Abstract
The photosynthetic and basal apicomplexan Chromera velia was recently described, expanding the membership of this otherwise nonphotosynthetic group of parasite protists. Apicomplexans are alveolates with secondary plastids of red algal origin, but the evolutionary history of their nuclear genes is still actively discussed. Using deep sequencing of expressed genes, we investigated the phylogenetic affinities of a stringent filtered set of 3,151 expressed sequence tag-contigs by generating clusters with eukaryotic homologs and constructing phylogenetic trees and networks. The phylogenetic positioning of this alveolate alga was determined and sets of phyla-specific proteins extracted. Phylogenetic trees provided conflicting signals, with 444 trees grouping C. velia with the apicomplexans but 354 trees grouping C. velia with the alveolate oyster pathogen Perkinsus marinus, the latter signal being reinforced from the analysis of shared genes and overall sequence similarity. Among the 513 C. velia nuclear genes that reflect a photosynthetic ancestry and for which nuclear homologs were available both from red and green lineages, 263 indicated a red photosynthetic ancestry, whereas 250 indicated a green photosynthetic ancestry. The same 1:1 signal ratio was found among the putative 255 nuclear-encoded plastid proteins identified. This finding of red and green signals for the alveolate mirrors the result observed in the heterokont lineage and supports a common but not necessarily single origin for the plastid in heterokonts and alveolates. The inference of green endosymbiosis preceding red plastid acquisition in these lineages leads to worryingly complicated evolutionary scenarios, prompting the search for other explanations for the green phylogenetic signal and the amount of hosts involved.
Keywords: Chromera, Apicomplexa, Alveolata, chromalveolata, apicoplast, protist evolution
Introduction
The Apicomplexa are a group of parasite protists that, with the exception of intestinal parasites from the genus Cryptosporidium, house a relict plastid known as the apicoplast (reviewed in McFadden 2010). The organelle does not perform photosynthesis but is nevertheless essential for ultimate parasite survival and propagation. This can probably be attributed to the number of biochemical pathways the apicoplast contains, which include parts of the fatty acid and isopentenyl diphosphatase synthesis (Waller et al. 1998; Jomaa et al. 1999), the assembly of iron–sulfur complexes (Seeber 2002), and segments of heme biosynthesis which is most likely carried out in conjunction with the mitochondria (Ralph et al. 2004). The recent discovery of Chromera velia has added the first nonparasitic autotroph with a photosynthetically active plastid to the base of the apicomplexan phylum (Moore et al. 2008). At least one more photosynthetic basal apicomplexan has since been described, and collectively, they are currently designated as “chromerids” (Janouskovec et al. 2010; Obornik et al. 2011). Chromerid algae are suspected to be a missing link, connecting the parasitic Apicomplexa with their evolutionary past and algal relatives (Moore et al. 2008).
Apicomplexa belong to the alveolates, a group that includes the dinoflagellates and the ciliates, as well as other less intensely studied lineages such as the Perkinsidae (Gould, Waller, et al. 2008, Zhang et al. 2011). The infrakingdom Alveolata is characterized by the presence of cortical alveolae, a one-membrane bound compartment lying below the plasma membrane and together with longitudinal microtubules and an electron-dense layer of mainly unknown composition (referred to as epiplasm or subpellicular network) forms a multilayered cell pellicle (Cavalier-Smith 1991; Gould, Tham, et al. 2008). The evolutionary history of photosynthesis among the alveolates involves the acquisition of red algal plastids via secondary endosymbiosis (Stoebe and Maier 2002; Gould, Waller, et al. 2008; Archibald 2009; Janouskovec et al. 2010; Keeling 2010). Most apicomplexans studied to date have highly reduced plastid genomes, whereas Cryptosporidium has lost the organelle entirely and only comparatively few genes have been retained in this case, which betray the previous existence of a photosynthetic organelle (Huang et al. 2004). No ciliate has yet been identified that possesses a plastid or a relict thereof, and to our knowledge, only one report claims to have identified phylogenetic evidence for the past presence of such an organelle based on 16 ciliate nuclear genes (Reyes-Prieto et al. 2008). Eisen et al. (2006) noticed a similar weak green signal in their earlier genome analysis manuscript of Tetrahymena thermophila, but in contrast concluded this signal is not above “background noise” that are expected at random in the analysis of 10,000 of genes. In conclusion, there is currently no credible evidence that ciliates ever were secondarily photosynthetic. Finally, Perkinsidae, like Apicomplexa, have nonphotosynthetic plastids that in addition seem to lack DNA and, hence, must have all of their required protein content encoded by nuclear genes (Matsuzaki et al. 2009).
The evolutionary origin of alveolates and their plastid(s) is furthermore coupled to the waning dispute over the “chromalveolate” hypothesis, which proposed that a single secondary endosymbiotic event gave rise to all protist lineages harboring a secondary plastid of red algal origin (Cavalier-Smith 1999). Some analyses supported the chromalveolate concept (Bachvaroff et al. 2005; Harper et al. 2005; Patron et al. 2007), but more current data indicates that it is wrong with respect to the prediction of a single secondary symbiosis (Sanchez-Puerta and Delwiche 2008; Baurain et al. 2010; Felsner et al. 2011). The monophyletic origin was challenged earlier on by others, who also proposed an alternative evolutionary model (Bodyl 2005; Bodyl et al. 2009). In conclusion, a clarification of the specific evolutionary relationships between these complex phlya remains to be provided (reviewed in Gould, Waller, et al. 2008; Sanchez-Puerta and Delwiche 2008; Archibald 2009; Keeling 2010).
A phylogenetic analysis of the two chromerid plastid genomes leaves no doubt that they are of red algal origin, highly reduced compared with red algal plastids in general and larger than those of dinoflagellate plastids and apicomplexan apicoplasts (Janouskovec et al. 2010). But plastid genomes reflect neither the phylogenetic position nor the evolutionary history of the host. A recent analysis on the two sequenced genomes of the diatoms (stramenopiles) Thalassiosira and Phaeodactylum has added a new twist to the chromalveolate conundrum (Moustafa et al. 2009). They found that approximately 16% of the proteins potentially encoded by the nuclear genome of stramenopiles were found to reflect a green algal origin, that is, they indicated a closer phylogenetic affinity to the green lineage of primary plastids than to the red. Not unreasonably, they interpreted that observation as evidence for a green photosynthetic ancestry of the diatom host prior to the acquisition of the red algal plastid, as predicted in theory earlier on (Häuber et al. 1994, Becker et al. 2008, Frommolt et al 2008), but with several caveats, in particular concerning lineage sampling (Dagan and Martin 2009). With the goal of better understanding the phylogenomic position and photosynthetic history of protists with red secondary plastids, we have employed deep sequencing to investigate the phylogeny of C. velia expressed nuclear genes.
