Abstract
We show that the centrosome- and microtubule-regulating protein γ-tubulin interacts with E2 promoter binding factors (E2Fs) to modulate E2F transcriptional activity and thereby control cell cycle progression. γ-Tubulin contains a C-terminal signal that results in its translocation to the nucleus during late G1 to early S phase. γ-Tubulin mutants showed that the C terminus interacts with the transcription factor E2F1 and that the E2F1–γ-tubulin complex is formed during the G1/S transition, when E2F1 is transcriptionally active. Furthermore, E2F transcriptional activity is altered by reduced expression of γ-tubulin or by complex formation between γ-tubulin and E2F1, E2F2, or E2F3, but not E2F6. In addition, the γ-tubulin C terminus encodes a DNA-binding domain that interacts with E2F-regulated promoters, resulting in γ-tubulin-mediated transient activation of E2Fs. Thus, we report a novel mechanism regulating the activity of E2Fs, which can help explain how these proteins affect cell cycle progression in mammalian cells.—Höög, G., Zarrizi, R., von Stedingk, K., Jonsson, K., Alvarado-Kristensson, M. Nuclear localization of γ-tubulin affects E2F transcriptional activity and S-phase progression.
Keywords: cell cycle progression, signal transduction, cell signaling
During cell division, the point of no return is when a cell commits to divide at the G1/S transition. At the molecular level, during G1/S, the CDK2–cyclin E complex hyperphosphorylates retinoblastoma protein (RB), which leads to the dissociation of E2 promoter binding factor (E2F) from RB and initiation of E2F transcriptional activity. This event marks the onset of cell division, as E2F initiates the transcription of a number of genes necessary for cell cycle progression. Mutations in this RB–E2F-dependent cell cycle checkpoint have been implicated in the achievement of growth factor independence in multiple malignancies (1).
γ-Tubulin is a ubiquitously expressed protein that regulates interphase αβ-tubulin nucleation (2), centrosomal duplication (3), and spindle formation (4). γ-Tubulin occurs in the cytosol (5), centrosomes (3), and nucleus (6–8); in the latter location it associates with Rad51 during recombination repair (6). However, neither the mechanism underlying nuclear localization of γ-tubulin nor the nuclear functions of this protein have been elucidated. The present study detected a pool of γ-tubulin that is actively translocated to the nucleus of U2OS and NIH3T3 cells, where it modulates E2F transcriptional activity and cell cycle progression.
MATERIALS AND METHODS
Cell culture, transfection, and cell cycle analysis
Murine NIH3T3 embryonic fibroblasts and human U2OS osteosarcoma cells were cultured as reported elsewhere (3) and were transfected with various cDNAs using Lipofectamine Plus (Gibco BRL, Gaithersburg, MD, USA) and Jet Pei (Q-Biogene, Carlsbad, Ca, USA). To obtain equal protein levels of the various ectopically expressed proteins, the following DNA amounts were used in transfection and cotransfection experiments: 960 ng of γ-tubulin-green fluorescent protein (γtubGFP), N-γ-tubulin1–333, or C-γ-tubulin334–452 and/or 40 ng of hemagglutinin (HA)-E2F1, HA-E2F2, HA-E2F3, HA-E2F6, GFP, or HA-database of rice transcription factor polypeptide 1 (Dp1) in U2OS cells and 875 ng of γtubGFP in NIH3T3 cells was cotransfected with 188 ng of the various E2Fs or GFP. To achieve phase synchronization, cells were arrested in the G0 phase (3) and released for different periods of time. Phase distribution was examined by using propidium iodide to measure cell DNA content, as described previously (3).
Cell fractionation
To isolate chromatin, cells (0.5×106) were treated as described previously (9). In brief, cells were first lysed in buffer A (9) containing 0.1% triton X-100. Nuclei in the first pellet were collected and lysed in buffer B (9). Insoluble chromatin was collected by centrifugation. The purified fractions were boiled in sample buffer (3) and analyzed by Western blotting using α-tubulin and histone as molecular markers for the cytosolic and nuclear fractions, respectively.
cDNA and reagents
Human (h)γ-tubulin pcDNA3-GFP was provided by Dr. Jiri Bartek (Institute of Cancer Biology, Danish Cancer Society, Copenhagen, Denmark; ref. 10); pcDNA3-hemagglutinin (HA)hE2F1 was furnished by Dr. Joseph R. Nevins (Duke University, Durham, NC, USA; ref. 11); pGL3-TATA-6xE2F-Luc, hE2F2, hE2F3 and hDp1 were from Dr. Kristian Helin (Biotech Research and Innovation Centre, University of Copenhagen, Copenhagen, Denmark; refs. 12, 13); hE2F6 was provided by Dr. David M. Livingston (Dana-Farber Cancer Institute, Boston, MA, USA; ref. 14), and cyclin E promoter (plasmid 8458; Addgene, Cambridge, MA, USA) was furnished by Dr. Robert A. Weinberg (Massachusetts Institute of Technology, Cambridge, MA, USA). Human γ-tubulin, N-γ-tubulin1–333, and C-γ-tubulin334–452 were amplified by PCR and subcloned in frame into pGEX2T (Amersham, Piscataway, NJ, USA) using the primer sets P1 (5′-GCGCCCGGGCATGCCGAGGGAAATC-3′) and P2 (5′-CGCGAATTCCTCTGGGTGCCCCAGGA-3′), P1 and 5′-CGCGAATTCCTGACCTGGGTGGGGT-3′, and 5′-CGCCCCGGGCCACAAGAGCTTGCAG-3′ and P2, respectively. N-γtubulin1–333 and C-γtubulin334–452 were subcloned in frame into pEGFPN1 (Clontech, Palo Alto, CA, USA) using the oligo sets 5′-GCGGCTAGCGATGCCGAGGGAAATCATC-3′ and 5′-CGCAAGCTTGACCTGGGTGGGGT3-′, and 5′-GCGGCTAGCGATGAGCTTGCAGAGGATCCGG-3′ and P2, respectively. Human E2F1, E2F1 (Δ360–426), and E2F1 (Δ194–426) were amplified by PCR and subcloned in frame into pET21d (Novagen, San Diego, CA, USA) using the primer sets P3 (5′-CGCGAATTCACCATGGCCTTGGCCGGGGCC-3′) and 5′-GCGAAGCTTGAAATCCAGGGGGGTGAGG-3′, P3 and 5′-GCGAAGCTTGAAATCCAGGGGGGTGAGGTCCCCAAAGTCCAACAGCGGTTCTTGC-3′, and P3 and 5′-GCGAAGCTTGAAATCCAGGGGGGTGAGGCTGCCCAGCCACTGGAT-3′, respectively. Human HAE2F1 (Δ409–426), HAE2F1 (Δ2–126), HAE2F1 (Δ360–426), and HAE2F1 (Δ193–426) were obtained by PCR and subcloned into HindIII/EcoRI sites of pcDNA3.1 using the primer sets P4 (5′-GCGAAGCTTACCATGGCCTTGGCCGGGGCC3′) and 5′-CGCGAATTCTCACGCGTAATCCGGGACATCGTAC GGGTACATGGATCCGAAATCCAGGGGGGTGAGGTCCCCAAAGTCGAACTCCTCAGGGAGG-3′, 5′-GCGAAGCTTACCATGCGCTATGAGACCTCACTG-3′ and 5′-CGCGAATTCTCACGCGTAATCCGGGACATCGTACGGGTACATGGATCCGAAATCCAGGGGGGTGAGG-3′, P4 and 5′-CGCGAATTCTCACGCGTAATCCGGGACATCGTACGGGTACATGGATCCGAAATCCAGGGGGGTGAGGTCCCCAAAGTCCAACAGCGGTTCTTGC-3′, and P4 and 5′-CGCGAATTCTCACGCGTAATCCGGGACATCGTACGGGTACATGGATCCGAAATCCAGGGGGGTGAGGTCCCCAAAGTCGCTGCCCAGCCACTGGAT-3′, respectively. FLAGE2F1 (Δ193–284), FLAGE2F1 (Δ284–359), and FLAGE2F1 (Δ193–359) were obtained by PCR and subcloned into HindIII/BamHI/EcoRI sites of pcDNA3.1 using the primer sets P4, 5′-GCGGGATCCCAGCCACTGGATGTGGTTC-3′, P5 (5′-GCGAAGCTTGGATCCCTTAAGAGCAAACAAGG-3′), and P6 (5′-CGCGAATTCTCAGATCTTATCGTCGTCATCCTTGTAATCGAAATCCAGGGGGGTGAG-3′); P4, 5′-GCGGGATCCCTGAAAGTTCTCCGAAGAGT-3′, P7 (5′-GCGAAG CTTGGATCCCGGATGGGCAGCCTGC-3′) and P6; and P4, P5, P7, and P6; respectively. Arginines and lysines in an sh-resistant γ-TUBULIN gene (3) were replaced using a Quikchange Site-directed Mutagenesis Kit (Stratagene, La Jolla, CA. USA) and the following primers (mutated bases underscored): 5′-CCAGTATGACAAGCTGGCTAAGCGGGAGGCCTTC-3′ and 5′-GAAGGCCTCCCGCTTAGCCAGCTTGTCATACTGG-3′ (R399>A); 5′-CAGTATGACAAGCTGCGTGCGCGGGAGGCCTTCCTGG-3′ and 5′-CCAGGAAGGCCTCCCGCGCACGCAGCTTGTCATACTG-3′ (K400>A); 5′-GCCTTCCTGGAGCAGTTCGCCAAGGAGGACATGTTCAAG-3′ and 5′-CTTGAACATGTCCTCCTTGGCGAACTGCTCCAGGAAGGC-3′ (R409>A); 5′-GGAGCAGTTCCGCGCGGAGGACATGTTC-3′ and 5′-GAACATGTCCTCCGCGCGGAACTGCTCC-3′ (K410>A); 5′-CAGTATGACAAGCTGGCTGCGCGGGAGGCCTTCCTGG-3′ and 5′-CCAGGAAGGCCTCCCGCGCAGCCAGCTTGTCATACTG-3′ (R399>A K400>A); and 5′-GCCTTCCTGGAGCAGTTCGCCGCGGAGGACATGTTCAAG-3′ and 5′-CTTGAACATGTCCTCCGCGGCGAACTGCTCCAGGAAGGC-3′ (R409>A K410>A). The mutations were verified by sequencing.
