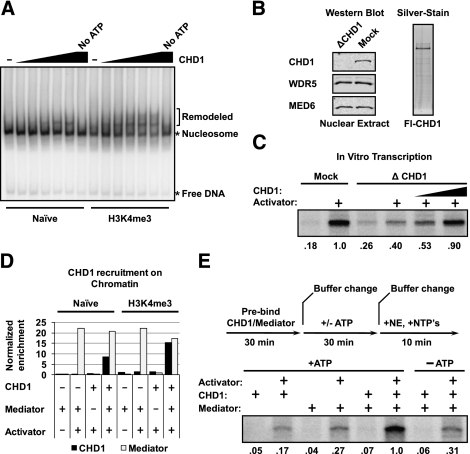
We next focused on the function of CHD1 at a promoter. To establish that recombinant CHD1 from the baculovirus system is functional, we performed a chromatin remodeling assay. In the presence of ATP, CHD1 is active in the in vitro remodeling assay, as illustrated by the shift in mobility of the nucleosome on a gel upon CHD1 treatment (Fig. 5A). Additionally, the remodeling is significantly more pronounced on H3K4me3 chromatin. This result agrees with the generally accepted view that one role of H3K4me3 is to enhance CHD1 remodeling of nucleosomes (Petesch and Lis 2008).
Figure 5.
Characterization of CHD1 during transcription initiation. (A) Autoradiograph of nucleosome remodeling assay demonstrating that recombinant CHD1 is enzymatically active on Naive and H3K4me3 32P-labeled 601 mononucleosomal templates. (B) Western blot of CHD1-depleted nuclear extract and a silver-stained gel of purified recombinant Flag-CHD1 expressed in baculovirus. (C) Autoradiograph of in vitro transcription primer extension assays using CHD1-depleted nuclear extracts as shown. Transcription was compared between CHD1-depleted (ΔCHD1) and mock-depleted nuclear extracts and rescued with the addition of purified CHD1 as shown. Signals were quantitated using Imagequant TL and normalized to Mock-treated + activator. (D) Graph showing CHD1 recruitment to H3K4me3 versus Naive immobilized chromatin templates with purified Mediator and activator. Trace levels of CHD1 were detected by immunoblotting in the purified Mediator alone. CHD1 recruitment in each lane was normalized to recruited Mediator. (E) Levels of transcription were measured in a rate-limiting assay performed as illustrated by the schematic. H3K4me3 chromatin templates were preincubated with activator, Mediator, and CHD1 as indicated. After 30 min, unbound protein was removed, and buffer with or without ATP was added. After a further 30-min incubation, the buffer was removed and beads were suspended in transcription buffer containing HeLa nuclear extract and nucleotides. After 10 min, the transcription reaction was subjected to primer extension analysis to measure mRNA. Signals were quantitated using Imagequant TL and normalized to the signal in lane 6.
To determine whether CHD1 contributes to transcriptional activation on H3K4me3 chromatin, we prepared extracts depleted of the protein using a native CHD1 antibody. Using this method, we depleted CHD1 from the HeLa extracts by >90% (Fig. 5B). The in vitro transcription experiment in Figure 5C shows that depletion of CHD1 decreases transcriptional activation, whereas addition of recombinant CHD1 restores the normal level of activated transcription. The effect is specific to the CHD1-depleted extracts, as addition of CHD1 to the mock-depleted extracts has no additional effect on transcription and even inhibits at high concentrations (Supplemental Fig. 4). Note that CHD1 depletion reduces but does not abolish transcription activation on the H3K4me3 chromatin template. This result could indicate that CHD1 is not absolutely required for, but does strongly contribute to, H3K4me3 chromatin transcription in our system. Collectively, the data in Figure 5, A–C, establish that CHD1 is active in remodeling and necessary for efficient activated transcription.
One possibility is that CHD1 recruitment to the PIC through the Mediator facilitates Pol II initiation on chromatin. If so, this could be a rate-limiting step that would be overcome by prebinding the Mediator and CHD1 to the template in the presence of ATP prior to adding HeLa extract and nucleotides. First, we tested binding of purified CHD1 and Mediator to chromatin. Figure 5D quantitates an immobilized template assay showing that purified CHD1 binds better in the presence of Mediator. Experiments on both naive unmodified and H3K4me3 chromatin revealed that the overall recruitment of CHD1 is further enriched on the H3K4me3 chromatin (Fig. 5D). We cannot say that this is a direct interaction, as neither protein is completely pure. Next, we tested the effect of prebinding Mediator and CHD1 on transcription of H3K4me3 templates. The data in Figure 5E establish that preincubation of Mediator and CHD1 with the chromatin template stimulates transcription in an ATP-dependent manner. Preincubation of Mediator and CHD1 in the absence of ATP failed to elicit a stimulatory effect within the short 10-min time frame of the in vitro transcription experiment. This result implies that the ATP-dependent activity shown in Figure 5A is necessary for transcription. We conclude that CHD1 enhances transcription in a Mediator- and ATP-dependent manner (Esnault et al. 2008; Boeing et al. 2010).
