Abstract
Widespread antimalarial resistance has been a barrier to malaria elimination efforts in Sri Lanka. Analysis of genetic markers in historic parasites may uncover trends in the spread of resistance. We examined the frequency of Plasmodium falciparum chloroquine transporter (pfcrt; codons 72–76) haplotypes in Sri Lanka in 1996–1998 and 2004–2006 using a high-resolution melting assay. Among 59 samples from 1996 to 1998, we detected the SVMNT (86%), CVMNK (10%), and CVIET (2%) haplotypes, with a positive trend in SVMNT and a negative trend in CVMNK frequency (P = 0.004) over time. Among 24 samples from 2004 to 2006, we observed only the SVMNT haplotype. This finding indicates selection for the SVMNT haplotype over time and its possible fixation in the population.
Introduction
Malaria affects over 2 billion people across the globe who live at risk of acquiring this infection.1 It primarily affects the poor populations in tropical and subtropical parts of the world, where the temperature and rainfall are conducive to the development of the malaria parasites in Anopheles mosquitoes. In Sri Lanka, malaria is traditionally considered to be endemic in two-thirds of the country, but annual incidence in recent years has been remarkably low, with a precipitous drop from 210,000 to 467 cases between the years 2000 and 2009.2 A national plan is currently in place for the elimination of malaria from this island over a 5-year period.3
Meanwhile, resistance to first-line antimalarials, particularly chloroquine, has become increasingly common in the Plasmodium falciparum population.4 The first chloroquine-resistant cases on the island were identified in 1984 in the Dambulla area of Matale district.5 Since that time, studies have found treatment failure rates ranging from 30% to 53.5% and in vitro resistance rates around 65% in different regions of the country.6,7
Chloroquine resistance has been linked to single-nucleotide polymorphisms (SNPs) in the P. falciparum chloroquine transporter (pfcrt) gene, and the K76T mutation specifically has been considered the best molecular marker for tracking resistance in parasite populations.8,9 Furthermore, the haplotypes observed in codons 72–76 have been associated with different fitness costs, coresistance to amodiaquine, and geographic origins.10–12
A previous study has investigated these markers in Sri Lanka, finding that, among 76 samples collected from patients in the Mannar district of Northern Province in 2002 and 2004, 76.3% had the T76 mutation.7 However, no published studies from outside the Mannar district have yet measured the change in haplotype frequencies in Sri Lanka over time.
Longitudinal data from past years have the potential to inform treatment and control efforts during the ongoing elimination program by providing a baseline for measuring changes in haplotype frequencies. This information may also provide clues to the evolutionary history of drug-resistant parasites in Sri Lanka. A sensitive, high-throughput method for detecting haplotypes is important for both observing past trends and monitoring current changes in resistance markers in the population. In recent years, genotyping technology using high-resolution melting assays has been found to be more sensitive, time-efficient, cost-efficient, and easy to use than traditional methods like restriction enzyme digestion and direct sequencing (Daniels R, unpublished data). This study uses a high-resolution melting assay to examine the changes in haplotype frequencies in codons 72–76 of the pfcrt gene in Sri Lankan parasites collected over two periods: the first period was between 1996 and 1998 from the Anadhurapura district, and the second period was between 2004 and 2006 from several endemic districts in Sri Lanka.
Materials and Methods
Sample collection.
Samples from 1996, 1997, and 1998 were collected from slide-positive patients who presented themselves at government health facilities in Anadhurapura district. In total, the 13, 16, and 30 samples collected in 1996, 1997, and 1998, respectively, were successfully analyzed for this study. A second, smaller set was collected between 2004 and 2006 among slide-positive patients attending one of the health posts/hospitals in the districts of Vavuniya, Trincomalee, Anuradhapura, Polonnaruwa, Ampara, Batticaloa, and Kurunegala as part of an earlier study13 (Figure 1). Ten, thirteen, and one samples collected from 2004, 2005, and 2006, respectively, were successfully analyzed in this study. All blood samples were collected on filter paper, and genomic DNA was extracted using a chelex-100 method (Bio-Rad Laboratories, Hercules, CA). Ethical clearance for this project was granted by the Committee on Research and Ethical Review at the Faculty of Medicine, Peradeniya, Kandy, and verbal consent was obtained from participants, parents, and/or guardians.
Figure 1.
