Abstract
Short interspersed nuclear elements (SINEs) are a type of class 1 transposable element (retrotransposon) with features that allow investigators to resolve evolutionary relationships between populations and species while providing insight into genome composition and function. Characterization of a Carnivora-specific SINE family, Can-SINEs, has, has aided comparative genomic studies by providing rare genomic changes, and neutral sequence variants often needed to resolve difficult evolutionary questions. In addition, Can-SINEs constitute a significant source of functional diversity with Carnivora. Publication of the whole-genome sequence of domestic dog, domestic cat, and giant panda serves as a valuable resource in comparative genomic inferences gleaned from Can-SINEs. In anticipation of forthcoming studies bolstered by new genomic data, this review describes the discovery and characterization of Can-SINE motifs as well as describes composition, distribution, and effect on genome function. As the contribution of noncoding sequences to genomic diversity becomes more apparent, SINEs and other transposable elements will play an increasingly large role in mammalian comparative genomics.
Keywords: carnivore, genome, SINE
Short interspersed nuclear elements (SINEs) are repetitive genomic sequences, members of class 1 transposable elements (retrotransposons), that are present in most eukaryotic organisms (Wicker et al. 2007), including monotreme, marsupial, and eutherian mammals, (Nishihara et al. 2006; Gu et al. 2007; Munemasa et al. 2008). Characterized by unique features in structure, proliferation, and genome distribution, SINEs are chimeras of transcribed RNA genes and simple repeats that proliferate via reverse transcription using the enzymatic machinery of autonomous elements, which recognize internal polymerase III promoter sequences (Collier and Largaespada 2007). Approximately 40 (transfer RNA) tRNA, 7SL RNA, and 5S ribosomal RNA derived SINE families have been described in mammals thus far, many of which are present in more than 104 copies (Miyamoto 1999; Gu et al. 2007). As a source of insertional mutagenesis, SINEs have been linked to unequal recombination events (Callinan et al. 2005) and genetic diseases (Deininger and Batzer 1999) as well as proven to be informative evolutionary markers across genomes. With the publication of whole-genome sequences from domestic dog (Canis familiaris) (Lindblad-Toh et al. 2005), domestic cat (Felis catus) (Pontius et al. 2007), and most recently giant panda (Ailuropoda melanoleuca) (Li et al. 2010), new insights are possible on the family of SINEs unique to the Carnivora order termed Can-SINEs. Here, we review the structural, functional, and evolutionary impact of Can-SINEs on carnivore genomes.
Discovery and Characterization of Can-SINEs
Can-SINEs were first described in the early 1990s when an interspersed repetitive element was discovered on the X chromosome of the American mink (Mustela vison) that shared 55% sequence similarity with tRNA-lysine derived rodent B2 SINEs (Lawrence et al. 1985; Lavrentieva et al. 1991). Subsequent studies of other caniform suborder species including harbor seal (Phoca vitulina concolor), dog (C. familiaris), wolf (C. lupus), coyote (C. latrans), and mink (M. vision) affirmed the presence of these sequences in high copy number (Coltman and Wright 1994). Initially not observed in the feliform suborder representative, F. catus, this newly characterized SINE family appeared caniform specific and therefore was named Can-SINE (Coltman and Wright 1994). However, subsequent hybridization studies with F. catus (van der Vlugt and Lenstra 1995; Vassetzky and Kramerov 2002) as well as a sequence study of 3 Y-chromosome genes among all members of the Felis genus, the bobcat (Lynx rufus), C. familiaris, and several bear species (Pecon-Slattery et al. 2000) conclusively demonstrated that Can-SINEs are ubiquitous across Carnivora.
Can-SINEs are defined by a tRNA-related region, which includes A and B promoter boxes, followed by a (CT)n microsatellite and terminate with a poly-A/T tail containing the polyadenylation signal AATAAA (Figure 1) (Lavrentieva et al. 1991). The tRNA-related region has primary sequence similarity of 70–79% to lysine-tRNAs (Vassetzky and Kramerov 2002), and most inserts are between 150 and 300 base pairs (bp) in length, depending on the size of the (CT)n and poly A/T regions (Vassetzky and Kramerov 2002; Pecon-Slattery et al. 2004). Each Can-SINE locus is flanked by target site duplications of 8–15 bp generated during retrotransposition. The number of SINE repeats within carnivore genomes, based on estimates of the C. familiaris genome, range from 1.1 × 106 (Lindblad-Toh et al. 2005) to 1.3 × 106 (Coltman and Wright 1994) copies.
Figure 1.
Alignment and minimum evolution phylogeny of Repbase Can-SINE voucher sequences. All Can-SINE vouchers begin with the designation “SINEC,” followed by species of first discovery where “Fc” = Felis catus, “CF” = Canis familiaris, “Ame” = Ailuropoda melanoleuca, and “Pv” = Phoca vitulina. (A) The initial global alignment of lysine-tRNA derived segments was estimated using the Geneious alignment module (Drummond et al. 2010) and refined by hand. Lines indicate RNA polymerase III promoter boxes A and B. (B) The neighbor-joining clustering algorithm was used to estimate phylogeny with the Tamura–Nei distance model, and branch support approximated using 1000 bootstrap replications. Feliform SINEs form a distinct clade.
Can-SINE Amplification
Although the specific retrotransposition mechanism has yet to be fully described, comparative evidence indicates that Can-SINEs proliferate in a manner similar to other mammalian SINEs (Gentles et al. 2005). In general, SINE amplification occurs through a “copy and paste” mechanism known as target-primed reverse transcription wherein a few SINE master or source copies are transcribed in high copy number (Cordaux et al. 2004; Tong et al. 2010). Lacking functional coding regions, SINE amplification, and integration is dependent on enzymes derived from the host genome and long interspersed nuclear elements (LINEs) (Kajikawa and Okada 2002). Proliferation is initiated by recognition of the promoter boxes residing in the RNA-related region by host-derived RNA-polymerase III and followed by cleavage of the genomic DNA at TTAAAA motifs between the T’s and A’s by LINE-derived enzymes (Gentles et al. 2005; Cordaux and Batzer 2009). This process allows the poly-A/T tail of the SINE transcript to bind to the single-stranded genomic DNA, creating a primer for LINE-derived reverse transcriptase to synthesize complementary SINE DNA (Christensen et al. 2006; Kurzynska-Kokorniak et al. 2007). A subsequent nick in the opposite genomic strand 6–10 bp downstream from the initial cut site results in a novel SINE insert flanked by target site duplications (Jurka 1997; Christensen et al. 2006) (Figure 2).