The following antibodies and reagents were used: anti-Histone (Chemicon/Millipore, Bi1lerica, MA, USA); anti-HA, anti-GFP, anti-RB, anti-pRB, anti-E2F1, anti-E2F2 anti-E2F3, and anti-Dp1 (Santa Cruz Biotechnology, Santa Cruz, CA, USA); rabbit anti-γ-tubulin T3320 and T5192 and mouse anti-γ-tubulin GTU-88 (Sigma, St. Louis, MO, USA) and anti-α-tubulin (Calbiochem, San Diego, CA, USA); anti-His (Abcam, Cambridge, MA, USA); protein G PLUS-agarose and anti-GST (Amersham); 32PγATP (Perkin-Elmer, Wellesley, MA, USA); an enhanced chemiluminescence (ECL) kit (Santa Cruz Biotechnology); SDS-PAGE reagents (Bio-Rad, Hercules, CA, USA). All other reagents were obtained from Sigma, unless otherwise indicated.
RNA interference
For human E2F1, E2F2, and E2F3 (15) shRNA, the annealed oligonucleotides 5′-GATCCCTATCTGTACTACGCAGCTGTTCAAGAGACAGCTGCGTAGTACAGATATTTTTGGAAA-3′ and 5′-AGCTTTTCCAAAAATATCTGTACTACGCAGCTGTCTCTTGAACAGCTGCGTAGTACAGATAGG-3′, 5′-GATCCCGACTCGGTATGACACTTCGTTCAAGAGACGAAGTGTCATACCGAGTCTTTTTGGAAA-3′ and 5′-AGCTTTTCCAAAAAGACTCGGTATGACACTTCGTCTCTTGAACGAAGTGTCATACCGAGTCGG-3′, and 5′-GATCCCGGCTGGAGCTAGGAGAAAGTTCAAGAGACTTTCTCCTAGCTCCAGCCTTTTTGGAAA-3′ and 5′-AGCTTTTCCAAAAAGGCTGGAGCTAGGAGAAAGTCTCTTGAACTTTCTCCTAGCTCCAGCCGG-3′, respectively, were cloned into the BglII and HindIII sites of the plasmid pTER (a gift from Dr. Marc van de Wetering (Hubrecht Laboratory, Centre for Biomedical Genetics, Utrecht, The Netherlands), as described elsewhere (3). Human γ-TUBULIN shRNA and the RNAi- resistant γ-TUBULIN-gene were prepared as previously reported (3). The pBS/U6 plasmid (a gift of Dr. Yang Shi, Harvard Medical School, Boston, MA, USA) was prepared as described previously (3), encoding the shRNA for mouse γ-Tubulin with the annealed oligonucleotides, 5′-GGCTCATGATGGCCAACCACCAAGCTTGGTGGTTGGCCATCATGAGCCCTTTTTC-3′ and 5′-AATTGAAAAAGGGCTCATGATGGCCAACCACCAAGCTTGGTGGTTGGCCATCATGAGCC-3′. Cells were transfected as reported earlier (3).
Expression and purification of recombinant proteins
The human GST fusion proteins (N-terminal-GST γ-tubulin GST-N-γtubulin1–333 and GST-C-γtubulin334–452) were expressed in Escherichia coli DH5a and C-terminal His6-tagged E2F1, E2F1 (Δ360–426), and E2F1 (Δ194–426) were expressed in E. coli BL21(DE3) (Stratagene; ref. 3). Exponentially growing bacteria bearing these plasmids were induced (37°C, 2 h) with 0.2 and 1 mM isopropyl-1-thio-b-d-galactopyranoside (IPTG), respectively. Recombinant proteins were purified under native conditions using glutathione-Sepharose 4B (Amersham Pharmacia Biotech) or Ni2+ affinity resin (Qiagen, Valencia, CA, USA), according to the manufacturer's instructions.
Immunoprecipitation and Western blot analysis
Using previously described methodology (16), cell extracts were obtained, the remaining supernatants were precleared, and immunoprecipitation was performed using anti-HA IgG2a monoclonal antibody (Santa Cruz Biotechnology) or anti-E2F1 or anti-γ-tubulin IgG1 monoclonal antibody. Western blotting was performed as reported elsewhere (17).
Microscopy
NIH3T3 or U2OS cells were cultured and fixed as described previously (3). Staining of endogenous γ-tubulin was performed with the following antibodies: rabbit anti-γ-tubulin T3320 or T5192 or mouse anti-γ-tubulin GTU-88. Fluorescence images were captured and processed using an Olympus Bx 51 microscope (Olympus, Tokyo, Japan). A minimum of 100 cells were examined in each sample.
Reconstitution of the E2F–γ-tubulin complex
GST-γtub, GST-C-γtub334–452, or GST-N-γtub1–333 (500 ng) was incubated for 45 min with or without His-E2F1 or His-E2F1Δ334–452 (500 ng); this was done on ice in a buffer (total volume of 1 ml) containing 20 mM Tris (pH 7.5), 0.1% Triton X-100, 1 mM MgCl2, 0.25 mM GTP, 5 mM β-mercaptoethanol, 137 mM NaCl, and 5% glycerol. The GST-tag proteins were purified by adsorption on 40 ml of glutathione-Sepharose 4B (18) and subsequently washed 4 times with buffer and analyzed by Western blotting.
Luciferase assays
U2OS cells were transfected with 170 ng of the desired luciferase reporter construct, 1 ng of PRL-CMV-Renilla luciferase reporter construct (Promega, Madison, WI, USA), 20 ng of an E2F construct, and 200 ng of a γ-tubulin construct. If the total amount of plasmid DNA did not reach 400 ng, pCDNA 3.1 or pEGFP empty vectors were added, and an equal amount of plasmid DNA was used in the transfections. At 1 d after transfection, cells were harvested for determination of luciferase and Renilla activity. Double determination of luciferase activity was achieved using a dual-luciferase reporter assay system (Promega).
Gene expression analysis
Total RNA from transfected U2OS cells was extracted as previous described (3), followed by a cleaning step using the RNeasy Mini Kit (Qiagen). mRNA expression array analysis was performed using the human Illumina platform. The gene set enrichment analysis (GSEA) program (http://www.broadinstitute.org/gsea/) was used to generate a ranked gene list according to differential expression between control-shRNA- and γ-TUBULIN-shRNA-transfected samples. Genes were ranked in descending order with regard to level of expression in the γ-TUBULIN-shRNA-transfected cells. GSEA (19) was performed on the generated ranked gene list using an E2F1-upregulated gene set generated from the publically available data set (accession number GSE1562; http://www.ncbi.nlm.nih.gov/geo/) consisting of control (LacZ) and exogenous E2F1 expressing melanoma SK-MEL-2 cells (20). E2F1-upregulated genes were defined as all genes displaying ≥5-fold higher expression in the E2F1-expressing cells compared to control (Table 1).
Table 1.