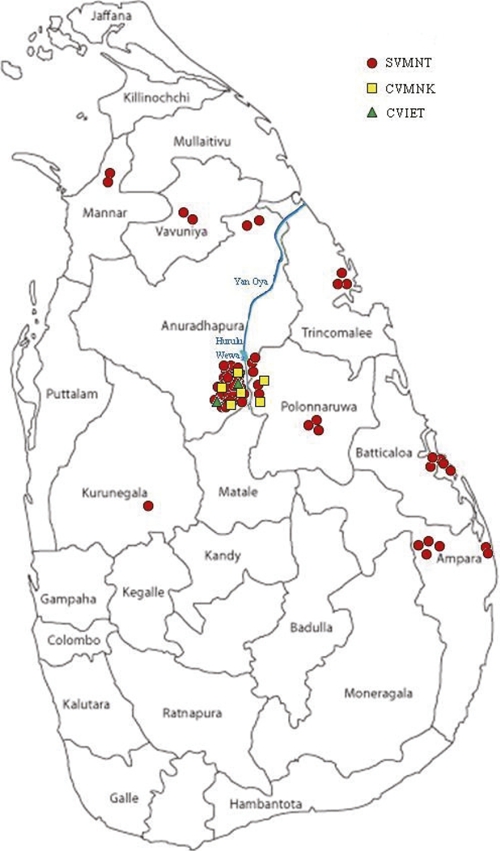
Map of Sri Lanka, showing the patient sample collection sites and haplotype distribution in each site.
Polymerase chain reaction.
A nested polymerase chain reaction (PCR) was performed on samples to obtain a 167-bp fragment containing pfcrt codons 72–76. The outer primers used were PFCRT1f: 5′-CCTTGTCGACCTTAACAGATG-3′ and PFCRT1r: 5′-GTTTTATATTGGTAGGTGGAATAGATTCTC-3′, and the inner primers were PfCRT2f: 5′-CTTGGTAAATGTGCTCATGTGTTTAAAC-3′ and PFCRT2r: 5′-GTTTCGGATGTTACAAAACTATAGTTACC-3′. For the outer reaction, 1 μL supernatant from the Chelex extraction was added to a 25-μL reaction mixture containing 2× goTaq Green master mix (Promega, Madison, WI) and 0.3 μM outer primers. The nested reaction was performed by adding 2 μL products from the first reaction to a 50-μL reaction mixture containing the inner primers. The PCR conditions for the outer PCR were initial activation for 3 minutes at 94°C followed by 30 cycles of 94°C for 30 seconds, 51°C for 30 seconds, 65°C for 45 seconds, and a final extension at 65°C for 3 minutes. The conditions for the inner PCR were initial activation at 94°C for 3 minutes followed by 30 cycles of 94°C for 30 seconds, 55°C for 30 seconds, 65°C for 30 seconds, and a final extension at 65°C for 3 minutes. Products were run on 2% agarose gels and visualized by ethidium bromide staining.
High-resolution melting analysis.
An aliquot of all PCR products were processed by Exo-sap-it (USB, Cleveland, OH) according to manufacturer protocols and quantified using a Nanodrop spectrophotometer (Thermo Scientific, Waltham, MA) before high-resolution melting (HRM) analysis.
These products were asymmetrically PCR-amplified using the internal primers described above in 5-μL reactions using 1 ng template, 2.5× LightScanner Master Mix (catalog #HRLS-ASY-0003; Idaho Technology, Inc., Salt Lake City, UT), 0.05 μM forward primer, 0.25 μM reverse primer, and 0.20 μM unlabeled, 3′-blocked probe (5′-GTGTAGTGTAATGAAAATTTTTG-3′ BLOCK) that was designed to be centered around the codons 72–76 region of pfcrt. The block is used to prevent extension of the probe during amplification; the purpose of the asymmetric primer concentrations is to drive the production of a single-stranded product that will bind to this blocked probe during HRM analysis. The combination of blocked probe and single-stranded product ensures that enough material is present for probe-based melting analysis.
The reactions were overlaid with mineral oil and amplified (95°C for 120 seconds denaturation and 45 cycles at 95°C and 68°C for 30 seconds each followed by one cycle each of 95°C and 28°C for 30 seconds).
After amplification, the samples were analyzed by HRM on a LightScanner-384 (Idaho Technology, Inc., Salt Lake City, UT) starting at 40°C and ending at 90°C. Sequencing-verified controls for haplotypes were run alongside the experimental samples. Samples were analyzed using LightScanner software according to Idaho Technology, Inc. protocol; sample melting plots were normalized and transformed to display differential plots of the melting peaks both for the entire amplicon and in the probe melting region.
Cloning and sequencing.
