Abstract
In this study, we characterized the anti-HIV activities of various R88-APOBEC3G (R88-A3G) mutant fusion proteins in which each A3G mutant was fused with a virus-targeting polypeptide (R14-88, hereafter named R88) derived from HIV-1 Vpr. Our results show that the introduction of the deaminase-defective mutant E259Q into R88-A3G did not affect the virion incorporation of this mutant but blocked the protein's ability to inhibit HIV-1 infection. Our data also reveal that the antiviral effect of A3GY124A, a previously described A3G virus-packaging mutant, was completely rescued when the mutant was fused with R88. In an attempt to identify the most potent R88-A3G fusion proteins against HIV-1 infection, we introduced two Vif-binding mutants (D128K and P129A) into the R88-A3G fusion protein and showed that both R88-A3GD128K and R88-A3GP129A possessed very potent anti-HIV activity. When R88-A3GP129A was transduced into CD4+ C8166 T cells, HIV-1 infection was completely abolished for at least 24 days. In an attempt to further test the anti-HIV effect of this mutant in primary human HIV susceptible cells, we introduced R88-A3GP129A into human peripheral blood mononuclear cells (PBMCs) and macrophages with a recombinant adeno-associated virus (rAAV2/5) vector. The results demonstrate that a significant inhibition of HIV-1 infection was observed in the transduced PBMCs and macrophages. These results provide evidence for the feasibility of an R88-A3G–based anti-HIV strategy. The further optimization of this system will contribute to the development of new anti-HIV gene therapy approaches.
Ao and colleagues generate a panel of R88-APOBEC3G (R88-A3G) mutant fusion proteins and characterize their anti-HIV properties. They demonstrate the ability to use AAV2/5 to express a protein with particularly potent anti-HIV activity (R88-A3GP129A) into CD4+ T cells and human peripheral blood mononuclear cells and macrophages and efficiently inhibit HIV-1 infection.
Introduction
The Apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 3G (APOBEC3G; hereafter referred to as A3G), is a cellular cytidine deaminase that can interfere with the replication of a broad range of retroviruses and hepatitis B viruses (Sheehy et al., 2002; Turelli et al., 2004). It is well known that in the absence of HIV Vif, A3G is capable of efficiently incorporating into viral particles during HIV assembly by interacting with the viral Gag precursor (Alce and Popik, 2004; Cen et al., 2004; Luo et al., 2004; Schafer et al., 2004). In the viral particle, A3G associates with the HIV-1 reverse transcription complex, and this association is enhanced by the presence of viral RNA (Khan et al., 2005). After virus entry into susceptible cells, the virion-incorporated A3G interrupts HIV infectivity by introducing dC-to-dU mutations into the minus strand of the viral DNA during reverse transcription (Harris et al., 2003; Lecossier et al., 2003; Mangeat et al., 2003; Mariani et al., 2003; Zhang et al., 2003). In addition to its deaminase activity, A3G has been shown to inhibit viral reverse transcription directly (Chiu et al., 2005; Newman et al., 2005). All of these studies clearly indicate that host A3G possesses multiple anti-HIV activities during HIV-1 replication.
During wild-type HIV-1 infection, however, the antiviral effects of A3G are counteracted by the presence of the viral protein Vif. It is well known that Vif can directly bind to A3G and significantly prevent the virion incorporation of A3G by accelerating A3G ubiquitination and proteasomal degradation, thus decreasing the intracellular concentration of A3G (Conticello et al., 2003; Kao et al., 2003; Mariani et al., 2003; Marin et al., 2003; Sheehy et al., 2003; Stopak et al., 2003; Yu et al., 2003, 2004). In addition, other studies suggest that Vif can prevent A3G incorporation into virions independent of the reduction in A3G intracellular levels (Kao et al., 2003, 2004; Mariani et al., 2003; Sheehy et al., 2003). Indeed, Opi et al. (2007) showed that HIV-1 Vif efficiently inhibits the virion packaging and antiviral activity of a degradation-resistant A3G mutant.
Given that A3G possesses potent anti-HIV activity and that its action is counteracted by HIV-1 Vif protein, considerable interest has been focused on restoring A3G's antiviral activity during wild-type HIV-1 infection because the antiviral activity of A3G represents a novel therapeutic strategy against HIV-1 infection. One way to overcome the inhibition of A3G by Vif is to disrupt the Vif-A3G interaction. Previous studies have shown that a single amino-acid substitution at position 128 of A3G (representing a change of an aspartic acid [D] to a lysine [K]) blocked the Vif interaction with A3G and rescued the antiviral activity of A3G (Bogerd et al., 2004; Mangeat et al., 2004; Schrofelbauer et al., 2004; Xu et al., 2004; Huthoff and Malim, 2007; Lavens et al., 2010). This finding demonstrated the feasibility of designing pharmaceutical agents that could block Vif binding to A3G. Interestingly, while the three amino acid (aa) motif comprised of aspartic acid, proline, and aspartic acid (DPD) at aa positions 128 to 130 of A3G was identified as a crucial region for the interaction between A3G and HIV-1 Vif (Huthoff and Malim, 2007), a YYFW (124 to 127) region that is adjacent to the N-terminus of the DPD motif of A3G was found to be critical for the virion packaging of A3G. Such close alignment of these two functional domains within A3G makes it difficult to target the DPD motif with a pharmaceutical agent without affecting A3G packaging. Thus, other approaches, including delivering A3G into HIV-1 particles independent of the blockage by Vif and subsequently inactivating HIV-1 infectivity, could hold promise as a potential HIV therapeutic approach.
We recently reported that the R88-A3G fusion protein, which was made by fusing A3G to a virus-targeting polypeptide (R14-88) derived from the HIV-1 Vpr protein (Yao et al., 1999), was able to overcome the HIV-1 Vif barrier and efficiently deliver A3G into the virus (Ao et al., 2008). Moreover, the study demonstrated that the expression of the R88-A3G fusion protein in Vif+ HIV-1–producing cells drastically inhibited progeny virus infection in different cell types, including CD4+ C8166 T cells and human primary peripheral blood mononuclear cells (PBMCs) (Ao et al., 2008). In the present study, we introduced different A3G mutants into the R88-A3G fusion system and characterized their anti-HIV properties. Furthermore, this study demonstrates that, through an AAV2/5 vector, we were able to introduce R88-A3GP129A into CD4+ T cells human PBMCs and macrophages and efficiently inhibit HIV-1 infection.
Material and Methods
Plasmids
CMVin-R14-88-pcs-A3Gwt (R88-A3Gwt), HA-A3Gwt, and pYEF1-R88-A3Gwt were constructed previously (Ao et al., 2008). The CMVin-R88-A3G mutants were generated by a polymerase chain reaction (PCR)-based method (Ao et al., 2007). Briefly, in a two-step PCR technique that has been previously described (Yao et al., 1995b), the primers used for generating different A3G mutants were the 5′-A3G-XbaI primer (5′-TCATCTAGAAAGCCTCACTTCAG-3′), 3′-BglII primer (5′-TAAGAGCTCATGCCTGCAGCC-3′), and the corresponding mutagenic oligonucleotides. Each of the amplified A3G mutant cDNAs was cloned into CMVin-R14-88-pcs as described previously (Ao et al., 2008). The presence of the corresponding mutations in A3G was confirmed by sequencing. The HIV-1 Vif expressor (pcDNA-hVif) (Nguyen et al., 2004) was obtained from the National Institutes of Health (NIH) AIDS Research and Reference Reagent Program. The HIV-1 proviral clones HxBru-Vif+ and pNL4.3-Nef+/GFP have been described previously (Ao et al., 2008). The AAV2CMVlacZ plasmid expressing the lacZ reporter gene from a cytomegalovirus (CMV) promoter and flanked by AAV2 inverted terminal repeats has been described before (Auricchio et al., 2001; Bello et al., 2009). The R88-A3GP129A gene was amplified by PCR from plasmid CMVin-R88-A3GP129A using the following primer pair: 5′-R88-NotI primer (5′-ATAGCGGCCGCCATGCCACACAATGAA-3′) and 3′-A3G-BamHI primer (5′-GCTCGGATCCTCAGTTTTCCTGATT-3′).
Cell lines, antibodies, and chemicals
Human embryonic kidney 293T cells and HeLa cells were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal calf serum (FCS) and 1% penicillin and streptomycin. The CD4+ C8166 T-cell line was maintained in RPMI-1640 medium containing 10% FCS and 1% penicillin and streptomycin. PBMCs were isolated from the blood of healthy adult volunteers by sedimentation on a Ficoll (Lymphoprep; Axis-Shield, Norton, MA) gradient. Isolated PBMCs were stimulated with 0.1% phytohemaglutinin for 3 days and maintained in RPMI 1640 supplemented with 5% interleukin-2. PBMC-derived macrophages were prepared by dispensing fresh PBMCs into 12-well plates at the desired density at 37°C for 2 hours. After gentle washing with DMEM, the adherent cells were cultured in DMEM containing 10% fetal bovine serum (FBS) and 10-ng/ml macrophage colony-stimulating factor (M-CSF; R&D Systems, Minneapolis, MN) for 7 days. The DNA transfection of 293T cells or HeLa cells was performed using a standard calcium phosphate DNA precipitation method.