Figure 2.
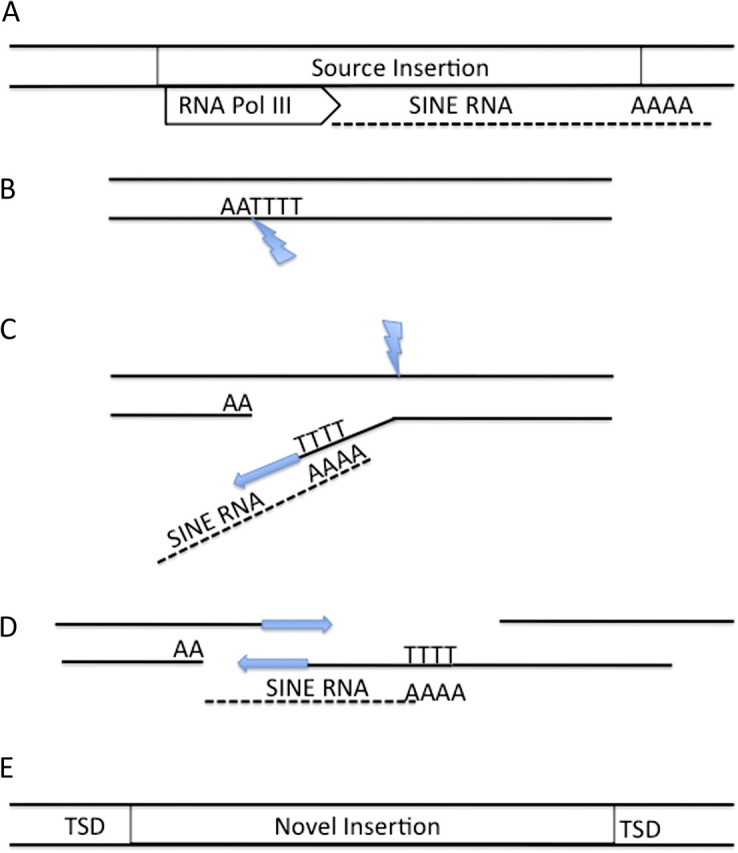
Retrotransposition of mammalian SINE insertions. (A) The master SINE copy is transcribed by RNA polymerase III. (B) LINE-derived enzyme nicks a chromosome strand at the motif AATTTT. (C) The poly-A tail of a transcribed SINE binds to the free TTTT and acts as a primer for LINE-derived reverse transcriptase. A nick on the opposite strand frees the complementary target site duplication sequence. (D) Reverse transcriptase synthesizes complementary strands. (E) A new SINE insertion with characteristic target site duplications. Adapted from (Cordaux and Batzer 2009). (This figure appears in color in the online version of Journal of Heredity.)
The involvement of LINEs in SINE proliferation has been attributed to structural similarities between the 3′ portions (microsatellite and poly A/T regions) of each transposable element. For example, monotreme Mon-1 and marsupial Ther-2 SINE families share 3′ end sequences with LINEs of the same genome, which are believed to facilitate SINE retrotransposition (Ohshima and Okada 2005). Conversely, the endogenous L1 family LINEs that facilitate rodent and primate 7SL-derived SINE proliferation do not share primary sequence homology (Ohshima and Okada 2005). Whole-genome analyses of C. familiaris and F. catus reveal that at least 19% and 8% of the genome are comprised of LINE sequences respectively, derived from the L1 and L2 families (Lindblad-Toh et al. 2005; Pontius et al. 2007). In addition, complete open reading frames may be found amongst carnivore L1s, indicating recent retrotransposition activity of this LINE family (Pontius et al. 2007). However, further comparative genomic analyses are essential in clarifying the functional and evolutionary associations between carnivore-specific SINEs and LINEs.
Can-SINE Voucher Sequences and Subfamilies
At any given time, only a few SINE insertions will act as the source of novel SINE transcripts during amplification (Cordaux et al. 2004). Gradual accumulation of mutations will eventually inhibit transcription of a given master copy, which then becomes dormant and is replaced by an alternate copy. This process results in SINE subfamilies with diagnostic nucleotide sequence that are classified into phylogenetic lineages (Ray et al. 2006). The 16 Can-SINE voucher sequences described within the Repbase library and RepeatMasker software to date (Jurka et al. 2005; Smit and Green 2005) serve as prototypes for subfamily classification schemes. Can-SINE sequences are collectively designated “SINEC” followed by putative subfamily and species of first discovery identifiers (Figure 1). For example, the first Can-SINE insertion sequence found in the A. melanoleuca genome is designated SINEC1_AMe (Li et al. 2010).
Phylogenetic analysis of existing Can-SINE voucher sequences defines 2 evolutionary lineages concordant with genome origin as either caniform or feliform (Figure 1). Amongst the caniform SINEs, divergent lineages represented by A. melanoeuca (family Ursidae) and P. vitulina (family Phocidae) are interspersed with putative Canis (family Canidae) specific sequences. This structure, unrelated to carnivore taxonomy, suggests SINE master copies that originated early in caniforms have remained active sources of SINE proliferation within independent lineages (Figure 1). Future comparative research will fully characterize the Can-SINE subfamilies and provide further insight to the carnivore genome diversity.
Biomedical Effects of Can-SINEs
Transposable elements, including SINEs, contribute broadly to the functional diversity of mammalian genomes. Although the vast majority of novel insertions that manage to evade purifying selection and genetic drift are functionally benign, a few confer neutral or deleterious phenotypic variants (Belancio et al. 2008). SINE insertions may be co-opted by host genomes as sites for nonhomologous genome rearrangement, as sources for coding sequence, and as regulatory elements. Numerous transposable element insertions have been implicated in human disease (Deininger and Batzer 1999; Batzer and Deininger 2002), and transgenic mice engineered for over expression of the transposable element type known as “Sleeping Beauty” experienced increased cancer development (Dupuy et al. 2005). Relative to primates and rodents, the phenotypic consequences of SINE insertions among Carnivores are less thoroughly documented. However, an increasing number of SINE-derived variants have been found amongst the domestic dog breeds, which provide well-defined populations ideal for studying morphological variation and disease susceptibility.