A subset of PCR reactions was also sequenced. Because of short amplicon length, PCR products were cloned using a TOPO TA cloning kit (Invitrogen, Carlsbad, CA) using the manufacturer's protocol before sequencing. Plasmids were isolated using a Qiagen Miniprep kit and Sanger sequenced by standard methods using the M13 forward and reverse primers (Genewiz, Boston, MA). Sequencing data were analyzed using the Seqman software package (Lasergene Version 7.1; DNA Star Inc., Madison, WI).
Statistical analysis.
Fisher–Freeman–Halton exact test was used to compare the frequency of the three haplotypes over the 3-year period (1996–1998). Cochran–Armitage exact trend test was used to evaluate whether frequencies of the two more common haplotypes had a linear increasing or decreasing trend across the 3 years. All analyses were performed in StatXact 8 (Cytel Inc., Cambridge, MA).
Results
HRM analysis of 59 samples from 1996 to 1998 in Anadhurapura found three distinct peaks for the codons 72–76 region of the pfcrt gene (Figure 2). The majority of the isolates showed a melting curve consistent with the mutant SVMNT haplotype (N = 51; 86.4%), and a small number of samples showed the wild-type CVMNK pattern (N = 6; 10.2%) or the mutant CVIET pattern (N = 2; 3.4%). Minor outlier peaks in four of the SVMNT curves may suggest mixed infections but more likely are the result of artifacts from the amplification process with variant bases added from enzyme error. Of 43 samples from the 1996–1997 set that were successfully cloned and sequenced, only one had a single synonymous nucleotide discrepancy compared with HRM results, which was most likely because of enzyme error during either the cloning or sequencing process.
Figure 2.

High-resolution melt curves. The three peaks correspond to the CVIET (blue), SVMNT (grey), and CVMNK (red) haplotypes. To better visualize the melting peak differences, the temperature is plotted against the negative rate of change of the fluorescence over time and normalized against the background fluorescence.
Among the 24 samples collected from seven districts in 2004–2006 (10 from 2004, 13 from 2005, and 1 from 2006), HRM peaks indicated that all isolates carried the SVMNT allele. Figure 1 shows the geographic distribution of haplotypes from both 1996–1998 and 2004–2006.
According to the Fisher exact test, there was a statistically significant change in the distribution of haplotypes over the 3-year period from 1996 to 1998 (P = 0.007) (Table 1). Cochran–Armitage exact test showed a linear trend in the increasing frequency of SVMNT mutants and decreasing frequency of the wild-type CVMNK over the 3-year period (P = 0.004). Because there were only two CVIET mutants observed with no distinguishable trend in frequency, these samples were excluded from the trend test.
Table 1.
Distribution pattern of P. falciparum pfcrt haplotypes over time
| Haplotype | Sequence | Frequency of haplotypes (%) | |||
|---|---|---|---|---|---|
| Anadhurapura | Seven districts | ||||
| 1996 | 1997 | 1998 | 2004–2006 | ||
| CVMNK | TGTGTAATGAATAAA (six samples sequenced) | 4 (31%) | 2 (13%) | 0 (0) | 0 (0) |
| SVMNT | AGTGTAATGAATACA (36 samples sequenced)* | 9 (69%) | 13 (81%) | 29 (97%) | 24 (100%) |
| CVIET | TGTGTAATTGAAACA (one sample sequenced) | 0 (0) | 1 (6%) | 1 (3%) | 0 (0) |
1996–1998 = Fisher–Freeman–Halton test (P = 0.007). 2004–2006 = Cochran–Armitage test (CVMNK and SVMNT; P = 0.004).
One sample was sequenced as AGTGTGATGAATACA.
Discussion
To our knowledge, this study is the first to examine changes in the distribution of pfcrt haplotype frequencies in Sri Lanka over an extended period of decreasing malaria prevalence. Although the sample size is small, the data indicates a significant trend in haplotype frequencies. HRM analysis was a sensitive and high-throughput method for detecting haplotypes that was less labor-intensive and more cost-efficient than cloning and direct sequencing of samples.
We found a significant change in haplotype frequencies between 1996 and 1998, suggesting that there was strong selection pressure in favor of the SVMNT haplotype and against the wild type. The replacement of wild-type parasites in the population with the SVMNT mutants corresponds to a period of continuous chloroquine use as first treatment and decreasing malaria incidence in the district of Anadhurapura. The total number of malaria cases in the district declined from 7,995 in 1996 to 5,528 cases in 1998 and continued to drop even more rapidly over the next 10 years, corresponding with nationwide trends. By 2004, the nationwide prevalence of P. falciparum in Sri Lanka was 520, and by 2006, only 27 cases were reported. The sole prevalence of the SVMNT parasites between 2004 and 2006 may indicate a fixation of the haplotype on the island.