E2F1-upregulated target gene signature
| No. | Gene | No. | Gene | No. | Gene | No. | Gene | No. | Gene |
|---|---|---|---|---|---|---|---|---|---|
| 1. | BMP2 | 17. | PCK1 | 33 | RORA | 49. | HPGD | 65. | INHBB |
| 2. | LRIG1 | 18. | BRF1 | 34. | CDKN2C | 50. | ZBTB1 | 66. | H2AFB1 |
| 3. | EIF4A1 | 19. | MAP3K5 | 35. | KIAA0495 | 51. | CCNE2 | 67. | FOXF2 |
| 4. | FOXD1 | 20. | NPTX2 | 36. | HOXD8 | 52. | ARR3 | 68. | MYB |
| 5. | RB1 | 21. | PEG10 | 37. | GATA2 | 53. | GZMB | 69. | NR2F1 |
| 6. | FST | 22. | KLF10 | 38. | PTK7 | 54. | AMY2B | 70. | CDKN1A |
| 7. | FOXA1 | 23. | GNRHR | 39. | CYP26A1 | 55. | ATP8A1 | 71. | BAI2 |
| 8. | VEGFC | 24. | FOXO1 | 40. | TCF7 | 56. | WIF1 | 72. | F3 |
| 9. | PTHLH | 25. | FGFR1 | 41. | PLAG1 | 57. | IRX5 | 73. | CDKN1B |
| 10. | DUSP2 | 26. | AKAP5 | 42. | DLX2 | 58. | C8G | 74. | SKP2 |
| 11. | NHLH2 | 27. | EFNB2 | 43. | GNAO1 | 59. | MPP6 | 75. | STX11 |
| 12. | ABCC4 | 28. | LOC652637 | 44. | KITLG | 60. | KIAA1107 | 76. | RBM38 |
| 13. | RFPL3S | 29. | TERF2 | 45. | CSPG5 | 61. | PKLR | 77. | FGF9 |
| 14. | ZIC3 | 30. | PLCL2 | 46. | LHX2 | 62. | CCNE1 | ||
| 15. | IRF1 | 31. | ESR2 | 47. | ABCA2 | 63. | SYNGR3 | ||
| 16. | FGFR3 | 32. | CHST1 | 48. | BCL2L11 | 64. | MAN1C1 |
All genes displaying a ≥5-fold higher expression in E2F1 overexpressing melanoma SK-MEL-2 cells compared to control SK-MEL-2 cells (data set NCBI GEO accession number GSE1562). Genes are displayed in order of most positively enriched (genes 1–35) to the most negatively enriched (genes 37–77) in the γ-Tubulin-shRNA-transfected cells. HOXD8 (gene 36) displayed no enrichment.
To examine γ-tubulin-correlated gene expression in tumor material, two publically available datasets were examined including 315 breast tumors, 121 lung tumors [National Center for Biotechnology (NCBI) Gene Expression Omnibus (GEO) accession number GSE2109] and 290 colon tumors (GEO accession number GSE14333). For each tumor type, all genes were ranked according to correlation to γ-tubulin expression. The top 100 correlated genes to γ-tubulin in each case were used to perform a Gene Ontology (GO) analysis, using the Functional Annotation tool in the online Database for Annotation, Visualization and Integrated Discovery (DAVID; refs.21, 22).
Electrophoretic mobility shift assay (EMSA)
The double-stranded oligonucleotides used for EMSA were derived from the dihydrofolate reductase promoter, an E2F1 consensus site, and the sequences were as follows (23): nonmutated 5′AGCTTCTAGTGCAATTTCGCGCCAAACTTGG3′ and 5′AATTCCAAGTTTGGCGCGAAATTGCACTAGA3′ (E2F site underscored); mutated 5′AGCTTCTAGTGCAATTGCTCGACCAACTTGG3′ and 5′AATTCCAAGTTGGTCGAGCAATTGCACTAGA3′ (mutated bases underscored). The oligos were aligned and subcloned into HindIII/EcoRI sites of pcDNA3.1.
A 20 μl reaction mixture containing 2 μg of each recombinant protein, 0.1 ng of the HindIII/EcoRI-fragmented 32P-labeled DNA probe, 0.25mg/ml poly(dI-dC)-poly(dI-dC), and DNA competitor (100-fold cold oligo; wild-type or mutated) or antibody (1.0 μl), was incubated in binding buffer (1.5 M Tris, pH 7.5; 1 M MgCl2; 20% triton X-100; 0.5 M EDTA; 5 M NaCl; 30 mM Na3VO4; 0.25 M PMSF; 3M KCl; and 500 mM NaF) at room temperature for 30 min and then analyzed on a 4% acrylamide gel in 0.5 TBE at 4°C.
Chromatin immnunoprecipitation (ChIP)
ChIP was described elsewhere (24). ChIPs were performed using the following rabbit polyclonal antibodies: anti-E2F1 C-20, anti-E2F2 C-20, anti-E2F3 N-20, and anti-Dp1 K-20 (Santa Cruz Biotechnology) and anti-γ-tubulin T3320 or affinity-purified anti-γ-tubulin T5192 (Sigma), on nontransfected aphidicolin treated U2OS cells (3). In re-ChIP experiments, endogenous E2Fs or γ-tubulin–chromatin complexes were first immunoprecipitated and then eluted by incubation for 30 min at 37°C in 10 mM DTT, 2 mM EDTA, and 10 mM Tris (pH 8). The eluted chromatin complexes were subjected to a second ChIP. Coprecipitated chromatin was analyzed by PCR for the presence of cyclin E promoter DNA between –76 to 86 bp using primers 5′-TCTGGGTCCCGCGCGGCCGCTGAGG-3′ and 5′-ATCCGCGCCTGCCCCCTACACCGCG-3′.
Microtubule regrowth assay
Cells were treated and analyzed as described previously (3).
Statistical evaluation
All data are expressed as means ± sd, and Student's paired t test was used to analyze the differences. Cell cycle profiles were assessed using FlowJo (Tree Star, Inc., Ashland, OR, USA). For gene expression array analyses, correlated ranked gene lists were generated based on Pearson's product moment correlation coefficient. Relative P values and 95% confidence intervals were also calculated for the Pearson's correlations. GO analyses performed on the generated ranked gene lists displayed overall cluster enrichment score (log of geometric mean of EASE scores of all GEO terms included in the functional cluster) and both EASE score (modified Fisher exact test ; ref. 25) P values for GEO term enrichment and Bonferoni corrected P values to account for multiple testing. GSEA displayed the enrichment score and corresponding nominal P value representing the significance of enrichment of the single tested gene set (E2F1 up-regulated; ref. 19).
RESULTS
During G1/S transition, γ-tubulin localizes to chromatin and interacts with E2F1
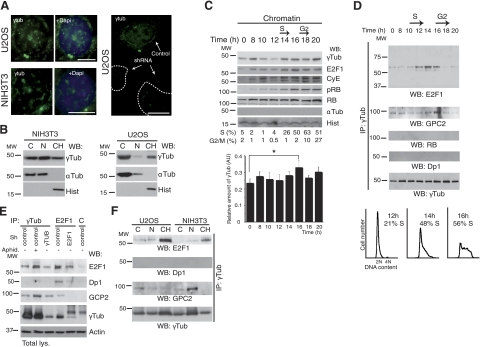
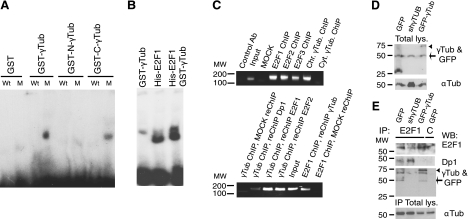
Immunofluorescence analysis using 3 different anti-γ-tubulin antibodies showed a constitutive localization of endogenous γ-tubulin to the nucleus of NIH3T3 and U2OS cells (6–8). In addition, the centrosomic, nuclear, and cytosolic immunofluorescence-signal decreased with selective shRNA (Fig. 1A). Analysis of biochemical fractionations showed that part of the endogenous γ-tubulin was found in the chromatin fraction (ref. 9 and Fig. 1B), and a densitometric analysis of the different fractions showed that 29.3 ± 2.6 and 25.8 ± 2.6% (n=3) of the total amount of endogenous γ-tubulin was present in the chromatin of U2OS and NIH3T3, respectively. To determine variations in localization during the cell cycle, we investigated distribution of endogenous γ-tubulin in synchronized NIH3T3 cells (3). This revealed accumulation in the chromatin fraction of cells in late G1 and early S phase (6), in the latter reaching a maximum at 16 h (Fig. 1C and Supplemental Fig. S1A). Translocation of γ-tubulin to the chromatin coincided with a rise in E2F1 protein levels and an increase in hyperphosphorylated RB (Fig. 1C and Supplemental Fig. S1A), events that mark the onset of cell division (1). Analysis of endogenous γ-tubulin immunoprecipitates disclosed a cell cycle-dependent association of γ-tubulin with endogenous E2F1, but not with cyclin E, Dp1, or RB (Fig. 1D). By comparison to other γ-tubulin-interacting proteins, such as GCP2 (2), the E2F1–γ-tubulin complex appeared transiently at G1/S entry and in early S phase (Fig. 1D). Additional characterization showed that levels of E2F1–γ-tubulin complex decreased by shRNAi-induced reduction of either E2F1 or γ-tubulin (Fig. 1E) and increased when U2OS cells were aphidicolin arrested (3, 6) in early S phase (Fig. 1E). Next, we investigated the cellular localization of the E2F1–γ-tubulin by immunoprecipitating endogenous γ-tubulin from fractions of U2OS and NIH3T3 cells. In comparison with the microtubule associated GCP2–γ-tubulin complex, E2F1–γ-tubulin occurred mainly in the chromatin fraction, suggesting that γ-tubulin may modulate the nuclear transcriptional activity of E2F1 (Fig. 1F). Furthermore, E2F heterodimerization partner Dp1 was found associated with E2F1 immunoprecipitates but not with γ-tubulin immunoprecipitates (Fig. 1D, E). These findings support a specific nuclear association between γ-tubulin and E2F1 during early S phase.