The merle coat pattern of C. familiaris is an incompletely dominant inherited phenotype noted by marbled coat pigmentation that is sometimes accompanied by symptoms analogous to those associated with human Waardenburg syndrome including auditory, visual, skeletal, cardiac, and reproductive impairments (Clark et al. 2006). This phenotype, common in the Corgi, Dachshund, Australian Shepard, and other domestic breeds, segregates with a region of the C. familiaris genome that includes the silver” gene, SILV, which is responsible for coat pigment in multiple mammalian species including mouse and horse (Kwon et al. 1995; Theos et al. 2005). In merle canines, the SILV gene harbors a Can-SINE insertion in reverse orientation at the boundary of intron 10 and exon 11, which causes alternate splicing of exon 11 and ultimately results in the merle phenotype (Clark et al. 2006).
The autosomal recessive version of spontaneous centronuclear myopathy (CNM), a muscular disorder that affects multiple mammalian species including humans, occurs in Labrador retrievers. The disease is characterized by muscle weakness, muscular atrophy, and other afflictions attributed to compromised muscle fibers (Hu et al. 1996; Laporte et al. 1996). Disease associated genotypes were mapped to the canine autosomal protein tyrosine phosphatase-like member A gene (PTPLA), the mouse and human homologs of which are expressed in myogenic precursors during embryogenesis and in adult skeletal muscles (Uwanogho et al. 1999; Breen et al. 2004). Characterization of canine PTPLA revealed a Can-SINE insertion in the reverse orientation within exon 2 that segregates with CNM disease. The disorder conforms to a recessive model of inheritance in which unaffected carriers are heterozygous, whereas affected individuals are homozygous for the insertion. Can-SINE presence results in 7 transcript isoforms of which only 2 are identical to wild-type PTPLA, whereas the remaining 5 are presumably ineffective or toxic (Pelé et al. 2005).
Narcolepsy is a sleep disorder that affects humans and other mammals including domestic dogs. A mutation within the hypocretin receptor 2 gene (HCRTR2) is among the multiple genetic causes of human early onset narcolepsy (Peyron et al. 2000). Doberman Pinschers with narcolepsy also have an HCRTR2 mutation in the form of a Can-SINE insertion prior to the fourth exon that results in inefficient pre- (messenger RNA) mRNA splicing. Other domestic dog breeds with increased incidence of narcolepsy, however, do not have the HCRTR2 insertion, and thus, similar to the human disease, there may be multiple causes of canine narcolepsy (Lin et al. 1999).
Domestic dogs are characterized by profound body size diversity, ranging from 2 kg Chihuahuas to 82 kg Mastiffs. Recent efforts to unveil genetic factors influencing canine body size have uncovered multiple Can-SINE insertion variants in linkage disequilibrium with size determining genes. Within the second intron of the insulin-like growth factor 1 (IGF1) gene, an SNP and Can-SINE segregate with a wide variety of small breeds (Sutter et al. 2007). Although SINE insertions are demonstrated factors in gene expression (Hasler and Strub 2006; Lin et al. 2008), the causative mutation in IGF1 has yet to be determined. From an evolutionary perspective, although the “small” haplotype is not observed in the domestic dog forbearer, the gray wolf (C. lupus), overall genetic similarity to the homologous locus in Middle Eastern wolves suggests a Middle Eastern origin for a small dog variant that occurred after the initial domestication (Gray et al. 2010).
Retrogenes are derived from processed mRNAs sequences that have been retrotransposed into the genome via reverse transcription and are usually not expressed due to the absence of regulatory elements (Brosius 1999a). However, in rare instances, retrogenes can employ the internal promoters of nearby LINEs or SINEs for transcription and expression (Brosius 1999b; Esnault et al. 2000). Chondrodysplasia (shortened limbs) is a feature of several domestic dog breeds linked to the fibroblast-growth-factor receptor 4 retrogene (fgfr4). This retrogene has been inserted into a LINE sequence that is in close proximity to multiple Can-SINE insertions suggesting that these elements provide promoter sequences that allow expression of the fgfr4 at critical time points in development (Parker et al. 2009).
The Can-SINE induced phenotypes described above, associated with dog domestication and breed development, serve as models for the role of SINEs in gene function and complex genetic diseases in natural populations. Increased genomic data and advances in bioinformatics tools will further elucidate the interplay between Can-SINEs, coding sequences, and regulatory regions (Spady and Ostrander 2008).
Evolutionary Insights
SINEs have been used as synapomorphic markers in the reconstruction of several mammalian phylogenies including the afrotherian, cetacean, and artiodactyls lineages (Nikaido et al. 1999, 2001; Nishihara et al. 2005, 2006). However, very few studies have investigated the utility of Can-SINEs in carnivore systematics. Prior to the availability of carnivore whole-genome sequences, Can-SINEs were primarily characterized as a by-product of other genetic studies. For example, a survey of microsatellite loci inadvertently uncovered a species-specific SINE insertion in the Mel08 locus of American mink (M. vison) (Lopez-Giraldez et al. 2006) that was subsequently used in conservation efforts to distinguish invasive M. vison from the ecologically imperiled European mink (M. lutreola) (Lopez-Giraldez et al. 2005) (Table 1). Similarly, sequence analysis of the transthyretin gene revealed an orthologous Can-SINE locus that is a synapomorphy amongst caniforms (Zehr et al. 2001). Comparative studies of the feliform species bay cat (Profelis temminckii) and Pallas cat (Otocolobus manul), as well as the caniform gray wolf (C. lupus), red wolf (C. rufus), otter (Lutra lutra), and striped skunk (Mephitis mephitis) unveiled multiple, independently-derived Can-SINE insertions in the ß-fibrinogen gene (Yu and Zhang 2005). Moreover, the M. mephitis insert is a chimera, illustrating the tendency for SINEs to be incorporated within or adjacent to existing SINEs (Yu et al. 2004, 2008). (Table 1).
