Abstract
The introduction of antibiotic therapy as first-line treatment of Buruli ulcer underlines the importance of laboratory confirmation of clinical diagnosis. Because smear microscopy has very limited sensitivity, the technically demanding and more expensive IS2404 diagnostic polymerase chain reaction (PCR) has become the main method for confirmation. By optimization of the release of mycobacteria from swab specimen and concentration of bacterial suspensions before smearing, we were able to improve the detection rate of acid-fast bacilli by microscopy after Ziehl–Neelsen staining. Compared with IS2404 PCR, which is the gold standard diagnostic method, the sensitivity and specificity of microscopy with 100 concentrated specimens were 58.4% and 95.7%, respectively. We subsequently evaluated a stepwise laboratory confirmation algorithm with detection of AFB as first-line method and IS2404 PCR performed only with those samples that were negative in microscopic analysis. This stepwise approach reduced unit cost by more than 50% to $5.41, and the total costs were reduced from $917 to $433.
Introduction
Buruli ulcer (BU) is a chronic necrotizing infection of subcutaneous tissue caused by Mycobacterium ulcerans. The disease has been identified in more than 30 tropical and subtropical countries in the world, with the majority of cases being identified in West Africa. In terms of numbers, BU is the third most important mycobacterial disease of public health importance globally; however, in endemic countries such as Ghana, it ranks second after tuberculosis.1 BU affects people from impoverished communities, where there are significant resource constraints, especially for diagnosis and treatment. A key element in the management of BU is the early diagnosis before the development of large ulcers. However, most cases are seen late at health facilities, resulting in increased patient suffering and prolonged hospital stays.2
Laboratory diagnostic tests available for confirmation of BU are smear microscopy for detection of acid-fast bacilli (AFB), M. ulcerans isolation by culture, histopathology, and polymerase chain reaction (PCR) for detection of the insertion sequence IS2404. Although both histopathology and cultivation are technically very demanding and time-consuming diagnostic approaches, the fast and less demanding smear microscopy has limited sensitivity. Therefore, IS2404 PCR has become the gold standard diagnostic tool,4 although its routine use in resource-poor settings is limited by the high costs and need for sophisticated laboratory infrastructure. Therefore, PCR tests are usually not performed at the treating facilities but in bulk at national reference laboratories or outside the endemic country at international reference laboratories.5 These shortcomings have reduced microbiological confirmation to a quality control tool for clinical diagnosis. Although the general perception is that diagnosis based on clinical judgment alone can be adequate,6 incidents of misdiagnosis have been reported. BU cases have been missed initially,7–9 but presumed BU lesions have proven to be other conditions.10–13
In the past, surgery was used as the only means of BU case management; however, the World Health Organization (WHO) currently recommends an 8-week combination therapy with rifampicin and streptomycin as first-line treatment.14 With the introduction of this antimycobacterial treatment, laboratory confirmation of clinically suspected cases became even more crucial for the clinical management of BU. We, therefore, evaluated the usefulness of microscopy for analyzing swab and fine-needle aspiration (FNA) samples to reduce cost of laboratory confirmation of clinical diagnosis.
Materials and Methods
Source and collection of specimen.
Pathological samples were collected from 100 suspected BU patients from eight districts and three regions in Ghana. The majority of the samples (69) were from the West and South districts in the Greater Accra region; 29 were from the Akwapim North and South, Atiwa, West Akim, and Manya Kobo districts, all in the Eastern region, and 2 were from Nkoranza North district in the Brong-Ahafo regions. Forty-seven of the cases were females; age range was between 3 and 82 years. Arithmetic mean age was 25.1 years, median was 14 years, and 10 years was the mode. The remaining 53 cases were males between 4 months and 82 years; the arithmetic mean was 26.1 years, the median was 15.5 years, and the mode was 10 years. Among the 100 cases, 99 were new cases, and 1 was recorded as a recurrent case. The majority of the suspects had ulcers (86): 74 had only ulcers, 9 had edema with ulcers, 1 had ulcers with oesteomylitis, and 2 had nodules and ulcers. Only 14 cases were pre-ulcerative: 12 nodules, 1 plaque, and 1 edema. The lesion category was recorded for 80 cases, and of these cases, 31 (38.75%), 22 (27.5%), and 29 (33.75%) were categories I, II, and III, respectively.
