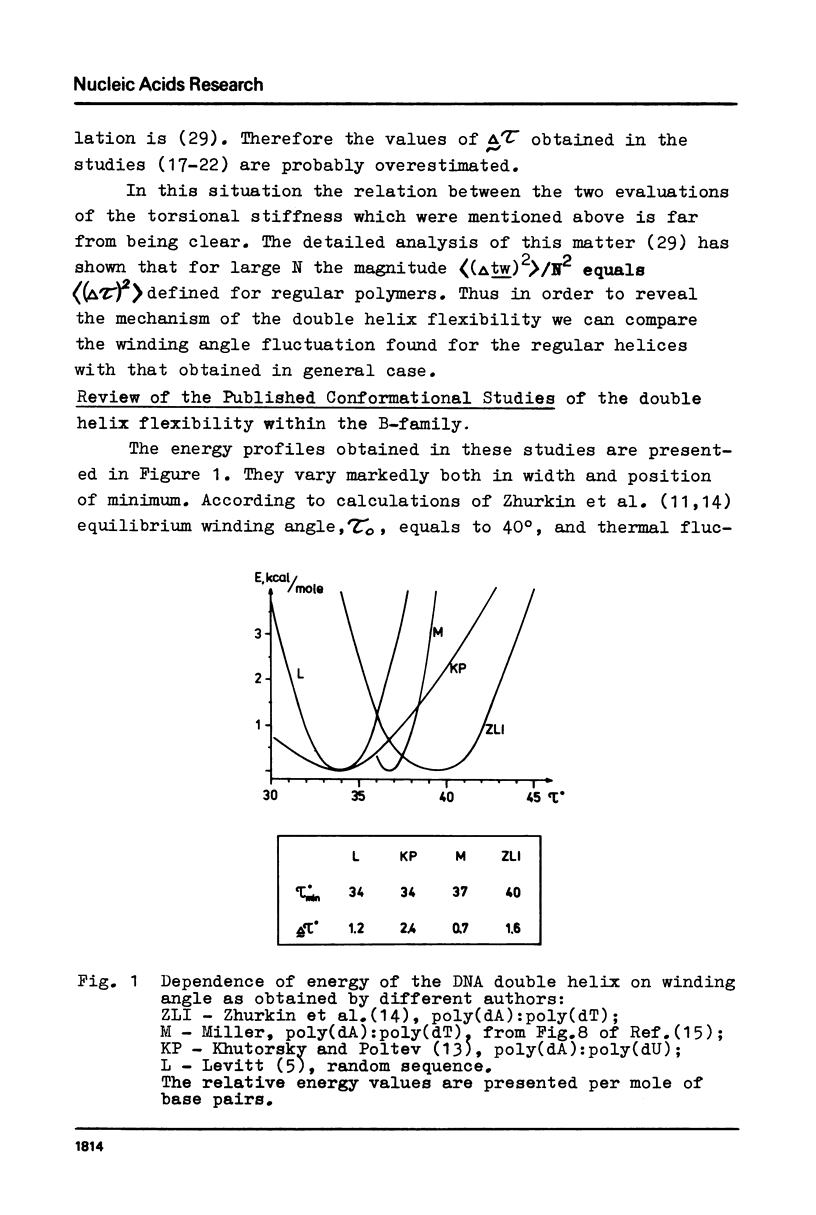
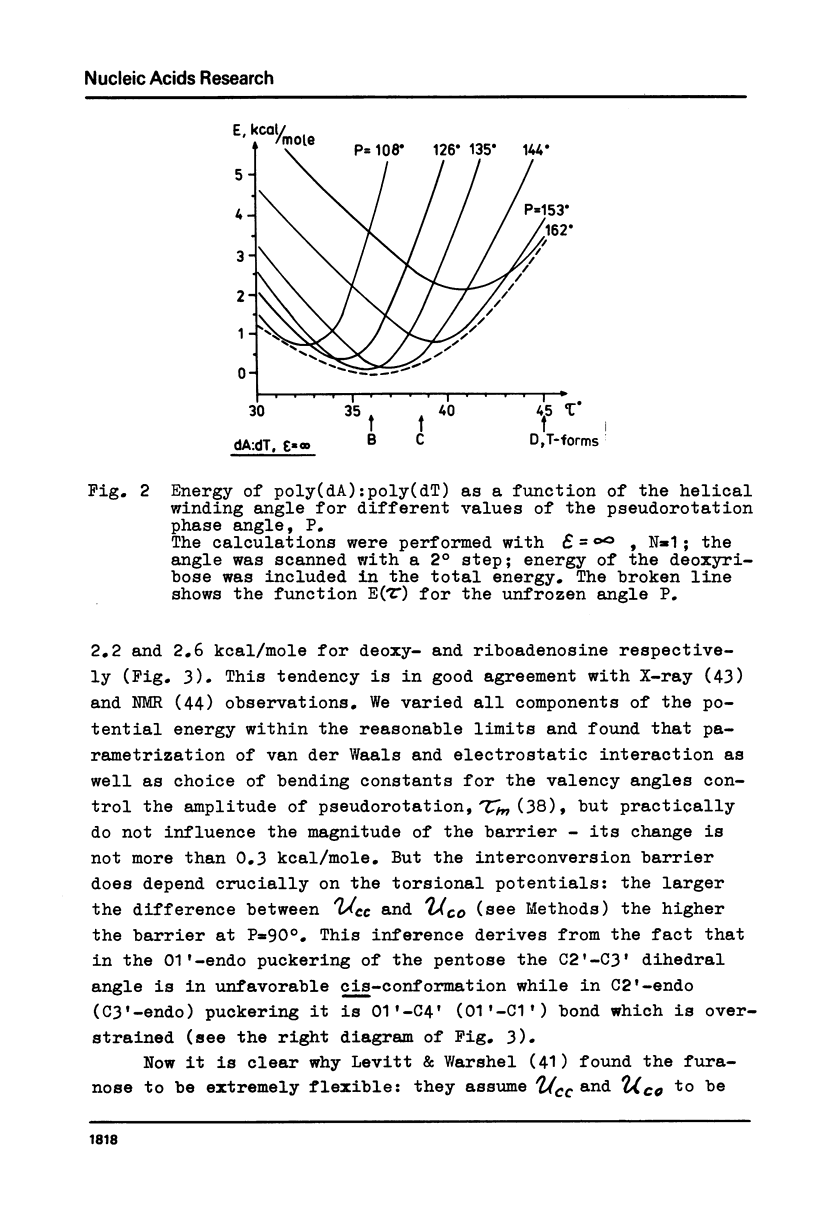
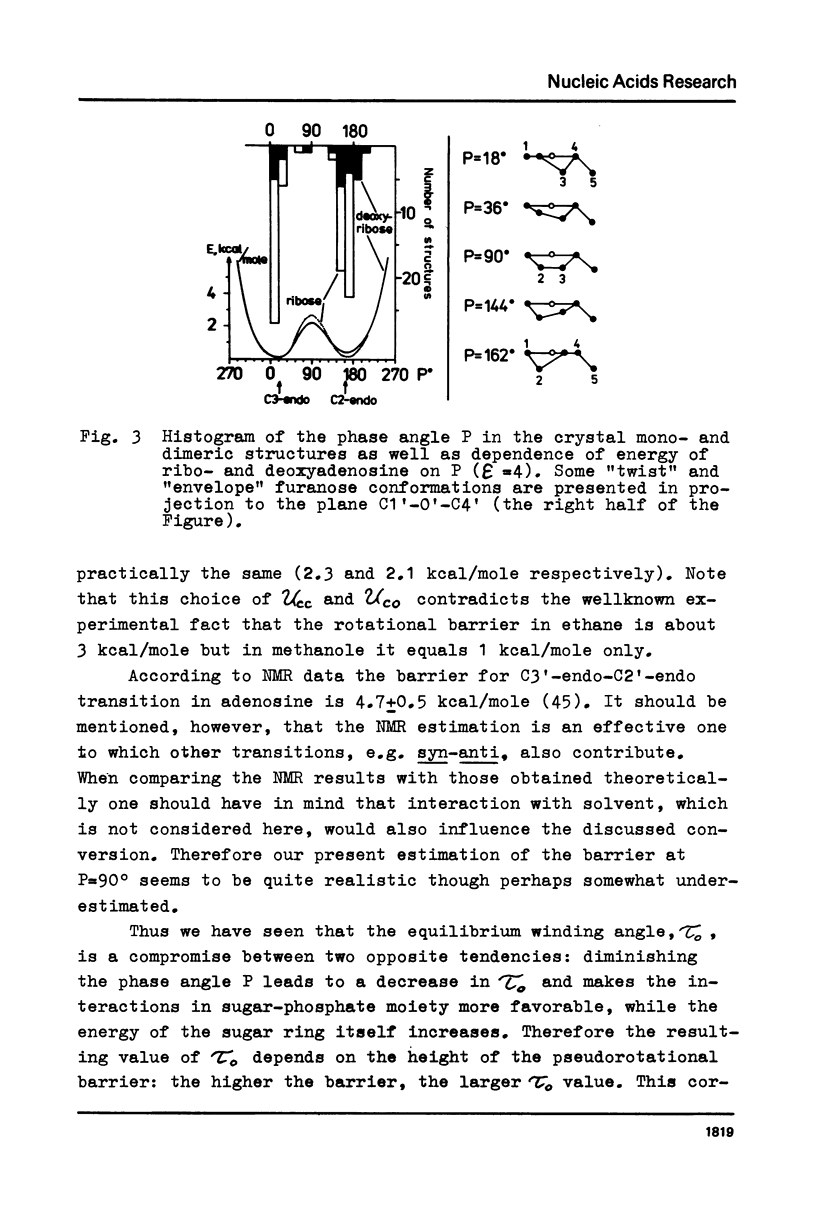
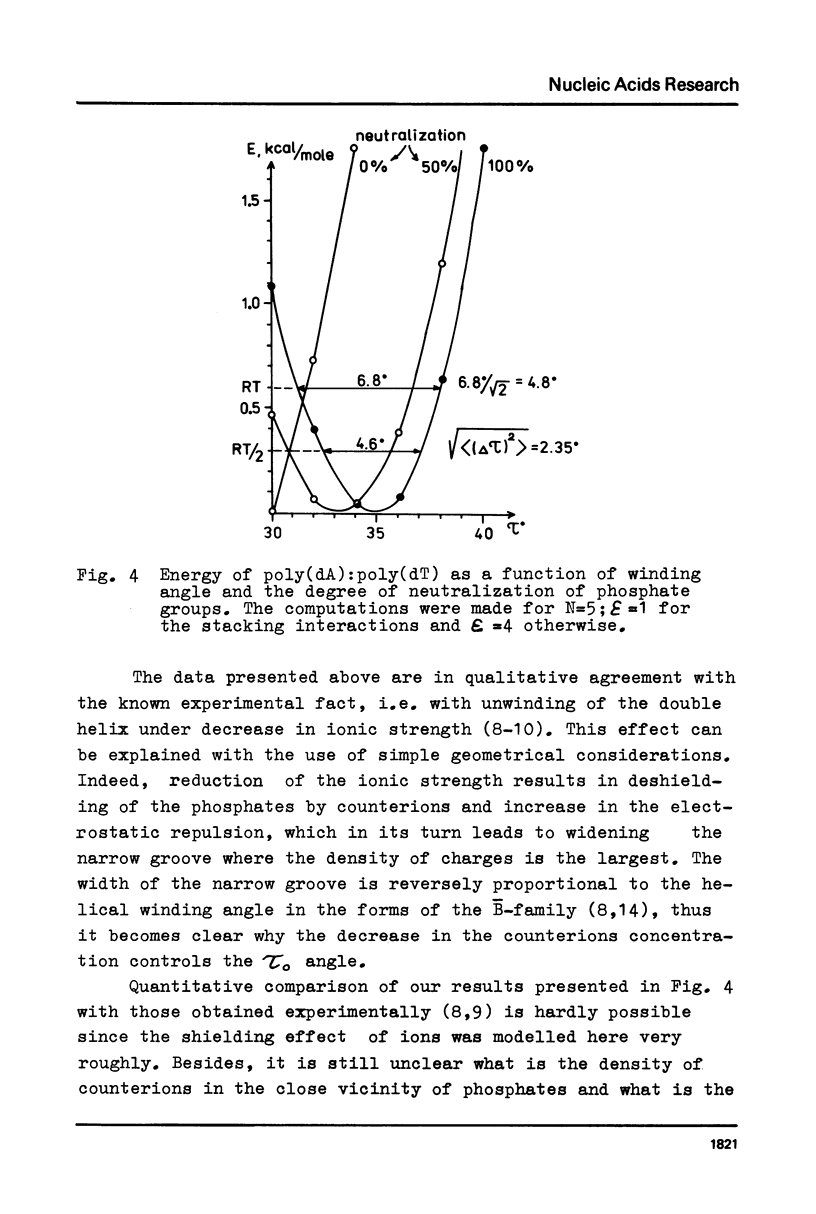
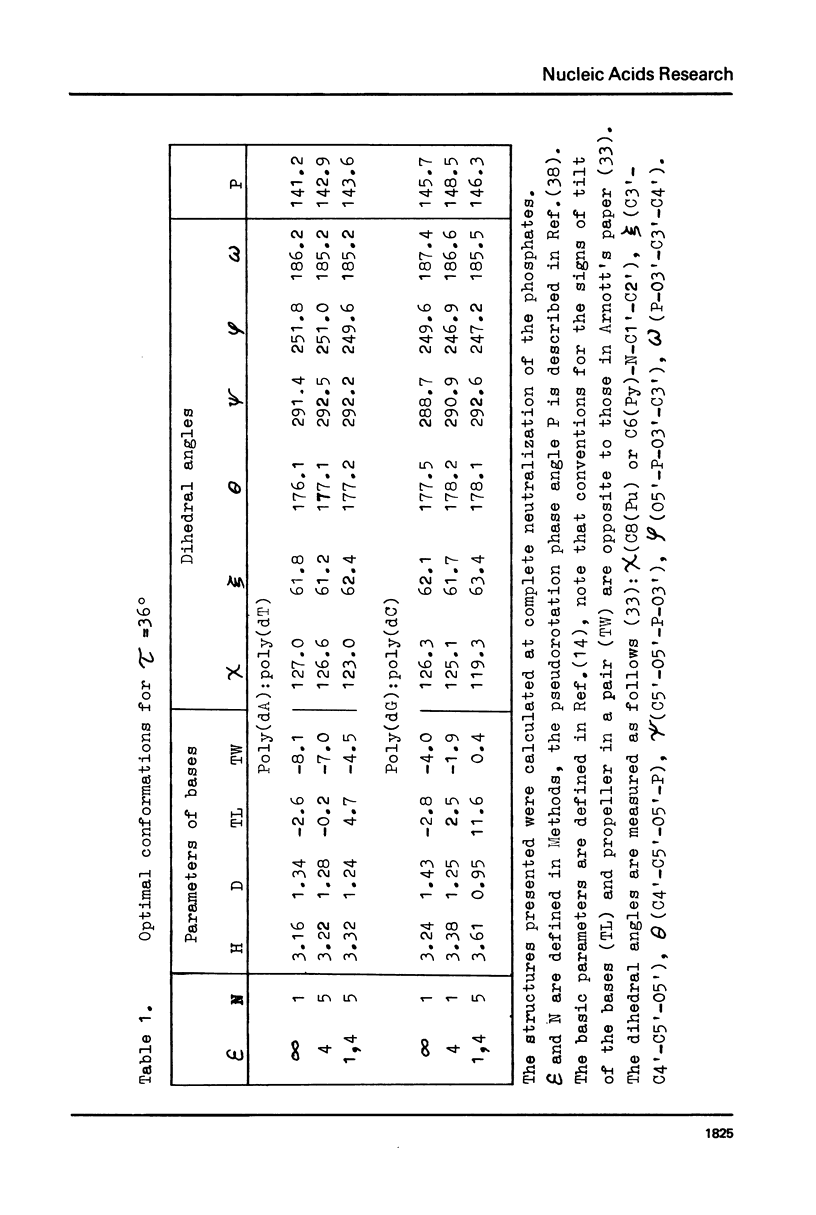
Abstract
The thermal fluctuations of a regular double helix belonging to the B-family were studied by means of atom-atomic potentials method. The winding angle fluctuation was found to be 2.4 degrees for poly(dA):poly(dT) and 3.0 degrees for poly(dG):poly(dC). The reasonable agreement of these estimations with those obtained experimentally reveals the essential role of the small-amplitude torsional vibrations of atoms in the mechanism of the double helix flexibility. The calculated equilibrium winding angle, tau 0, essentially depends on the degree of neutralization of phosphate groups, being about 35.5 degrees for the full neutralization. The deoxyribose pucker is closely related to the tau angle: while tau proceeds from 30 degrees to 45 degrees the pseudorotation phase angle, P, increases from 126 degrees to 164 degrees. Fluctuations of the angles TL and TW, which specify inclination of the bases to the helix axis, were evaluated to be 5 degrees-10 degrees. Possible correlation between conformational changes in the adjacent nucleotides is discussed.
Full text
PDF














Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Altona C., Sundaralingam M. Conformational analysis of the sugar ring in nucleosides and nucleotides. A new description using the concept of pseudorotation. J Am Chem Soc. 1972 Nov 15;94(23):8205–8212. doi: 10.1021/ja00778a043. [DOI] [PubMed] [Google Scholar]
- Arnott S., Bond P. J., Chandrasekaran R. Visualization of an unwound DNA duplex. Nature. 1980 Oct 9;287(5782):561–563. doi: 10.1038/287561a0. [DOI] [PubMed] [Google Scholar]
- Arnott S., Chandrasekaran R., Birdsall D. L., Leslie A. G., Ratliff R. L. Left-handed DNA helices. Nature. 1980 Feb 21;283(5749):743–745. doi: 10.1038/283743a0. [DOI] [PubMed] [Google Scholar]
- Arnott S. The geometry of nucleic acids. Prog Biophys Mol Biol. 1970;21:265–319. doi: 10.1016/0079-6107(70)90027-1. [DOI] [PubMed] [Google Scholar]
- Baase W. A., Johnson W. C., Jr Circular dichroism and DNA secondary structure. Nucleic Acids Res. 1979 Feb;6(2):797–814. doi: 10.1093/nar/6.2.797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belintsev B. N., Gagua A. V., Nedospasov S. A. The effect of the superhelicity on the double helix twist angle in DNA. Nucleic Acids Res. 1979 Mar;6(3):983–992. doi: 10.1093/nar/6.3.983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies D. B., Danyluk S. S. Nuclear magnetic resonance studies of 2'- and 3'-ribonucleotide structures in solution. Biochemistry. 1975 Feb 11;14(3):543–554. doi: 10.1021/bi00674a013. [DOI] [PubMed] [Google Scholar]