Table 1.
Clade and species-specific Can-SINE insertion sites published in previous evolutionary studies.
| Taxonomic group | Locus name | Publication |
| Suborder | ||
| Caniformia | Ttr intron 1 | Zehr et al. (2001)/Yu et al. (2011) |
| Caniformia | CF_L002d | Schröder et al. (2009) |
| Caniformia | CF_L003c | Schröder et al. (2009) |
| Caniformia | CF_L006a | Schröder et al. (2009) |
| Caniformia | CF_L010 | Schröder et al. (2009) |
| Caniformia | CF_L013 | Schröder et al. (2009) |
| Superfamily | ||
| Arctiodea | CF_L003b | Schröder et al. (2009) |
| Musteloidea | CF_L002b | Schröder et al. (2009) |
| Musteloidea | Ccng2 intron 2 | Yu et al. (2011) |
| Pinnipedia (2) | Ssr1 intron 5 | Yu et al. (2011) |
| Pinnipedia | Wasf1 intron 3 | Yu et al. (2011) |
| Family | ||
| Ursidae | Zfy | Pecon-Slattery et al. (2000) |
| Canidae | CF_L001 | Schröder et al. (2009) |
| Canidae* | CF_L003a | Schröder et al. (2009) |
| Canidae | CF_L004 | Schröder et al. (2009) |
| Canidae | CF_L007a | Schröder et al. (2009) |
| Canidae | CF_L007b | Schröder et al. (2009) |
| Canidae | CF_L008 | Schröder et al. (2009) |
| Canidae | CF_L011 | Schröder et al. (2009) |
| Canidae | CF_L014 | Schröder et al. (2009) |
| Canidae | CF_L015 | Schröder et al. (2009) |
| Odobenidae/Otariidae | CF_L006b | Schröder et al. (2009) |
| Mustelidae (except Meles) | CF_L004b | Schröder et al. (2009) |
| Canidae* | CF_L016 | Schröder et al. (2009) |
| Mustelidae | Ccng2 intron 2 | Yu et al. (2011) |
| Otaeiidae | Ccng2 intron 2 | Yu et al. (2011) |
| Ursidae | Cidea1 | Yu et al. (2011) |
| Mustelidae | Cidea1 | Yu et al. (2011) |
| Mustelidae (2) | Impa1 intron 6 | Yu et al. (2011) |
| Procyonidae | Plod2 intron 14 | Yu et al. (2011) |
| Subfamily | ||
| Caninae | CF_L002e | Schröder et al. (2009) |
| Caninae* | CF_L009b | Schröder et al. (2009) |
| Genus | ||
| Felis | Zfy | Pecon-Slattery et al. (2000) |
| Canis | ß-fibrinogen intron 7 | Yu and Zhang (2005) |
| Ursus | Coro1c intron 5 | Yu et al. (2011) |
| Ursus | Impa1 intron 6 | Yu et al. (2011) |
| Species | ||
| Mustela vison | Mel08 locus | Lopez-Giraldez et al. (2005) |
| Otocolobus manul | Ube1y | Pecon-Slattery et al. (2000) |
| O. manul | ß-fibrinogen intron 7 | Yu and Zhang (2005) |
| Profelis temminckii | ß-fibrinogen intron 7 | Yu and Zhang (2005) |
| M. erminea | Zfy | Yamada and Masuda (2010) |
| Meles anakuma | Zfy | Yamada and Masuda (2010) |
| Mephitis mephitis | ß-fibrinogen intron 7 | Yu and Zhang (2008) |
| Lutra lutra | ß-fibrinogen intron 4 | Yu and Zhang (2008) |
| Ursus arctos | Smcy | Pecon-Slattery et al. (2000) |
| Lynx rufus | Smcy | Pecon-Slattery et al. (2004) |
| Felis margarita/silvestris | Smcy | Pecon-Slattery et al. (2004) |
| Ursus arctos* | CF_L002a | Schröder et al. (2009) |
| Mep. mephitis* | CF_L002c | Schröder et al. (2009) |
| Mep. mephitis* | CF_L008a | Schröder et al. (2009) |
| Canis familiaris* | CF_L005 | Schröder et al. (2009) |
| C. familiaris* | CF_L012 | Schröder et al. (2009) |
| Procyon lotor* | CF_L009a | Schröder et al. (2009) |
| Zalophus californianus | CF_L017 | Schröder et al. (2009) |
| U. arctos* | CF_L018 | Schröder et al. (2009) |
| Proc. lotor* | CF_L021 | Schröder et al. (2009) |
| C. familiaris/lupus* | Atp5d intron 2 | Yu et al. (2011) |
| C. familiaris/lupus* | Fgb7 | Yu et al.(2011) |
| C. familiaris/lupus* (2) | Ssr1 intron 5 | Yu et al. (2011) |
| Arctonyx collaris* | Cidea1 | Yu et al. (2011) |
| Proc. lotor* (2) | Ccng2 intron 6 | Yu et al. (2011) |
| Mep. mephitis* | Cidea1 | Yu et al. (2011) |
| Mep. mephitis* | Impa1 intron 6 | Yu et al. (2011) |
| M. kathiah* | Impa1 intron 6 | Yu et al. (2011) |
| Mep. mephitis* (2) | Fgb7 | Yu et al. (2011) |
| Ailuropoda melanoleuca* | Guca1b intron 3 | Yu et al. (2011) |
| A. melanoleuca* | Impa1 intron 6 | Yu et al. (2011) |
| A. melanoleuca* | Ssr1 intron 5 | Yu et al. (2011) |
| Mep. mephitis* | Tbk1 intron 8 | Yu et al. (2011) |
| Martes flavigula* | Ociad1 intron 4 | Yu et al. (2011) |
*Phylogenetic range is not yet conclusive as data is not available from all related taxa. Numbers in parentheses indicate adjacent independent insertions within the same taxa.