Materials and Methods
Cell Culture, mRNA Processing, and Library Assembly
Cells were grown at 25 °C with a 16 h light and 8 h dark cycle in Tropic Marin PRO-REEF (Tropic Marin, Germany) supplemented with f/2 AlgaBoost (AusAqua, Australia). Cells of 800 ml culture (about 5 × 105cells/ml) from three different time points (every 8 h starting 1 h before the light turned on) were harvested by centrifugation at 3,000 × g for 20 min. RNA of those three samples was isolated separately with TRIzol (Invitrogen, Germany) following the manufactures protocol with the following modification: the cell pellet was grinded in the presence of liquid nitrogen for 5–10 min before TRIzol was added. After RNA quantification, the samples were pooled so that an equal amount of each was present and send on dry ice for further processing to GATC-Biotech (Germany). At GATC, the RNA was amplified using their standard protocol for “True-Full-Length cDNA” and then additionally normalized before sequencing 2 million reads on a Titanium GS FLX (Roche). Trimming of adapter sequences, primary clustering, and assembly of the reads was performed by GATC-Biotech. Sequencing resulted in 2502269 reads with an average length of 239 bases, which were assembled into 29,856 contigs. Additionally, we included 2,854 C. velia expressed sequence tags (ESTs) from GenBank (Benson et al. 2009). Multiple copy proteins were unified and EST-contigs shorter than 100 nt removed. Furthermore, such EST-contigs with BlastN hits to the plastidal genome of C. velia (e value cutoff 10−10, downloaded from RefSeq, Pruitt et al. 2007) or the Rfam database (Gardner et al. 2009) were deleted in order to remove remnants of chloroplast-encoded transcripts and noncoding RNA families. All sequences have been deposited under JO786643–JO814452.
Database Preparation
The protein database sequences were obtained from either EuPathDB (Aurrecoechea et al. 2007) RefSeq or in the case of Cyanidioschyzon merolae (Matsuzakiet al. 2004), Ectocarpus siliculosus (Cock et al. 2010), and Emiliania huxleyi (http://genome.jgi-psf.org/Emihu1/Emihu1.download.ftp.html) from their corresponding genome project homepages. From the downloaded files, we removed C-terminal stop codons and replaced selenocysteins by Xs. In cases where no adequate number of protein sequences was available, EST-contigs were used instead or in addition. For this purpose, we created an EST-contig database by downloading ESTs for all lineages with >1,000 entries from GenBank, with exception of the Galdieria ESTs, which were downloaded from the Galdieria sulphuraria genome project homepage (Weber et al. 2004). For further information and a list of organisms, see supplementary information (Supplementary Material online). The EST-contigs were translated into proteins by the method described below and merged with the protein database.
Chromera EST-contigs were translated in a protein sequence similarly to the method described in Min et al. (2005). The EST sequences were blasted (BlastX; Altschul et al. 1997), using e value threshold ≤ 1 × 10−5 to the protein database and SwissProt database (Boeckmann et al. 2003). For sequences with blast hits, we translated the EST-contigs using the reading frame of the best blast hit (BBH). Sequences lacking a blast hit were predicted de novo by searching for the open reading frame (ORF) yielding the longest polypeptide (using both sense and antisense). In ORFs lacking an N-terminal methionine, the first codon in the EST-contig was translated into the first amino acid. When a C-terminal STOP codon was missing, the last codon in the EST-contig was translated into the last amino acid. Translated EST-contigs of C. velia were clustered into cognates of nearly identical EST-contigs by CDHIT (Weizhong and Godzik 2006) with a 95% amino acid sequence identity as a threshold, using the slow mode (–g 1). For the remaining EST-contigs, a search for reciprocal BBH (rBBH; Tatusov et al. 1997) with an e value cutoff of <1 × 10−10 was performed against the protein/EST data set of each species/genus. In case of multiple BBH having identical e values, all hits were retained. In this case, the rBBH approach was used to reduce redundant hits within the ESTs of the same gene. Pairwise alignments of Chromera EST-contigs and their rBBH were reconstructed with Needleman and Wunsch alignment algorithm (Needleman and Wunsch 1970) using Needle (EMBOSS; Rice et al. 2000). Pairs with a global amino acid identity ≥25% (excluding external gapped positions) were retained for further analysis. In case of multiple equally similar hits per one Chromera EST-contig or per one protein within the Chromera EST-contigs, the rBBH with the highest global similarity was used. Clusters of homologous proteins were constructed for Chromera EST-contigs and their homologs in all species data sets. An exclusion of 359 clusters comprising only EST-contigs yielded 3,151 clusters in total.
Phylogenetic Trees and Splits Networks
To reconstruct phylogenetic trees, all “nonchromalveolate” sequences except for one outgroup (the one showing the higher sequence similarity to the Chromera EST-contigs) were excluded from the clusters. Clusters having <4 remaining members were omitted. A total of 3,151 clusters of homologous proteins were aligned by MAFFT (Katoh and Toh 2008) using the default parameters. Multiple alignment quality was assessed using Guidance (Penn et al. 2010). Gapped alignment positions were removed and 86 short alignments (<10 positions) were excluded from further analysis. Phylogenetic trees were reconstructed from 2,258 multiple sequence alignments with PhyML (Guindon and Gascuel 2003) using the best fit model as inferred by ProtTest 3 (Darriba et al. 2011) using the Akaike information criterion (Akaike 1974) measure. For the reconstruction of a splits network, all splits within the phylogenetic trees were extracted using a Perl script and converted into a binary pattern that included 37 digits. If the split contained taxon i then digit xi in the corresponding pattern was set to “1,” otherwise it was “0.” Taxa that were missing in a tree were indicated by a “?.” The resulting patterns were summarized in a splits network using SplitsTree (Huson and Bryant 2006).
To find Chromera sequences of green or red origin, only1,174 clusters including proteins from Rhodophyta and Chloroplastida were used. All nonrhodophyta and nonchloroplastida sequences were removed from the clusters, except for those of Chromera. As an outgroup for each tree, the BBH to C. velia was used, which did not belong to Rhodophyta, Chloroplastida, a translated EST-contig or any organisms with a red algae as secondary endosymbiont. Phylogenetic trees were reconstructed from the resulting alignments (having ≥50 positions) using the same methodology described above, yielding 813 trees with an outgroup in total. The nearest neighbor to Chromera within each tree was determined by searching for the smallest clade that included C. velia and either only rhodophyta (red signal) or chloroplastida (green signal) and did not include the outgroup. For the determination of the position of C. velia in the trees as sister group or inside the red or green clades, we rooted the trees by the outgroups and searched for the second nearest neighbors using Newick Utilities package (Junier and Zdobnov 2010). Extraction of the longest branches to assess long-branch attraction was performed by the same package. Additional two split networks were reconstructed from trees sorted into red or green nearest neighbor using a composite outgroup regardless of the outgroup identity in each single tree.