The purified rabbit anti-hA3G (cat. no. 10201) was obtained through the NIH AIDS Research and Reference Reagent Program. The rabbit anti-Vpr antibody has been described previously (Yao et al., 1995b). The mouse anti-HIVp24 and anti-CD4 monoclonal antibodies used in this study have been described previously (Yao et al., 1995a; Ao et al., 2005).
Virus production and infection
To generate viruses that incorporated different R88-A3G fusions, 293T cells were co-transfected with the corresponding HIV-1 proviral DNA and R88-A3Gwt/mut plasmids. Virus-containing supernatants were collected at 48 hr after transfection and subjected to ultracentrifugation (32,000 rpm for 1 hr at 4°C) to pellet the virus. The quantification of virus stocks was determined by Gag-p24 measurements using an HIV-1 p24 ELISA Kit (purchased from the AIDS Vaccine Program of the Frederick Cancer Research and Development Center, Ft. Detrick, MD). To infect CD4+ C8166 T cells, equal amounts of virus were incubated with susceptible cells at 37°C for 2 hr. The cells were then washed and incubated with fresh medium. At different time points, HIV replication levels in each infected culture were monitored by measurement of HIV-1 Gag-p24 antigen by HIV-1 Gag-p24 ELISA. The infection level mediated by the pNL4.3-GFP HIV-1 virus was monitored using a fluorescence-activated cell sorter (FACS; Becton Dickinson FACS Calibur, Franklin Lakes, NJ) or observed under fluorescence microscopy after the infected cells were fixed with phosphate-buffered saline (PBS)-4% paraformaldehyde.
Establishment of R88-A3Gwt/mutant-expressing stable C8166 cell lines
The C8166 cell lines expressing R88-A3Gwt or R88-A3GP129A were generated as described previously (Ao et al., 2008). Briefly, pseudotyped lentiviral vector stocks were produced by co-transfecting 293T cells with pYEF1-mcs or pYEF1-R88-A3Gwt/P129A-puro vectors and the HIV-packaging pCMV ΔR8.2 vector (Kobinger et al., 2001) and VSV-G expression plasmids. The lentiviral vector produced (300 ng of Gag-p24) was then incubated with 0.5 × 106 C8166 T cells overnight. Forty-eight hours after transduction, the transduced C8166 T cells were placed under selection with puromycin (0.5 μg/ml) for at least 10 days before analysis. The cell growth, cell cycle profiles, and CD4 receptor expression levels of the various cell lines were analyzed as described previously (Ao et al., 2008).
AAV production and transduction
The AAV2/5 vectors were produced by triple transfection of HEK 293 cells with AAV2/5 CMVlacZ or AAV2/5CMVR88-A3GP129A, the pACK2/5 packaging plasmid, which comprises rep from AAV2 and cap from AAV5 (Bello et al., 2009) and the pAd.DELTA F6 helper plasmid containing adenoviral genes necessary to drive AAV replication (E2a, E4, VARNA). Transfections were performed with calcium phosphate co-precipitation (Bello et al., 2009). Recombinant AAV2/5 vectors were all purified by the standard cesium chloride sedimentation method (Xiao et al., 1998). The titers of AAV2/5 vector preparations were determined by TaqMan PCR with primers for the bGH polyA (Bello et al., 2009).
The cells were transduced with recombinant AAV2/5CMVLacZ or AAV2/5-CMVR88-A3GP129A at 1 × 106 virus particles (vp)/cell in serum-free medium and incubated for 2 hr at 37°C with gentle agitation every 15 min followed by the addition of fresh media with 10% FBS and overnight incubation.
Quantitative detection of R88-A3GP129A mRNA in transduced cells by reverse transcription and real-time PCR analysis
Human PBMCs and macrophages were transduced with AAV2/5CMVLacZ or AAV2/5CMVR88-A3GP129A vectors at 106 vp/cell. Seventy-two hours after transduction, total RNA from 1 × 106 cells was extracted using the TRIZOL reagent (Invitrogen, Carlsbad, CA) following the manufacturer's instructions. RNA was subjected to reverse-transcription PCR (RT-PCR) with Moloney murine leukemia virus reverse transcriptase (Promega, Madison, WI). The cDNA was then PCR-amplified with Taq DNA polymerase using the following primers: 5′-R88-NotI-ATAAGCGGCCGCCATGCCACACAATGAA, 3′-A3GP129A-TGGTAATCTGTCCCAGAA. The 5′ and 3′ primers targeted Vpr and A3G, respectively. The mRNA content was normalized by amplifying the human β-globin gene as described previously (Simon and Malim, 1996). All PCR products were separated by electrophoresis on 1.2% agarose gels. To quantitatively determine the expression levels of R88-A3GP129A mRNA and the total cellular A3G mRNA (including endogenous A3G mRNA) in PBMC and in macrophages, the extracted RNA was first subjected to RT-PCR with Moloney murine leukemia virus reverse transcriptase (Promega). Then the reverse-transcripted DNA from each sample was quantified by real-time PCR analysis by using two sets of primers separately: one set for R88-A3GP129A (Vpr-5, 5′-GATACTTGGGCAGGAGTGG-3′ and A3G-3; 5′-CACCTGGCCTCGAAAGAT-3′) and another set for A3G (A3G-5; 5′-CTTCAGAAACACAGTGGAGC-3′ and A3G-3; 5′-CACCTGGCCTCGAAAGAT-3′). The R88-A3GP129A and the total cellular A3G levels were quantified in Mx3000P real-time PCR system (Stratagene, Santa Clara, CA). The reaction was carried out in a final volume of 20 μl, consisting 1 × FastStart DNA Master SYBR Green I (Roche Diagnostics, Mannheim, Germany) and 0.2 μM of each sense and antisense primers.
Radiolabeling, immunoprecipitation, and Western blot analyses
To perform pulse-chase radiolabeling experiments, 293T cells were co-transfected with wild-type R88-A3G or the mutants and a pcDNA-hVif expressor. After 48 hr, cells were incubated for 30 min with starvation medium (DMEM without methionine, plus 10% dialyzed FBS) followed by pulse-labeling for 30 min with 250 μCi of [35S]-methionine (PerkinElmer Life Science, Boston, MA). After labeling, the radiolabeled medium was replaced with medium containing an excess of unlabeled methionine, and the cells were incubated at 37°C. Labeled cells were collected at 0, 3.5, or 5 hr and subjected to immunoprecipitation using an anti-Vpr antibody. Immunoprecipitates were resolved by SDS–PAGE in a 12% gel followed by autoradiography.
To analyze protein expression in viral particles, lysed viral samples were directly loaded into a 12% SDS–PAGE gel, and different proteins were detected by Western blot analysis with the corresponding antibodies. The horseradish peroxidase–conjugated donkey anti-rabbit IgG and sheep anti-mouse IgG (Amersham Biosciences, Piscataway, NJ) were used as secondary antibodies, and the protein bands were visualized using an enhanced chemiluminescence kit (PerkinElmer Life Science).
Immunofluorescence
HeLa cells were grown on glass coverslips (12 mm2) in 24-well plates and transfected with CMVin-HA-A3Gwt or different R88-A3Gwt/mutant plasmids. After 48 hr, the cells were washed with PBS and fixed and permeabilized with methanol/acetone (1:1 ratio) for 30 min at room temperature. The coverslips were incubated with a rabbit anti-A3G antibody (1:500) for 2 hr at 37°C followed by incubation with a 1:500 dilution of fluorescein isothiocyanate–conjugated anti-rabbit antibody for 1 hr. Nuclei were stained with 4′,6-diamidino-2-phenylindole. The coverslips were mounted with Mowiol 4-88, and the cells were visualized under an Axiovert 200 microscope (Carl Zeiss, Jena, Germany) with a × 20 objective.
Results
Expression of different R88-A3G variants and their resistance to Vif-mediated degradation
Our previous study demonstrated that the fusion protein R88-A3Gwt comprised of an A3G and a virion-targeting polypeptide (R14-88) significantly inhibited HIV-1 infection even in the presence of Vif (Ao et al., 2008). To develop a more potent anti-HIV R88-A3G fusion molecule and to test the requirement of A3G enzymatic activity for the anti-HIV effect of R88-A3G fusion proteins, we introduced several A3G mutants into the CMVin-R88-A3G fusion protein, including R88-A3GY124A, R88-A3GD128K, R88-A3GP129A, and R88-A3GE259Q (Fig. 1A). Among these mutants, D128K and P129A, which are located in the previously described HIV Vif-binding region 128DPD130, were intended to block the Vif-A3G interaction (Xu et al., 2004; Huthoff and Malim, 2007). The A3GY124A has a mutation located in a 124YYFW127 region that is important for the packaging of A3G into HIV-1 particles. This mutant has been previously shown to lose the virion incorporation ability (Huthoff and Malim, 2007). We also included a C-terminal cytidine deaminase domain mutant, R-A3GE259Q (Shindo et al., 2003; Schumacher et al., 2008; Browne et al., 2009), to test the importance of the enzymatic function of A3G in the antiviral activity of R-A3G.