Genomic Chd1 binding overlaps with Mediator in vivo
Our hypothesis is that CHD1 binding to the PIC is controlled by the Mediator, possibly through bridging factors. CHD1 can then act on the H3K4me3 nucleosomes that are found at gene promoters. This hypothesis predicts that CHD1 occupancy would correlate with Mediator predominantly at active genes. To address this issue, we first examined the recruitment of CHD1 along with Mediator in a U2OS cell line bearing a doxycycline-inducible TetR-VP16-activated reporter gene (Fig. 6A). CHD1 recruitment closely follows Mediator recruitment in a time course of doxycycline induction at the promoter, but is not observed at the 3′ end of the reporter gene at the time points tested (Supplemental Fig. 5A). These data suggest that Mediator and CHD1 join the VP16-stimulated PIC as it is assembled in vivo.
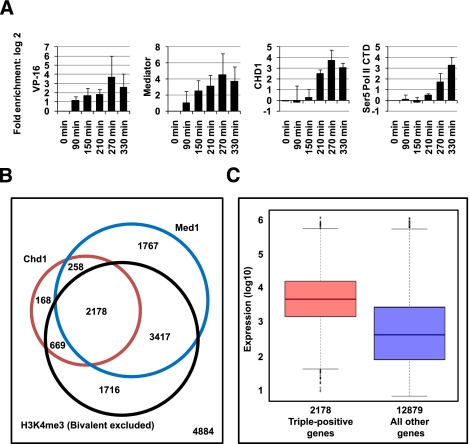
Figure 6.
Chd1 and Mediator binding correlate in vivo. (A) ChIP analysis of doxycycline-induced enrichment of VP16, Med1-containing Mediator, CHD1, and Pol II on a stably integrated doxycycline-inducible Luciferase reporter in U2OS cells during a time course. (B) Venn diagram showing distribution and overlap of genes for Chd1 and Med1 binding and H3K4me3 (excluding H3K27me3) occupancy across the mouse ES cell genome. P-values are as follows: Chd1-Med1, P-value = 1.42 × 10−216; Chd1-H3K4me3 (excluding H3K27me3), P-value < 9.34 × 10−322; Med1-H3K4me3 (excluding H3K27me3), P-value < 1.05 × 10−321. (C) Box plot of gene expression levels of Chd1-positive, Med1-positive, and H3K4me3 (excluding H3K27me3)-positive genes and all other genes, including singly and doubly bound as well as unbound genes. Chd1-, Med1-, and H3K4me3-cobound genes show an overall higher level of transcription. For triple-positive versus all other genes: D = 0.4276, P-value < 2.2 × 10−16.
We next examined two previously published genome-wide data sets in mouse ES cells (Gaspar-Maia et al. 2009; Sridharan et al. 2009; Kagey et al. 2010). We analyzed binding events for the Med1 Mediator subunit on the same promoter regions that were analyzed on promoter arrays for Chd1 and H3K4me3. Previous studies have reported that Chd1, which is required for ES cell self-renewal, localizes to only 12% of the promoter regions of bivalent genes and is highly enriched at active H3K4me3 genes. We found a significant genome-wide overlap for Chd1, Med1, and H3K4me3 (Fig. 6B; Supplemental Fig. 5B,C). As shown previously, Chd1 localizes predominantly to promoters occupied by H3K4me3. This correlation is even stronger when bivalent promoters are excluded from the pool of H3K4me3-positive promoters (Supplemental Fig. 5B). Similarly, Med1 occupancy is significantly reduced across bivalent promoters (Supplemental Fig. 5C). These data imply that the mechanism to “poise/prime” bivalent genes for future activation is not PIC-dependent. Although not all Med1-bound, H3K4me3-occupied genes are Chd1 targets, both Med1 and H3K4me3 are found at the vast majority of Chd1-bound promoters. Binding of Med1 and Chd1, along with H3K4me3, is indicative of highly active transcription as shown in Figure 6C (see also Supplemental Fig. 5D). We suggest, based on the data above, that the Mediator coordinates H3K4me3-stimulated initiation of transcription by Chd1.
Med1 is required for coimmunoprecipitation of Mediator and CHD1
One problem with studying cross-talk between Mediator and CHD1 in cells is that indiscriminate disruption of the Mediator is likely to have widespread indirect consequences. It was therefore necessary to identify specific Mediator subunits that might be involved in the recruitment. Using in vitro transcription and translation, we synthesized individual Mediator subunits and studied their affinity for Flag-CHD1 bound to the Flag antibody resin (Supplemental Fig. 6). In this type of analysis, binding does not necessarily indicate a direct interaction because transcription factors are known to be present in reticulocyte lysates and the baculovirus-synthesized CHD1 may contain contaminants from insect cells. Nevertheless, this approach represents the most logical way to determine which Mediator subunit is involved in recruitment of CHD1. Our assay identified Med1 as the subunit with the highest affinity among the 29 subunits tested. This result, however, does not preclude the possibility that other Mediator subunits may also be involved.