Figure 1.
γ-Tubulin localizes to chromatin and interacts with E2F1. A) Localization of endogenous γ-tubulin (eγTub) was examined by immunofluorescence staining with γTub (green), and nuclei were detected with DAPI (blue) in NIH3T3 and U2OS cells expressing human γ-TUBULIN -shRNA. Scale bars = 10 μm. B) Cells (0.5×106) were biochemically divided into cytosolic (C), nuclear (N), and chromatin (CH) fractions and analyzed by Western blotting (WB) with an anti-γ-tubulin antibody (γTub), α-tubulin (αTub) and histone (Hist) (n=5 or 6). C) Extracts from synchronous NIH3T3 cells were prepared as in A and examined by WB using antibodies against E2F1, cyclin E (CyE), phosphoretinoblastoma (pRB), total RB, γ- and α-tubulin, and histone (n=6). DNA content was determined by flow cytometry (percentage of S- or G2/M-phase cells, indicated beneath blots). Graph illustrates densitometric analysis of γTub content in the chromatin fraction (means±sd; n=6). *P < 0.05. D) Using extracts from synchronous NIH3T3 cells, γ-tubulin was immunoprecipitated with an anti-γ-tubulin antibody, developed by WB with an anti-E2F1 antibody (top), and reprobed with GCP2, RB, Dp1, and γTub (bottom). Bottom panels illustrate DNA content of the cells detected by flow cytometry (percentage S-phase cells indicated, n=5). E) Immunoprecipitation with the indicated antibodies or with a control antibody (C) was performed using U2OS cells expressing human γ-TUBULIN- or E2F1-shRNA, or control plasmids. Some control cells were treated with aphidicolin (Aphid.). Total lysate was run as loading control (bottom panel; n=4), and WBs were analyzed with the indicated antibodies. F) NIH3T3 and U2OS cells were biochemically divided into cytosolic, nuclear membrane, and chromatin fractions. Each fraction was subjected to immunoprecipitation with an anti-γ-tubulin antibody and examined by WB.
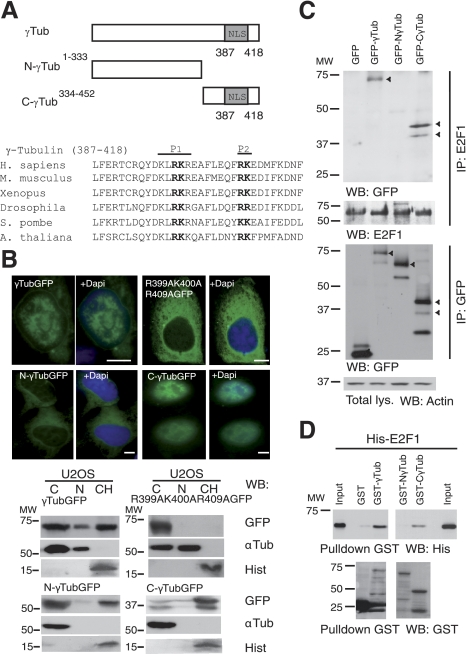
γ-Tubulin C terminus contains a nuclear localization signal that translocates γ-tubulin to the nucleus and regulates γ-tubulin association with E2F1
Determinants of γ-tubulin nuclear targeting were elucidated using 2 fragments of human γ-tubulin fused to GFP: N-γtubGFP1–333 and C-γtubGFP334–452 (Fig. 2A). Both wild-type γtubGFP and the C-γtubGFP334–452 constructs exhibited stronger nuclear localization than N-γtubGFP1–333, although a pool of N-γtubGFP1–333 was still found in the chromatin fraction (Fig. 2B). The γ-tubulin sequence included a putative nuclear localization signal (NLS) located close to the C terminus that contained two short stretches of basic amino acids, one comprising residues K397, R399, K400, and R401, and the other encompassing R409 and K410. Sequence alignments showed that the residues in the NLS are highly conserved among species (Fig. 2A). Single or double mutations of R399A, K400A, R409A, and K410A partially blocked nuclear and chromatin accumulation of γtubGFP (Fig. 2B and Supplemental Fig. S1B). The amount of γtubGFP in the chromatin decreased significantly when R399A, K400A, and R409A were mutated (Fig. 2B and Supplemental Fig. S1B), confirming the implication of both stretches in the γ-tubulin NLS. Immunoprecipitation of E2F1 from cells transiently expressing γtubGFP, N-γtubGFP1–333, or C-γtubGFP334–452 showed that E2F1 associated with γtubGFP and C-γtubGFP334–452, but not with N-γtubGFP1–333 (Fig. 2C). Furthermore, bacterially produced GST-γtub and GST-C-γtub334–452, but not GST-N-γtub1–333, formed a complex with His-E2F1 (Fig. 2D), which further strengthens that the C terminus of γ-tubulin interacts directly with E2F1.
Figure 2.
γ-Tubulin C terminus contains a nuclear localization signal. A) Top panel: structure of wild-type γ-tubulin and various γ-tubulin constructs, depicting the nuclear localization signal (NLS). Bottom panel: bold type indicates the larger (P1) and the smaller (P2) basic patch of mutated amino acids in the γ-tubulin NLS. B) Top panels: immunofluorescence analysis of U2OS cells transfected with GFP-γ-tubulin (γTubGFP), γtubGFP-R399A-K400A-R409A (R399A-K400A-R409A), N-γtubGFP1–333 (N-γTubGFP), or C-γtubGFP334–452 (C-γTubGFP). Nuclei were detected using DAPI (blue). Scale bars = 10 μm. Bottom panels: cells (0.5×106) were analyzed biochemically, as summarized in Fig. 1A, and examined by WB (n=3–6). C) U2OS cells expressing the various GFP-γ-tubulin constructs, as indicated, were separately immunoprecipitated with first anti-E2F1 (top panels), and then the same lysate was immunoprecipitated a second time with anti-GFP antibodies (bottom panels) and analyzed by WB. Part of the lysate used for the immunoprecipitations was run as loading control (total lys.). Cellulose membranes containing immunoprecipitated E2F1 were first analyzed by WB with an anti-GFP antibody and reprobed with an anti-E2F1. Arrowheads indicate the various GFP-fused proteins. D) Purified full-length GST-γ-tubulin was incubated with His-E2F1, and the GST-γ-tubulin was subsequently retrieved by adsorption to glutathione-Sepharose 4B and examined by WB (n=3). Total amounts of loaded His-E2F1 were analyzed by WB (input).
γ-Tubulin moderates E2F1, E2F2, and E2F3 transcriptional activities in a RB-binding-independent manner
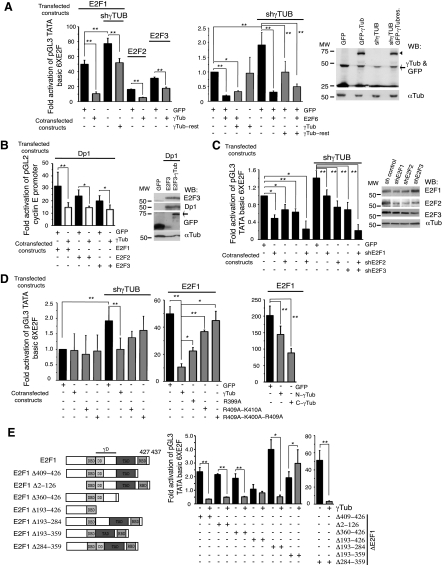
Depending on their transcriptional effects, the human transcription factor E2F family can be divided into 2 groups: transcriptional activators, E2F1, E2F2, and E2F3; and transcriptional repressors, E2F4, E2F5, E2F6, E2F7, and E2F8 (26). To analyze the effect of γ-tubulin heterodimerization on E2F transcriptional activity, we performed an assay using luciferase reporter plasmids containing E2F binding sites (12) or the cyclin E promoter (27). The luciferase activity was measured in U2OS cells transfected with E2F activators E2F1, E2F2, and E2F3 or the transcriptional repressor E2F6 (1). Also, cells were transfected with γ-tubulin and/or the E2F heterodimerization partner, Dp1 (1). The luciferase activity attributed to E2F1, E2F2, or E2F3 expression in the presence or absence of Dp1 was decreased when γ-tubulin was coexpressed with these E2Fs; however, the repressive effect of E2F6 was not reverted on coexpression with γ-tubulin (Fig. 3A, B). Furthermore, γ-TUBULIN-shRNA increased the transcriptional activity of both endogenous and recombinant E2F1, and this increase was reverted on introduction of an RNAi- resistant γ-TUBULIN gene (Fig. 3A). The transcriptional activity in control shRNA- or γ-TUBULIN-shRNA-transfected cells was decreased by shRNAi-induced reduction of the endogenous levels of E2F1, E2F2, or E2F3 (Fig. 3C). The moderating effect of γ-tubulin on E2F1 transcriptional activity was reversed when Ala399-γtubGFP, Ala409-Ala410-γtubGFP, Ala399-Ala400-Ala409-γtubGFP, or Dp1 was coexpressed with E2F1 (Fig. 3A, B, D). Expression of C-γ-tubulin334–452 remarkably reduced the transcriptional activity of E2F1, compared to the effect of N-γ-tubulin1–333 on E2F1 transcriptional activity (Fig. 3D). Considering this, we hypothesized that γ-tubulin C-terminal domain interaction with E2F moderates E2F transcriptional activities.