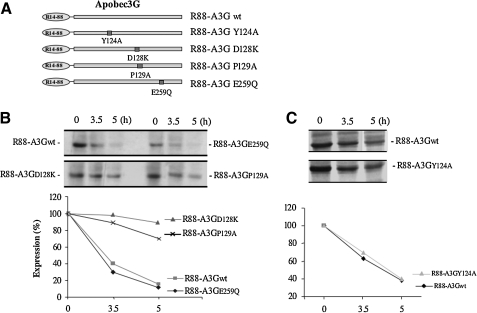
FIG. 1.
The expression of R88-APOBEC3G (R88-A3G) mutants and their resistance to Vif-mediated degradation. (A) Schematic representation of R88-A3G mutants at the cytidine deaminase activity site (E259Q), viral packaging site (P124A), and Vif-response site (D128K, P129A). (B and C) R88-A3G wild-type or mutant expressors were co-transfected with a pcDNA-hVif expressor in 293T cells. At 48-hr post-transfection, cells were pulse-radiolabeled with [35S]-methionine for 30 min and collected and lysed at 0, 3.5, and 5 hr. The level of R88-A3Gwt/mutant in each lysed cell sample was evaluated by anti-Vpr immunoprecipitation (upper panel). The level of degradation of wild-type and mutant R88-A3G was quantified by laser densitometry (lower panel). The result shown is representative of two independent experiments.
We next tested the expression of different A3G mutants and their resistance to Vif-mediated degradation. It is known that HIV Vif targets A3G to the proteasome for degradation, and that this targeting results in reduced intracellular levels of A3G, consequently inhibiting A3G packaging into virions (Kao et al., 2003; Mariani et al., 2003; Marin et al., 2003; Sheehy et al., 2003; Stopak et al., 2003). In this study, we transfected each of the R88-A3G expressors with an HIV-1 Vif plasmid (pcDNA-hVif) (Nguyen et al., 2004) into 293T cells. At 48 hr post-transfection, the expression of each protein in 293T cells was analyzed by [35S]methionine pulse-chase radiolabeling and immunoprecipitation with anti-Vpr antibodies as described previously (Ao et al., 2008). Our results show that, consistent with a previous study (Ao et al., 2008), R88-A3Gwt was rapidly degraded in the presence of Vif and had a half-life of approximately 3 hr (Fig. 1B). Similarly, R88-A3GE259Q and R88-A3GY124A were also sensitive to Vif-mediated degradation (Fig. 1B and C). By contrast, the R88-A3GD128K and R88-A3GP129A mutants were found to be resistant to degradation and had half-lives of more than 5 hr (Fig. 1B). These results are consistent with previous observations for wild-type A3G (Xu et al., 2004; Schrofelbauer et al., 2006; Huthoff and Malim, 2007) and imply that the Vif-resistant properties of R88-A3GD128K and R88-A3GP129A may affect their antiviral activities.
Inhibitory effects of different R88-A3G mutants on Vif+ HIV-1 infectivity
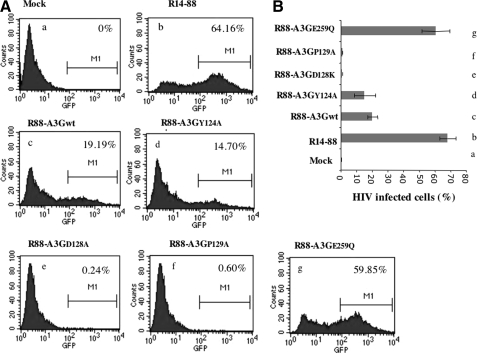
To investigate the inhibitory effect of each R88-A3G mutant on Vif+ HIV-1 infectivity, we produced virus stocks by co-transfecting pNL4.3/GFP+/Vif+ provirus with either wild-type R88-A3G or different mutant expression vectors in 293T cells. Forty-eight hours after transfection, progeny viruses were pelleted by ultracentrifugation and quantified by Gag-p24 measurements using an HIV-1 p24 ELISA kit. The virus production from different R88-A3G mutant-transfected cells did not differ significantly according to the p24 levels (data not shown). Equal amounts of the viruses (adjusted by HIV-1 Gag-p24 levels) were used to infect CD4+ C88166 T cells, and the level of infection was determined in CD4+ C88166 T cells by FACS analysis of the percentage of infected (green fluorescent protein [GFP]-positive) cells (Fig. 2). In the absence of any R88-A3G, approximately 64% of the cells were infected (Fig. 2A and B, b), while in the presence of R88-A3Gwt, only 19% of cells were found to be positive (Fig. 2A and B, c). Interestingly, R88-A3GE259Q, a catalytically defective mutant, did not exhibit significant antiviral activity compared with R88-A3Gwt (Fig. 2A and B, g). These results indicate that, in the context of the R88-A3G fusion, the cytidine deaminase mutant A3GE259Q was not active against HIV-1, even though it was efficiently incorporated into the virus (Fig. 3B, left panel). In contrast, the packaging-defective mutant R88-A3GY124A (Huthoff and Malim, 2007) possessed anti-HIV activity as efficiently as the wild-type R88-A3G (Fig. 2A and B, compare d with c). This result provides evidence that the fusion of a virion-targeting signal (R88) to A3GY124A can rescue the antiviral activity of A3GY124A.
FIG. 2.
The inhibitory effects of R88-A3G mutants on Vif+ virus infectivity. Viruses were produced from 293T cells transfected with 3 μg of HIV-1 pNL4.3-GFP or co-transfected with 4 μg of a R88-A3Gwt/mutant expressor. Equal amounts of produced viruses (as adjusted by the Gag-p24 level) were used to infect CD4+ C8166 T cells. At 72 hr post-infection, the percentage of infected (green fluorescent protein [GFP]-positive) cells was measured by fluorescence-activated cell sorter (FACS) analysis. (A) Representative FACS profiles. (B) Mean percentages of infected cells from each cell line (a–g) with standard errors are the results for duplicated samples from the experiment.
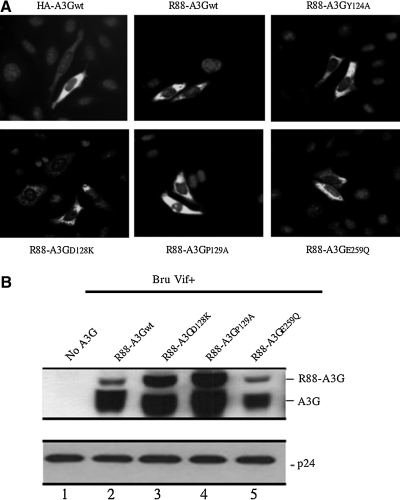
FIG. 3.
Intracellular localization and virion incorporation of different R88-A3G mutants. (A) HeLa cells were transfected with equal amounts of HA-A3G or R88-A3Gwt/mutant plasmid DNA. Forty-eight hours post-transfection, cells were fixed and treated with rabbit anti-A3G polyclonal antibody followed by incubation with fluorescein isothiocyanate (FITC)-conjugated anti-rabbit antibody and observed by fluorescence microscopy ( × 20 objective). (B) 293T cells were transfected with HxBru-Vif+ (3 μg; lane 2) or co-transfected with HxBru-Vif+ and 2 μg of R88-A3Gwt/mutant (D128K, P129A, or E259Q; lanes 3 to 6). After 48 hr, the produced virus particles were collected from the supernatant by ultracentrifugation through a 20% sucrose cushion. The virus lysate samples were loaded onto a 12% SDS-PAGE gel and analyzed by Western blotting with rabbit anti-A3G and anti-p24 antibodies, as indicated. The result shown is representative of two independent experiments.
Two R88-A3G mutants, R88-A3GD128K and R88-A3GP129A, showed significantly enhanced anti-HIV activities compared with R88-A3Gwt. The infection was almost completely blocked when the infecting virus was produced from R88-A3GD128K– or R88-A3GP129A–expressing cells (Fig. 2A and B, e and f). This result was expected because the two mutants have been shown to be resistant to inhibition by Vif (Mangeat et al., 2004; Schrofelbauer et al., 2004; Xu et al., 2004; Huthoff and Malim, 2007). Moreover, our data indicate that wild-type R88-A3G is not fully Vif resistant, and that introducing D128K or P129A into R-A3G could almost completely inhibit HIV-1 replication by overcoming the blockage of Vif. These results indicate that R88-A3GD128K and R88-A3GP129A are potent antiviral molecules.
Intracellular localization and virion incorporation of different R88-A3G mutants
The results discussed above show that different R88-A3G mutants possess varying anti-HIV activities. To better understand the mechanism underlying the actions of these mutants, we investigated their intracellular localization and virion incorporation. Previous studies showed that A3G localized predominantly to the cytoplasm and that the single aa substitutions within the region from aa 113 to 128 did not affect the cytoplasmic localization of A3G (Stenglein et al., 2008). Because A3G was fused to a HIV-1 Vpr peptide, a karyophilic protein that is predominantly localized in the nucleus (Lu et al., 1993; Yao et al., 1995b), we examined whether this fusion strategy changed the intracellular distribution of different A3G mutants. Each R88-A3G mutant was transfected into HeLa cells, and their intracellular localization was analyzed by an anti-A3G immunofluorescence assay. In parallel, an HA-tagged A3G expressor (Ao et al., 2008) was also transfected into HeLa cells as a control. The results show that HA-A3G was evenly distributed in the cytoplasm (Fig. 3A). Similarly, wild-type R88-A3G and all R88-A3G mutants were predominantly localized in the cytoplasm (Fig. 3A). These results indicate that fusing R14-88 with A3G did not affect the intracellular localization of the fusion protein.