The Y chromosome is described as a “sink” for transposable elements where SINEs can accumulate in the nonrecombining region (Krzywinskia et al. 2004; Jurka et al. 2007). Can-SINE insertions have been found within the Zfy gene of the Felis genus, the bear (Ursidae) family, the Japanese badger (Meles anakuma), and the stoat (M. erminea) (Pecon-Slattery et al. 2000; Yamada and Masuda 2010). The Smcy gene hosts an orthologous insertion in the sand cat (F. margarita) and the wildcat (F. silvestris) species complex. Species-specific insertions are also found in the Smcy gene of Ursus arctos and Ly. rufus (Pecon-Slattery et al. 2004). Otocolobus manul has a species-specific insertion in the Ube1y gene (Pecon-Slattery et al. 2000). (Table 1).
As with all mammalian SINEs, Can-SINE insertion distributions are generally congruent with existing hypotheses of speciation (Ray et al. 2006). However, the intronic insertions found in all Felis species and L. rufus (described above) occur at identical genomic coordinates (Pecon-Slattery et al. 2000, 2004), which if interpreted as a single synapomorphy, would unite distantly related species. However, phylogenetic reconstruction with sequences adjacent to the insertion site and differences in the poly A/T tails indicate that the 2 insertions are the result of independent SINE integration events at identical loci that occurred after the divergence of the major cat lineages (Pecon-Slattery et al. 2004; Johnson et al. 2006).
Comparative Genomics Fosters Can-SINEs Analyses
Whole-genome sequencing technologies enable inclusion of SINEs and other transposable elements in comparative analyses. Computational tools for analysis of noncoding regions littered with SINE insertions are becoming more accessible while sequences from model organisms provide a point of reference for the pursuit of informative SINEs in closely related and divergent taxa Wang et al. 2006; Liu et al. 2009; Schröder et al. 2009).
Whole-genome sequences derived from a Standard Poodle and Boxer indicate Can-SINEs account for approximately 8% of the C. familiaris genome (Kirkness et al. 2003; Lindblad-Toh et al. 2005). Nearly, all Can-SINE sequences that most closely align to the SINEC_Cf2 voucher sequence are homozygous and conserved between the 2 individuals, indicating that this subfamily is inactive. However, approximately 7% of sequences that most closely resemble the SINEC_Cf voucher sequence are unfixed, either within or between the 2 canines, which suggests recent proliferation of the corresponding subfamily (Kirkness et al. 2003). In contrast, recently acquired insertions account for only 0.5% of the Alu content in the human genome of which only 25% are unfixed (Batzer and Deininger 2002). Approximately 7.9% of the giant panda (A. melanoleuca) genome is comprised of SINE insertions that are <10% diverged from Repbase voucher sequences (Li et al. 2010). Further, de novo characterization of transposable elements identified an additional 0.1% of sequence belonging to SINE elements that were not yet identified in the Repbase, which suggests the presence of panda-specific subfamilies (Li et al. 2010). Initial estimates of the transposable element content in F. catus found that 11% of the genome is comprised of Can-SINEs similar to SINEC_FC1, SINEC_FC2, and SINEC_C1 voucher sequences and mammalian-wide MIR SINEs. However, novel Can-SINE sequences were also identified (Pontius et al. 2007), suggesting feliform-specific subfamilies in addition to those in Repbase.
Whole-genome sequences may be used as references for comparative analyses, allowing large-scale identification of Can-SINE insertion sites in both coding and noncoding regions that are population, species, or lineage specific. Within C. familiaris, 64 unfixed SINEC_Cf insertions have been localized that segregate with breed (Wang and Kirkness 2005). In addition, 17 intronic parsimony informative Can-SINE loci, distributed among a collection of anonymous introns, were located among 21 caniform species, all of which are congruent with other molecular data (Schröder et al. 2009). Whole-genome sequencing also facilitated identification of 31 diagnostic Can-SINE insertions amongst 14 intronic regions (Yu et al. 2011). Within Feliformia, comparative genomics analysis resulted in the identification of over 90 informative loci that can discern suborder, familial, and species relationships (Walters-Conte KB, unpublished data).
Conclusions
Our understanding of mammalian transposable elements has accelerated in the last 20 years as a consequence of advances in sequencing and computational technologies. Through these advances, we find that carnivore-specific Can-SINEs have a significant impact on genome content and gene function. These abundant retroelement insertions can directly alter phenotypes by becoming incorporated into coding regions and by providing promoter sequences that disrupt the transcriptional regulation of adjacent genes. As sources of genetic diversity, Can-SINEs have proven to be highly informative markers to differentiate protected and invasive species. In addition, Can-SINEs can define domestic dog breeds as well as diagnose species, genus, familial, and suborder relationships throughout Carnivora. Whole-genome sequences provide references to investigate the plethora of Can-SINEs that are clustered within intergenic regions. Future advancements in sequencing and bioinformatics technologies will provide further insights into Can-SINE biology, which will add to our general knowledge of the function and evolution of mammalian SINEs.
Funding
This project has been supported in part with federal funds from the National Cancer Institute, National Institutes of Health, under contract N01-CO-12400. The Robert Weintraub Program in Systematics and Evolution administered by The George Washington University also provided financial support.