- Depew D. E., Wang J. C. Conformational fluctuations of DNA helix. Proc Natl Acad Sci U S A. 1975 Nov;72(11):4275–4279. doi: 10.1073/pnas.72.11.4275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickerson R. E., Drew H. R. Structure of a B-DNA dodecamer. II. Influence of base sequence on helix structure. J Mol Biol. 1981 Jul 15;149(4):761–786. doi: 10.1016/0022-2836(81)90357-0. [DOI] [PubMed] [Google Scholar]
- Finch J. T., Lutter L. C., Rhodes D., Brown R. S., Rushton B., Levitt M., Klug A. Structure of nucleosome core particles of chromatin. Nature. 1977 Sep 1;269(5623):29–36. doi: 10.1038/269029a0. [DOI] [PubMed] [Google Scholar]
- Frank-Kamenetskii M. D., Lazurkin Y. S. Conformational changes in DNA molecules. Annu Rev Biophys Bioeng. 1974;3(0):127–150. doi: 10.1146/annurev.bb.03.060174.001015. [DOI] [PubMed] [Google Scholar]
- Fuller F. B. The writhing number of a space curve. Proc Natl Acad Sci U S A. 1971 Apr;68(4):815–819. doi: 10.1073/pnas.68.4.815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogan M. E., Jardetzky O. Effect of ethidium bromide on deoxyribonucleic acid internal motions. Biochemistry. 1980 May 13;19(10):2079–2085. doi: 10.1021/bi00551a012. [DOI] [PubMed] [Google Scholar]
- Hogan M., Dattagupta N., Crothers D. M. Transient electric dichroism of rod-like DNA molecules. Proc Natl Acad Sci U S A. 1978 Jan;75(1):195–199. doi: 10.1073/pnas.75.1.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Il'icheva I. A., Kister A. E., Dashevskii V. G., Esipova N. G., Tumanian V. G. B-forma kak absoliutnyi énergeticheskii minimum DNK. Entropiinaia priroda B--A-perekhoda. Biofizika. 1978 Sep-Oct;23(5):947–951. [PubMed] [Google Scholar]