Specimens were collected using standard procedures as prescribed by the WHO. From cases with ulcerative lesions, two swab samples were collected by swabbing the surface beneath the entire undermined edges per each lesion to make sure that the areas with the highest load of bacteria are sampled. In the case of pre-ulcerative lesions, one FNA specimen was collected at the center of the lesion.15 We used a 21/23-gauge needle attached to a 5-mL syringe. Swabs were either transported dry or inserted into 15-mL falcon tubes containing transport medium to cover the entire tip of the swab, whereas FNA specimens were immediately drained into an Epperndorf tube containing 500 µL phosphate buffered saline (PBS). A specimen collection form was used to obtain detailed information about the specimen, and each pair of specimens and matching specimen collections were packed in a zip-locked bag and then transported to the Noguchi Memorial Institute for Medical Research (NMIMR) for processing and analysis. The procedures used for sample collection and analysis conform to the WHO standards, and they form part of the national Buruli ulcer control program system of laboratory confirmation of BU cases. Ethical approval of the study was obtained from the Institutional Review Committee of the NMIMR. In addition, consent was obtained from all adult participants and legal guardians of all minors.
Laboratory analysis.
Optimization of bacteria release from swab specimen.
Swab samples that are received in our laboratory usually come as dry swabs and have spent between 2 days and 1 month in transit. To optimize bacteria release from the swab sample, two procedures were evaluated using 20 paired samples. The first process, swab specimen(s) from the same lesion were pooled together and soaked in sterile PBS for 30 minutes, which was followed by vortexing for 2 minutes. In the second process, swab specimen(s) from the same lesion were pooled together and soaked for 30 minutes in a tube containing sterile PBS and 10 3-mm-diameter undrilled glass beads (Merck, Darmstadt, Germany). The specimen was then vortexed at full speed for 2 minutes (Figure 1).
Figure 1.
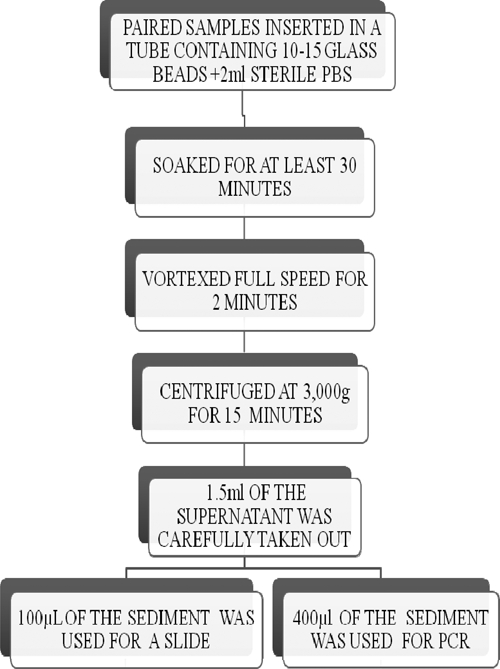
Work flow.
The effectiveness of the two procedures was evaluated semiquantitatively by microscopy and real-time PCR. Thereafter, the better of the two procedures was used for subsequent sample processing for analysis.
Processing of swab and FNA specimens.
The bacteria suspensions obtained as already described from swab specimens were concentrated by centrifuging at 3,000 × g for 15 minutes. About 1.5 mL supernatant were carefully taken out, and 100 and 400 µL sediment were used to prepare slides for Ziehl–Neelson (ZN) microscopy and IS2404 PCR, respectively. DNA was extracted using the Qiagen DNA mini prep kit (Qiagen, Hilden, Germany), and the manufacturer's instruction was followed.
Smear microscopy.