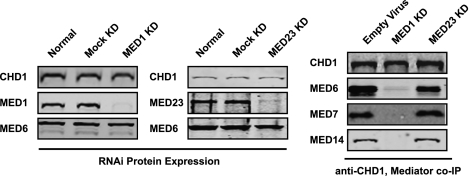
Med1 was then knocked down in 293T cells along with a control subunit, Med23, using lentiviruses encoding shRNAs targeting the two subunits. Both Med1 and Med23 are known to be targeted by specific activators in vivo (Yuan et al. 1998; Boyer et al. 1999). Mediators lacking Med1 and Med23 have been isolated (Ito et al. 2000; Stevens et al. 2002). Thus, disruption of Med1 or Med23 is unlikely to have a strong impact on Mediator structure. We then performed coimmunoprecipitation experiments with CHD1 as bait. We chose Med6, Med7, and Med14 as subunits indicative of the three known structural modules of Mediator (Conaway et al. 2005). The data in Figure 7 show that CHD1 coimmunoprecipitated with Mediator subunits in extracts from cells treated with the virus alone and with the Med23 shRNA knockdown virus but not in the Med1 shRNA knockdown. The data suggest that select disruption of the Mediator can abolish its ability to coimmunoprecipitate with CHD1.
Figure 7.
CHD1 associates with Med1-containing Mediator in vivo. Immunoblots showing specific shRNA knockdown of Med1 and Med23 subunits of Mediator in 293T cells (left panels) and Med1-dependent coimmunoprecipitation of Mediator by targeted immunoprecipitation of CHD1 (right panel). Subunits representing the head (Med6), middle (Med7), and tail (Med14) modules of Mediator are blotted.
Discussion
Our data support the concept that activator-stimulated PIC assembly is physically and mechanistically linked with the subsequent effects of H3K4me3. The use of MuDPIT and immunoblotting to interrogate the PICs formed on chromatin, coupled with the purification and analysis of CHD1, allowed us to determine that the Mediator coordinates assembly of the PIC and recruitment of CHD1. Our data provide a basis for the specificity of CHD1 for active genes.
Our major finding was that CHD1 binding was largely coupled to activator-mediated PIC assembly but was also weakly stimulated by H3K4me3 alone. Indeed, the PIC and H3K4me3 together caused the most efficient recruitment of CHD1. Previous studies have shown that CHD1 binds to H3K4me3, an observation highlighted by recent proteomic analyses (Sims et al. 2005; Bartke et al. 2010; Vermeulen et al. 2010). However, this event alone seems to contribute only a small amount of CHD1 binding to an active gene. Indeed, recent studies have demonstrated that in Drosophila, the CHD1 chromodomains, which recognize H3K4me3, are not required for its colocalization to active genes (Morettini et al. 2011). Moreover, Kornberg and colleagues (Ehrensberger and Kornberg 2011) have recently found that S. cerevisiae CHD1, whose chromodomains do not recognize H3K4me3, selectively removes nucleosomes at the promoter in vitro and in vivo in an activator-dependent manner. In mammalian cells, the close correlation between localization of Mediator, Chd1, and H3K4me3 and active transcription in mouse ES cells reinforces the notion that the major mechanism for Chd1 localization is PIC assembly. Dual interactions between the transcriptional machinery and the SET1-catalyzed H3K4me3 modification reinforce the specificity of CHD1.
Among our findings was that the SET1 complex is also recruited to the PIC (as shown in Supplemental Fig. 1A). It has been reported that some activators interact with subunits of the SET1 complex and that some promoters of inactive genes are bound by activators. CHD1, however, is found only at genes that are actively transcribed. Inactive genes are typically di- or trimethylated at H3K9 or H3K27. Hence, bivalent domains might be a natural by-product of PIC absence. In such cases, where full PIC assembly might be limited by an incomplete complement of activators, CHD1 occupancy may be prevented by both the absence of Mediator and the presence of repressive complexes like PRC1 or HP1 that bind the modifications that correlate with inactive genes.
MuDPIT analysis of proteins bound to CHD1 led to the discovery that it associates with several proteins normally found near or immediately downstream from the start site, including the PAF1 complex, INO80, P-TEFb, NELF, TAFIIs, TFIIH, and Mediator, among others. P-TEFb releases the stalled Pol II found 30–50 bp downstream from many genes by phosphorylating DSIF and NELF (Peterlin and Price 2006). Similarly, numerous TAFII subunits and the TFIIH XPB subunit cross-link immediately downstream from the start site in vitro (Dvir et al. 2001). These data, along with the observation that many genes contain a nucleosome positioned near the start site (Schones et al. 2008), reinforce the notion that CHD1 action is functionally coupled to regulatory events associated with the promoter, including initiation and early elongation. The role of INO80 is unclear, but studies have implicated this ATP remodeling complex in promoter-dependent transcription (Cai et al. 2007). We also found small amounts of PAF1 associated with CHD1 as previously observed by others, suggesting the possibility that the two factors interact directly (Sims et al. 2007; Warner et al. 2007).
Analysis of higher-stringency CHD1 pull-downs suggested that among the interacting proteins, TFIIH has a high affinity (Fig. 4B). TFIIH has previously been shown to bind Mediator in work done by others (Esnault et al. 2008; Boeing et al. 2010). This suggests that TFIIH may contribute to the docking of CHD1 with Mediator. Indeed, our unpublished immobilized template data support a Mediator–TFIIH–CHD1 interaction. However, it remains a possibility that this may be enabled by factors copurifying with Mediator or with CHD1 from the insect cell expression system. These issues notwithstanding, our data suggest that Mediator controls CHD1 recruitment.