Figure 3.
γ-Tubulin moderates E2F1, E2F2, and E2F3 transcriptional activity. Assay of the luciferase activity driven by 6 E2F promoter binding sites (A, C, E) or cyclin E promoter (B, D) on transient transfection of U2OS cells with a Renilla reporter construct and the following constructs: GFP, γ-TUBULIN-shRNA (shγTub), HA-E2F1, HA-E2F2, HA-E2F3, HA-E2F6, HA-Dp1, GFP-γ-tubulin (γTub), RNAi-resistant γ-TUBULIN gene (γTub-rest), E2F1-shRNA (shE2F1), E2F2-shRNA (shE2F2), E2F3-shRNA (shE2F3), Ala399-γtubGFP (R399A), Ala409-Ala410-γtubGFP (R409A-K410A), Ala399-Ala400-Ala409-γtubGFP (R399A-K400A-R409A), N-γtubGFP1–333 (N-γTub), and C-γtubGFP334–452 (C-γTub), or various E2F1 mutants: HA-E2F1Δ409–426 (Δ409–426), HA-E2F1Δ2–126, HA-E2F1Δ360–426, HA-E2F1Δ193–426, Flag-E2F1Δ193–284, Flag-E2F1Δ193–359 or Flag-E2F1Δ284–359, as indicated. Luciferase activity of cells transfected with control construct was set as 1, and relative activities were calculated (means±sd, n=3–10). A–C) Total lysates of transfected U2OS cells were analyzed by WB with the indicated antibodies. Arrowheads and arrows indicate GFP-γ-tubulin and endogenous γ-tubulin, respectively. E) Structure of wild-type E2F1 and various E2F1 constructs comprising the DNA-binding (DBD), dimerization (DD), transactivating (TAD), RB-binding (RBD), and γ-tubulin-interacting (γD) domains. *P < 0.05; **P < 0.01.
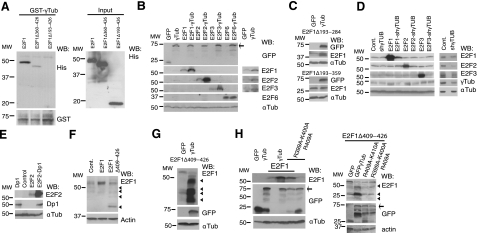
To identify the E2F1 domain important in the E2F1–γ-tubulin complex, we tested various E2F1 constructs (Fig. 3E). The effect of γ-tubulin on E2F1 transcriptional activity was not blocked by deletion of the E2F1 RB-binding domain (RBD; HA-E2F1Δ409–426), N-terminal domain (HA-E2F1Δ2–126), or transactivating domain (TAD; HA-E2F1Δ360–426). However, E2F1 transcriptional activity was reduced by deletion of the E2F1 region (HA-E2F1Δ193–426; Fig. 3E) that includes the predicted dimerization domain (DD; residues 195 to 284) (28–30). To determine whether E2F1Δ193–426 interacts with γ-tubulin in a cell-free system, we tested bacterially produced His-E2F1, His-E2F1Δ360–426, and His-E2F1Δ193–426 and GST-γtub in vitro. E2F1–γ-tubulin complex formation occurred in the presence of His-E2F1Δ360–426 but not with His-E2F1Δ193–426 (Fig. 4A), showing that residues 193 to 360 are involved in the interaction. To ascertain the involvement of the DD (28–30); we deleted residues Ser193 to Ile284, Ile284 to Leu359, or Ser193 to Leu359 in E2F1 and tested each of the constructs in a luciferase reporter assay. The luciferase activity due to Flag-E2F1Δ193–284 or Flag-E2F1Δ284–359 expressions was decreased when γ-tubulin was coexpressed with these mutants (Fig. 3E). The repressor effect of γ-tubulin on E2F1 activity was abolished when residues 193–359 where deleted. However, it is worth noting that the luciferase activity of Flag-E2F1Δ193–359 was enhanced by increased levels of γ-tubulin, which is probably caused by the presence of additional sequence on E2F1 that are necessary for γ-tubulin–E2F complex formation (Fig. 3E). Together, these data demonstrate that the C terminus of γ-tubulin interacts directly with the γ-tubulin interacting domain (γD; Ser193 to Leu359) of E2F1.
Figure 4.
γ-Tubulin affects the expression levels of E2F1, E2F2, and E2F3. A) Purified full-length GST-γ-tubulin was incubated with His-E2F1, His-E2F1Δ349–426, or His-E2F1Δ193–426, and the GST-γ-tubulin was subsequently retrieved by adsorption to glutathione-Sepharose 4B and examined by WB (n=3). Total amounts of loaded His-tagged proteins were analyzed by WB (input). B–H) Total lysates of transfected U2OS cells with the following constructs: GFP, HA-E2F1, -E2F2, -E2F3, -E2F6, -Dp1, GFP-γ-tubulin (γTub), γ-TUBULIN-shRNA (shγTub), HA-E2F1Δ409–426 (Δ409–426), Flag-E2F1Δ193–284, Flag-E2F1Δ193–359, Ala399-Ala400-Ala409-γtubGFP (R399A-K400A-R409A), γtubGFP-R409A-K410A (R409A-K410AGFP), or empty vector (Cont.) were analyzed by WB with the indicated antibodies. Arrows and arrowheads indicate GFP-fused proteins and E2F1 degradation products, respectively (n=3). D) Overexposed WBs of the endogenous expression of E2Fs are shown at right.
To rule out nonspecific transcriptional variations that could cause the observed effect of γ-tubulin on E2F activity, we studied variations in the protein levels of E2F. In the studied cells, the protein levels of endogenous E2F or ectopically expressed E2F varied, dependent on the γ-tubulin protein levels. Unexpectedly, increased protein levels of E2Fs were detected when γ-tubulin was coexpressed with E2F1, E2F1Δ193–284, E2F2, and E2F3, but not E2F6 (Fig. 4B) or E2F1Δ193–359 (Fig. 4C), suggesting that γ-tubulin protected E2F1, E2F1Δ193–284, E2F2, and E2F3 from degradation. Accordingly, γ-TUBULIN-shRNA-transfected cells showed decreased E2F1, E2F2, and E2F3 levels (Fig. 4D), but the protective effect of γ-tubulin on E2Fs was impaired in the presence of Dp1 (Fig. 3B and data not shown). Nonetheless, coexpression of Dp1 with E2Fs induced an increase in expression of E2Fs (Fig. 4E and data not shown). Interestingly, the E2F γD (residues 193–359) encloses the DD (residues 195–284), which suggests that Dp1 and γ-tubulin compete for the same E2F-interacting site and that the interaction of γ-tubulin with the DD of E2F protects E2F from degradation. These results indicate the existence of two different E2F complexes: an active complex form of Dp1–E2F and a nonactive complex composed of γ-tubulin–E2F.
Association of E2Fs with RB regulates activities and turnover of E2Fs within cells (ref. 1 and Fig. 4F); consequently, we analyzed the effects of γ-tubulin on expression of an RBD-deficient E2F1 mutant (HA-E2F1Δ409–426). Indeed, the absence of RBD did not influence the γ-tubulin-induced increase in protein levels of HA-E2F1Δ409–426; instead, HA-E2F1Δ409–426 was rapidly degraded (Fig. 4G). To ascertain that the effect on E2F levels was dependent on the C terminus of γ-tubulin, we coexpressed HA-E2F1 or HA-E2F1Δ409–426 with γ-tubulin mutants and found that the γ-tubulin mutants did not affect HA-E2F1 or HA-E2F1Δ409–426 expression (Fig. 4H). Considering these findings, we concluded that γ-tubulin's regulatory effect on E2Fs protein levels depends on the γ-tubulin C terminus in a RB-binding-independent manner.