Slides were allowed to dry and were fixed by heat. The smears were then stained as previously described using the ZN procedure and graded by the International Union against Tuberculosis and Lung Diseases grading system. A slide is graded as negative only after reading at least 100 high fields without the detection of AFB under oil immersion and confirmation by a second reader.
Quality control.
One-tenth of all the slides were blindly evaluated by an independent technician. A discordant reading was resolved by a second assessor.
TaqMan real-time PCR.
The primers and TaqMan MGB probes (Applied Biosystems, California) for detecting the M. ulcerans-specific sequence, IS2404, were as optimized in the work by Fyfe and others16 with some modifications in reaction conditions. IS2404 real-time PCR mixtures contained 1× Qiagen master mix (containing HotstarTaq plus DNA polymerase, deoxyribonucleotide triphosphate (dNTP) mix, and PCR buffer), 1 μL extracted template DNA, 0.5 µM concentrations of each primer, and 0.2 µM probe, 1× TaqMan exogenous internal positive control (IPC), and probe reagents (Applied Biosystems) in a total volume of 20 µL. Amplification and detection were performed with the Rotor-Gene (Qiagen, Hilden, Germany) using the following program: 1 cycle of 95°C for 5 minutes and 40 cycles of 95°C for 15 seconds and 60°C for 15 seconds. Each PCR run contains two non-template controls and an IS2404 positive control.
Conventional PCR.
The IS2404 sequence was amplified in a 20-µL reaction volume using the Qiagen Fast cycling PCR kit. The primer sets used for the amplification were as described previously.17 The reaction mixture contained a 0.5-µM concentration of each primer, a 200-µM concentration of each deoxynucleoside triphosphate, an optimized concentration of MgCl2, 1× PCR buffer, 1 U HotstarTaq plus DNA polymerase, and approximately 50 ng DNA. Thermocycling parameters were 95°C for 5 minutes and 35 cycles of 5 seconds at 96°C, 5 seconds at 60°C, 15 seconds at 60°C, and 1 minute at 72°C; 10 μL amplified DNA were subjected to electrophoresis in a 2% agarose gel and detected by ethidium bromide staining and ultraviolet (UV) transillumination. To ensure the quality of diagnostic procedure, all batches of extraction included controls, and amplification procedures included negative and positive controls. The amplicons were sized by comparing with a 1-kb ladder.
Stepwise confirmation of BU.
In this case, a case was confirmed by first analyzing with ZN microscopy, which is cheaper and technologically less expensive, and PCR was done only when microscopy was negative (approach B), which contrasts the previous approach of analyzing all samples by PCR (approach A). We assumed that, in already described endemic areas, the specificity of microscopy will be very high, which is in accordance with the diagnosis of other mycobacterial disease in Ghana.
Data handling and analysis.
The positivity rates of each of the two methods were determined by dividing the number of positive tests by the total number of tests. The sensitivity and specificity of the microscopy test were calculated using IS2404 PCR as the gold standard.
Cost analysis.
The definition used to calculate the cost was defined as the value of resources used to produce something (hence, the costs incurred by smear microscopy and PCR), and the cost was calculated based on the direct cost of reagents and materials used to arrive at the result. We compared the cost of the routine PCR analysis of all samples (approach A) with the cost of our proposing strategy of first microscopy followed by PCR only on smear negative samples (approach B) using another batch of received specimen. P value for statistical significance of difference in cost of the two approaches was determined by normal approximation to binomial test calculator.
Results
Optimization of the release of bacteria from swab samples.
Efficacy of two procedures for the release of mycobacteria from dry swab samples was compared. Swabs soaked in PBS were vortexed with or without glass beads. Suspensions were subsequently evaluated by microscopy after ZN staining and semiquantitative IS2404 real-time (RT) PCR. Included in the comparative study were 19 BU lesions testing positive by RT-PCR (cycle threshold [Ct] ≤ 39) after sample processing with glass beads (Table 1). Although 12 of 13 (92%) suspensions with a Ct < 31 in RT-PCR were also positive by microscopy, only 3 of 25 (12%) suspensions with a Ct > 31 were microscopy positive, showing the overall superior sensitivity of PCR.