The idea that the Mediator serves as a platform for other proteins and conveys signals to the general transcription machinery was originally proposed by Young (Chao et al. 1996). The concept that Mediator coordinates events at chromatin is emerging and makes sense given that the machineries coevolved in almost all eukaryotes. In addition to Shilatifard's work on COMPASS (Krogan et al. 2003; Wu et al. 2008), a previous study in our laboratory (Black et al. 2006) linked p300-mediated acetylation with PIC assembly. We also observed in our MuDPIT analysis of PICs the enrichment of other chromatin factors—such as GCN5, NuA4, and other CHD family members—in an activator-stimulated fashion. Most importantly, Mediator has been linked directly to recruitment of Pol II SEC via the Med26 subunit (Takahashi et al. 2011). It would be interesting if this event was linked to the action of CHD1 at the start site. Boyer and colleagues (Ding et al. 2008) have shown that the connectivity also extends to silencing factors, as G9a links directly with the Cdk8 module of the Mediator.
Our current view of the protein–protein interactions identified by proteomic and mechanistic studies is that CHD1 is recruited only to active genes by its interaction with the PIC via the Mediator. Activators are known to interact directly with Mediator and TFIID. The VP16 activation domain docks with Mediator via the MED25 subunit (Yang et al. 2004; Uhlmann et al. 2007) and with TFIID via TAF9 (Goodrich et al. 1993). VP16 is known to recruit both complexes to DNA in vitro and in vivo (Berk et al. 1998). TFIID and Mediator also interact to form a coactivator complex (Johnson and Carey 2003), which is necessary in vitro for binding of the GTFs and Pol II. Pol II binds tightly to the active form of Mediator in a somewhat mutually exclusive manner with the Cdk8 module (Paoletti et al. 2006).
In conclusion, our data suggest that the Mediator not only serves as a docking platform for activator-stimulated PIC assembly of the GTFs, but coordinates the recruitment of CHD1 during active transcription as well. As the Mediator signifies active genes, the specificity of CHD1 recruitment can best be described as cooperative protein–protein interactions between the PIC, CHD1, and H3K4me3.
Materials and methods
Methyl-lysine histone octamer preparation
Lys 4 of histone H3 was mutated to a cysteine by site-directed mutagenesis of Xenopus H3.1 bearing a C110A mutation, expressed and purified from Escherichia coli inclusion bodies, and subjected to chemical alkylation by (2-bromoethyl) trimethylammonium bromide (Simon et al. 2007) before assembly into histone octamers (Luger et al. 1997).
Chromatin preparation
A 602-bp biotinylated PCR fragment that directly encompasses G5E4T (Johnson and Carey 2003) was assembled into chromatin by salt dilution as described previously (Steger et al. 1997) and was validated by EMSA in native PAGE. Chromatin was immobilized on M280 streptavidin beads (Dynal) in chromatin-binding buffer (20 mM HEPES at pH 8.0, 150 mM KCl, 10% glycerol, 4 mM MgCl2, 1 mM DTT, 200 μg/mL BSA).
Immobilized template recruitment assay
The 40-μL immobilized template recruitment assays contained 80 μg of HeLa nuclear extract, and 50 ng of chromatin or naked DNA template in immobilized template binding buffer (120 mM KCl, 10 mM HEPES at pH 8.0, 5% glycerol, 0.2 μg/mL BSA, 0.05% NP-40). After the indicated time periods, the beads were washed three times in immobilized template buffer. Captured PICs were incubated with 500 μM ATP in immobilized template binding buffer where indicated in Supplemental Figure 1B for Pol II phosphorylation assays. Bound protein was eluted in 10 μL of 2× Laemmli buffer, fractionated by SDS-PAGE, and immunoblotted. For proteomic analysis, the 1-h time point was scaled up and subjected to MuDPIT. Antibodies used in immunoblotting included MED23 (BD Pharmingen), Pol II CTD 8WG16 (QED Bioscience), TFIIB (Tantin et al. 1996), WDR5 (Upstate Biotechnology), ASH2L, RBBP5, PAF1, and CHD1 (Bethyl Laboratories). All other antibodies were purchased from Santa Cruz Biotechnologies.
Extract and protein preparation
HeLa nuclear extract (Dignam et al. 1983) and GAL4-VP16 were prepared as previously described. Immunodepletion of TFIID from 1 mL of HeLa nuclear extract was performed using 200 μg each of antibodies against TAF4, TAF1, and TAF3 (Santa Cruz Biotechnologies). Mediator was depleted from 1 mL of extract with antibodies against MED1, MED6, MED7, MED25, and CDK8. CHD1 was depleted with an antibody against CHD1 (Bethyl). Antibodies were cross-linked to protein A and G paramagnetic beads (Invitrogen) using 20 mM dimethylpimelimidate in 0.1 M sodium borate buffer (pH 9) and washed extensively with 50 mM glycine (pH 2.5). The cross-linked beads were equilibrated in buffer D (20 mM HEPES at pH 7.9, 0.1 mM EDTA, 20% glycerol, 0.1 M KCl) and incubated with HeLa nuclear extract in buffer D for 4 h at 4°C. The supernatant was isolated and used for immobilized template analysis as described above.