- Il'icheva I. A., Tumanian V. G., Kister A. E., Dashevskii V. G. Izuchenie zavisimosti vtorichnoi struktury DNK ot posledovatel'nosti osnovanii metodom teoreticheskogo konformatsionnogo analiza. Biofizika. 1978 Mar-Apr;23(2):201–207. [PubMed] [Google Scholar]
- Ivanov V. I., Minchenkova L. E., Minyat E. E., Frank-Kamenetskii M. D., Schyolkina A. K. The B to A transition of DNA in solution. J Mol Biol. 1974 Aug 25;87(4):817–833. doi: 10.1016/0022-2836(74)90086-2. [DOI] [PubMed] [Google Scholar]
- Ivanov V. I., Minchenkova L. E., Schyolkina A. K., Poletayev A. I. Different conformations of double-stranded nucleic acid in solution as revealed by circular dichroism. Biopolymers. 1973;12(1):89–110. doi: 10.1002/bip.1973.360120109. [DOI] [PubMed] [Google Scholar]
- Khutorsky V. E., Poltev V. I. Conformations of double-helical nucleic acids. Nature. 1976 Dec 2;264(5585):483–484. doi: 10.1038/264483a0. [DOI] [PubMed] [Google Scholar]
- Klevan L., Hogan M., Dattagupta N., Crothers D. M. Electric dichroism studies of the size and shape of nucleosomal particles. Cold Spring Harb Symp Quant Biol. 1978;42(Pt 1):207–214. doi: 10.1101/sqb.1978.042.01.023. [DOI] [PubMed] [Google Scholar]