Table 1.
Release of M. ulcerans cells from swab samples
| Ct RT-PCR with beads | Ct RT-PCR without beads | Microscopy with beads | Microscopy without beads |
|---|---|---|---|
| 23.9 | Neg | + | − |
| 23.9 | 25.9 | + | + |
| 27.2 | 29.4 | + | − |
| 27.4 | 25.2 | + | + |
| 29.8 | Neg | + | − |
| 29.9 | 30.2 | + | + |
| 30.4 | 35.5 | + | − |
| 30.7 | 31.7 | + | − |
| 30.9 | 31.2 | + | − |
| 33.4 | Neg | − | − |
| 33.7 | 38.5 | − | − |
| 33.8 | 34.7 | + | + |
| 33.9 | 36.1 | − | − |
| 34.5 | 35.3 | + | − |
| 36.5 | Neg | − | − |
| 36.9 | 35.5 | − | − |
| 37.1 | 37.1 | − | − |
| 37.6 | Neg | − | − |
| 39.0 | Neg | − | − |
Swabs were soaked in PBS and vortexed with or without glass beads. After concentration, aliquots were used for microscopic analysis after Ziehl–Neelsen staining and IS2404 real-time PCR after DNA extraction. Results for individual patients are shown and ordered by RT-PCR Ct values after vortexing of samples with glass beads.
The addition of glass beads increased the microscopic detection of AFBs from 21% (4/19) to 58% (11/19); 6 of 19 (32%) lesions rated negative by RT-PCR when swabs were vortexed without glass beads turned positive when glass beads were used. To increase sensitivity of microscopy, we subsequently concentrated supernatants by centrifugation. For all subsequent analyses, glass beads were added, and suspensions were concentrated.
Sensitivity and specificity of the optimized procedure for microscopic detection of AFB in swab and FNA samples.
In the next step, we analyzed 105 swab samples from 86 suspected BU cases with ulcerative lesions; 68.6% of the samples (72/105) were positive by conventional IS2404 PCR. From 19 lesions, two samples were analyzed; in six cases, both samples were positive, in five cases, both were negative, and in eight cases, PCR results were diverging. Testing more than one sample, thus, increased lesion positivity from 58% to 66% for PCR. Microscopy testing of more than one sample increased the positivity from 32% to 39%.
In addition, FNA samples collected from 14 suspected BU patients with non-ulcerative lesions were analyzed. Of these patients, 11 (78.6%) were positive by PCR, and 6 (43%) of 11 were positive by microscopy.
Taken together, of 100 suspected BU cases, 77 cases were confirmed by IS2404 PCR; 45 of 77 PCR positives (58%) were also confirmed by microscopy, and 1 of 23 PCR-negative samples was microscopy-positive. Combining the two methods, 78 of 100 suspected BU cases were confirmed. Taking IS2404 PCR as the gold standard diagnostic method, the sensitivity and specificity of microscopy were 57.1% (44/77) and 95.7% (22/23), respectively.
Reduction of costs for laboratory diagnosis of BU by using the optimized microscopic procedures as first-line test.
In the next step, we evaluated whether microscopic analysis with the optimized procedures as first-line diagnostic test can substantially reduce costs for BU diagnosis. Monitoring the materials needed to perform the two tests, the unit cost of ZN smear microscopy was $0.22, and the unit cost of IS2404 PCR was $11.24. We analyzed 80 samples from 80 patients using an approach in which PCR was done only when microscopy was negative. By microscopy, 53.8% (43/80) of samples were positive, and costs of materials for the microscopic examination of all 80 samples were $17.60. PCR was performed only on the 37 microscopy-negative samples, bringing material costs for PCR of $415.88. Of these samples, 17 of 37 (43.6%) samples were PCR-positive. The total material costs of analyses were $433.48, reflecting a unit cost of $5.41. Costs for PCR analysis of all samples would have been more than two times as high ($899.20), with a unit cost of $11.24.