Absence/Presence of Homologs in Other Species
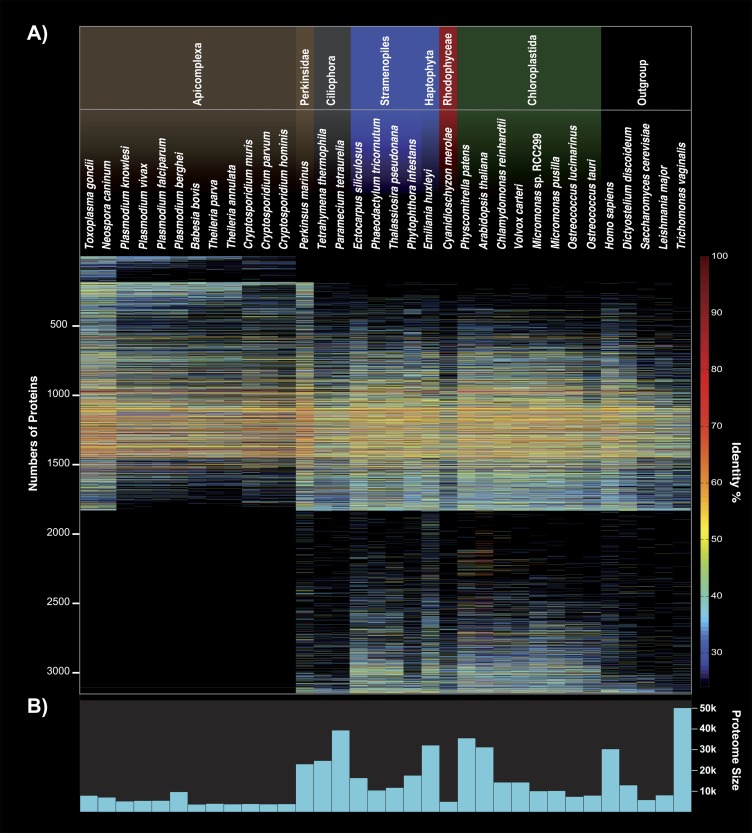
In addition to the rBBH approach, homologs to Chromera EST-contigs within each species were identified by Blasting the clustered Chromera EST-contigs against the species data set. BBHs with an e value ≤ 1 × 10−10 were aligned with their Chromera homolog using Needle (EMBOSS; Rice et al. 2000). Global pairwise alignments resulting in ≥25% amino acid identity after removal of external gapped positions were classified as a present homolog. The global amino acid identities presented in figure 2 were extracted from the pairwise alignments. The clusters that are shown along the y axis are sorted as follows: 1) all clusters specific for the apicomplexan phylum, 2) clusters of all members, 3) clusters that, except for C. velia, do have members just outside of apicomplexa. Within the three categories, the clusters were sorted by ascending number of present homologs within the Apicomplexa and descending number of present homologs within the non-Apicomplexa.
FIG. 2.—
Presence/absence pattern and identity of the nuclear-encoded Chromera velia ESTs compared with 34 organisms. (A) The 3,151 sequences are sorted by their specificity and frequency to other Apicomplexa sequences. One hundred and fifty-one sequences have homologs only in Apicomplexa, whereas 1,316 sequences had homologs only in organisms other than Apicomplexa. Note that outside the Apicomplexa, C. velia shares the highest amount of overall identity with Perkinsus marinus. In (B), the potential amount of proteins encoded within the genomes used in the analysis.
Prediction of Plastidal and Secretory Proteins
For the prediction of a signal peptide, only EST-contigs that were translated into a protein that started with a methionine were used. SignalP V3.0 (Emanuelsson et al. 2007) was used to find sequences with potential plastidal signal peptides. Chromera sequences having homologs (see “Database Preparation“) that were annotated as plastid targeted were classified as plastidal proteins as well. All 657 detected sequences were then manually inspected, and an analysis including BlastP, SignalP, and TargetP (Emanuelsson et al. 2007) was used to determine the cleavage sites and distinguish plastidal from other secretory proteins. A sequence logo of the targeting signal was created using Weblogo (Crooks et al. 2004) from positions −20 to +20 in respect to the predicted cleavage site.
Annotation of Sequences
KEGG annotations were determined by using KAAS (Moriya et al. 2007) using translated Chromera sequences as query against the KEGG maps of 27 eukaryotes including (for the complete species name, see http://www.genome.ad.jp/tools/kaas/): hsa, dme, cel, ath, osa, olu, cme, sce, ddi, ehi, pfa, pyo, pkn, tan, tpv, bbo, cpv, cho, tgo, tet, ptm, tbr, tcr, lma, tva, pti, and tps. Protein functional categories were summarized as follows: KOs were mapped to the corresponding annotations obtained from KEGG FTP Server (http://www.genome.jp/kegg/download/). The main categories “Cellular Processes” and “Environmental Information Processing” were merged into “Cellular Processing and Signaling.” Proteins in the “Unclassified, poorly characterized” category were classified as “Unclassified.” All other “Unclassified” categories were added to subcategory “Other” of the corresponding main classification. Genes potentially associated with photosynthetic were identified by searching for the KEGG categories “Photosynthesis” and “Photosynthetic.”
Results and Discussion
To obtain a broad sample of expressed genes, we isolated the RNA from exponentially growing cells every 8 h from three different time points, covering light and dark cycle. The culture contained mainly nonflagellated immotile cells, although motile cells were also observed. The RNA was enriched for full-length transcripts and normalized before library sequencing. After assembly and filtering, 32,020 contigs with a balanced GC content of 50.76% and an average length of 827 bases were used to predict the protein sequences by BlastX, using a database containing the swissprot database and 34 selected genomes (for details, see Materials and Methods). To reduce redundancy, the predicted proteins were clustered by 95% identity, and homologous clusters formed by reciprocal blast to the protein database that additionally included predicted proteins from ESTs of lineages from which no genomic data were available, such as dinoflagellates. As a result, we obtained 3,151 clusters encoding on average 239 amino acids, which were then used for all subsequent analyses shown and discussed below. The predicted ORFs use all regular codons to encode the 20 standard amino acids, and no significant preference for certain codons was observed (supplementary table 1, Supplementary Material online).
Using Conserved Targeting to Identify Plastid Proteins
In order to screen for nuclear-encoded plastid proteins, we analyzed whether the targeting signal of these proteins—having to cross four membranes to reach the stroma—is as conserved as in many other organisms harboring a plastid of red algal origin (Patron and Waller 2007; Gould, Waller, et al. 2008). The plastid targeting signals of these organisms are well conserved, the translocon components involved are potential drug targets in Apicomplexa, and they have, hence, been a central topic of research. Furthermore, do they provide a molecular nontree-based evidence for the common ancestry—though not necessarily single origin—of the secondary plastids in the group (Gould, Sommer, Hadfi, et al. 2006; Patron and Waller 2007; Sommer et al. 2007; Lim et al. 2009; Spork et al. 2009; McFadden 2010).
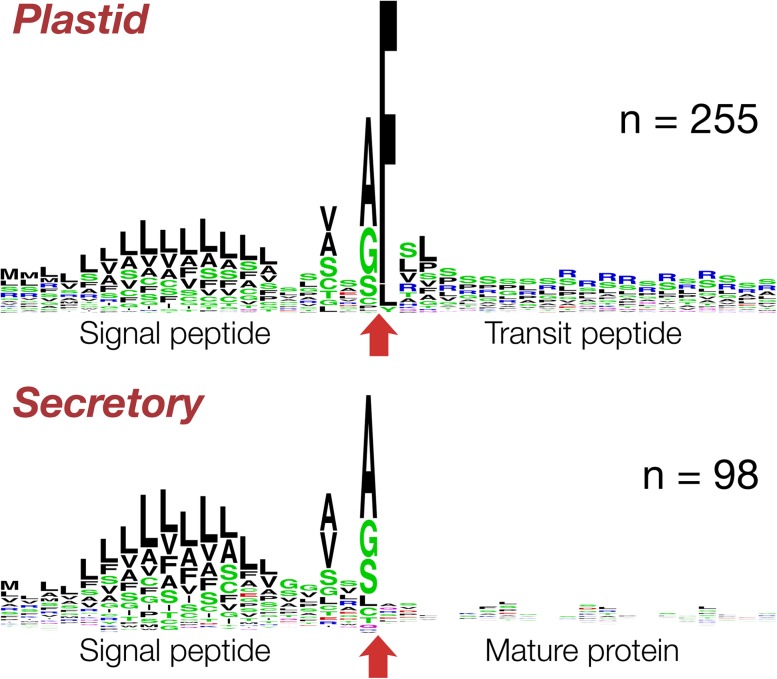
First, we collected all contigs retrieving homologs with keywords such as plastid, chloroplast, or apicoplast within their annotation and analyzed their 5′ end for an encoded signal peptide and its predicted cleavage site. From more than a 100 initial sequences, it became apparent that Chromera encodes a bipartite targeting signal (BTS) with a conserved cleavage motif (Ala-Phe) between signal and transit peptide. This position is crucial for correct targeting across the second innermost membrane of the plastids and present in cryptophytes, heterokontophytes, haptophytes, many dinoflagellates, and to a certain degree in the apicomplexan Toxoplasma (Gould, Sommer, Kroth, et al. 2006; Gruber et al. 2007; Patron and Waller 2007). It varies only a little, allowing to a lesser degree other bulky aromatic amino acids such as leucine, tyrosine, or tryptophane at the +1 position (Gruber et al. 2007; Patron and Waller 2007). The features of the subsequent transit peptide vary significantly more, even among apicomplexa themselves, but generally the level of phosphorylatable amino acids (serine/threonine) and positively charged amino acids (lysine and arginine) are elevated (Patron and Waller 2007).