- Le Bret M. Monte Carlo computation of the supercoiling energy, the sedimentation constant, and the radius of gyration of unknotted and knotted circular DNA. Biopolymers. 1980 Mar;19(3):619–637. doi: 10.1002/bip.1980.360190312. [DOI] [PubMed] [Google Scholar]
- Levitt M. How many base-pairs per turn does DNA have in solution and in chromatin? Some theoretical calculations. Proc Natl Acad Sci U S A. 1978 Feb;75(2):640–644. doi: 10.1073/pnas.75.2.640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar D. P., Robbins R. J., Zewail A. H. Direct observation of the torsional dynamics of DNA and RNA by picosecond spectroscopy. Proc Natl Acad Sci U S A. 1980 Oct;77(10):5593–5597. doi: 10.1073/pnas.77.10.5593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller K. J. Interactions of molecules with nucleic acids. I. An algorithm to generate nucleic acid structures with an application to the B-DNA structure and a counterclockwise helix. Biopolymers. 1979 Apr;18(4):959–980. doi: 10.1002/bip.1979.360180415. [DOI] [PubMed] [Google Scholar]
- Peck L. J., Wang J. C. Sequence dependence of the helical repeat of DNA in solution. Nature. 1981 Jul 23;292(5821):375–378. doi: 10.1038/292375a0. [DOI] [PubMed] [Google Scholar]
- Pulleyblank D. E., Shure M., Tang D., Vinograd J., Vosberg H. P. Action of nicking-closing enzyme on supercoiled and nonsupercoiled closed circular DNA: formation of a Boltzmann distribution of topological isomers. Proc Natl Acad Sci U S A. 1975 Nov;72(11):4280–4284. doi: 10.1073/pnas.72.11.4280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhodes D., Klug A. Sequence-dependent helical periodicity of DNA. Nature. 