To examine the encapsidation of R-A3G mutants into the virus in the presence of HIV Vif, we transfected 293T cells with the HxBru-Vif+ provirus and either the wild-type or mutant (D128K, P129A, or E259Q) CMVin-R88-A3G plasmids. At 48 hr post-transfection, viral particles were collected and concentrated by ultracentrifugation through a 20% sucrose cushion. Similar amounts of each virus (adjusted by the amount of HIV p24) were lysed and directly loaded onto a 12% SDS-PAGE gel. The virion-associated R88-A3G and Gag-p24 proteins were detected by Western blotting with rabbit anti-A3G and mouse anti-p24 antibodies. Consistent with our previous observations (Ao et al., 2008), a significant amount of R88-A3Gwt was shown to be packaged into Vif+ HIV-1 virions (Fig. 3B, left panel, lane 2). Interestingly, the R88-A3GD128K and R88-A3GP129A mutants that are resistant to Vif-mediated degradation (Fig. 1B) were incorporated into viruses at much higher levels compared with wild-type R88-A3Gwt (Fig. 3B, left panel, compare lanes 3 and 4 with lane 2). The virion-incorporation level of the cytidine deaminase–deficient mutant R88-A3GE259Q was similar to that of wild-type R88-A3G (Fig. 3B, left panel, lane 5). These data indicate that one explanation for the remarkable anti-HIV activities of R88-A3GD128K and R88-A3GP129A (as shown in Fig. 2B) arises from their high levels of virion incorporation.
Expression of R88-A3GP129A in CD4+ C8166 T cells effectively blocks HIV-1 replication and spread
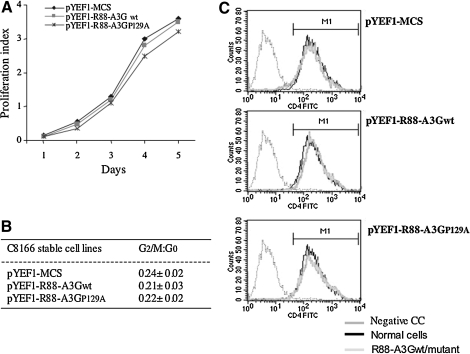
The results from the above-mentioned experiments showed that R88-A3GD128K and R88-A3GP129A exhibited the most effective antiviral activity when they were transiently over-expressed in HIV-producing 293T cells. To determine how these mutants affect HIV replication and spread when expressed in CD4+ T cells, we generated CD4+ T-cell lines that stably expressed a low level of R88-A3G and R88-A3GP129A through a lentiviral vector system as previously described (Ao et al., 2008). Briefly, the CD4+ C8166 T cells were subsequently transduced with an equal amount of R88-A3G– and R88-A3GP129A–expressing vector particles (300 ng of Gag-p24), as described in Materials and Methods, and selected with puromycin (0.5 μg/ml). Two C8166 T-cell lines that stably expressed R88-A3Gwt or R88-A3GP129A were obtained. The expression of the fusion protein was detected by monitoring virion-incorporated R88-A3Gwt or R88-A3GP129A protein from highly concentrated HxBru-Vif+ viruses produced in these two cell lines (data not shown). We were unable to obtain cells containing R88-A3GD128K due to the extremely strong inhibitory effect of R88-A3GD128K on lentiviral vector transduction efficiency. Prior to examining the effects of the R-A3Gwt/mutant on HIV replication, we evaluated the cell growth, cell cycle profile, and CD4 receptor expression on the cell surface as described previously (Ao et al., 2008). The results showed that cell growth and the cell cycle profiles did not significantly differ in R88-A3Gwt, R88-A3GP129A, or empty vector–transduced C8166 T-cell lines (Fig. 4A and B). In addition, equivalent levels of the surface CD4 receptor were detected on these stable cell lines (Fig. 4C).
FIG. 4.
Generation and biological characterization of CD4+ T cells stably expressing R88-A3GP129A. C8166 T cells (0.5 × 106) were transduced with an empty vector (pYEF1-MCS) or lentiviral vectors containing either R88-A3Gwt (pYEF1-R88-A3Gwt) or R88-A3GP129A (pYEF1-R88-A3GP129A) and selected with 0.5 μg/ml puromycin to produce puromycin-resistant cell populations. (A) To assess the growth of vector- and R88-A3Gwt/mut-transduced C8166 T cells, a cell proliferation reagent WST assay was performed to determine the cell viability at several time points. (B) The cell-cycle profiles of vector- and R88-A3G–transduced C8166 T cells were analyzed by staining with 30 μg/ml of propidium iodide followed by flow cytometry to measure cellular DNA content. (C) CD4 receptor expression levels in vector- and R88-A3Gwt/mut–transduced C8166 T cells were analyzed by anti-CD4 staining and a flow cytometry assay.
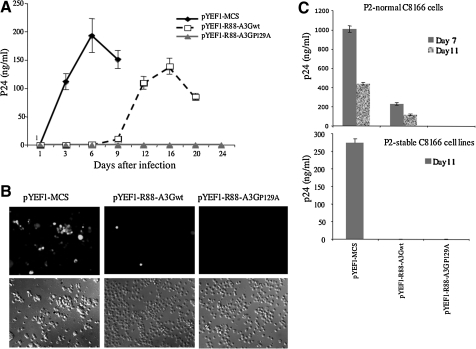
To determine whether the R88-A3GP129A–transduced C8166 T cells exerted inhibitory effects on HIV-1 infection, we infected R88-A3Gwt, R88-A3GP129A, and empty vector–transduced C8166 T-cell lines in parallel with equal amounts of pNL4.3-GFP virus (Ao et al., 2008). Viral replication was monitored by measuring the level of the HIV-1 Gag-p24 antigen in the supernatant for a 24-day period (Fig. 5A). In addition, the presence of HIV-1–infected cells (GFP-positive) at day 6 was determined using fluorescence microscopy (Fig. 5B). The results showed that the empty vector control cell line supported Vif+ pNL4.3-GFP virus replication, which peaked after 6 days of infection. In the R-A3Gwt–transduced cell line, Vif+ pNL4.3-GFP robust virus replication began at day 12 and peaked at day 16 (Fig. 5A and B). Interestingly, no HIV-1 replication was detected during the 24 days of observation in cells expressing R88-A3GP129A (Fig. 5A and B). These data indicate that R88-A3GP129A expressed in highly susceptible CD4+ T cells was capable of providing long-term inhibition of Vif+ HIV-1 replication.
FIG. 5.
The R88-A3GP129A–transduced CD4+ C8166 T cells significantly inhibited HIV-1 infection. Equal amounts of pNL4.3-GFP virus were used to infect vector-, R88-A3Gwt-, or R88-A3GP129A–transduced C8166 cell lines. (A) At different time points, virion-associated Gag-p24 antigen levels in the supernatant were measured by anti-p24 ELISA. (B) After 6 days of infection, the numbers of infected (GFP-positive) cells were visualized under fluorescence microscopy. (C) Identical volumes of 72-hr infectious supernatants were used to infect nontransduced (upper panel) or transduced C8166 cells (lower panel; passage 2). Levels of HIV-1 Gag-p24 antigen in supernatants from passage 2 were then measured by an HIV p24 ELISA assay at days 7 and 11 post-infection. The result shown is representative of two independent experiments. Means with standard errors were calculated from the duplicated samples from one experiment.
To further test the effect of R88-A3Gwt and R88-A3GP129A on HIV transmission, the supernatant from the initial infection culture (passage 1) was collected at 72 hr post-infection. Similar volumes (1 ml) of infectious supernatants were used to infect normal (Fig. 5C, upper panel) or fresh R88-A3Gwt– or R88-A3GP129A–expressing C8166 T cells (Fig. 5C, lower panel). After infection, we performed an HIV-1 p24 ELISA to determine the infectivity of the progeny virus by measuring HIV-1 Gag-p24 levels in the supernatants from the corresponding cultures at days 7 and 11 post-infection. The results show that the infectivity of the progeny virus from R88-A3Gwt–transduced cells was reduced approximately fivefold compared with the virus from empty vector–transduced cells in normal C8166 cells (passage 2; Fig. 5C, upper panel). However, when the viral infection was carried out in freshly R88-A3Gwt–transduced cells (passage 2; Fig. 5, lower panel), no infection was detected until day 11. Interestingly, there were no escaped infectious viruses produced in R88-A3GP129A–expressing cells at passage 1. Therefore, no infection was observed at passage 2 post-infection in either normal or freshly R88-A3GP129A–transduced cells (passage 2; Fig. 5C). Taken together, these results further demonstrate that the presence of R88-A3G in CD4+ T cells significantly inhibited HIV infection, and that R88-A3GP129A was able to completely block HIV infection and spread in CD4+ C8166 T cells.