In total, we collected 255 nuclear-encoded plastid proteins from our data set with a full-length 5′ end encoding a BTS, from which we generated a sequence logo (fig. 1; supplementary table 2, Supplementary Material online). In 88.6% of them, the +1 position—that is, the first amino acid of the transit peptide—was a phenylalanine, in 7% a leucine, and in 2% a tyrosine. Compared with other transit peptides targeting to secondary red plastids, the current sample from C. velia represents a mix of features individually found in the transit peptides of other lineages with red plastids. They feature both enriched level of serine residues at the beginning and a stretch of positively charged amino acids that follows (fig. 1). For the latter, arginine instead of lysine residues are used when compared with Plasmodium. The latter can most likely be attributed to the high AT content of the Plasmodium genome. We found only one secretory nonplastid protein (a cathepsin homolog) with an Ala-Phe cleavage site. In this case, though, no transit peptide was predicted to succeed the signal peptide. Only a minor amount, less than 2% of likely plastid proteins such as a thylakoid lumen protein or a uroporphyrinogen III synthase (con11984 and con06800, respectively), encode amino acids other than F, L, or Y at the +1 position of the transit peptide. Apart from a wrong targeting signal prediction, there is, furthermore, the possibility that some of those proteins are translocated across only the first 2 of the 4 membranes into the periplastidal compartment, present also in C. velia (Moore et al. 2008). These proteins harbor a BTS but different amino acids at the +1 transit peptide position (Gould, Sommer, Kroth, et al. 2006; Gruber et al. 2007). Of the latter, two ubiquitin-conjugating enzymes (con13687 and con23963) are of special interest as such enzymes are involved in protein translocation across the secondary plastids of red algal origin (McFadden 2010; Felsner et al. 2011; Moog et al. 2011).
FIG. 1.—
Sequence logo of the BTS of nuclear-encoded plastid proteins. The logo was curated based on 255 sequences, which encode an N-terminal signal peptide followed by a transit peptide. The −20/+20 positions relative to the cleavage site (red arrow) between the two parts of the BTS are shown. Secretory and plastid proteins both encode an almost identical signal peptide but only in the latter case a transit peptide follows. The N-terminal part of the transit peptide is enriched in serine residues and the C-terminal end with positively charged arginine residues.
The transit peptides of nuclear-encoded apicoplast proteins are generally characterized by a simple set of parameters, of which an overall positive charge is important (Tonkin et al. 2008). The chromerid BTS is an extraordinary example with chimeric characters, individually conserved in the different phyla and genera housing a red algal endosymbiont. The nature of the apicomplexan transit peptide holds the key to ultimately understanding how the proteins are selected from other secretory proteins in order to be transported to the apicoplast. Chromera velia, with its wealth of new sequences and a conserved targeting motif, offers a chance to commence a new search for the components involved once the entire nuclear genome becomes available.
Phyla Affinity and Phylogenetic Positioning
Evolution of protists with secondary plastids has generated a smorgasbord of organisms whose genomes show phylum-specific expansion of certain protein families and reduction of others, in Apicomplexa often reflecting the specialization of parasite–host interactions (Martens et al. 2008). Chromera velia is a nonparasitic phototrophic and basal apicomplexan and allows to investigate the question of what degree photosynthesis loss has in fact shaped apicomplexan parasites and their genomes compared with their photosynthetic relative.
Using 25% amino acid sequence identity as a cutoff, we found 151 C. velia EST-contigs that are unique to apicomplexa, and on the opposite almost 42% of our filtered EST-contigs retrieved homologs only outside the Apicomplexa (fig. 1 and supplementary information, Supplementary Material online). Twenty sequences are exclusively shared with Perkinsus marinus and 11 with ciliates. Thirty-five C. velia EST-contigs are exclusively shared with P. marinus and Apicomplexa and 13 with P. marinus and dinoflagellates, 80 with Apicomplexa, P. marinus, and dinoflagellates. In sum, 367 sequences of Chromera are exclusively shared with at least one other alveolate and five sequences were found unique to all alveolates. Expanding onto other phyla with secondary plastids of red origin (Haptophyta, Stramenopiles, and Cryptophyta), we find 143 EST-contigs exclusively shared with these. One hundred and ninety-nine EST-contigs of Chromera find homologs only outside of the alveolate, haptophyte, heterokont, and cryptophyte phyla.
As expected, the vast majority of the phyla-specific hits are proteins of unknown function (supplementary table 3, Supplementary Material online). Hence, an interpretation of what protein families might have expanded early within the apicomplexan phylum based on our EST-contig data would be unreasonable. Nevertheless, the amount of Chromera-encoded proteins that identify homologs only in organisms other than Apicomplexa, with 1,316 of 3,151, is huge. Martens et al. (2008) noticed a massive loss of genes encoding proteins involved especially in amino acid, carbohydrate, and lipid metabolisms and attributed this to the parasitic lifestyle of apicomplexa. Indeed, approximately one-third of our 1,316 EST-contigs with a KEGG annotation retrieve KEGG annotations belonging to the three metabolic categories mentioned above and only 44 of them were classified in categories associated with photosynthesis. This confirms that losing photosynthesis (not the plastid) and giving up a host-independent lifestyle has had massive impact on the parasitic apicomplexan coding capacity.
The overall identity of the nuclear-encoded EST-contigs was compared with 34 organisms and summarized in a quantifying presence/absence pattern (fig. 2). The highest overall identity is shared with nonphotosynthetic Apicomplexa; by far with Toxoplasma gondii and another sarcocystidae, Neospora canium. The loss of the plastid in the genus Cryptosporidium has not affected the overall amount of sequence identity with Chromera as much as expected, when compared with the apicoplast bearing Aconoidasida, such as Plasmodium, Babesia, or Theileria. Generally, the amount of sequence identity of expressed Chromera genes can neither be directly linked to photosynthesis ability nor genome size (fig. 2). Notably and unexpectedly, the highest fraction of overall identity outside of the apicomplexan phylum is shared with P. marinus—a nonphotosynthetic but plastid bearing oyster pathogen.