Recombinant Flag-tagged human CHD1 was purified from SF9 cells using a baculovirus overexpression system (Invitrogen). Briefly, cells were resuspended in 0.3 M buffer F (0.3 M NaCl, 20% glycerol, 20 mM HEPES at pH 7.9, 4 mM MgCl2, 0.2% Triton X-100, 0.1% NP-40) and sonicated. Lysates were then treated with DNase I (1 U/mL) and heparin (12.5 μg/mL) and cleared by centrifugation at 30,000g. The resulting lysate was bound to M2 anti-Flag resin (Sigma), washed extensively in 0.5 M buffer F, and eluted using 3× Flag peptide (0.25 mg/mL; Sigma). Mediator was purified from HeLa cells expressing Flag-tagged human Intersex (Med29) as described previously (Sato et al. 2003). The HeLa Intersex cell line was a gift from Joan and Ron Conaway.
MuDPIT analysis of immobilized templates and purified proteins
For MuDPIT analysis, the equivalent of 300 immobilized template reactions were pooled together. Samples were eluted in 50 mM Tris (pH 8.0) and 6 M urea. For CHD1, ∼250 μg of Flag-CHD1 was immobilized on Flag antibody beads and incubated with 0.45 mL of HeLa nuclear extract in buffer containing 100 mM KCl in the presence of either 1 U/mL DNase and 12.5 μg/mL heparin (low stringency) or 75 μg/mL heparin and 10 U/mL DNase I in the presence of 2 mM CaCl2 (high stringency). Bound proteins were washed in 0.3 M buffer F, and F-CHD1 and interacting proteins were then eluted using a 3× Flag peptide (0.25 mg/mL; Sigma). Protein samples were precipitated in 20% TCA, washed with cold acetone, and digested with trypsin. The digested peptide samples were then fractionated with sequential cation exchange and reverse-phase chromatography and eluted directly into a LTQ-Orbitrap mass spectrometer (Thermo Fisher). MS/MS spectra were collected as described (Law et al. 2010). Data analysis was performed with the SEQUEST and DTASelect2 algorithms and filtered with at least two peptides per protein and a peptide-level false-positive rate of <5% as estimated by a decoy database strategy (Law et al. 2010). NSAF values were calculated as described in Law et al. (2010). Proteins were assigned into complexes using the online resource CORUM (Ruepp et al. 2008) and Mediator submodules as published previously (Bourbon 2008).
In vitro transcription assays
The 40-μL standard reactions in Figure 1 contained 50 ng of linear chromatin template, nuclear extract, and GAL4-VP16 as described previously (Black et al. 2006). After 60 min, RNA was harvested and analyzed by primer extension as described previously. Transcription of immobilized chromatin templates in Figure 5 was carried out on 50 ng of linear H3K4me3 chromatin templates, which were bound with saturating levels of GAL4-VP16, followed by removal of excess activator. Templates were then bound with saturating amounts of Mediator and CHD1, as determined by immobilized template assays, and incubated for 30 min at 30°C in the presence or absence of ATP, after which unbound protein was removed. Eighty micrograms of HeLa extract and NTPs were added, and transcription was allowed to proceed for 10 min under standard conditions as described previously (Black et al. 2006).
Cross-correlation of genomic chromatin immunoprecipitation (ChIP) data sets
ChIP–chip data sets from experiments performed on mouse ES cells were used to determine whether gene promoters were enriched for Chd1 (Gaspar-Maia et al. 2009) and H3K4me3 or H3K27me3 (Sridharan et al. 2009). ChIP-seq data for Med1-occupied genes were from Supplemental Table 5 of the study by Young and colleagues (Kagey et al. 2010). The ChIP–chip experiments were performed on Agilent promoter arrays with probes covering the region −5.5 kb to +2.5 kb relative to the transcription start site. Hence, only those Med1-enriched regions that had significant peaks within this 8-kb region, based on the location of enriched regions from Supplemental Table 4 in Kagey et al. (2010), were used for further overlap analysis. The overlap of genes whose promoters were enriched for each feature (Med1, Chd1, and H3K4me3 without H3K27me3) was determined pairwise, and the significance of the overlap was evaluated using the hypergeometric test. Box plots were used to visualize the distribution of expression levels in mouse ES cells for the set of genes exhibiting co-occupancy by all three features as compared with those for gene sets with different combinations of the three features in their promoter regions or their complete absence. Gene expression levels were obtained from Supplemental Table 2 in Gaspar-Maia et al. (2009). A two-sample Kolmogorov-Smirnov test was used to compare the distributions of expression values between the two sets of genes.
shRNA knockdown of Mediator subunits and coimmunoprecipitation
293T cells were grown to 30% confluency in 10-cm dishes and infected with lentiviruses expressing shRNAs targeting MED23 (TTGTGAGTGTCATCAGCAGCC) and MED1 (GTCATGGAGAAGAGGGTTGTG). After 96 h, cell lysates were prepared and immunoblotted to determine the extent of MED1 and MED23 knockdown using the MED6 antibody as a control. The lysates were subjected to immunoprecipitation with a CHD1 antibody (Bethyl Laboratories). The immunoprecipitates were blotted with antibodies to CHD1, MED6, MED7, and MED14 (Santa Cruz Biotechnologies) to determine the amount of coimmunoprecipitation.
ChIP
ChIP from the U2OS cells, various times after doxycyline treatment, was performed using antibodies to CHD1, MED1, VP16, and Ser 5 Pol II (Abcam) as previously described (Black et al. 2006).