R88-A3GP129A inhibits HIV-1 replication in human PBMCs and macrophages
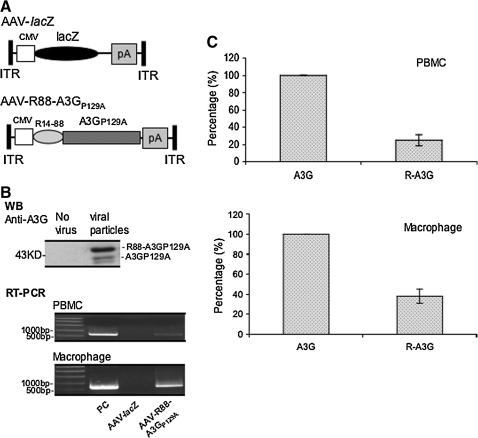
Human CD4+ T cells and macrophages are the main HIV-1 infection target cells in vivo. Thus, it is important to introduce R88-A3GP129A in these primary cells and test its inhibitory effect on HIV infection. Because the low efficient lentiviral vector-mediated introduction of R88-A3GP129A into these primary cells poses a significantly obstacle, we utilized an adeno-associated viral (AAV2/5) vector to introduce the R88-A3GP129A gene into primary PBMCs and macrophages because previous studies showed that the production and transduction of the AAV vector were not inhibited by A3G (Chen et al., 2006; Narvaiza et al., 2009). To produce AAV2/5-expressing R88-A3GP129A, we first replaced the lacZ gene in an AAV2-lacZ vector (Auricchio et al., 2001; Bello et al., 2009) with a cDNA encoding R88-A3GP129A (Fig. 6A). AAV2/5-lacZ- and AAV2/5-R88-A3GP129A particles were subsequently produced by transfecting 293T cells with AAV2-lacZ or AAV2-R88-A3GP129A, the pACK2/5 packaging plasmid, and the helper plasmid pAd.DELTA F6. The recombinant AAV2/5 vector particles were collected, purified by cesium chloride gradients and titrated by TaqMan PCR as previously described in the Material and Methods section. To test whether AAV2/5 could transduce C8166 T cells, human PBMCs, and macrophages, we transduced these cells with the AAV2/5-R88-A3GP129A vector (106 vp/cell). After 3 days, R88-A3GP129A and the total cellular A3G mRNA levels were quantified by reverse transcription followed by PCR (Fig. 6B) and by the real-time PCR analysis (Fig. 6C). The results showed that R88-A3GP129A–specific mRNA was detected in PBMCs and in macrophages (Fig. 6B, middle and lower panels), and the data from quantitative real-time PCR revealed that the R88-A3GP129A mRNA level was approximately 23% of the total A3G mRNA level in PBMCs (Fig. 6C, upper panel). Interestingly, the R88-A3GP129A mRNA level in macrophages reached nearly 40% of the total A3G mRNA level (Fig. 6C, lower panel). These results indicate that by using AAV2/5 vector transduction, R88-A3GP129A can be efficiently introduced into primary PBMCs and macrophages. Moreover, by using Western blot with an anti-A3G antibody, we could directly detect R88-A3GP129A fusion protein and its cleaved product, A3GP129A, in HIV progeny viruses produced from AAV2/5-R88-A3GP129A–transduced C8166 T cells infected with HIV-1 (Fig. 6B, upper panel).
FIG. 6.
AAV2/5 vectors deliver R88-A3GP129A into C8166 T cell, human PBMCs, and macrophages. (A) Structures of AAV2/5CMV-transgene vectors. AAV2/5CMV-R88-A3GP129A vector plasmids were constructed by substituting the lacZ gene with R88-A3GP129A between the cytomegalovirus (CMV) promoter and the poly(A) sequence. The entire vector DNA is flanked by the AAV2 inverted terminal repeats (ITRs). (B) C8166 cells were transduced with AAV2/5CMV-lacZ or AAV2/5CMV-R88-A3GP129A vectors at 106 vp/cell. Seventy-two hours after transduction, cells were infected with the pNL4.3-GFP HIV virus (multiplicity of infection of 1). Seven days after infection, viruses were pelleted from the supernatant, lysed, and loaded onto a 10% SDS-PAGE gel and analyzed by Western blotting using an anti-A3G polyclonal antibody (upper panel). Total RNA was isolated from AAV2/5CMV-lacZ or AAV2/5CMV-R88-A3GP129A–transduced human PBMCs and macrophages 72 hr after transduction. The samples were analyzed by reverse-transcription polymerase chain reaction (RT-PCR) for the presence of R88/A3GP129A mRNA. As a positive control (PC), the AAV2/5CMV-R88/A3GP129A plasmid was used as a template. (C) Quantitative real-time PCR analysis to determine R88-A3GP129A mRNA expression level. Cellular mRNA was extracted from PBMCs and macrophages, and subjected to reverse transcription. The reverse-transcribed DNA from each sample was then amplified by real-time PCR to quantify R88-A3GP129A (R-A3G) and the total A3G (A3G) mRNA levels. The total A3G mRNA level was arbitrarily set at 100%. The result is a representative of two independent experiments.
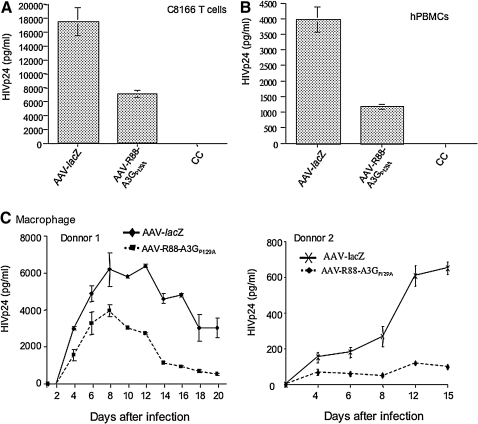
We next tested whether AAV2/5-R88-A3GP129A–transduced C8166 T cells and human PBMCs were resistant against HIV-1 infection. First, we transduced C8166 T cells and phytohemaglutinin-stimulated human PBMCs with AAV2/5-R88-A3GP129A or AAV2/5-lacZ particles (106 vp/cell) for 3 days. The cells were subsequently infected with equal amounts (adjusted by amounts of HIV Gagp24) of T-tropic HIV-1 (pNL4.3–GFP). At days 3 (for C8166 T cells) or 6 (for PBMCs), supernatants were collected and the HIV-1 replication level was monitored by measuring HIV-1 Gagp24 antigen levels. The results show that in both R88-A3GP129A–expressing C8166 T cells and human PBMCs, HIV-1 replication was reduced approximately two- to threefold compared with that in AAV2/5-lacZ–transduced cells (Fig. 7A and B).
FIG. 7.
AAV2/5CMV-R88-A3GP129A–transduced CD4+ C8166 T cells, human PBMCs (hPBMCs), and macrophages were significantly resistant to HIV-1 replication. (A) C8166 cells, (B) phytohemaglutinin (PHA)-stimulated human PBMCs, and (C) macrophage colony-stimulating factor (M-CSF)-stimulated human macrophages (donor 1 and 2) were transduced with AAV2/5CMV-lacZ or AAV2/5CMV-R88-A3GP129A vectors for 72 hr and infected with pNL4.3-GFP+, nevirapine-resistant HIV-1, or the pNL4.3-Bal virus strain. At different time points after infection, the supernatants from the infected cell cultures were collected, and viral replication was monitored by measuring the level of the HIV-1 Gag-p24 antigen. The result shown is representative of two independent experiments. Means with standard errors were calculated from the results of duplicated samples from one experiment.
To further test the effect of R88-A3GP129A on HIV replication in macrophages, we transduced macrophages from two different donors with the same AAVs. After 3 days, the transduced cells were infected with a macrophage-tropic pNL4.3-Bal virus as previously describe (Bishop et al., 2006; Miyagi et al., 2007; Ao et al., 2010). At different time points after infection, supernatants from each infected cell culture were collected, and the viral infection level was monitored using an anti-p24 ELISA. The data from Fig. 7C show that HIV-1 replication was inhibited in AAV2/5-R88-A3GP129A–transduced macrophages. From donor 1, HIV-1 replication in AAV2/5-lacZ–transduced macrophages peaked at days 8 to 12, while in AAV2/5-R88-A3GP129A–transduced macrophages, HIV-1 infection peaked at day 8, while viral replication was reduced by approximately 40%. Of interest, after 8 days of infection, the HIV-1 infection level in AAV2/5-R88-A3GP129A–transduced macrophages rapidly decreased to a low level (Fig. 7C, left panel), suggesting that either HIV-1 infection could not persist or the progeny virus were unable to initiate efficient subsequent infections. In macrophages from donor 2, HIV-1 infection was significantly less productive (approximately 10-fold decrease in HIV Gagp24 production) than that in macrophages from donor 1 (Fig. 7C, compare right panel with the left panel). Interestingly, in AAV2/5-R88-A3GP129A–transduced macrophages, it appeared that HIV-1 could not initiate a productive infection up to 15 days (Fig. 7C, right panel). All of these results indicate that AAV2/5-mediated introduction of R88-A3GP129A in primary PBMCs and macrophages are sufficient to inhibit HIV-1 infection.