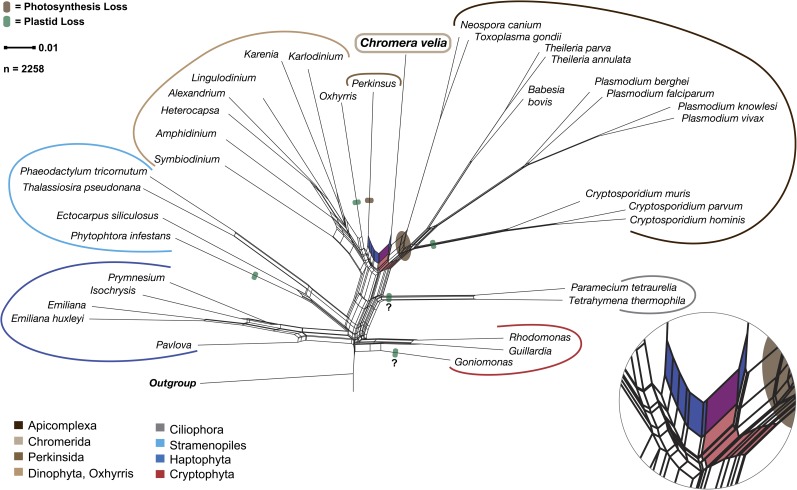
A split network of distances derived from a matrix representation of all splits from the 2,258 homolog cluster trees with at least four members supports the monophyletic origin of alveolates and positions C. velia as the most basal apicomplexan (red split in fig. 3), but there is a conflicting split that links C. velia with P. marinus. Hence, the phylogenomic analysis is consistent with the presence/absence and sequence similarity of genes shown in figure 2. This, furthermore, raises the question as to whether chromerids should not only be pigeonholed as a basal apicomplexan but might need to be treated as a separate lineage, sitting between the Perkinsidae and Apicomplexa, as already suggested by Moore et al. (2008). This is supported by our results, but should only be answered with confidence once more chromerid sequences, such as those of CCMP3155, become available. Our results, furthermore, suggest the possibility that a eukaryote–eukaryote endosymbiosis involving a red alga occurred after the ciliate phyla branched off independently, which included maybe a red alga phylogentically linked but not identical to the one engulfed by the heterokont ancestor.
FIG. 3.—
Splits network of distances derived from a matrix representation of all splits from the 2,258 homolog cluster trees generated. The net places the apicomplexan Chromera velia between nonphotosynthetic organisms. Bottom right shows an enlargement of the two splits that on the one side unites C. velia’s nuclear gene phylogeny with the Apicomplexa (light red)—whereby C. velia shows a basal position—and on the other highlights the signal linking it with the nonphotosynthetic Perkinsus marinus (blue split). Not only is this seen in the phylogeny above but also clearly in the gene distribution pattern in figure 1. The question marks indicate the two cases (Ciliates and Goniomonas), where it is disputed whether they lost or never had a plastid.
Green and Red Phylogenetic Signals among Nuclear-Encoded Proteins
The chromalveolate hypothesis posits that all members of this superphylum are united by the monophyletic origin of their secondary plastid from a red alga (Cavalier-Smith 1999). The plastid genome of the two chromerids supports a monophyletic rise of the currently present plastid in alveolates and heterokonts (Janouskovec et al. 2010) but that analysis did not include nuclear-encoded genes. Genome-sequencing projects of the two diatoms Thalassiosira pseudonana and Phaeodactylum tricornutum and the oomycete Phytophtora had noticed a green signal in their phylogenetic analyses (Armbrust et al. 2004; Tyler et al. 2006; Bowler et al. 2008), but it was not until a genome-wide search for a green signal in diatom genomes, that the idea of a more complex evolutionary scenario specifically involving green endosymbionts was formulated explicitly (Moustafa et al. 2009). The authors claim 16% of the diatoms nuclear genome could be of green origin and suggest that more than 1,700 genes were introduced into the diatom genomes by a green algal endosymbiont preceding the red one.
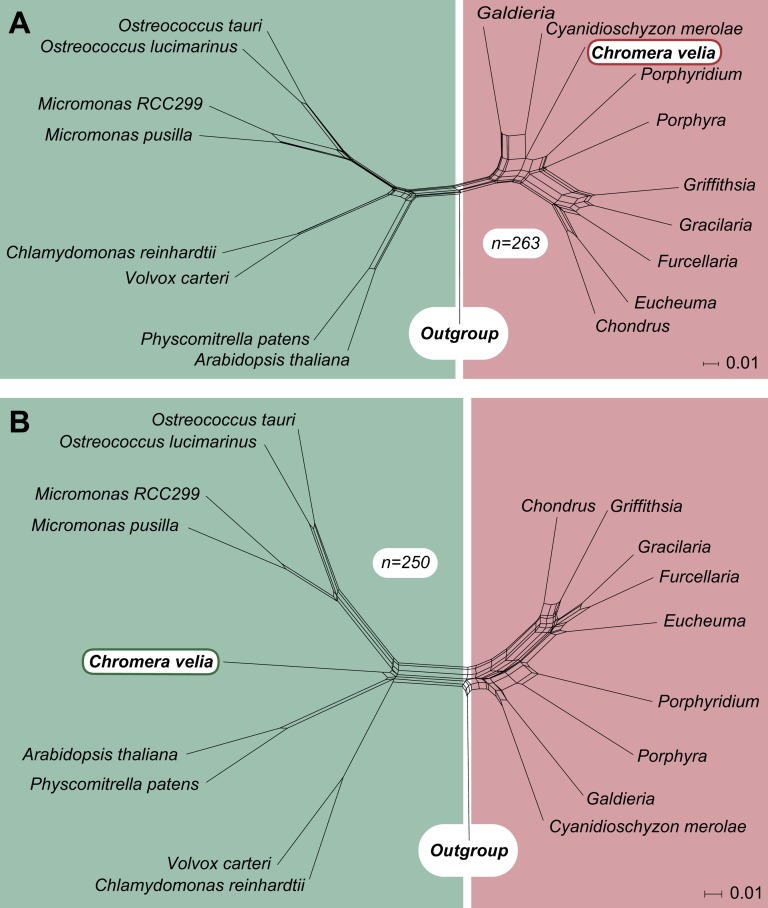
From our set of Chromera clusters, 831 sequences had homologs only to the chlorophytes (chloroplastida/viridiplantae), whereas 176 retrieve a phylogenetic association with only the rhodophytes. As this result could strongly be influenced by the difference in gene sample size available for chlorophytes and rhodophytes, we compared only those EST-contigs, for which homologs were present in both. From those sequence clusters, we generated 1,053 individual alignments (minimum of 50 amino acid positions) and maximum likelihood trees that contain a red as well as a green homologue, whereby 813 of them—comprising in total 93,745 aligned amino acid positions—furthermore, contained an outgroup. From those with an outgroup, 263 nearest neighbors were of the red, 250 of the green lineage. Furthermore, 55 of the 263 red and 86 of 250 green signals were positioned inside the red or green clades, respectively. A ratio of 1:1 was also found for the nuclear-encoded plastid proteins, where 16 proteins have a red and 16 proteins have a green affiliation. Based on the nearest neighbor trees, we generated two splits networks that reflect the position of Chromera within either the red or the green group, which themselves are clearly separated (fig. 4).