The change in frequency might be explained by SVMNT parasites' relative fitness over both the wild-type CVMNK and the mutant CVIET parasites under the standard treatment regimen at the time. Before May 2008, chloroquine (CQ) was the standard first-line treatment of malaria cases in Sri Lanka. Furthermore, a Sri Lankan study conducted several years ago suggested that recrudescent CQ-resistant malaria strains had a lower detectability rate and a higher gametocyte rate that may have conferred an additional survival advantage over CQ-sensitive strains.6 Recent studies also suggest that CVIET mutants may incur a higher fitness cost than SVMNT mutants, which was indicated by the recovery of CQ-sensitive strains after the cessation of drug pressure in regions where the CVIET haplotype was prevalent but not in regions of high SVMNT prevalence.11 This finding may be informative in planning future treatment policies, because artemisinin-based combination therapy (ACT) has replaced CQ as first-line therapy for P. falciparum-infected patients in Sri Lanka as of 2008.3 Furthermore, several recent studies indicate that the SVMNT mutations may be sufficient to confer resistance to amodiaquine.11,12 Although the current ACT regimen used in Sri Lanka is artemether-lumefantrine, the high prevalence of the SVMNT haplotype found in Sri Lanka cautions against the widespread adoption of amodiaquine-based ACTs in the country.
The highly prevalent SVMNT type strain in Sri Lanka may be related to parasites in India, where the SVMNT haplotype has also reached near fixation.14 There is evidence the Indian strains are related to strains from Papua New Guinea.14 In contrast, the CVIET-type parasites have generally been traced to a common origin at the Thai–Cambodian border, which subsequently spread throughout Southeast Asia as well as Africa.10 Recent research suggests importation of cases may play a significant role in hindering elimination efforts on islands.15 As the malaria elimination program progresses, future tracking of pfcrt haplotypes, along with other data, will be informative in determining the frequency and origin of imported cases.
ACKNOWLEDGMENTS
The authors would like to thank Dr. G. Galappaththy and the staff of Anti-Malaria Campaign, Ministry of Health, Sri Lanka, for facilitating the sample collection; Kayla Barnes, Harvard School of Public Health (HSPH), and Sharmini Gunawardana, Faculty of Medicine, University of Colombo (UCFM), for coordinating the transfer of samples; Chamika de Silva, UCFM, for assistance with graphics; and Sarah Volkman, HSPH, for valuable comments on the draft manuscript.
Footnotes
Financial support: This study received financial support from National Institutes of Health Grants 5R03TW007966-02 and 1R01AI075080-01A1.
Authors' addresses: Jenny J. Zhang, Tharanga N. Senaratne, Rachel Daniels, Clarissa Valim, and Dyann F. Wirth, Department of Immunology and Infectious Diseases, Harvard School of Public Health, Boston, MA, E-mails: zhang33@post.harvard.edu, nsenarat@hsph.harvard.edu, rdaniels@broadinstitute.org, cvalim@hsph.harvard.edu, and dfwirth@hsph.harvard.edu. Michael Alifrangis and Flemming Konradsen, Centre for Medical Parasitology and International Health Unit, Institute for International Health, Immunology and Microbiology, Copenhagen, Denmark, E-mails: micali@sund.ku.dk and flko@sund.ku.dk. Priyanie Amerasinghe and Rupika Rajakaruna, Department of Zoology, University of Peradeniya, Peradeniya, Sri Lanka, E-mails: P.Amerasinghe@cgiar.org and rupikar@pdn.ac.lk. Nadira D. Karunaweera, Department of Parasitology, Faculty of Medicine, University of Colombo, Colombo, Sri Lanka, E-mail: nadira@parasit.cmb.ac.lk.