Acknowledgments
We thank Joan and Ron Conaway for gifts of TFIIH and the Flag-Med29 cell line, and Matteo Pellegrini for advice on statistical analyses. This work was supported by NIH grants R01-GM74701 (to M.C.) and R01-GM089778 (to J.A.W.); funds from the Jonsson Cancer Center at UCLA (to J.A.W.); the USPHS National Research Service Award GM07104 (to G.B.); the Jonsson Cancer Center Foundation at UCLA (to R.S.); the NIH Director's Young Innovator Award DP2OD001686 (to K.P.); a CIRM Young Investigator Award RN1-00564 (to K.P.); and the Eli and Edythe Broad Center of Regenerative Medicine and Stem Cell Research at UCLA (to K.P.).
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.17554711.
References
- Bartke T, Vermeulen M, Xhemalce B, Robson SC, Mann M, Kouzarides T 2010. Nucleosome-interacting proteins regulated by DNA and histone methylation. Cell 143: 470–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berk AJ, Boyer T, Kapanidis AN, Ebright RH, Kobayashi NN, Horn PJ, Sullivan SM, Koop R, Surby MA, Triezenberg SJ 1998. Mechanism of viral activators. Cold Spring Harb Symp Quant Biol 63: 243–252 [DOI] [PubMed] [Google Scholar]
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. 2006. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 125: 315–326 [DOI] [PubMed] [Google Scholar]
- Black JC, Choi JE, Lombardo SR, Carey M 2006. A mechanism for coordinating chromatin modification and preinitiation complex assembly. Mol Cell 23: 809–818 [DOI] [PubMed] [Google Scholar]
- Boeing S, Rigault C, Heidemann M, Eick D, Meisterernst M 2010. RNA polymerase II C-terminal heptarepeat domain Ser-7 phosphorylation is established in a Mediator-dependent fashion. J Biol Chem 285: 188–196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourbon H-M 2008. Comparative genomics supports a deep evolutionary origin for the large, four-module transcriptional mediator complex. Nucleic Acids Res 36: 3993–4008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer TG, Martin MED, Lees E, Ricciardi RP, Berk AJ 1999. Mammalian Srb/Mediator complex is targeted by adenovirus E1A protein. Nature 399: 276–279 [DOI] [PubMed] [Google Scholar]
- Cai Y, Jin J, Yao T, Gottschalk AJ, Swanson SK, Wu S, Shi Y, Washburn MP, Florens L, Conaway RC, et al. 2007. YY1 functions with INO80 to activate transcription. Nat Struct Mol Biol 14: 872–874 [DOI] [PubMed] [Google Scholar]
- Chao DM, Gadbois EL, Murray PJ, Anderson SF, Sonu MS, Parvin JD, Young RA 1996. A mammalian SRB protein associated with an RNA polymerase II holoenzyme. Nature 380: 82–85 [DOI] [PubMed] [Google Scholar]
- Conaway RC, Sato S, Tomomori-Sato C, Yao T, Conaway JW 2005. The mammalian Mediator complex and its role in transcriptional regulation. Trends Biochem Sci 30: 250–255 [DOI] [PubMed] [Google Scholar]
- Dignam JD, Lebovitz RM, Roeder RG 1983. Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acids Res 11: 1475–1489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding N, Zhou H, Esteve PO, Chin HG, Kim S, Xu X, Joseph SM, Friez MJ, Schwartz CE, Pradhan S, et al. 2008. Mediator links epigenetic silencing of neuronal gene expression with X-linked mental retardation. Mol Cell 31: 347–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drapkin R, Reardon JT, Ansari A, Huang J-C, Zawel L, Ahn K, Sancar A, Reinberg D 1994. Dual role of TFIIH in DNA excision repair and in transcription by RNA polymerase II. Nature 368: 769–772 [DOI] [PubMed] [Google Scholar]
- Dvir A, Conaway JW, Conaway RC 2001. Mechanism of transcription initiation and promoter escape by RNA polymerase II. Curr Opin Genet Dev 11: 209–214 [DOI] [PubMed] [Google Scholar]
- Ehrensberger AH, Kornberg RD 2011. Isolation of an activator-dependent, promoter-specific chromatin remodeling factor. Proc Natl Acad Sci 108: 10115–10120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esnault C, Ghavi-Helm Y, Brun S, Soutourina J, Van Berkum N, Boschiero C, Holstege F, Werner M 2008. Mediator-dependent recruitment of TFIIH modules in preinitiation complex. Mol Cell 31: 337–346 [DOI] [PubMed] [Google Scholar]
- Gaspar-Maia A, Alajem A, Polesso F, Sridharan R, Mason MJ, Heidersbach A, Ramalho-Santos J, McManus MT, Plath K, Meshorer E, et al. 2009. Chd1 regulates open chromatin and pluripotency of embryonic stem cells. Nature 460: 863–868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodrich JA, Hoey T, Thut CJ, Admon A, Tjian R 1993. Drosophila TAFII40 interacts with both a VP16 activation domain and the basal transcription factor TFIIB. Cell 75: 519–530 [DOI] [PubMed] [Google Scholar]
- Ito M, Yuan C-X, Okano HJ, Darnell RB, Roeder RG 2000. Involvement of the TRAP220 component of the TRAP/SMCC coactivator complex in embryonic development and thyroid hormone action. Mol Cell 5: 683–693 [DOI] [PubMed] [Google Scholar]