FIG. 4.—
Comparison of the red and green signal of nuclear-encoded Chromera genes. Five hundred and thirteen phylogenetic trees contained genes of green and red origin and also an outgroup. Those part almost 50–50 into trees, in which the nearest neighbor of the Chromera velia homolog is either of red or green origin. In two splits networks combining all the red and green trees separately, the apicomplexan alga is unambiguously positioned among either the rhodophytes (A) or the chloroplastida (B).
Thus, we can confirm the presence of green and red phylogenetic signals in chromalveolate genomes, as found by Moustafa et al. (2009), but the interpretation of that observation becomes very complex. The single origin of a secondary red algal plastid in the common ancestor of haptophytes, heterokonotphytes, and cryptophytes and alveolates is rejected by the most recent molecular data (Stiller et al. 2009; Baurain et al. 2010; Felsner et al. 2011) and because the ciliates lack both green and red signals in their nuclear genes, a single origin of the green signal in the common ancestor of diatoms (Moustafa et al. 2009) and Chromera (this paper) can be excluded. Thus, if we interpret the green signal as evidence for a symbiosis and gene transfer, then two independent origins of the green signal must be postulated. In the simplest scenarios, this could entail 1) independent secondary symbioses of green algal symbionts in the ancestors of the Chromera and diatom lineages followed by replacement of the green plastid with additional independent red secondary symbioses (four secondary symbioses total) or 2) origin of the green signal via secondary symbiosis in a common ancestor of the red plastid donor for the diatom and Chromera lineages, in which case these would be tertiary plastids, counter to conventional wisdom (and three symbioses at the minimum are required, two of which entail closely related endosymbionts).
In general, that seems to be quite a bit of symbiosis and gene transfer in parallel, so it is prudent to question the premise that the green signal does in fact represent evidence for a biological event rather than being a manifestation of sampling, random, or other bias in the data. Because the red signal can be readily attributed to the origin of the red plastid, it is the green signal that is suspect, as it is the only reason to entertain the possibility of a large number of inferred symbioses that are otherwise not supported by any independent data. We looked to see if there was a tendency for the green alignments to be shorter, less reliable, or more poorly conserved, such that these factors might generate spurious phylogenetic signal. No such tendency was detected. We looked to see if amino acid content of the green versus red genes was significantly different and again no such tendency was detected (supplementary table 4, Supplementary Material online). We looked to see whether a strong skew existed with respect to functional categories, but we observed none (supplementary table 5, Supplementary Material online). To test for a possible long-branch attraction caused by using only one outgroup sequence, we checked if the tree root is located between the two longest branches in the tree. Long branch attraction was observed in only 10 red and 14 green phylogenies. Furthermore, we tested for differences regarding organism distribution, which were used as an outgroup, and found no significant differences.
Could the green signal both in diatoms and in Chromera simply be a random phylogenetic error? This is a possibility. How so? If we go back to Moustafa et al. (2009), what they reported was a collection of green phylogenetic signals corresponding to diatom nuclear genes that branch with chlorophyte, streptophyte, and prasinophyte homologues. At face value, their data indicated three independent green secondary endosymbiotic events (at least), but the simplest and most reasonable interpretation—and the one that they favored—was that it was in fact only one green event with an endosymbiont (donor) of probably prasinophyte-like phylogenetic identity, whereby the streptophyte and chlorophyte signals represent, by inference, random phylogenetic error. But only one branch removed from the green lineage resides the red lineage. In other words, in the interpretation of Moustafa et al. (2009) regarding diatoms, one green endosymbiosis gave rise to three different green signals, two of which are the result of phylogenetic error (and very implicitly, the later red endosymbiosis for which we have evidence in the form of the plastid gave rise to no error at all). In our current interpretation of the diatom and the Chromera data, one red endosymbiosis each gave rise to the red signal in those lineages, but each red signal also contained error, namely all three green signals that Moustafa et al. (2009) observe (not just the two that they assume to be in error). Accordingly, in Chromera, the green signal is best interpreted as a phylogenetic error, in toto. Indeed, Moustafa et al. (2009) found about 1,700 green and about 400 red genes (a ratio of 4:1) in diatoms, and in our analysis, with slightly improved sampling, we see a ratio of about 1:1 (250:263). When we performed the same analysis with just the red algal genome of C. merolae, as Moustafa et al. (2009) did, the green signal increased and the red signal decreased by about 7% (56% and 44% vs. 49% and 51%). So, the green signal is attributable to sampling. A report by Stiller et al. (2009), which focused on red signals within the organisms having potentially lost their red algal endosymbiont, describes a similar correlation and they conclude: “to move away from a posteriori data interpretations and toward direct tests of explicit predictions from standing and future evolutionary hypothesis.” Hence, we expect that with improved sampling—especially more than one red algal genome available—and with more refined phylogenetic methods, the green signal in both the diatoms and Chromera should continue to decline. Whether the green signal is then reduced to nothing more but “background noise” remains to be seen.
In general, the more genes that are investigated to explain the origin of complex plastids, the more conflict is observed in the data (reviewed, e.g., in Gould, Waller, et al. 2008; Sanchez-Puerta and Delwiche 2008; Archibald 2009; Keeling 2010).The more organisms and genomic data are studied, the more apparent it becomes that a monophyletic scenario summarized in the chromalveolate hypothesis—although maybe attractive—must be rejected. The origin of organisms with secondary red plastids might entail similar but nonidentical hosts (that of heterokonts, haptophytes, and cryptophytes) and similar but nonidentical endosymbionts (that of heterokonts and alveolates). Untangling these branches, keeping random phylogenetic errors in mind, remains a substantial challenge.
Supplementary Material
supplementary information and supplementary tables 1–5 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
We thank Jan Slapeta for discussing RNA isolation methods. This work is funded by Fit for Excellence (grant number 38700018). S.B.G. was furthermore funded by the Strategischer Forschungsfonds (grant number 3702008), both of the HH-University (HHU) Dusseldorf. Computational support and infrastructure was provided by the Zentrum für Informations- und Medientechnologie of the HHU Dusseldorf.