Acknowledgments
The authors thank Dr. Stephen J. O'Brien for providing research support. We also acknowledge Dr. Warren Johnson and Dr. Diana Lipscomb for their helpful suggestions. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
References
- Batzer MA, Deininger PL. Alu repeats and human genomic diversity. Nat Rev Genet. 2002;3:370–379. doi: 10.1038/nrg798. [DOI] [PubMed] [Google Scholar]
- Belancio VP, Hedges DJ, Deininger P. Mammalian non-LTR retrotransposons: for better or worse, in sickness and health. Genome Res. 2008;18:343–358. doi: 10.1101/gr.5558208. [DOI] [PubMed] [Google Scholar]
- Breen M, Hitte C, Lorentzen T, Thomas R, Cadieu E, Sabacan L, Scott A, Evanno G, Parker H, Kirkness E, et al. An integrated 4249 marker FISH/RH map of the canine genome. BMC Genomics. 2004;5(1):65. doi: 10.1186/1471-2164-5-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brosius J. Many G-protein-coupled receptors are encoded by retrogenes. Trends Genet. 1999a;15(8):304–305. doi: 10.1016/s0168-9525(99)01783-7. [DOI] [PubMed] [Google Scholar]
- Brosius J. Transmutation of tRNA. Nat Genet. 1999b;22:8–9. doi: 10.1038/8711. [DOI] [PubMed] [Google Scholar]
- Callinan PA, Wang J, Herke SW, Garber RK, Liang P, Batzer MA. Alu retrotransposition-mediated deletion. J Mol Biol. 2005;348:791–800. doi: 10.1016/j.jmb.2005.02.043. [DOI] [PubMed] [Google Scholar]
- Christensen S, Ye J, Eickbush T, et al. RNA from the 5′ end of the R2 retrotransposon controls R2 protein binding to and cleavage of its DNA target site. Proc Natl Acad Sci U S A. 2006;103(47):17602–17607. doi: 10.1073/pnas.0605476103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark LA, Wahl JM, Rees CA, Murphy KE, et al. Retrotransposon insertion in SILV is responsible for merle patterning of the domestic dog. Proc Natl Acad Sci U S A. 2006;103(5):1376–1381. doi: 10.1073/pnas.0506940103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collier LS, Largaespada DA. Transposable elements and the dynamic somatic genome. Genome Biol. 2007;8(Suppl 1):S5. doi: 10.1186/gb-2007-8-s1-s5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coltman DW, Wright JM. Can SINEs: a family of tRNA-derived retroposons specific to the superfamily Canoidea. Nucleic Acids Res. 1994;22(14):2726–2730. doi: 10.1093/nar/22.14.2726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordaux R, Batzer M. The impact of retrotransposons on human genome evolution. Nat Rev Genet. 2009;10:691–703. doi: 10.1038/nrg2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordaux R, Hedges DJ, Batzer MA. Retrotransposition of Alu elements: how many sources? Trends Genet. 2004;20(10):464–467. doi: 10.1016/j.tig.2004.07.012. [DOI] [PubMed] [Google Scholar]
- Deininger P, Batzer M. Alu repeats and human disease. Mol Genet Metab. 1999;67(3):183–193. doi: 10.1006/mgme.1999.2864. [DOI] [PubMed] [Google Scholar]
- Dupuy AJ, Akagi K, Largaespada DA, Copeland NG, Jenkins NA. Mammalian mutagenesis using a highly mobile somatic Sleeping Beauty transposon system. Nature. 2005;436:221–226. doi: 10.1038/nature03691. [DOI] [PubMed] [Google Scholar]
- Esnault C, Maestre J, Heidmann T. Human LINE retrotransposons generate processed pseudogenes. Nat Genet. 2000;24:363–367. doi: 10.1038/74184. [DOI] [PubMed] [Google Scholar]
- Gentles AJ, Kohany O, Jurka J. Evolutionary diversity and potential recombinogenic role of integration targets of non-LTR retrotransposons. Mol Biol Evol. 2005;22:1983–1991. doi: 10.1093/molbev/msi188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray M, Sutter N, Ostrander E, Wayne RK. The IGF1 small dog haplotype is derived from Middle Eastern grey wolves. BMC Biol. 2010;8:16. doi: 10.1186/1741-7007-8-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu W, Ray DA, Walker JA, Barnes EW, Gentles AJ, Samollow PB, Jurka J, Batzer MA, Pollock DD. SINEs, evolution and genome structure in the opossum. Gene. 2007;396(1):46–58. doi: 10.1016/j.gene.2007.02.028. [DOI] [PubMed] [Google Scholar]
- Hasler J, Strub K. Survey and summary—Alu elements as regulators of gene expression. Nucleic Acids Res. 2006;34:5491–5497. doi: 10.1093/nar/gkl706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu L-J, Laporte J, Kioschis P, Heyberger S, Kretz C, Poustka A, Mandel J-L, Dahl N. X-linked myotubular myopathy: refinement of the gene to a 280-kb region with new and highly informative microsatellite markers. Hum Genet. 1996;98(2):178–181. doi: 10.1007/s004390050185. [DOI] [PubMed] [Google Scholar]
- Johnson WE, Eizirik E, Pecon-Slattery J, Murphy WJ, Antunes A, Teeling E, O'Brien SJ. The late Miocene radiation of modern Felidae: a genetic assessment. Science. 2006;311(5757):73–77. doi: 10.1126/science.1122277. [DOI] [PubMed] [Google Scholar]
- Jurka J. Sequence patterns indicate an enzymatic involvement in integration of mammalian retroposons. Proc Natl Acad Sci U S A. 1997;94:1872–1877. doi: 10.1073/pnas.94.5.1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurka J, Kapitonov V, Kohany O, Jurka MV. Repetitive sequences in complex genomes: structure and evolution. Annu Rev Genomics Hum Genet. 2007;8:241–259. doi: 10.1146/annurev.genom.8.080706.092416. [DOI] [PubMed] [Google Scholar]
- Jurka J, Kapitonov VV, Pavlicek A, Klonowski P, Kohany O, Walichiewicz J. Repbase Update, a database of eukaryotic repetitive elements. Cytogenet Genome Res. 2005;110:462–467. doi: 10.1159/000084979. [DOI] [PubMed] [Google Scholar]
- Kajikawa M, Okada N. LINEs mobilize SINEs in the eel through a shared 3' sequence. Cell. 2002;111(3):433–444. doi: 10.1016/s0092-8674(02)01041-3. [DOI] [PubMed] [Google Scholar]