References
- 1.Snow RW, Guerra CA, Noor AM, Myint HY, Hay SI. The global distribution of clinical episodes of Plasmodium falciparum malaria. Nature. 2005;434:214–217. doi: 10.1038/nature03342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Epidemiology Unit, Ministry of Health, Sri Lanka . Quarterly Reports, Epidemiology Unit, Ministry of Health, Colombo, Sri Lanka, 2000–2009. Sri Lanka: Ministry of Health; 2009. [Google Scholar]
- 3.Ministry of Health, Sri Lanka . Strategic Plan for Elimination of Malaria 2008–2012. Anti-Malaria Campaign, Ministry of Health, Colombo, Sri Lanka. Sri Lanka: Ministry of Health; 2008. [Google Scholar]
- 4.Rajakaruna R, Amerasinghe P, Galappaththy G, Konradsen F, Briet O, Alifrangis M. Current status of malaria and anti-malarial drug resistance in Sri Lanka. Ceylon J Sci (Bio Sci) 2008;37:15–22. [Google Scholar]
- 5.Ratnapala R, Subramaniam K, Yapabandara MG, Fernando WP. Chloroquine resistant Plasmodium falciparum in Sri Lanka. Ceylon Med J. 1984;29:135–145. [PubMed] [Google Scholar]
- 6.Handunnetti SM, Jayasinghe S, Pathirana PP, Fernando R, Sheriff MH, Mendis KN. Sulphadoxine-pyrimethamine and chloroquine resistant Plasmodium falciparum infection in Sri Lanka. Ceylon Med J. 1994;39:45–46. [PubMed] [Google Scholar]
- 7.Hapuarachchi HC, Abeysundara S, Dayanath MY, Manamperi A, Abeyewickreme W, de Silva NR. Molecular markers of chloroquine resistance in Plasmodium falciparum in Sri Lanka: frequency before revision of the antimalarial drug policy. Ann Trop Med Parasitol. 2009;103:351–356. doi: 10.1179/136485909X435067. [DOI] [PubMed] [Google Scholar]
- 8.Fidock DA, Nomura T, Talley AK, Cooper RA, Dzekunov SM, Ferdig MT, Ursos LMB, Sidhu AS, Naudé B, Deitsch KW, Su X, Wootton JC, Roepe PD, Wellems TE. Mutations in the P. falciparum digestive vacuole transmembrane protein PfCRT and evidence for their role in chloroquine resistance. Mol Cell. 2000;6:861–871. doi: 10.1016/s1097-2765(05)00077-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Djimde A, Doumbo OK, Cortese JF, Kayentao K, Doumbo S, Diourte Y, Dicko A, Su X, Nomura T, Fidock D, Wellems T, Plowe C, Coulibaly D. A molecular marker for chloroquine-resistant falciparum malaria. N Engl J Med. 2001;344:257–263. doi: 10.1056/NEJM200101253440403. [DOI] [PubMed] [Google Scholar]
- 10.Mita T, Tanabe K, Kita K. Spread and evolution of Plasmodium falciparum drug resistance. Parasitol Int. 2009;58:201–209. doi: 10.1016/j.parint.2009.04.004. [DOI] [PubMed] [Google Scholar]
- 11.Sá JM, Twu O, Hayton K, Reyes S, Fay MP, Ringwald P, Wellems TE. Geographic patterns of Plasmodium falciparum drug resistance distinguished by differential responses to amodiaquine and chloroquine. Proc Natl Acad Sci USA. 2009;106:18883–18889. doi: 10.1073/pnas.0911317106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Beshir K, Sutherland CJ, Merinopoulos I, Durrani N, Leslie T, Rowland M, Hallett R. Amodiaquine resistance in Plasmodium falciparum malaria in Afghanistan is associated with the Pfcrt SVMNT allele at codons 72 to 76. Antimicrob Agents Chemother. 2010;54:3714–3716. doi: 10.1128/AAC.00358-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schousboe ML, Rajakaruna RS, Salanti A, Hapuarachchi HC, Galappaththy GN, Bygbjerg IC, Amerasinghe PH, Konradsen F, Alifrangis M. Island-wide diversity in single nucleotide polymorphisms of the Plasmodium vivax dihydrofolate reductase and dihydropteroate synthetase genes in Sri Lanka. Malar J. 2007;6:28. doi: 10.1186/1475-2875-6-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mixson-Hayden T, Jain V, McCollum AM, Poe A, Nagpal AC, Dash AP, Stiles JK, Udhayakumar V, Singh N. Evidence of selective sweeps in genes conferring resistance to chloroquine and pyrimethamine in Plasmodium falciparum isolates in India. Antimicrob Agents Chemother. 2010;54:997–1006. doi: 10.1128/AAC.00846-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rebaudet S, Bogreau H, Silai R, Lepere JF, Bertaux L, Pradines B, Delmont J, Gautret P, Parola P, Rogier C. Genetic structure of Plasmodium falciparum and elimination of malaria, Comoros Archipelago. Emerg Infect Dis. 2010;16:1686–1694. doi: 10.3201/eid1611.100694. [DOI] [PMC free article] [PubMed] [Google Scholar]