Discussion
In this study, we investigated the anti-HIV activity of different R88-A3G mutant fusion proteins including the A3G virus-packaging mutant Y124A, the deaminase-defective mutant E259Q, and the Vif-binding mutants D128K and P129A. We showed that, while all R-A3G mutants could be efficiently incorporated into Vif+ HIV-1 particles, the highest virion-incorporation levels were observed for R88-A3GD128K and R88-A3GP129A mutants because of their resistance to Vif-mediated degradation. In addition, the results show that A3G deaminase activity was required for R88-A3G antiviral activity. Most importantly, we demonstrated that introducing R88-A3GP129A into CD4+ C8166 T cells, PBMCs, and macrophages through AAV2/5-mediated transduction could significantly inhibit HIV-1 infection. All of these data provide further evidence for a potent A3G-based gene therapy approach to inhibit HIV-1 infection.
In the absence of HIV Vif, A3G can be efficiently incorporated into viral particles and interrupt HIV infectivity by introducing dC-to-dU mutations in the minus viral DNA strand during reverse transcription in target cells (Harris et al., 2003; Lecossier et al., 2003; Mangeat et al., 2003; Mariani et al., 2003; Zhang et al., 2003). Although the cytidine deaminase activity mediated by A3G plays an important role in restricting HIV-1 infection, the deaminase-independent antiviral activity of A3G remains controversial. Several studies have suggested that the antiviral function of A3G can be dissociated from its cytidine deaminase activity (Shindo et al., 2003; Newman et al., 2005; Bishop et al., 2006). However, other studies have shown that a specific deaminase-defective mutant of A3G, E259Q, has little or no antiviral activity (Navarro et al., 2005; Miyagi et al., 2007; Schumacher et al., 2008; Browne et al., 2009). To exclude the possibility that E259Q could impair the virus-packaging ability of A3G, we introduced E259Q into R88-A3G and tested its virus incorporation and antiviral activity. Our results indicate that although this fusion protein was effectively incorporated into viruses (Fig. 3), it did not exhibit any antiviral effect (Fig. 2). These data suggest that the anti-HIV activity of R88-A3G is deaminase dependent.
A previous study identified a 4-aa region (residues 124YYFW127) immediately adjacent to the Vif-binding site (128DPD130) as being a critical region for A3G virion incorporation (Huthoff and Malim, 2007). Moreover, the authors showed that the mutant Y124A located in this region lost its anti-HIV activity, presumably due to its inability to be targeted into the virus. In the present study, we fused this A3G Y124A to the R88 peptide and found that the anti-HIV activity of this fusion protein was rescued to a similar level as the wild-type R88-A3G (Fig. 2). These data indicate that R88 is sufficient to compensate for the virus-packaging defect of A3G (Fig. 2B), and that the Y124A mutation does not affect the antiviral property of R88-A3G. Of note, approximately 275 Vpr molecules were estimated to be incorporated into each HIV-1 particle (Muller et al., 2000), but only 0.3 to 0.8 active A3G molecules were present in each Vif+ particle and four to nine A3G molecules were present in Vif– particles (Xu et al., 2007; Nowarski et al., 2008; Browne et al., 2009). Thus, it is expected that R88 is capable of delivering R88-A3GY124A into HIV particles and fully rescuing its antiviral effect. Another interesting observation in this study was that, while the R88-mediated virion incorporation pathway of R88-A3Gwt could overcome the blocking effect by Vif, the encapsidation of R88-A3Gwt was still significantly affected by the presence of Vif (Fig. 3B). This is due to R88-A3Gwt remaining sensitive to Vif-mediated degradation, which results in a relatively low level of intracellular R88-A3G that is available for viral encapsidation. Fortunately, this defect in R-A3Gwt can be compensated for by introducing the Vif-binding mutations D128K or P129A into this fusion protein. Our results demonstrate that the virion incorporation levels of R88-A3GD128K and R88-A3GP129A were significantly higher than that of R88-A3Gwt (Fig. 3B). In the present study, we also compared the antiviral activities of wild-type R88-A3Gwt and the different mutants and found that fusing R88 to A3Gwt could not completely inhibit viral replication, and R88-A3GD128K and R88-A3GP129A exhibited a much stronger anti-HIV effect compared with R88-A3Gwt (Figs. 2 and 4). Although these two Vif-binding–defective mutants were efficiently incorporated into the virus, we still could not conclude that their remarkable anti-HIV activity could be attributed to higher levels of virion incorporation. If HIV Vif located within the virus could directly interfere with A3G's biological activity, then one would expect that the action of R88-A3GD128K and R88-A3GP129A would not be affected by Vif. Indeed, a previous study provided evidence that the production of infectious Vif+ virus does not depend on the reduced level of A3G from virus-producing cells (Kao et al., 2004), suggesting that the Vif present in the virus may directly interfere with A3G's biological activity. Interestingly, Santa-Marta et al. (2005) also showed that HIV Vif can directly inhibit A3G-mediated hypermutation in bacteria. Because no ubiquitin-proteasome system is present in bacteria, these observations argue in favor of a direct effect of Vif on the enzymatic activity of A3G.
To further explore the feasibility of using this approach as a gene therapy strategy, we first introduced R88-A3Gwt and R88-A3GP129A into CD4+ C8166 T cells using a lentiviral vector followed by puromycin selection. The selected cell lines were subsequently used to test the effect of these fusions on HIV-1 replication and spread. Our results show that the presence of R88-A3Gwt was sufficient to attenuate virus replication; however, after 12 days, the virus started to replicate again (Fig. 4). Intriguingly, in R88-A3GP129A–expressing CD4+ T cells, HIV-1 infection was completely blocked, and no sign of viral infection during the 24-day period could be detected. These results clearly demonstrate that introducing R88-A3GP129A in CD4+ T cells efficiently inhibits HIV replication, although the long-term effect of the expression of this mutant requires further investigation. We still cannot exclude the possibility that HIV-1 could develop resistance against the antiviral effect of R88-A3GP129A because previous work has shown that changes in aa 14 to 17 of Vif could modify its interaction with A3GD128K and inhibit the activity of the mutant (Schrofelbauer et al., 2006). However, it would be very difficult for the HIV to develop simultaneous resistance against the actions of both R88 and A3GP129A. This is a very interesting question and is currently under further investigation.
Another important question is how to introduce the R88-A3G fusion protein into human PBMCs and macrophages to investigate its impact on HIV replication. Lentiviral vectors cannot be used for this purpose because vector transduction efficiency is extremely low in the presence of R88-A3G. Therefore, we used an AAV vector system, which is currently the only nonpathogenic viral vector that can transduce genes into a number of tissues and cells, including dendritic cells, PBMCs, and macrophages, without causing toxicity. In addition, the AAV vector system may direct long-term expression (Flotte et al., 1993; Herzog et al., 1997; During et al., 1998; Fan et al., 1998; Lewin et al., 1998; Ponnazhagan et al., 2001; Xin et al., 2002). Importantly, two previous studies have shown that the production and transduction of AAV are not affected by the presence of A3G (Chen et al., 2006; Narvaiza et al., 2009), which makes it an ideal vector to deliver R88-A3G into primary cells. Indeed, our results show that AAV2/5 was able to quite efficiently introduce R88-A3GP129A into human PBMCs and macrophages. The quantitative real-time PCR analysis revealed that the R88-A3GP129A mRNA level was approximately 23% of total A3G mRNA level in PBMCs and reached nearly 40% of the total A3G mRNA level in macrophages (Fig. 6B and C). Therefore, it is conceivable that an AAV2/5-R88-A3GP129A vector system can provide protection against HIV-1 infection. Interestingly, a recent study has shown that AAV5, rather than AAV2, can more effectively transduce human primary CD4+ T cells, macrophages, and dendritic cells (Xin et al., 2006). Thus, it will be interesting to test whether AAV5 could also be more efficient than AAV2/5 in transducing R88-A3G into primary PBMCs and macrophages and providing effective protection against HIV infection. Overall, this study has provided evidence for the feasibility of using an AAV-R88-A3G vector system as a gene therapy approach to inhibit HIV infection and spread, particularly against HIV-1 mucosal infection during sexual transmission. More studies are required to characterize and optimize this anti-HIV gene therapy approach.
Acknowledgements
We thank Drs. J. Lingappa, S. Bour, and K. Strebel for providing the anti-A3G antibodies and HIV-1 pcDNA-hVif expressor, which were obtained through the AIDS Research Reference Reagent Program, Division of AIDS, NIAID, NIH. Z.-J. Ao and X.-X. Wang are recipients of a postdoctoral fellowship from the Canadian Institutes of Health Research (CIHR) International Infectious Disease & Global Health Training Program (IID&GHTP) and a studentship from the China Scholarship Council (CSC), respectively. K. Danappa Jayappa is a recipient of Manitoba Health Research Council/Manitoba Institute of Child Health (MHRC/MICH) and IID&GHTP scholarships. X.-J. Yao is a recipient of the Basic Science Career Development Research Award from The Manitoba Medical Service Foundation. This work was supported by a CIHR HIV prevention operating grant (HPR85525) and the Leaders Opportunity Fund Award from Canadian Foundation of Innovation (CFI) to X.-J. Yao.
Author Disclosure Statement
No competing financial interests exist.