- Kirkness EF, Bafna V, Halpern AL, Levy S, Remington K, Rusch DB, Delcher AL, Pop M, Wang W, Fraser CM. The dog genome: survey sequencing and comparative analysis. Science. 2003;301:1898–1903. doi: 10.1126/science.1086432. [DOI] [PubMed] [Google Scholar]
- Krzywinskia J, Nusskernb DR, Kerna MK, Besanskya NJ. Isolation and characterization of Y chromosome sequences from the African malaria mosquito Anopheles gambiae. Genetics. 2004;166:1291–1302. doi: 10.1534/genetics.166.3.1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurzynska-Kokorniak A, Jamburuthugoda V, Bibillo A, Eickbush T. DNA-directed DNA polymerase and strand displacement activity of the reverse transcriptase encoded by the R2 retrotransposon. J Mol Biol. 2007;374(2):322–333. doi: 10.1016/j.jmb.2007.09.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon B, Halaban R, Ponnazhagan S, Kim K, Chintamaneni C, Bennett D, Pickard R. Mouse silver mutation is caused by a single base insertion in the putative cytoplasmic domain of Pmel 17. Nucleic Acids Res. 1995;23(1):154–158. doi: 10.1093/nar/23.1.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laporte J, Hu L, Kretz C, Mandel J, Kioschis P, Coy J, Klauck S, Poustka A, Dahl N. A gene mutated in X-linked myotubular myopathy defines a new putative tyrosine phosphatase family conserved in yeast. Nat Genet. 1996;13(2):175–182. doi: 10.1038/ng0696-175. [DOI] [PubMed] [Google Scholar]
- Lavrentieva MV, Rivkin MI, Shilov AG, Kobetz ML, Rogozin IB, Serov OL. B2-like repetitive sequence from the X chromosome of the American mink (Mustela vison) Mamm Genome. 1991;1(3):165–170. doi: 10.1007/BF00351063. [DOI] [PubMed] [Google Scholar]
- Lawrence C, McDonnell D, Ramsey WJ. Analysis of repetitive sequence elements containing tRNA-like sequences. Nucleic Acids Res. 1985;13(12):4239–4252. doi: 10.1093/nar/13.12.4239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R, Fan W, Tian G, Zhu H, He L, Cai J, Huang Q, Cai Q, Li B, Bai Y, et al. The sequence and de novo assembly of the giant panda genome. Nature. 2010;463(7279):311–317. doi: 10.1038/nature08696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin L, Faraco J, Li R, Kadotani H, Rogers W, Lin X, Qui X, de Jon PJ, Nishino S, Maignot E. The sleep disorder canine narcolepsy is caused by a mutation in the hypocretin (Orexin) receptor 2 gene. Cell. 1999;98:365–376. doi: 10.1016/s0092-8674(00)81965-0. [DOI] [PubMed] [Google Scholar]
- Lin L, Shen S, Tye A, Cai J, Jiang P, Davidson B, Xing Y. Diverse splicing patterns of exonized Alu elements in human tissues. PLoS Genet. 2008;4:e10000225. doi: 10.1371/journal.pgen.1000225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindblad-Toh K, Wade CM, Mikkelsen TS, Karlsson EK, Jaffe DB, Kamal M, Clamp M, Chang JL, Kulbokas EJ, 3rd, Zody MC, et al. Genome sequence, comparative analysis and haplotype structure of the domestic dog. Nature. 2005;438(7069):803–819. doi: 10.1038/nature04338. [DOI] [PubMed] [Google Scholar]
- Liu G, Alkan C, Jiang L, Zhao S, Eichler EE. Comparative analysis of Alu repeats in primate genomes. Genome Res. 2009;19:876–885. doi: 10.1101/gr.083972.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Giraldez F, Andres O, Domingo-Roura X, Bosch M. Analyses of carnivore microsatellites and their intimate association with tRNA-derived SINEs. BMC Genomics. 2006;7:269. doi: 10.1186/1471-2164-7-269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Giraldez F, Gomez-Moliner BJ, Marmi J, Domingo-Roura X. Genetic distinction of American and European mink (Mustela vison and M. lutreola) and European polecat (M. putorius) hair samples by detection of species-specific SINE and RFLP assay. J Zool London. 2005;265:405–410. [Google Scholar]
- Miyamoto MM. Molecular systematics: perfect SINEs of evolutionary history? Curr Biol. 1999;9:816–819. doi: 10.1016/s0960-9822(99)80498-9. [DOI] [PubMed] [Google Scholar]
- Munemasa M, Nikaido M, Nishihara H, Donnellan S, Austin CC, Okada N. Newly discovered young CORE-SINEs in marsupial genomes. Gene. 2008;407(1–2):176–185. doi: 10.1016/j.gene.2007.10.008. [DOI] [PubMed] [Google Scholar]
- Nikaido M, Matsuno F, Hamilton H, Brownell RL, Jr, Cao Y, Ding W, Zuoyan Z, Shedlock AM, Fordyce, et al. Retroposon analysis of major cetacean lineages: the monophyly of toothed whales and the paraphyly of river dolphins. Proc Natl Acad Sci U S A. 2001;98(13):7384–7389. doi: 10.1073/pnas.121139198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikaido M, Rooney AP, Okada N. Phylogenetic relationships among cetartiodactyls based on insertions of short and long interspersed elements: hippopotamuses are the closest extant relatives of whales. Proc Natl Acad Sci U S A. 1999;96(18):10261–10266. doi: 10.1073/pnas.96.18.10261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishihara H, Hasegawa M, Okada N. Pegasoferae, an unexpected mammalian clade revealed by tracking ancient retroposon insertions. Proc Natl Acad Sci U S A. 2006;103(26):9929–9934. doi: 10.1073/pnas.0603797103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishihara H, Satta Y, Nikaido M, Thewissen JGM, Stanhope M, Okada N. Retrotransposon analysis of Afrotherian phylogeny. Mol Biol Evol. 2005;9:1823–1833. doi: 10.1093/molbev/msi179. [DOI] [PubMed] [Google Scholar]
- Ohshima K, Okada N. SINEs and LINEs: symbionts of eukaryotic genomes with a common tail. Cytogenet Genome Res. 2005;110(1–4):475–490. doi: 10.1159/000084981. [DOI] [PubMed] [Google Scholar]