References
- Akaike H. A new look at the statistical model identification. IEEE Trans Automat Contr. 1974;19:716–723. [Google Scholar]
- Altschul SF, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Archibald JM. The puzzle of plastid evolution. Curr Biol. 2009;19:81–88. doi: 10.1016/j.cub.2008.11.067. [DOI] [PubMed] [Google Scholar]
- Armbrust EV, et al. The genome of the diatom Thalassiosira pseudonana: ecology, evolution, and metabolism. Science. 2004;306:79–86. doi: 10.1126/science.1101156. [DOI] [PubMed] [Google Scholar]
- Aurrecoechea C, et al. ApiDB: integrated resources for the apicomplexan bioinformatics resource center. Nucleic Acids Res. 2007;35:427–430. doi: 10.1093/nar/gkl880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachvaroff TR, Puerta MVS, Delwiche CF. Chlorophyll c-containing plastid relationships based on analyses of a multigene data set with all four chromalveolate lineages. Mol Evol Biol. 2005;22:1772–1782. doi: 10.1093/molbev/msi172. [DOI] [PubMed] [Google Scholar]
- Baurain D, et al. Phylogenomic evidence for separate acquisition of plastids in cryptophytes, haptophytes, and stramenopiles. Mol Evol Biol. 2010;27:1698–1709. doi: 10.1093/molbev/msq059. [DOI] [PubMed] [Google Scholar]
- Becker B, Hoef-Emden K, Melkonian M. Chlamydial genes shed light on the evolution of photoautotrophic eukaryotes. BMC Evol Biol. 2008;8:203. doi: 10.1186/1471-2148-8-203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Sayers EW. GenBank. Nucleic Acids Res. 2009;37:26–31. doi: 10.1093/nar/gkn723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodyl A. Do plastid-related characters support the chromalveolate hypothesis? J Phycol. 2005;41:712–719. [Google Scholar]
- Bodyl A, Stiller JW, Mackiewicz P. Chromalveolate plastids: direct descent or multiple endosymbioses? Trends Ecol Evol. 2009;24:119–121. doi: 10.1016/j.tree.2008.11.003. [DOI] [PubMed] [Google Scholar]
- Boeckmann B, et al. The SWISS-PROT protein knowledgebase and its supplement TrEMBL in 2003. Nucleic Acids Res. 2003;31:365–370. doi: 10.1093/nar/gkg095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowler C, et al. The Phaeodactylum genome reveals the evolutionary history of diatom genomes. Nature. 2008;456:239–244. doi: 10.1038/nature07410. [DOI] [PubMed] [Google Scholar]
- Cavalier-Smith T. Cell diversification in heterotrophic flagellates. In: Patterson D, Larsen J, editors. The biology of free-living heterotrophic flagellates. Oxford: Clarendon Press; 1991. pp. 113–131. [Google Scholar]
- Cavalier-Smith T. Principles of protein and lipid targeting in secondary symbiogenesis: euglenoid, dinoflagellate, and sporozoan plastid origins and the eukaryote family tree. J Eukaryot Microbiol. 1999;46:347–366. doi: 10.1111/j.1550-7408.1999.tb04614.x. [DOI] [PubMed] [Google Scholar]
- Cock J, et al. The Ectocarpus genome and the independent evolution of multicellularity in brown algae. Nature. 2010;465:617–621. doi: 10.1038/nature09016. [DOI] [PubMed] [Google Scholar]
- Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dagan T, Martin W. Seeing green and red in diatom genomes. Science. 2009;324:1651–1652. doi: 10.1126/science.1175765. [DOI] [PubMed] [Google Scholar]
- Darriba D, Taboada GL, Doallo R, Posada D. ProtTest 3: fast selection of best-fit models of protein evolution. Bioinformatics. 2011;27:1164–1175. doi: 10.1093/bioinformatics/btr088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen JA, et al. Macronuclear genome sequence of the ciliate Tetrahymena thermophila, a model eukaryote. PLoS Biol. 2006;4:e286. doi: 10.1371/journal.pbio.0040286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuelsson O, Brunak S, von Heijne G, Nielsen H. Locating proteins in the cell using TargetP, SignalP and related tools. Nat Protoc. 2007;2:953–971. doi: 10.1038/nprot.2007.131. [DOI] [PubMed] [Google Scholar]
- Felsner G, et al. ERAD components in organisms with complex red plastids suggest recruitment of a preexisting protein transport pathway for the periplastid membrane. Genome Biol Evol. 2011;3:140–150. doi: 10.1093/gbe/evq074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frommolt R, et al. Ancient recruitment by chromists of green algal genes encoding enzymes for carotenoid biosynthesis. Mol Biol Evol. 2008;25:2653–2667. doi: 10.1093/molbev/msn206. [DOI] [PubMed] [Google Scholar]
- Gardner PP, et al. Rfam: updates to the RNA families database. Nucleic Acids Res. 2009;37:136–140. doi: 10.1093/nar/gkn766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould SB, Sommer MS, Hadfi K, et al. Protein targeting into the complex plastid of cryptophytes. J Mol Evol. 2006;62:674–681. doi: 10.1007/s00239-005-0099-y. [DOI] [PubMed] [Google Scholar]
- Gould SB, Sommer MS, Kroth PG, et al. Nucleus-to-nucleus gene transfer and protein retargeting into a remnant cytoplasm of cryptophytes and diatoms. Mol Biol Evol. 2006;23:2413–2422. doi: 10.1093/molbev/msl113. [DOI] [PubMed] [Google Scholar]
- Gould SB, Tham WH, Cowman AF, McFadden GI, Waller RF. Alveolins, a new family of cortical proteins that define the protist infrakingdom Alveolata. Mol Biol Evol. 2008;25:1219–1230. doi: 10.1093/molbev/msn070. [DOI] [PubMed] [Google Scholar]
- Gould SB, Waller RF, McFadden GI. Plastid evolution. Annu Rev Plant Biol. 2008;59:491–517. doi: 10.1146/annurev.arplant.59.032607.092915. [DOI] [PubMed] [Google Scholar]
- Gruber A, et al. Protein targeting into complex diatom plastids: functional characterisation of a specific targeting motif. Plant Mol Biol. 2007;64:519–530. doi: 10.1007/s11103-007-9171-x. [DOI] [PubMed] [Google Scholar]
- Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- Harper JT, Waanders E, Keeling PJ. On the monophyly of chromalveolates using a six-protein phylogeny of eukaryotes. Int J Syst Evol Microbiol. 2005;55:487–496. doi: 10.1099/ijs.0.63216-0. [DOI] [PubMed] [Google Scholar]
- Häuber MM, Müller SB, Speth V, Maier UG. How to evolve a complex plastid?—A hypothesis. Bot Acta. 1994;107:383–386. [Google Scholar]
- Huang J, et al. Phylogenomic evidence supports past endosymbiosis, intracellular and horizontal gene transfer in Cryptosporidium parvum. Genome Biol. 2004;5:R88. doi: 10.1186/gb-2004-5-11-r88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huson DH, Bryant D. Application of phylogenetic networks in evolutionary studies. Mol Biol Evol. 2006;23:254–267. doi: 10.1093/molbev/msj030. [DOI] [PubMed] [Google Scholar]
- Janouskovec J, Horak A, Obornik M, Lukes J, Keeling PJ. A common red algal origin of the apicomplexan, dinoflagellate, and heterokont plastids. Proc Natl Acad Sci U S A. 2010;107:10949–10954. doi: 10.1073/pnas.1003335107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jomaa H, et al. Inhibitors of the non-mevalonate pathway of isoprenoid biosynthesis as antimalarial drugs. Science. 1999;285:1573–1576. doi: 10.1126/science.285.5433.1573. [DOI] [PubMed] [Google Scholar]
- Junier T, Zdobnov EM. The Newick utilities: high-throughput phylogenetic tree processing in the UNIX shell. Bioinformatics. 2010;26:1669–1670. doi: 10.1093/bioinformatics/btq243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Toh H. Recent developments in the MAFFT multiple sequence alignment program. Brief Bioinform. 2008;9:286–298. doi: 10.1093/bib/bbn013. [DOI] [PubMed] [Google Scholar]