γ-Tubulin binds to DNA on the same DNA-binding motif as E2F
To determine whether γ-tubulin, similar to Dp1 (1), binds to the E2F DNA binding site, we performed an electrophoretic mobility shift assay (EMSA; ref. 31). Surprisingly, GST-γtub and GST-C-γtub334–452, but not GST-N-γtub1–333, displayed binding to the radioactively labeled probe representing the E2F binding site, which was inhibited by an unlabeled competing wild-type E2F binding site (Fig. 5A). In addition, incubating His-E2F1 with GST-γtub increased the DNA-binding activity of GST-γtub relative to the amount of bound probe detected when the different components were incubated separately (Fig. 5B). To study γ-tubulin's ability to bind to E2F-regulated promoters, a ChIP assay using γ-tubulin antibodies was performed (24). Endogenous γ-tubulin was present on the cyclin E promoter in aphidicolin-treated U2OS cells, and sequential ChIP assay (E2F ChIP followed by γ-tubulin re-ChIP or vice versa) showed that endogenous E2F1 and E2F2, but not Dp1, colocalized with endogenous γ-tubulin on cyclin E promoter (Fig. 5C). These results indicated that the C terminus of γ-tubulin may interact directly with the γD of E2Fs and thereby negatively modulate E2F transcriptional activity. All the above-mentioned experiments suggested that γ-tubulin and Dp1 compete for the same binding site on E2Fs. To confirm the hypothesis, we analyzed the Dp1 content on E2F1 immunoprecipitates from cells expressing variable protein levels of γ-tubulin (Fig. 5D). We detected that lower γ-tubulin levels were associated with higher levels of Dp1 in E2F1 immunoprecipitates (Fig. 5E). This result revealed that the mechanism by which γ-tubulin moderates E2F activity is by competing out Dp1 in the E2F–Dp1 complex.
Figure 5.
E2F–γ-tubulin complex binds to cyclin E promoter. A, B) DNA-binding activity of GST-γ-tubulin or γ-tubulin fragments in the presence or absence of His-E2F1 was assessed by an electrophoretic mobility assay. A 100-fold excess of unlabeled competing probe containing a wild-type (Wt) or a mutant (M) E2F site was added to the assay as indicated. C) Aphidicolin-treated U2OS cells were analyzed by single or sequential ChIP using E2F or γ-tubulin antibodies. PCR primers amplified the cyclin E promoter region from −76 to 86 bp. D, E) U2OS cells expressing GFP, γ-TUBULIN-shRNA, or GFP-γ-tubulin were lysed; total lysates were analyzed by WB (total lys.; D) or were used for immunoprecipitation with anti-E2F1 antibody or with a control (C) antibody (E, top panel). Part of the total lysates used for immunoprecipitations were run in a gel as loading control (IP Total lys.; E, bottom panel), and WBs were analyzed with the indicated antibodies. Arrowheads and arrows indicate GFP-γ-tubulin and endogenous γ-tubulin, respectively (n=3).
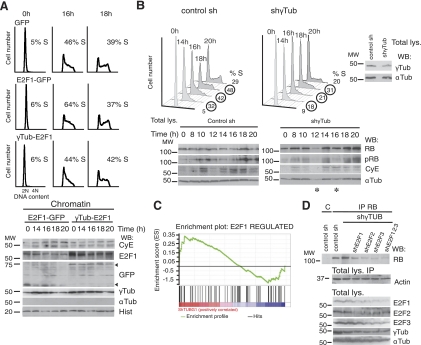
γ-Tubulin levels affect cell cycle progression and the expression of E2F-regulated genes
To further confirm the effect of γ-tubulin on E2Fs, we altered γ-tubulin levels in cells expressing HA-E2F1 in a synchronized cell population. Increased γ-tubulin levels impaired the S-phase entry effect caused by HA-E2F1 by negatively regulating the expression of E2F transcriptional targets, such as cyclin E expression (Fig. 6A). Moreover, alterations of the γ-tubulin levels during cell cycle in U2OS cells expressing GFP, γtubGFP, γTUBULIN-shRNA, and NLS-γtubGFP mutants showed that a larger number of control cells expressing GFP or γtubGFP were in S phase compared to the cells with various γtubGFP mutants (Supplemental Fig. S2A). In a synchronized cell population, S-phase progression was delayed in NIH3T3 cells transfected with γTubulin-shRNA or any of the NLS-γtubGFP mutants, suggesting that γ-tubulin levels and localization determine optimal cell cycle progression (Fig. 6B and Supplemental Fig. S2B).
Figure 6.
Variations in γ-tubulin levels affect cell cycle progression. A) Cell cycle distribution of synchronous control (GFP)-, E2F1 and GFP (E2F1-GFP)-, or E2F1 and γtubGFP (γTub-E2F1)-transfected NIH3T3 cells. Cells were synchronized in G0 and released for different time periods, and DNA content of the samples was determined by flow cytometry. Percentage of S-phase cells is indicated in each panel (n=4). Extracts from synchronous NIH3T3 cells transfected with E2F1 and GFP or E2F1 and γtubGFP were prepared as in Fig. 1A and examined by WB using the indicated antibodies (n=4). Arrowheads indicate various GFP-fused proteins. B) Cell cycle distribution of synchronous control shRNA- or γ-Tubulin-shRNA-transfected NIH3T3 cells. Cells were synchronized in G0 as in A and released for different time periods, and DNA content of the samples was determined by flow cytometry. Percentage of S-phase cells is indicated in each panel (n=3). Synchronous control shRNA- or γ-Tubulin-shRNA-transfected NIH3T3 cells were analyzed using the indicated antibodies (n=3). Total lysate was run in parallel as control for γ-tubulin levels and as loading control (indicated). Asterisks indicate lanes with variation in protein loading (n=3). C) GSEA of E2F1-upregulated gene set performed on a ranked gene list of differentially expressed genes between γ-TUBULIN-shRNA-transfected (n=4) and control-shRNA-transfected (n=5) cells. Genes were ranked in descending order with regard to level of expression in γ-TUBULIN-shRNA-transfected cells. (enrichment score 0.34, nominal P=0.026). D) U2OS cells expressing indicated constructs were used for immunoprecipitation of RB with anti-RB or control (C) antibody. Total lysate was run as loading control (bottom panels; n=3).
Intriguingly, low expression or mutations of γ-tubulin elevated E2F activity but delayed S-phase entry. To further elucidate the effect of reduced γ-tubulin levels on E2F activity, an mRNA expression array was performed on control shRNA- and γ-TUBULIN-shRNA-transfected cells, and the effect of γ-TUBULIN reduction on an E2F1 target gene signature was examined (Table 1). GSEA revealed that on reduced mRNA expression levels of γ-TUBULIN, a significant enrichment of E2F1 positively regulated target genes was found (enrichment score 0.34, nominal P=0.026; Fig. 6C). It can also be noted that RB lay among the most positively enriched E2F1 targets (Table 1). In accordance with this, we found increased protein levels of RB in γTUBULIN-shRNA- and γTubulin-shRNA-transfected cells (Fig. 6B, D), which provides a potential explanation for the observed delay in S-phase entry (Fig. 6B). These findings further support the moderating effect of γ-tubulin on E2F activities, as the increase in RB levels concurred with a decrease in levels of phosphorylated RB.
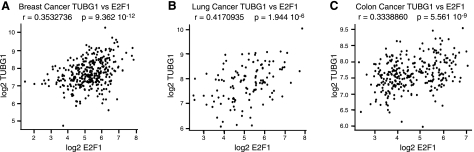
High expression of γ-tubulin correlates with high expression of E2F1 and cell cycle-associated gene signatures in various tumor types
To identify underlying biological processes associated with γ-tubulin expression in various tumor types, we made use of publically available expression array datasets, including 351 breast cancers, 121 lung cancers (NCBI GEO accession number GSE2109), and 290 colon cancers (NCBI GEO GSE14333; ref. 32). For each tumor type, all genes were ranked according to correlation to γ-tubulin expression. As expected from our results, γ-tubulin expression was significantly positively correlated with E2F1 expression in all three-tumor types (Fig. 7). In addition, GO analysis was performed using the top 100 positively correlated genes to γ-tubulin expression. In each tumor type, γ-tubulin-correlated gene expression displayed highly significant enrichment of cell cycle-related processes (Supplemental Tables S1 and S2). The fact that high levels of γ-tubulin coincide with high levels of cell cycle-regulated genes supports the hypothesis of γ-tubulin acting as a cell cycle regulator and also suggests that γ-tubulin may be involved in cell cycle regulation in multiple malignancies.