- Parker HG, VonHoldt B, Quignon P, Margulies E, Shao S, Mosher D, Spady T, Elkaloun A, Michele C, Jones PG, et al. An expressed fgfr retrogene is associated with breed-defining chondrodyplasia in domestic dogs. Science. 2009;325(5943):995–998. doi: 10.1126/science.1173275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pecon-Slattery J, Murphy WJ, O'Brien SJ. Patterns of diversity among SINE elements isolated from three Y-chromosome genes in carnivores. Mol Biol Evol. 2000;17(5):825–829. doi: 10.1093/oxfordjournals.molbev.a026361. [DOI] [PubMed] [Google Scholar]
- Pecon-Slattery J, Wilkerson AJ, Murphy WJ, O'Brien SJ. Phylogenetic assessment of introns and SINEs within the Y chromosome using the cat family Felidae as a species tree. Mol Biol Evol. 2004;21(12):2299–2309. doi: 10.1093/molbev/msh241. [DOI] [PubMed] [Google Scholar]
- Pelå M, Tiret L, Kessler J-L, Blot S, Panthier J-J. SINE exonic insertion in the PTPLA gene leads to multiple splicing defects and segregates with the autosomal recessive centronuclear myopathy in dogs. Hum Mol Genet. 2005;14(11):1417–1427. doi: 10.1093/hmg/ddi151. [DOI] [PubMed] [Google Scholar]
- Peyron C, Faraco J, Rogers W, Ripley B, Overeem S, Charnay Y, Nevsimalova S, Aldrich M, Reynolds D, Albin R, et al. A mutation in a case of early onset narcolepsy and a generalized absence of hypocretin peptides in human narcoleptic brains. Nat Med. 2000;6(9):991–997. doi: 10.1038/79690. [DOI] [PubMed] [Google Scholar]
- Pontius J, Mullikin J, Smith D, Lindblad-Toh K, Gnerre S, Clamp M, Chang J, Stephens R, Neelam B, Volfovsky N, et al. Initial sequence and comparative analysis of the cat genome. Genome Res. 2007;17(11):1675–1689. doi: 10.1101/gr.6380007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray DA, Xing J, Salem AH, Batzer MA. SINEs of a nearly perfect character. Syst Biol. 2006;55(6):928–935. doi: 10.1080/10635150600865419. [DOI] [PubMed] [Google Scholar]
- Schröder C, Bleidorn C, Hartmann S, Tiedemann R. Occurrence of Can-SINEs and intron sequence evolution supports robust phylogeny of pinniped carnivores and their terrestrial relatives. Gene. 2009;448(2):221–226. doi: 10.1016/j.gene.2009.06.012. [DOI] [PubMed] [Google Scholar]
- Smit A, Green P. Repeat Masker Website and Server Seattle, WA: Institute for Systems Biology. 2005. [Google Scholar]
- Spady T, Ostrander E. Canine behavioral genetics: pointing out the phenotypes and herding up the genes. Am J Hum Genet. 2008;82:10–18. doi: 10.1016/j.ajhg.2007.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutter N, Bustamante C, Chase K, Gray M, Zhao K, Lan Z, Padhukasahasram B, Karlins E, Davis S, Jones PG, et al. A single IGF1 allele is a major determinant of small size in dogs. Science. 2007;316(5821):112–115. doi: 10.1126/science.1137045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theos AC, Truschel ST, Raposo G, Marks MS. The Silver locus product Pmel17/gp100/Silv/ME20: controversial in name and in function. Pigment Cell Res. 2005;18(5):322–336. doi: 10.1111/j.1600-0749.2005.00269.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tong C, Gan X, He S. Multiple source genes of HAmo SINE actively expanded and ongoing retroposition in cyprinid genomes relying on its partner LINE. BMC Evol Biol. 2010;10(115):115. doi: 10.1186/1471-2148-10-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uwanogho DA, Hardcastle Z, Balogh P, Mirza G, Thornburg KL, Ragoussis J, Sharpe PT. Molecular cloning, chromosomal mapping, and developmental expression of a novel protein tyrosine phosphatase-like gene. Genomics. 1999;62(3):406–416. doi: 10.1006/geno.1999.5950. [DOI] [PubMed] [Google Scholar]
- van der Vlugt HH, Lenstra JA. SINE elements of carnivores. Mamm Genome. 1995;6(1):49–51. doi: 10.1007/BF00350894. [DOI] [PubMed] [Google Scholar]
- Vassetzky NS, Kramerov DA. CAN—a pan-carnivore SINE family. Mamm Genome. 2002;13(1):50–57. doi: 10.1007/s00335-001-2111-1. [DOI] [PubMed] [Google Scholar]
- Wang J, Song L, Gonder MK, Azrak S, Batzer MA, Tishkoff SA, Liang P. Whole genome computational comparative genomics: a fruitful approach for ascertaining Alu insertion polymorphisms. Gene. 2006;365:11–20. doi: 10.1016/j.gene.2005.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Kirkness EF. Short interspersed elements (SINEs) are a major source of canine genomic diversity. Genome Res. 2005;15:1798–1808. doi: 10.1101/gr.3765505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicker T, Sabot F, Hua-Van A, Bennetzen JL, Capy P, Chalhoub B, Flavell A, Leroy P, Morgante M, Panaud, et al. A unified classification system for eukaryotic transposable elements. Nat Rev Genet. 2007;8(12):973–982. doi: 10.1038/nrg2165. [DOI] [PubMed] [Google Scholar]
- Yamada C, Masuda R. Molecular phylogeny and evolution of sex-chromosomal genes and SINE sequences in the family Mustelidae. Mamm Study. 2010;35:17–30. [Google Scholar]
- Yu L, Li Q-w, Ryder O, Zhang Y-p. Phylogenetic relationships within mammalian order Carnivora indicated by sequences of two nuclear DNA genes. Mol Phylogenet Evol. 2004;33:694–705. doi: 10.1016/j.ympev.2004.08.001. [DOI] [PubMed] [Google Scholar]
- Yu L, Liu J, Luan P-t, Lee H, Lee M, Min M-s, Ryder O, Chemnick L, Davis H, Zhang Y-p. New insights into the evolution of intronic sequences of the Beta-fibrinogen gene and their application in reconstructing mustelid phylogeny. Zool Sci. 2008;25(6):622–672. doi: 10.2108/zsj.25.662. [DOI] [PubMed] [Google Scholar]
- Yu L, Luan P-t, Jin W, Ryder OA, Chemnick LG, Davis HA, Zhang Y-p. Phylogenetic utility of nuclear introns in interfamilial relationships of Caniformia (order Carnivora) Syst Biol. 2011;60(2):175–187. doi: 10.1093/sysbio/syq090. [DOI] [PubMed] [Google Scholar]
- Zehr SM, Nedbal MA, Flynn JJ. Tempo and mode of evolution in an orthologous Can SINE. Mamm Genome. 2001;12(1):38–44. doi: 10.1007/s003350010224. [DOI] [PubMed] [Google Scholar]