1981 Jul 23;292(5821):378–380. doi: 10.1038/292378a0. [DOI] [PubMed] [Google Scholar]
- Robinson B. H., Lerman L. S., Beth A. H., Frisch H. L., Dalton L. R., Auer C. Analysis of double-helix motions with spin-labeled probes: binding geometry and the limit of torsional elasticity. J Mol Biol. 1980 May 5;139(1):19–44. doi: 10.1016/0022-2836(80)90113-8. [DOI] [PubMed] [Google Scholar]
- Schellman J. A. Flexibility of DNA. Biopolymers. 1974 Jan;13(1):217–226. doi: 10.1002/bip.1974.360130115. [DOI] [PubMed] [Google Scholar]
- Shindo H., Wooten J. B., Pheiffer B. H., Zimmerman S. B. Nonuniform backbone conformation of deoxyribonucleic acid indicated by phosphorus-31 nuclear magnetic resonance chemical shift anisotropy. Biochemistry. 1980 Feb 5;19(3):518–526. doi: 10.1021/bi00544a020. [DOI] [PubMed] [Google Scholar]
- Sobell H. M., Tsai C. C., Gilbert S. G., Jain S. C., Sakore T. D. Organization of DNA in chromatin. Proc Natl Acad Sci U S A. 1976 Sep;73(9):3068–3072. doi: 10.1073/pnas.73.9.3068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tumanian V. G., Miannik Ia Kh, Il'icheva I. A. Struktura B-formy DNK na primere gomopolimera poly(dA) . poly(dT). Utochnenie po rentgenograficheskim i énergeticheskim kriteriiam. Biofizika. 1979 Jul-Aug;24(4):765–769. [PubMed] [Google Scholar]