Figure 7.
γ-Tubulin expression in tumor material displays a positive correlation to E2F1 expression. Scatter plots of log2 γ-tubulin expression vs. log2 E2F1 expression in breast tumors (A; n=351, r=0.353, 95% CI 0.258–0.442), lung tumors (B; n=121, r=0.417, 95% CI 0.258–0.554), and colon tumors (C; n=290, r=0.334, 95% CI 0.227–0.432); r represents the Pearson product moment correlation coefficient between γ-tubulin and E2F1 expression, with respective P values.
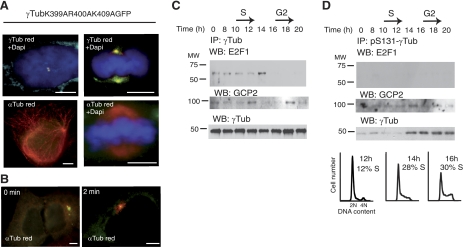
NLS-γ-tubulin mutants do not affect centrosome or αβ-tubulin dynamics and E2F1 does not associate with γ-tubulin phosphorylated on Ser131
To establish that the observed effect of γtubGFP mutants on E2F activities and cell cycle was not caused by defective centrosomes or altered αβ-tubulin dynamics, we examined the effect of γtubGFP-R399A-K400A-R409A on centrosome and αβ-tubulin regulation. Ectopic expression of γtubGFP-R399A-K400A-K409A in U2OS cells did not influence the number of centrosomes or mitotic spindles formed or altered interphase microtubules or astral microtubule regrowth (Fig. 8A, B). This finding suggests that the effects of γtubGFP mutants on cell cycle progression and E2F activity are not due to defective centrosomes or altered αβ-tubulin dynamics.
Figure 8.
γ-Tubulin-GFP-R399A-K400A-R409A does not alter centrosome replication or microtubule nucleation. A, B) Effect of γtubGFP-R399A-K400A-K409A on centrosome function was examined by immunofluorescence analysis of transfected U2OS cells. A) Tubulin in transfected cells was stained using anti-γ-tubulin (γTub; red) or anti-α-tubulin antibody (red), as indicated; nuclei were detected with DAPI (blue). B) α-Tubulin (red) in U2OS cells stained immediately following cold treatment (0 min) or 2 min after adding warm medium. Scale bars = 10 μm. C, D) γ-Tubulin phosphorylated on Ser131 does not associate with E2F1. Synchronous NIH3T3 cells (2.106/time point) were lysed and divided into 2 samples. C) In one sample, γ-tubulin was immunoprecipitated from extracts using γTub, then detected by Western blotting (WB) using anti-E2F1 antibody, and finally stripped and reprobed with γTub (bottom panel), as described in Fig. 1A. D) In the second sample, endogenous pSer131-γ-tubulin was immunoprecipitated with anti-pSer131-γ-tubulin antibody, subsequently developed by WB using anti-E2F1 antibody, and then stripped and reprobed with γTub (bottom blot panel). Bottom panels illustrate DNA content of the cells detected by flow cytometry (percentage of S-phase cells indicated, n=3).
We have previously shown that γ-tubulin is phosphorylated on Ser131 (pSer131), and pSer131 enhances microtubule polymerization in the centriole (3). Thus, in search for the mechanism that regulates nuclear translocation of γ-tubulin, we examined the amount of E2F1 associated with immunoprecipitates of pSer131–γ-tubulin from synchronized NIH3T3 cells (Fig. 8C, D). E2F1 interacted with total γ-tubulin immunoprecipitates in a cell-cycle-dependent manner (Fig. 8C) but did not associate with endogenous pSer131–γ-tubulin immunoprecipitates (Fig. 8D), implying the existence of an alternative mechanism that translocates γ-tubulin to the nucleus.
DISCUSSION
We describe an NLS that mediates γ-tubulin translocation to the nucleus during S phase (6–8). In the nucleus, γ-tubulin governs E2F1, E2F2, and E2F3 transcriptional activities. We found that decreased γ-tubulin levels or expression of NLS–γ-tubulin mutants increase E2F transcriptional activity, while elevated γ-tubulin levels reduce E2F activity. Furthermore, the protein levels of recombinant E2F1, E2F2, and E2F3 vary depending on the γ-tubulin and Dp1 expression. Low E2F protein levels were found in cells with reduced γ-tubulin levels, despite the high measured E2F transcriptional activities. Increased expression of γ-tubulin caused high protein levels of E2F, but the transcriptional activity of E2F was low. Also, we found that high expression levels of Dp1 increased the total levels of E2Fs; considering that low protein levels of γ-tubulin decreased E2Fs expression, this finding suggest that the transient increase of E2Fs during G1 and S phase could be caused by the dynamic formation of the Dp1- and γ-tubulin–E2F complexes.
γTUBULIN-shRNA induced an increase in E2F transcriptional activity and caused an E2F-dependent increase in the protein levels of RB. The RB signal transduction pathways can trigger a G1 arrest (33, 34); consequently, an increase in RB may be responsible for causing the observed S-phase entry delay in cells with reduced γ-tubulin protein levels. In G1/S entry, γ-tubulin binds to cyclin E promoter to ensure a transient expression of cyclin E, and, in doing so, γ-tubulin coordinates the cell cycle. In addition, the chromatin-associated pool of γ-tubulin was not bound to microtubules or GCP2-containing fractions, suggesting that γ-tubulin has centrosome-independent functions. Altogether, these observations indicate that γ-tubulin plays a role in the regulation of E2F transcriptional activity during the cell cycle.
Our finding of an E2F-mediated increase of RB in response to reduced γ-tubulin levels implicates γ-tubulin as a regulator in the G1/S phase transition. In line with this view, the presence or absence of centrosomes promotes either proliferation or a G1 arrest (35). The mechanism by which centrosomes regulate the G1/S transition is unclear, but it remains possible that the triggering signal that translocates γ-tubulin to the nucleus may originate at the centrosomes. Once centrosome duplication is initiated and cyclin E activity is no longer needed, the translocation of γ-tubulin to the nucleus will inhibit the transcription of cyclin E, and in doing so, will prevent a cyclin E-dependent reduplication of the centrosomes (36). In this way, a timely regulation of cell cycle progression can be achieved.
Alteration in the RB-E2F signal transduction pathways are connected to cancer development in various tumors types, as uncontrolled E2F signaling may trigger cell division (1). Consequently, in these tumors, uncontrolled cell division coincides with expression of cell cycle-regulated genes, which should also include γ-tubulin. To test this possibility, we examined γ-tubulin-correlated gene expression in tumor material and found that γ-tubulin displayed significant positive correlation to E2F1 expression, as well as significant enrichment of cell cycle-associated processes on GO analysis. These trends did not seem to be tumor type specific, as similar patterns were observed in multiple tumor types, including breast, lung, and colon cancers. This may suggest a role for γ-tubulin in cell cycle regulation in not only normal cells but also in malignancies, however further investigations are required.
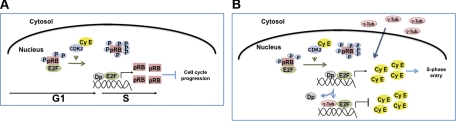
In summary, the mechanism that allows γ-tubulin to moderate E2F transcriptional activity involves direct binding to E2Fs. This, in turn, moderates E2F transcriptional activity during S-phase entry. In the absence of γ-tubulin, increased E2F activity elevates E2F-mediated expression of RB and delays S-phase entry (Fig. 9A). At the G1/S phase transition, RB releases E2Fs, which may induce E2F–γ-tubulin complex formation, ensuring a transient transcription of genes necessary for S-phase entry (Fig. 9B).
Figure 9.
γ-Tubulin protein levels regulate E2F transcriptional activity. A) In the absence of γ-tubulin, at G1/S phase transition, the released E2F-Dp1 complex transcribes RB. B) In the presence of γ-tubulin, at G1/S transition, the released E2F-Dp1 complex transcribes gene products necessary for S-phase entry, such as cyclin E. As nuclear γ-tubulin levels increase, the E2F–γ-tubulin complex forms and puts an end to the transcriptional activity of E2Fs.
Supplementary Material
Acknowledgments
The authors thank Jiri Bartek (Institute of Cancer Biology, Danish Cancer Society, Yang Shi (Harvard Medical School, Boston, MA, USA), Kristian Helin (Biotech Research and Innovation Centre, University of Copenhagen, Copenhagen, Denmark), Marc van de Wetering (Hubrecht Laboratory, Centre for Biomedical Genetics, Utrecht, The Netherlands), Joseph R. Nevins (Duke University, Durham, NC, USA), Robert A. Weinberg (Massachusetts Institute of Technology, Cambridge, MA, USA), and David M. Livingston (Dana-Farber Cancer Institute, Boston, MA, USA) for reagents, and P. Ödman for editorial assistance.