- Johnson KM, Carey M 2003. Assembly of a Mediator/TFIID/TFIIA complex bypasses the need for an activator. Curr Biol 13: 772–777 [DOI] [PubMed] [Google Scholar]
- Kagey MH, Newman JJ, Bilodeau S, Zhan Y, Orlando DA, van Berkum NL, Ebmeier CC, Goossens J, Rahl PB, Levine SS, et al. 2010. Mediator and cohesin connect gene expression and chromatin architecture. Nature 467: 430–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TH, Barrera LO, Zheng M, Qu C, Singer MA, Richmond TA, Wu Y, Green RD, Ren B 2005. A high-resolution map of active promoters in the human genome. Nature 436: 876–880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Guermah M, McGinty RK, Lee J-S, Tang Z, Milne TA, Shilatifard A, Muir TW, Roeder RG 2009. RAD6-mediated transcription-coupled H2B ubiquitylation directly stimulates H3K4 methylation in human cells. Cell 137: 459–471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konev AY, Tribus M, Park SY, Podhraski V, Lim CY, Emelyanov AV, Vershilova E, Pirrotta V, Kadonaga JT, Lusser A, et al. 2007. CHD1 motor protein is required for deposition of histone variant H3.3 into chromatin in vivo. Science 317: 1087–1090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornberg RD 2005. Mediator and the mechanism of transcriptional activation. Trends Biochem Sci 30: 235–239 [DOI] [PubMed] [Google Scholar]
- Kornberg RD 2007. The molecular basis of eukaryotic transcription. Proc Natl Acad Sci 104: 12955–12961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogan NJ, Dover J, Wood A, Schneider J, Heidt J, Boateng MA, Dean K, Ryan OW, Golshani A, Johnston M, et al. 2003. The Paf1 complex is required for histone H3 methylation by COMPASS and Dot1p: linking transcriptional elongation to histone methylation. Mol Cell 11: 721–729 [DOI] [PubMed] [Google Scholar]
- Lagrange T, Kim TK, Orphanides G, Ebright YW, Ebright RH, Reinberg D 1996. High-resolution mapping of nucleoprotein complexes by site-specific protein–DNA photocrosslinking: organization of the human TBP-TFIIA-TFIIB-DNA quaternary complex. Proc Natl Acad Sci 93: 10620–10625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law JA, Ausin I, Johnson LM, Vashisht AA, Zhu J-K, Wohlschlegel JA, Jacobsen SE 2010. A protein complex required for polymerase V transcripts and RNA-directed DNA methylation in Arabidopsis. Curr Biol 20: 951–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Carey M, Workman JL 2007. The role of chromatin during transcription. Cell 128: 707–719 [DOI] [PubMed] [Google Scholar]
- Luger K, Rechsteiner TJ, Flaus AJ, Waye MMY, Richmond TJ 1997. Characterization of nucleosome core particles containing histone proteins made in bacteria. J Mol Biol 272: 301–311 [DOI] [PubMed] [Google Scholar]
- Martinez E 2002. Multi-protein complexes in eukaryotic gene transcription. Plant Mol Biol 50: 925–947 [DOI] [PubMed] [Google Scholar]
- Morettini S, Tribus M, Zeilner A, Sebald J, Campo-Fernandez B, Scheran G, Wörle H, Podhraski V, Fyodorov DV, Lusser A 2011. The chromodomains of CHD1 are critical for enzymatic activity but less important for chromatin localization. Nucleic Acids Res 39: 3103–3115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paoletti AC, Parmely TJ, Tomomori-Sato C, Sato S, Zhu D, Conaway RC, Conaway JW, Florens L, Washburn MP 2006. Quantitative proteomic analysis of distinct mammalian Mediator complexes using normalized spectral abundance factors. Proc Natl Acad Sci 103: 18928–18933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterlin BM, Price DH 2006. Controlling the elongation phase of transcription with P-TEFb. Mol Cell 23: 297–305 [DOI] [PubMed] [Google Scholar]
- Petesch SJ, Lis JT 2008. Rapid, transcription-independent loss of nucleosomes over a large chromatin domain at Hsp70 loci. Cell 134: 74–84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranish JA, Yi EC, Leslie DM, Purvine SO, Goodlett DR, Eng J, Aebersold R 2003. The study of macromolecular complexes by quantitative proteomics. Nat Genet 33: 349–355 [DOI] [PubMed] [Google Scholar]
- Roeder RG 1998. Role of general and gene-specific cofactors in the regulation of eukaryotic transcription. Cold Spring Harb Symp Quant Biol 63: 201–218 [DOI] [PubMed] [Google Scholar]
- Ruepp A, Brauner B, Dunger-Kaltenbach I, Frishman G, Montrone C, Stransky M, Waegele B, Schmidt T, Doudieu ON, Stümpflen V, et al. 2008. CORUM: the comprehensive resource of mammalian protein complexes. Nucleic Acids Res 36: D646–D650 doi: 10.1093/nar/gkm936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruthenburg AJ, Allis CD, Wysocka J 2007. Methylation of lysine 4 on histone H3: intricacy of writing and reading a single epigenetic mark. Mol Cell 25: 15–30 [DOI] [PubMed] [Google Scholar]
- Sato S, Tomomori-Sato C, Banks CAS, Parmely TJ, Sorokina I, Brower CS, Conaway RC, Conaway JW 2003. A mammalian homolog of Drosophila melanogaster transcriptional coactivator intersex is a subunit of the mammalian Mediator complex. J Biol Chem 278: 49671–49674 [DOI] [PubMed] [Google Scholar]