Discussion
At the onset of global BU initiative, recommendations by the WHO required that a case of BU must be confirmed by at least two laboratory tests.3 However, despite that recommendation, most cases, especially in Africa, were treated by surgical excision based on clinical judgment. Although there was little error by experienced clinicians in endemic areas of ulcers, there were a number of reports of misdiagnosis, which could be as high as 30%.10–14 The WHO now puts a lot of emphasis on laboratory confirmation, because the introduction of antibiotic therapy results in the formation of a network of laboratories that confirms BU cases. Nevertheless, more emphasis on laboratory support for case management is focusing on IS2404 PCR15–18 and sidelining smear microscopy, which is the cornerstone of TB control.19 IS2404 PCR is another mycobacterial disease that is known to be cheaper, faster, and very specific. The overreliance on PCR, a technique that requires elaborate infrastructure, expertise, and costly reagents that are not available at the peripheral health facilities treating BU, brings about reliance on research laboratories (in some countries, even at international level) and therefore, affects availability of research projects.
We have previously reported a sensitivity of more than 70% for detection of AFB in stained smears of biopsy specimen collected during surgical excision. An important step was the concentrating of specimen before smearing. Our findings using swab and FNA samples indicate that smear microscopy can be used to confirm more than 50% of cases that would have been confirmed by PCR alone. We believe that this finding was achieved through (1) a good sample collection procedure using recommended guidelines, (2) at least two swabs per lesion analyzed, (3) concentration of specimen before smearing, and (4) good microscopy practice of taking care to read at least 100 high-power fields before declaring a slide negative.
By combining direct smear microscopy with PCR stepwise, the cost of analysis was reduced significantly to less than 50% of the cost if the samples were analyzed by PCR alone. However, it must be mentioned here that the cost analysis done involved only the cost of materials needed to perform the experiment, without taking into consideration the technician's time. We are of the opinion that, because all districts/subdistrict health facilities have the capacities to do ZN smear microscopy, facilities in known endemic communities treating BU should first perform microscopy on all lesions before beginning treatment; it will be of value to increase the number of samples collected per case to improve the sensitivity of microscopy. This process is used for TB, where three sputum samples are analyzed before a case is confirmed. That case will be treated only after microscopy-negative results show a strong clinical indication through epidemiological information that the case is BU. This process will definitely reduce the number of misdiagnoses as well as the number of specimen sent to central laboratories for case confirmation to ensure adequate treatment of cases. More importantly, the implementation of this system of laboratory confirmation will lead to significant reduction in cost of diagnosis, giving the opportunity for the different sectors of healthcare to be involved in BU control and reduce the overreliance on externally funded projects. The WHO guidelines require a case to be confirmed by at least two laboratory assays; however, with the focal distribution of BU, we believe that a suspected case can only be confirmed by ZN microscopy. This assertion is supported by the high specificity of ZN in this report using IS2404 by PCR as the gold standard detection method. The specificity of our study was 95.7, and this finding compares very well with a similar study conducted by Bretzel and others4 in 2006 that had a specificity of 96.6%.4 PCR will be required as a second confirmation test only if the case involves a new focus. Nevertheless, the fact that one of the microscopy-positive samples was found to be IS2404 PCR-negative could be a cause for concern, because there are other skin conditions that are caused by AFB-positive bacteria other than M. ulcerans. However, we believe that the possibility of this misdiagnosis is rare if clinical judgment is done well.
The acceptance of FNA should make it feasible for specimen collection from pre-ulcerative lesion for confirmation microscopy. Therefore, much effort needs to be put into making laboratory diagnosis a part of the routine case management of BU, and this goal can only be achieved in Africa if the role of ZN is clarified and reinforced.