References
- Alce T.M. Popik W. APOBEC3G is incorporated into virus-like particles by a direct interaction with HIV-1 Gag nucleocapsid protein. J. Biol. Chem. 2004;279:34083–34086. doi: 10.1074/jbc.C400235200. [DOI] [PubMed] [Google Scholar]
- Ao Z. Fowke K.R. Cohen E.A. Yao X. Contribution of the C-terminal tri-lysine regions of human immunodeficiency virus type 1 integrase for efficient reverse transcription and viral DNA nuclear import. Retrovirology. 2005;2:62. doi: 10.1186/1742-4690-2-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ao Z. Huang G. Yao H., et al. Interaction of human immunodeficiency virus type 1 integrase with cellular nuclear import receptor importin 7 and its impact on viral replication. J. Biol. Chem. 2007;282:13456–13467. doi: 10.1074/jbc.M610546200. [DOI] [PubMed] [Google Scholar]
- Ao Z. Jayappa K.D. Wang B., et al. Importin {alpha}3 interacts with HIV-1 integrase and contributes to HIV-1 nuclear import and replication. J. Virol. 2010;84:8650–8663. doi: 10.1128/JVI.00508-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ao Z. Yu Z. Wang L., et al. Vpr14-88-Apobec3G fusion protein is efficiently incorporated into Vif-positive HIV-1 particles and inhibits viral infection. PLoS ONE. 2008;3:e1995. doi: 10.1371/journal.pone.0001995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auricchio A. Hildinger M. O'Connor E., et al. Isolation of highly infectious and pure adeno-associated virus type 2 vectors with a single-step gravity-flow column. Hum. Gene Ther. 2001;12:71–76. doi: 10.1089/104303401450988. [DOI] [PubMed] [Google Scholar]
- Bello A. Tran K. Chand A., et al. Isolation and evaluation of novel adeno-associated virus sequences from porcine tissues. Gene Ther. 2009;16:1320–1328. doi: 10.1038/gt.2009.82. [DOI] [PubMed] [Google Scholar]
- Bishop K.N. Holmes R.K. Malim M.H. Antiviral potency of APOBEC proteins does not correlate with cytidine deamination. J. Virol. 2006;80:8450–8458. doi: 10.1128/JVI.00839-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogerd H.P. Doehle B.P. Wiegand H.L., et al. A single amino acid difference in the host APOBEC3G protein controls the primate species specificity of HIV type 1 virion infectivity factor. Proc. Natl. Acad. Sci. U. S. A. 2004;101:3770–3774. doi: 10.1073/pnas.0307713101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browne E.P. Allers C. Landau N.R. Restriction of HIV-1 by APOBEC3G is cytidine deaminase-dependent. Virology. 2009;387:313–321. doi: 10.1016/j.virol.2009.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cen S. Guo F. Niu M., et al. The interaction between HIV-1 Gag and APOBEC3G. J. Biol. Chem. 2004;279:33177–33184. doi: 10.1074/jbc.M402062200. [DOI] [PubMed] [Google Scholar]
- Chen H. Lilley C.E. Yu Q., et al. APOBEC3A is a potent inhibitor of adeno-associated virus and retrotransposons. Curr. Biol. 2006;16:480–485. doi: 10.1016/j.cub.2006.01.031. [DOI] [PubMed] [Google Scholar]
- Chiu Y.L. Soros V.B. Kreisberg , et al. Cellular APOBEC3G restricts HIV-1 infection in resting CD4+ T cells. Nature. 2005;435:108–114. doi: 10.1038/nature03493. [DOI] [PubMed] [Google Scholar]
- Conticello S.G. Harris R.S. Neuberger M.S. The Vif protein of HIV triggers degradation of the human antiretroviral DNA deaminase APOBEC3G. Curr. Biol. 2003;13:2009–2013. doi: 10.1016/j.cub.2003.10.034. [DOI] [PubMed] [Google Scholar]
- During M.J. Xu R. Young D., et al. Peroral gene therapy of lactose intolerance using an adeno-associated virus vector. Nat. Med. 1998;4:1131–1135. doi: 10.1038/2625. [DOI] [PubMed] [Google Scholar]
- Fan D.S. Ogawa M. Fujimoto , et al. Behavioral recovery in 6-hydroxydopamine-lesioned rats by cotransduction of striatum with tyrosine hydroxylase and aromatic L-amino acid decarboxylase genes using two separate adeno-associated virus vectors. Hum. Gene Ther. 1998;9:2527–2535. doi: 10.1089/hum.1998.9.17-2527. [DOI] [PubMed] [Google Scholar]
- Flotte T.R. Afione S.A. Conrad C., et al. Stable in vivo expression of the cystic fibrosis transmembrane conductance regulator with an adeno-associated virus vector. Proc. Natl. Acad. Sci. U. S. A. 1993;90:10613–10617. doi: 10.1073/pnas.90.22.10613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris R.S. Bishop K.N. Sheehy A.M., et al. DNA deamination mediates innate immunity to retroviral infection. Cell. 2003;113:803–809. doi: 10.1016/s0092-8674(03)00423-9. [DOI] [PubMed] [Google Scholar]
- Herzog R.W. Hagstrom J.N. Kung S.H., et al. Stable gene transfer and expression of human blood coagulation factor IX after intramuscular injection of recombinant adeno-associated virus. Proc. Natl. Acad. Sci. U. S. A. 1997;94:5804–5809. doi: 10.1073/pnas.94.11.5804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huthoff H. Malim M.H. Identification of amino acid residues in APOBEC3G required for regulation by human immunodeficiency virus type 1 Vif and Virion encapsidation. J. Virol. 2007;81:3807–3815. doi: 10.1128/JVI.02795-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao S. Khan M.A. Miyagi E., et al. The human immunodeficiency virus type 1 Vif protein reduces intracellular expression and inhibits packaging of APOBEC3G (CEM15), a cellular inhibitor of virus infectivity. J. Virol. 2003;77:11398–11407. doi: 10.1128/JVI.77.21.11398-11407.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao S. Miyagi E. Khan M.A., et al. Production of infectious human immunodeficiency virus type 1 does not require depletion of APOBEC3G from virus-producing cells. Retrovirology. 2004;1:27. doi: 10.1186/1742-4690-1-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan M.A. Kao S. Miyagi E., et al. Viral RNA is required for the association of APOBEC3G with human immunodeficiency virus type 1 nucleoprotein complexes. J. Virol. 2005;79:5870–5874. doi: 10.1128/JVI.79.9.5870-5874.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobinger G.P. Weiner D.J. Yu Q.C. Wilson J.M. Filovirus-pseudotyped lentiviral vector can efficiently and stably transduce airway epithelia in vivo. Nat. Biotechnol. 2001;19:225–230. doi: 10.1038/85664. [DOI] [PubMed] [Google Scholar]
- Lavens D. Peelman F. Van Der Heyden J., et al. Definition of the interacting interfaces of Apobec3G and HIV-1 Vif using MAPPIT mutagenesis analysis. Nucleic Acids Res. 2010;38:1902–1912. doi: 10.1093/nar/gkp1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecossier D. Bouchonnet F. Clavel F. Hance A.J. Hypermutation of HIV-1 DNA in the absence of the Vif protein. Science. 2003;300:1112. doi: 10.1126/science.1083338. [DOI] [PubMed] [Google Scholar]
- Lewin A.S. Drenser K.A. Hauswirth W.W., et al. Ribozyme rescue of photoreceptor cells in a transgenic rat model of autosomal dominant retinitis pigmentosa. Nat. Med. 1998;4:967–971. doi: 10.1038/nm0898-967. [DOI] [PubMed] [Google Scholar]
- Lu Y.L. Spearman P. Ratner L. Human immunodeficiency virus type 1 viral protein R localization in infected cells and virions. J. Virol. 1993;67:6542–6550. doi: 10.1128/jvi.67.11.6542-6550.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo K. Liu B. Xiao Z., et al. Amino-terminal region of the human immunodeficiency virus type 1 nucleocapsid is required for human APOBEC3G packaging. J. Virol. 2004;78:11841–11852. doi: 10.1128/JVI.78.21.11841-11852.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangeat B. Turelli P. Caron G., et al. Broad antiretroviral defence by human APOBEC3G through lethal editing of nascent reverse transcripts. Nature. 2003;424:99–103. doi: 10.1038/nature01709. [DOI] [PubMed] [Google Scholar]
- Mangeat B. Turelli P. Liao S. Trono D. A single amino acid determinant governs the species-specific sensitivity of APOBEC3G to Vif action. J. Biol. Chem. 2004;279:14481–14483. doi: 10.1074/jbc.C400060200. [DOI] [PubMed] [Google Scholar]
- Mariani R. Chen D. Schrofelbauer B., et al. Species-specific exclusion of APOBEC3G from HIV-1 virions by Vif. Cell. 2003;114:21–31. doi: 10.1016/s0092-8674(03)00515-4. [DOI] [PubMed] [Google Scholar]
- Marin M. Rose K.M. Kozak S.L. Kabat D. HIV-1 Vif protein binds the editing enzyme APOBEC3G and induces its degradation. Nat. Med. 2003;9:1398–1403. doi: 10.1038/nm946. [DOI] [PubMed] [Google Scholar]