- Schones DE, Cui K, Cuddapah S, Roh TY, Barski A, Wang Z, Wei G, Zhao K 2008. Dynamic regulation of nucleosome positioning in the human genome. Cell 132: 887–898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon MD, Chu F, Racki LR, de la Cruz CC, Burlingame AL, Panning B, Narlikar GJ, Shokat KM 2007. The site-specific installation of methyl-lysine analogs into recombinant histones. Cell 128: 1003–1012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims RJ, Chen C-F, Santos-Rosa H, Kouzarides T, Patel SS, Reinberg D 2005. Human but not yeast CHD1 binds directly and selectively to histone H3 methylated at lysine 4 via its tandem chromodomains. J Biol Chem 280: 41789–41792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims RJ, Millhouse S, Chen C-F, Lewis BA, Erdjument-Bromage H, Tempst P, Manley JL, Reinberg D 2007. Recognition of trimethylated histone H3 lysine 4 facilitates the recruitment of transcription postinitiation factors and pre-mRNA splicing. Mol Cell 28: 665–676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith E, Lin C, Shilatifard A 2011. The super elongation complex (SEC) and MLL in development and disease. Genes Dev 25: 661–672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sridharan R, Tchieu J, Mason MJ, Yachechko R, Kuoy E, Horvath S, Zhou Q, Plath K 2009. Role of the murine reprogramming factors in the induction of pluripotency. Cell 136: 364–377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steger DJ, Owen-Hughes T, John S, Workman JL 1997. Analysis of transcription factor-mediated remodeling of nucleosomal arrays in a purified system. Methods 12: 276–285 [DOI] [PubMed] [Google Scholar]
- Stevens JL, Cantin GT, Wang G, Shevchenko A, Shevchenko A, Berk AJ 2002. Transcription control by E1A and MAP kinase pathway via Sur2 Mediator subunit. Science 296: 755–758 [DOI] [PubMed] [Google Scholar]
- Stoller JZ, Huang L, Tan CC, Huang F, Zhou DD, Yang J, Gelb BD, Epstein JA 2010. Ash2l interacts with Tbx1 and is required during early embryogenesis. Exp Biol Med (Maywood) 235: 569–576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi H, Parmely TJ, Sato S, Tomomori-Sato C, Banks CA, Kong SE, Szutorisz H, Swanson SK, Martin-Brown S, Washburn MP, et al. 2011. Human Mediator subunit MED26 functions as a docking site for transcription elongation factors. Cell 146: 92–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan CC, Sindhu KV, Li S, Nishio H, Stoller JZ, Oishi K, Puttreddy S, Lee TJ, Epstein JA, Walsh MJ, et al. 2008. Transcription factor Ap2δ associates with Ash2l and ALR, a trithorax family histone methyltransferase, to activate Hoxc8 transcription. Proc Natl Acad Sci 105: 7472–7477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tantin D, Chi T, Hori R, Pyo S, Carey M 1996. Biochemical mechanism of transcriptional activation by GAL4-VP16. Methods Enzymol 274: 133–149 [DOI] [PubMed] [Google Scholar]
- Uhlmann T, Boeing S, Lehmbacher M, Meisterernst M 2007. The VP16 activation domain establishes an active Mediator lacking CDK8 in vivo. J Biol Chem 282: 2163–2173 [DOI] [PubMed] [Google Scholar]
- Vermeulen M, Eberl HC, Matarese F, Marks H, Denissov S, Butter F, Lee KK, Olsen JV, Hyman AA, Stunnenberg HG, et al. 2010. Quantitative interaction proteomics and genome-wide profiling of epigenetic histone marks and their readers. Cell 142: 967–980 [DOI] [PubMed] [Google Scholar]
- Warner MH, Roinick KL, Arndt KM 2007. Rtf1 is a multifunctional component of the Paf1 complex that regulates gene expression by directing cotranscriptional histone modification. Mol Cell Biol 27: 6103–6115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood A, Schneider J, Dover J, Johnston M, Shilatifard A 2003. The Paf1 complex is essential for histone monoubiquitination by the Rad6–Bre1 complex, which signals for histone methylation by COMPASS and Dot1p. J Biol Chem 278: 34739–34742 [DOI] [PubMed] [Google Scholar]
- Wu M, Wang PF, Lee JS, Martin-Brown S, Florens L, Washburn M, Shilatifard A 2008. Molecular regulation of H3K4 trimethylation by Wdr82, a component of human Set1/COMPASS. Mol Cell Biol 28: 7337–7344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang F, DeBeaumont R, Zhou S, Näär AM 2004. The activator-recruited cofactor/Mediator coactivator subunit ARC92 is a functionally important target of the VP16 transcriptional activator. Proc Natl Acad Sci 101: 2339–2344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan C-X, Ito M, Fondell JD, Fu Z-Y, Roeder RG 1998. The TRAP220 component of a thyroid hormone receptor-associated protein (TRAP) coactivator complex interacts directly with nuclear receptors in a ligand-dependent fashion. Proc Natl Acad Sci 95: 7939–7944 [DOI] [PMC free article] [PubMed] [Google Scholar]