ACKNOWLEDGMENTS
This study was funded by the Stop Buruli Consortium and Volkswagen Foundation. We are grateful to Kwaku Bio, William Opare, Dr. Benjamin Anku, and Isaac Lamptey of Ghana Health Service for providing us with clinical specimens for the analysis. We also appreciate the contribution of Inna Ibrahim, Grace Kpeli, David Mensah, and Zuliehatu Nakobu.
Footnotes
Authors' address: Dorothy Yeboah-Manu, Adwoa Asante-Poku, Kobina Asan-Ampah, Emelia Danso Edwin Ampadu, and Gerd Pluschke, Noguchi Memorial Institute for Medical Research, University of Ghana, Accra, Ghana, E-mails: dyeboah-manu@noguchi.mimcom.org, aasante-poku@noguchi.mimcom.org, kampah@noguchi.mimcom.org, edanso@noguchi.mimcom.org, ghanbu@4u.com.gh, and gerd.pluschke@unibas.ch.
References
- 1.Asiedu K, Sherpbier R, Raviglione MC. WHO Global Buruli Ulcer Initiative. Report 2000. Geneva, Switzerland: World Health Organization; 2000. (Buruli Ulcer Mycobacterium ulcerans Infection). [Google Scholar]
- 2.Wansbrough-Jones M, Phillips R. Buruli ulcer: emerging from obscurity. Lancet. 2006;367:1849–1858. doi: 10.1016/S0140-6736(06)68807-7. [DOI] [PubMed] [Google Scholar]
- 3.Portaels F, Johnson P, Meyers WM. Diagnosis of Mycobacterium ulcerans Disease. Geneva, Switzerland: World Health Organization; 2001. [Google Scholar]
- 4.Bretzel G, Siegmund V, Nitschke J, Herbinger KH, Thompson W, Klutse E, Crofts K, Massavon W, Etuaful S, Thompson R, Asamoah-Opare K, Racz P, Vloten F, Berberich VC, Kruppa T, Ampadu E, Fleischer B, Adjei O. A stepwise approach to the laboratory diagnosis of Buruli ulcer disease. Trop Med Int Health. 2007;12:89–96. doi: 10.1111/j.1365-3156.2006.01761.x. [DOI] [PubMed] [Google Scholar]
- 5.Eddyani M, Debacker M, Martin A, Aguiar J, Johnson CR, Uwizeye C, Fissette K, Portaels F.2008Primary culture of Mycobacterium ulcerans from human tissue specimens after storage in semisolid transport medium J Clin Microbiol 4669– 72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.van der Werf TS, van der Graaf WT, Tappero JW, Asiedu K. Mycobacterium ulcerans infection. Lancet. 1999;354:1013–1018. doi: 10.1016/S0140-6736(99)01156-3. [DOI] [PubMed] [Google Scholar]
- 7.Semret M, Koromihis G, MacLean JD, Libman M, Ward BJ. Mycobacterium ulcerans infection (Buruli ulcer): first reported case in a traveler. Am J Trop Med Hyg. 1999;61:689–693. doi: 10.4269/ajtmh.1999.61.689. [DOI] [PubMed] [Google Scholar]
- 8.Evans MR, Phillips R, Etuaful SN, Amofah G, Adomako J, Adjei O, Dennis-Antwi J, Lucas SB, Wansbrough-Jones MH. An outreach education and treatment project in Ghana for the early stage of Mycobacterium ulcerans disease. Trans R Soc Trop Med Hyg. 2003;97:159–160. doi: 10.1016/s0035-9203(03)90105-2. [DOI] [PubMed] [Google Scholar]
- 9.Coloma JN, Navarrete-Franco G, Iribe P, Lopez-Cepeda LD. Ulcerative cutaneous mycobacteriosis due to Mycobacterium ulcerans: report of two Mexican cases. Int J Lepr Other Mycobact Dis. 2005;73:5–12. doi: 10.1489/1544-581X(2005)73[5:UCMDTM]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 10.Revill WD, Morrow RH, Pike MC, Ateng J. A controlled trial of the treatment of Mycobacterium ulcerans infection with clofazimine. Lancet. 1973;2:873–877. doi: 10.1016/s0140-6736(73)92005-9. [DOI] [PubMed] [Google Scholar]