- Miyagi E. Opi S. Takeuchi H., et al. Enzymatically active APOBEC3G is required for efficient inhibition of human immunodeficiency virus type 1. J. Virol. 2007;81:13346–13353. doi: 10.1128/JVI.01361-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller B. Tessmer U. Schubert U. Krausslich H.G. Human immunodeficiency virus type 1 Vpr protein is incorporated into the virion in significantly smaller amounts than gag and is phosphorylated in infected cells. J. Virol. 2000;74:9727–9731. doi: 10.1128/jvi.74.20.9727-9731.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narvaiza I. Linfesty D.C. Greener B.N., et al. Deaminase-independent inhibition of parvoviruses by the APOBEC3A cytidine deaminase. PLoS Pathog. 2009;5:e1000439. doi: 10.1371/journal.ppat.1000439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro F. Bollman B. Chen H., et al. Complementary function of the two catalytic domains of APOBEC3G. Virology. 2005;333:374–386. doi: 10.1016/j.virol.2005.01.011. [DOI] [PubMed] [Google Scholar]
- Newman E.N. Holmes R.K. Craig H.M., et al. Antiviral function of APOBEC3G can be dissociated from cytidine deaminase activity. Curr. Biol. 2005;15:166–170. doi: 10.1016/j.cub.2004.12.068. [DOI] [PubMed] [Google Scholar]
- Nguyen K.L. Llano M. Akari H., et al. Codon optimization of the HIV-1 vpu and vif genes stabilizes their mRNA and allows for highly efficient Rev-independent expression. Virology. 2004;319:163–175. doi: 10.1016/j.virol.2003.11.021. [DOI] [PubMed] [Google Scholar]
- Nowarski R. Britan-Rosich E. Shiloach T. Kotler M. Hypermutation by intersegmental transfer of APOBEC3G cytidine deaminase. Nat. Struct. Mol. Biol. 2008;15:1059–1066. doi: 10.1038/nsmb.1495. [DOI] [PubMed] [Google Scholar]
- Opi S. Kao S. Goila-Gaur R., et al. Human immunodeficiency virus type 1 Vif inhibits packaging and antiviral activity of a degradation-resistant APOBEC3G variant. J. Virol. 2007;81:8236–8246. doi: 10.1128/JVI.02694-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponnazhagan S. Curiel D.T. Shaw D.R., et al. Adeno-associated virus for cancer gene therapy. Cancer Res. 2001;61:6313–6321. [PubMed] [Google Scholar]
- Santa-Marta M. da Silva F.A. Fonseca A.M. Goncalves J. HIV-1 Vif can directly inhibit apolipoprotein B mRNA-editing enzyme catalytic polypeptide-like 3G-mediated cytidine deamination by using a single amino acid interaction and without protein degradation. J. Biol. Chem. 2005;280:8765–8775. doi: 10.1074/jbc.M409309200. [DOI] [PubMed] [Google Scholar]
- Schafer A. Bogerd H.P. Cullen B.R. Specific packaging of APOBEC3G into HIV-1 virions is mediated by the nucleocapsid domain of the gag polyprotein precursor. Virology. 2004;328:163–168. doi: 10.1016/j.virol.2004.08.006. [DOI] [PubMed] [Google Scholar]
- Schrofelbauer B. Chen D. Landau N.R. A single amino acid of APOBEC3G controls its species-specific interaction with virion infectivity factor (Vif) Proc. Natl. Acad. Sci. U. S. A. 2004;101:3927–3932. doi: 10.1073/pnas.0307132101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrofelbauer B. Senger T. Manning Landau N.R. Mutational alteration of human immunodeficiency virus type 1 Vif allows for functional interaction with nonhuman primate APOBEC3G. J. Virol. 2006;80:5984–5991. doi: 10.1128/JVI.00388-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumacher A.J. Hache G. MacDuff D.A., et al. The DNA deaminase activity of human APOBEC3G is required for Ty1, MusD, and human immunodeficiency virus type 1 restriction. J. Virol. 2008;82:2652–2660. doi: 10.1128/JVI.02391-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheehy A.M. Gaddis N.C. Choi J.D. Malim M.H. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature. 2002;418:646–650. doi: 10.1038/nature00939. [DOI] [PubMed] [Google Scholar]
- Sheehy A.M. Gaddis N.C. Malim M.H. The antiretroviral enzyme APOBEC3G is degraded by the proteasome in response to HIV-1 Vif. Nat. Med. 2003;9:1404–1407. doi: 10.1038/nm945. [DOI] [PubMed] [Google Scholar]
- Shindo K. Takaori-Kondo A. Kobayashi M., et al. The enzymatic activity of CEM15/Apobec-3G is essential for the regulation of the infectivity of HIV-1 virion but not a sole determinant of its antiviral activity. J. Biol. Chem. 2003;278:44412–44416. doi: 10.1074/jbc.C300376200. [DOI] [PubMed] [Google Scholar]
- Simon J.H.M. Malim M.H. The human immunodeficiency virus type 1 Vif protein modulates the postpenetration stability of viral nucleoprotein complexes. J. Virol. 1996;70:5297–5305. doi: 10.1128/jvi.70.8.5297-5305.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenglein M.D. Matsuo H. Harris R.S. Two regions within the amino-terminal half of APOBEC3G cooperate to determine cytoplasmic localization. J. Virol. 2008;82:9591–9599. doi: 10.1128/JVI.02471-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stopak K. de Noronha C. Yonemoto W. Greene W.C. HIV-1 Vif blocks the antiviral activity of APOBEC3G by impairing both its translation and intracellular stability. Mol. Cell. 2003;12:591–601. doi: 10.1016/s1097-2765(03)00353-8. [DOI] [PubMed] [Google Scholar]
- Turelli P. Mangeat B. Jost S., et al. Inhibition of hepatitis B virus replication by APOBEC3G. Science. 2004;303:1829. doi: 10.1126/science.1092066. [DOI] [PubMed] [Google Scholar]
- Xiao W. Berta S.C. Lu M.M., et al. Adeno-associated virus as a vector for liver-directed gene therapy. J. Virol. 1998;72:10222–10226. doi: 10.1128/jvi.72.12.10222-10226.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin K.Q. Mizukami H. Urabe M., et al. Induction of robust immune responses against human immunodeficiency virus is supported by the inherent tropism of adeno-associated virus type 5 for dendritic cells. J. Virol. 2006;80:11899–11910. doi: 10.1128/JVI.00890-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin K.Q. Ooki T. Mizukami H., et al. Oral administration of recombinant adeno-associated virus elicits human immunodeficiency virus-specific immune responses. Hum. Gene Ther. 2002;13:1571–1581. doi: 10.1089/10430340260201662. [DOI] [PubMed] [Google Scholar]
- Xu H. Chertova E. Chen J., et al. Stoichiometry of the antiviral protein APOBEC3G in HIV-1 virions. Virology. 2007;360:247–256. doi: 10.1016/j.virol.2006.10.036. [DOI] [PubMed] [Google Scholar]
- Xu H. Svarovskaia E.S. Barr R., et al. A single amino acid substitution in human APOBEC3G antiretroviral enzyme confers resistance to HIV-1 virion infectivity factor-induced depletion. Proc. Natl. Acad. Sci. U. S. A. 2004;101:5652–5657. doi: 10.1073/pnas.0400830101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao X.J. Friborg J. Checroune F., et al. Degradation of CD4 induced by human immunodeficiency virus type 1 Vpu protein: a predicted alpha-helix structure in the proximal cytoplasmic region of CD4 contributes to Vpu sensitivity. Virology. 1995a;209:615–623. doi: 10.1006/viro.1995.1293. [DOI] [PubMed] [Google Scholar]
- Yao X.J. Kobinger G. Dandache S., et al. HIV-1 Vpr-chloramphenicol acetyltransferase fusion proteins: sequence requirement for virion incorporation and analysis of antiviral effect. Gene Ther. 1999;6:1590–1599. doi: 10.1038/sj.gt.3300988. [DOI] [PubMed] [Google Scholar]
- Yao X.J. Subbramanian R.A. Rougeau N., et al. Mutagenic analysis of human immunodeficiency virus type 1 Vpr: role of a predicted N-terminal alpha-helical structure in Vpr nuclear localization and virion incorporation. J. Virol. 1995b;69:7032–7044. doi: 10.1128/jvi.69.11.7032-7044.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X. Yu Y. Liu B., et al. Induction of APOBEC3G ubiquitination and degradation by an HIV-1 Vif-Cul5-SCF complex. Science. 2003;302:1056–1060. doi: 10.1126/science.1089591. [DOI] [PubMed] [Google Scholar]
- Yu Y. Xiao Z. Ehrlich E.S., et al. Selective assembly of HIV-1 Vif-Cul5-ElonginB-ElonginC E3 ubiquitin ligase complex through a novel SOCS box and upstream cysteines. Genes Dev. 2004;18:2867–2872. doi: 10.1101/gad.1250204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H. Yang B. Pomerantz R.J., et al. The cytidine deaminase CEM15 induces hypermutation in newly synthesized HIV-1 DNA. Nature. 2003;424:94–98. doi: 10.1038/nature01707. [DOI] [PMC free article] [PubMed] [Google Scholar]