- Vologodskii A. V., Anshelevich V. V., Lukashin A. V., Frank-Kamenetskii M. D. Statistical mechanics of supercoils and the torsional stiffness of the DNA double helix. Nature. 1979 Jul 26;280(5720):294–298. doi: 10.1038/280294a0. [DOI] [PubMed] [Google Scholar]
- Wang A. H., Quigley G. J., Kolpak F. J., Crawford J. L., van Boom J. H., van der Marel G., Rich A. Molecular structure of a left-handed double helical DNA fragment at atomic resolution. Nature. 1979 Dec 13;282(5740):680–686. doi: 10.1038/282680a0. [DOI] [PubMed] [Google Scholar]
- Worcel A., Strogatz S., Riley D. Structure of chromatin and the linking number of DNA. Proc Natl Acad Sci U S A. 1981 Mar;78(3):1461–1465. doi: 10.1073/pnas.78.3.1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhurkin V. B., Lysov Y. P., Ivanov V. I. Anisotropic flexibility of DNA and the nucleosomal structure. Nucleic Acids Res. 1979 Mar;6(3):1081–1096. doi: 10.1093/nar/6.3.1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhurkin V. B., Lysov Y. P., Ivanov V. I. Computer analysis of conformational possibilities of double-helical DNA. FEBS Lett. 1975 Nov 1;59(1):44–47. doi: 10.1016/0014-5793(75)80337-1. [DOI] [PubMed] [Google Scholar]
- Zhurkin V. B., Poltev V. I., Florent'ev V. L. Atom-atomnye potentsial'nye funktsii dlia konformatsionnykh raschetov nukleinovykh kislot. Mol Biol (Mosk) 1980 Sep-Oct;14(5):1116–1130. [PubMed] [Google Scholar]