- Keeling PJ. The endosymbiotic origin, diversification and fate of plastids. Philos Trans R Soc Lond B Biol Sci. 2010;365:729–748. doi: 10.1098/rstb.2009.0103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim L, Kalanon M, McFadden GI. New proteins in the apicoplast membranes: time to rethink apicoplast protein targeting. Trends Parasitol. 2009;25:197–200. doi: 10.1016/j.pt.2009.02.001. [DOI] [PubMed] [Google Scholar]
- Martens C, Vandepoele K, Van de Peer Y. Whole-genome analysis reveals molecular innovations and evolutionary transitions in chromalveolate species. Proc Natl Acad Sci U S A. 2008;105:3427–3432. doi: 10.1073/pnas.0712248105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuzaki M, et al. Genome sequence of the ultrasmall unicellular red alga Cyanidioschyzon merolae 10D. Nature. 2004;428:653–657. doi: 10.1038/nature02398. [DOI] [PubMed] [Google Scholar]
- Matsuzaki M, et al. A DNA-lacking plastid in the oyster pathogen Perkinsus marinus. Phycologia. 2009;48:82–83. [Google Scholar]
- McFadden GI. The apicoplast. Protoplasma. 2010 doi: 10.1007/s00709-010-0250-5. doi: 10.1007/s00709-010-0250-5. [DOI] [PubMed] [Google Scholar]
- Min XJ, Butler G, Storms R, Tsang A. OrfPredictor: predicting protein-coding regions in EST-derived sequences. Nucleic Acids Res. 2005;33:677–680. doi: 10.1093/nar/gki394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moog D, Stork S, Zauner S, Maier UG. In silico and in vivo investigations of proteins of a minimized eukaryotic cytoplasm. Genome Biol Evol. 2011;3:375–382. doi: 10.1093/gbe/evr031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore RB, et al. A photosynthetic alveolate closely related to apicomplexan parasites. Nature. 2008;451:959–963. doi: 10.1038/nature06635. [DOI] [PubMed] [Google Scholar]
- Moriya Y, Itoh M, Okuda S, Yoshizawa A, Kanehisa M. KAAS: an automatic genome annotation and pathway reconstruction server. Nucleic Acids Res. 2007;35:182–185. doi: 10.1093/nar/gkm321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moustafa A, et al. Genomic footprints of a cryptic plastid endosymbiosis in diatoms. Science. 2009;324:1724–1726. doi: 10.1126/science.1172983. [DOI] [PubMed] [Google Scholar]
- Needleman SB, Wunsch CD. A general method applicable to the search for similarities in the amino acid sequence of two proteins. J Mol Biol. 1970;48:443–453. doi: 10.1016/0022-2836(70)90057-4. [DOI] [PubMed] [Google Scholar]
- Obornik M, et al. Morphology and ultrastructure of multiple life cycle stages of the photosynthetic relative of Apicomplexa, Chromera velia. Protist. 2011;162:115–130. doi: 10.1016/j.protis.2010.02.004. [DOI] [PubMed] [Google Scholar]
- Patron NJ, Inagaki Y, Keeling PJ. Multiple gene phylogenies support the monophyly of cryptomonad and haptophyte host lineages. Curr Biol. 2007;17:887–891. doi: 10.1016/j.cub.2007.03.069. [DOI] [PubMed] [Google Scholar]
- Patron NJ, Waller RF. Transit peptide diversity and divergence: a global analysis of plastid targeting signals. Bioessays. 2007;29:1048–1058. doi: 10.1002/bies.20638. [DOI] [PubMed] [Google Scholar]
- Penn O, Privman E, Landan G, Graur D, Pupko T. An alignment confidence score capturing robustness to guide-tree uncertainty. Mol Biol Evol. 2010;27:1759–1767. doi: 10.1093/molbev/msq066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruitt KD, Tatusova T, Maglott DR. NCBI reference sequence (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2007;35:61–65. doi: 10.1093/nar/gki025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ralph SA, et al. Tropical infectious diseases: metabolic maps and functions of the Plasmodium falciparum apicoplast. Nat Rev Microbiol. 2004;2:203–216. doi: 10.1038/nrmicro843. [DOI] [PubMed] [Google Scholar]
- Reyes-Prieto A, Moustafa A, Bhattacharya D. Multiple genes of apparent algal origin suggest ciliates may once have been photosynthetic. Curr Biol. 2008;18:956–962. doi: 10.1016/j.cub.2008.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice P, Longden I, Bleasby A. EMBOSS: the European molecular biology open software suite. Trends Genet. 2000;16:276–277. doi: 10.1016/s0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- Sanchez-Puerta MV, Delwiche CF. A hypothesis for plastid evolution in chromalveolates. J Phycol. 2008;44:1097–1107. doi: 10.1111/j.1529-8817.2008.00559.x. [DOI] [PubMed] [Google Scholar]
- Seeber F. Biogenesis of iron-sulphur clusters in amitochondriate and apicomplexan protists. Int J Parasitol. 2002;32:1207–1217. doi: 10.1016/s0020-7519(02)00022-x. [DOI] [PubMed] [Google Scholar]
- Sommer MS, et al. Der1-mediated preprotein import into the periplastid compartment of chromalveolates? Mol Biol Evol. 2007;24:918–928. doi: 10.1093/molbev/msm008. [DOI] [PubMed] [Google Scholar]
- Spork S, et al. An unusual ERAD-like complex is targeted to the apicoplast of Plasmodium falciparum. Eukaryot Cell. 2009;8:1134–1145. doi: 10.1128/EC.00083-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiller JW, Huang J, Ding Q, Tian J, Goodwillie C. Are algal genes in nonphotosynthetic protists evidence of historical plastid endosymbioses? BMC Genomics. 2009;10:484. doi: 10.1186/1471-2164-10-484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoebe B, Maier UG. One, two, three: nature’s tool box for building plastids. Protoplasma. 2002;219:123–130. doi: 10.1007/s007090200013. [DOI] [PubMed] [Google Scholar]
- Tatusov RL, Koonin EV, Lipman DJ. A genomic perspective on protein families. Science. 1997;278:631–637. doi: 10.1126/science.278.5338.631. [DOI] [PubMed] [Google Scholar]
- Tonkin CJ, et al. Evolution of malaria parasite plastid targeting sequences. Proc Natl Acad Sci U S A. 2008;105:4781–4785. doi: 10.1073/pnas.0707827105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyler BM, et al. Phytophthora genome sequences uncover evolutionary origins and mechanisms of pathogenesis. Science. 2006;313:1261–1266. doi: 10.1126/science.1128796. [DOI] [PubMed] [Google Scholar]
- Waller RF, et al. Nuclear-encoded proteins target to the plastid in Toxoplasma gondii and Plasmodium falciparum. Proc Natl Acad Sci U S A. 1998;95:12352–12357. doi: 10.1073/pnas.95.21.12352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber AP, et al. EST-analysis of the thermo-acidophilic red microalga Galdieria sulphuraria reveals potential for lipid A biosynthesis and unveils the pathway of carbon export from rhodoplasts. Plant Mol Biol. 2004;55:17–32. doi: 10.1007/s11103-004-0376-y. [DOI] [PubMed] [Google Scholar]
- Weizhong L, Godzik A. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics. 2006;22:1658–1659. doi: 10.1093/bioinformatics/btl158. [DOI] [PubMed] [Google Scholar]
- Zhang H, Campbell DA, Sturm NR, Dungan CF, Lin S. Spliced leader RNAs, mitochondrial gene frameshifts and multi-protein phylogeny expand support for the genus Perkinsus as a unique group of alveolates. PLoS One. 2011;6:e19933. doi: 10.1371/journal.pone.0019933. [DOI] [PMC free article] [PubMed] [Google Scholar]