- 11.Guarner J, Bartlett J, Whitney EA, Raghunathan PL, Stienstra Y, Asamoa K, Etuaful S, Klutse E, Quarshie E, van der Werf TS, van der Graaf WTA, King CH, Ashford DA. Histopathologic features of Mycobacterium ulcerans infection. Emerg Infect Dis. 2003;9:651–656. doi: 10.3201/eid0906.020485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stienstra Y, van der Werf TS, Guarner J, Raghunathan RL, Whitney EAS, van der Graaf WTA, Asamoa K, Tappero JW, Ashford DA, King CH. Analysis of an IS2404-based nested PCR for diagnosis of Buruli ulcer disease in regions of Ghana where the disease is endemic. J Clin Microbiol. 2003;41:794–797. doi: 10.1128/JCM.41.2.794-797.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Debacker M, Aguiar J, Steunou C, Zinsou C, Meyers WM, Guedenon A, Scott JT, Dramaix M, Portaels F. Mycobacterium ulcerans disease (Buruli ulcer) in rural hospital, Southern Benin, 1997–2001. Emerg Infect Dis. 2004;10:1391–1398. doi: 10.3201/eid1008.030886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Etuaful S, Carbonnelle B, Grosset J, Lucas S, Horsfield C, Phillips R, Evans M, Ofori-Adjei D, Klustse E, Owusu-Boateng J, Amedofu GK, Awuah P, Ampadu E, Amofah G, Asiedu K, Wansbrough-Jones M. Efficacy of the combination rifampin-streptomycin in preventing growth of Mycobacterium ulcerans in early lesions of Buruli ulcer in humans. Antimicrob Agents Chemother. 2005;49:3182–3186. doi: 10.1128/AAC.49.8.3182-3186.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Phillips RO, Sarfo FS, Osei-Sarpong F, Boateng A, Tetteh I, Lartey A, Adentwe W, Opare W, Asiedu KB, Wansbrough-Jones M. Sensitivity of PCR targeting Mycobacterium ulcerans by use of fine-needle aspirates for diagnosis of Buruli ulcer. J Clin Microbiol. 2009;47:924–926. doi: 10.1128/JCM.01842-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fyfe JAM, Lavender CJ, Johnson PDR, Globan M, Sievers A, Azuolas J, Stinear TP. Development and application of two multiplex real-time PCR assays for the detection of Mycobacterium ulcerans in clinical and environmental samples. Appl Environ Microbiol. 2007;73:4733–4740. doi: 10.1128/AEM.02971-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mensah-Quainoo E, Yeboah-Manu D, Asebi C, Patafuor F, Ofori-Adjei D, Junghanss T, Pluschke G. Diagnosis of Mycobacterium ulcerans infection (Buruli ulcer) at a treatment centre in Ghana: a retrospective analysis of laboratory results of clinically diagnosed cases. Trop Med Int Health. 2008;13:191–198. doi: 10.1111/j.1365-3156.2007.01990.x. [DOI] [PubMed] [Google Scholar]
- 18.Siegmund V, Adjei O, Nitschke J, Thompson W, Klutse E, Herbinger KH, Thompson R, van Vloten F, Racz P, Fleischer B, Loescher T, Bretzel G. Dry reagent-based polymerase chain reaction compared with other laboratory methods available for the diagnosis of Buruli ulcer disease. Clin Infect Dis. 2007;45:68–75. doi: 10.1086/518604. [DOI] [PubMed] [Google Scholar]
- 19.Cattamanchi A, Dowdy DW, Davis JL, Worodria W, Yoo S, Joloba N, Matovu J, Hopewell PC, Huang L. Sensitivity of direct versus concentrated sputum smear microscopy in HIV-infected patients suspected of having pulmonary tuberculosis. BMC Infect Dis. 2009;9:53. doi: 10.1186/1471-2334-9-53. [DOI] [PMC free article] [PubMed] [Google Scholar]