This work was supported by grants from the Royal Physiographic Society (Lund, Sweden); the Åke Wiberg, Thelma Zoegas, Per-Eric and Ulla Schyberg, O. E. and Edla Johansson, Gradfordska, H. and G. Jeassons, and O. and E. Ericsson Foundations; Gyllenstiernska Krapperupsstiftelsen; Universitetssjukhuset Malmö Allmäna Sjukhus (U-MAS) and the U-MAS Cancer Research Fund; and the Swedish Research Council. A fellowship from the Swedish Society for Medical Research provided additional support. The authors declare no conflicts of interest.
Footnotes
This article includes supplemental data. Please visit http://www.fasebj.org to obtain this information.
REFERENCES
- 1. Trimarchi J. M., Lees J. A. (2002) Sibling rivalry in the E2F family. Nat. Rev. Mol. Cell. Biol. 3, 11–20 [DOI] [PubMed] [Google Scholar]
- 2. Schiebel E. (2000) gamma-Tubulin complexes: binding to the centrosome, regulation and microtubule nucleation. Curr. Opin. Cell Biol. 12, 113–118 [DOI] [PubMed] [Google Scholar]
- 3. Alvarado-Kristensson M., Rodriguez M. J., Silio V., Valpuesta J. M., Carrera A. C. (2009) SADB phosphorylation of gamma-tubulin regulates centrosome duplication. Nat. Cell Biol. 11, 1081–1092 [DOI] [PubMed] [Google Scholar]
- 4. Muller H., Fogeron M. L., Lehmann V., Lehrach H., Lange B. M. (2006) A centrosome-independent role for gamma-TuRC proteins in the spindle assembly checkpoint. Science 314, 654–657 [DOI] [PubMed] [Google Scholar]
- 5. Moudjou M., Bordes N., Paintrand M., Bornens M. (1996) gamma-Tubulin in mammalian cells: the centrosomal and the cytosolic forms. J. Cell Sci. 109(Pt. 4), 875–887 [DOI] [PubMed] [Google Scholar]
- 6. Lesca C., Germanier M., Raynaud-Messina B., Pichereaux C., Etievant C., Emond S., Burlet-Schiltz O., Monsarrat B., Wright M., Defais M. (2005) DNA damage induce gamma-tubulin-RAD51 nuclear complexes in mammalian cells. Oncogene 24, 5165–5172 [DOI] [PubMed] [Google Scholar]
- 7. Korver W., Guevara C., Chen Y., Neuteboom S., Bookstein R., Tavtigian S., Lees E. (2003) The product of the candidate prostate cancer susceptibility gene ELAC2 interacts with the gamma-tubulin complex. Int. J. Cancer 104, 283–288 [DOI] [PubMed] [Google Scholar]
- 8. Andersen J. S., Lyon C. E., Fox A. H., Leung A. K., Lam Y. W., Steen H., Mann M., Lamond A. I. (2002) Directed proteomic analysis of the human nucleolus. Curr. Biol. 12, 1–11 [DOI] [PubMed] [Google Scholar]
- 9. Mendez J., Stillman B. (2000) Chromatin association of human origin recognition complex, cdc6, and minichromosome maintenance proteins during the cell cycle: assembly of prereplication complexes in late mitosis. Mol. Cell. Biol. 20, 8602–8612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kramer A., Mailand N., Lukas C., Syljuasen R. G., Wilkinson C. J., Nigg E. A., Bartek J., Lukas J. (2004) Centrosome-associated Chk1 prevents premature activation of cyclin-B-Cdk1 kinase. Nat. Cell Biol. 6, 884–891 [DOI] [PubMed] [Google Scholar]
- 11. Black E. P., Hallstrom T., Dressman H. K., West M., Nevins J. R. (2005) Distinctions in the specificity of E2F function revealed by gene expression signatures. Proc. Natl. Acad. Sci. U. S. A. 102, 15948–15953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Muller H., Bracken A. P., Vernell R., Moroni M. C., Christians F., Grassilli E., Prosperini E., Vigo E., Oliner J. D., Helin K. (2001) E2Fs regulate the expression of genes involved in differentiation, development, proliferation, and apoptosis. Genes Dev. 15, 267–285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lees J. A., Saito M., Vidal M., Valentine M., Look T., Harlow E., Dyson N., Helin K. (1993) The retinoblastoma protein binds to a family of E2F transcription factors. Mol. Cell. Biol. 13, 7813–7825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gaubatz S., Wood J. G., Livingston D. M. (1998) Unusual proliferation arrest and transcriptional control properties of a newly discovered E2F family member, E2F-6. Proc. Natl. Acad. Sci. U. S. A. 95, 9190–9195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Liu K., Luo Y., Lin F. T., Lin W. C. (2004) TopBP1 recruits Brg1/Brm to repress E2F1-induced apoptosis, a novel pRb-independent and E2F1-specific control for cell survival. Genes Dev. 18, 673–686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Alvarado-Kristensson M., Melander F., Leandersson K., Ronnstrand L., Wernstedt C., Andersson T. (2004) p38-MAPK signals survival by phosphorylation of caspase-8 and caspase-3 in human neutrophils. J. Exp. Med. 199, 449–458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Alvarado-Kristensson M., Porn-Ares M. I., Grethe S., Smith D., Zheng L., Andersson T. (2002) p38 Mitogen-activated protein kinase and phosphatidylinositol 3-kinase activities have opposite effects on human neutrophil apoptosis. FASEB J. 16, 129–131 [DOI] [PubMed] [Google Scholar]
- 18. Hiebert S. W., Chellappan S. P., Horowitz J. M., Nevins J. R. (1992) The interaction of RB with E2F coincides with an inhibition of the transcriptional activity of E2F. Genes Dev. 6, 177–185 [DOI] [PubMed] [Google Scholar]
- 19. Subramanian A., Tamayo P., Mootha V. K., Mukherjee S., Ebert B. L., Gillette M. A., Paulovich A., Pomeroy S. L., Golub T. R., Lander E. S., Mesirov J. P. (2005) Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U. S. A. 102, 15545–15550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Jamshidi-Parsian A., Dong Y., Zheng X., Zhou H. S., Zacharias W., McMasters K. M. (2005) Gene expression profiling of E2F-1-induced apoptosis. Gene 344, 67–77 [DOI] [PubMed] [Google Scholar]
- 21. Dennis G., Jr., Sherman B. T., Hosack D. A., Yang J., Gao W., Lane H. C., Lempicki R. A. (2003) DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 4, P3 [PubMed] [Google Scholar]
- 22. Huang da W., Sherman B. T., Lempicki R. A. (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57 [DOI] [PubMed] [Google Scholar]
- 23. Qin X. Q., Chittenden T., Livingston D. M., Kaelin W. G., Jr. (1992) Identification of a growth suppression domain within the retinoblastoma gene product. Genes Dev. 6, 953–964 [DOI] [PubMed] [Google Scholar]
- 24. Weinmann A. S., Farnham P. J. (2002) Identification of unknown target genes of human transcription factors using chromatin immunoprecipitation. Methods 26, 37–47 [DOI] [PubMed] [Google Scholar]
- 25. Hosack D. A., Dennis G., Jr., Sherman B. T., Lane H. C., Lempicki R. A. (2003) Identifying biological themes within lists of genes with EASE[b]. Genome Biol. 4, R70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wan Z., Zhi N., Wong S., Keyvanfar K., Liu D., Raghavachari N., Munson P. J., Su S., Malide D., Kajigaya S., Young N. S. (2010) Human parvovirus B19 causes cell cycle arrest of human erythroid progenitors via deregulation of the E2F family of transcription factors. J. Clin. Invest. 120, 3530–3544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Geng Y., Eaton E. N., Picon M., Roberts J. M., Lundberg A. S., Gifford A., Sardet C., Weinberg R. A. (1996) Regulation of cyclin E transcription by E2Fs and retinoblastoma protein. Oncogene 12, 1173–1180 [PubMed] [Google Scholar]
- 28. Q01094 (E2F1_HUMAN) UniProt Protein Knowledgebase. Retrieved December 2, 2010, from http://www.uniprot.org/uniprot/Q01094
- 29. Helin K., Wu C. L., Fattaey A. R., Lees J. A., Dynlacht B. D., Ngwu C., Harlow E. (1993) Heterodimerization of the transcription factors E2F-1 and DP-1 leads to cooperative trans-activation. Genes Dev. 7, 1850–1861 [DOI] [PubMed] [Google Scholar]
- 30. Vidal M., Braun P., Chen E., Boeke J. D., Harlow E. (1996) Genetic characterization of a mammalian protein-protein interaction domain by using a yeast reverse two-hybrid system. Proc. Natl. Acad. Sci. U. S. A. 93, 10321–10326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Mudryj M., Hiebert S. W., Nevins J. R. (1990) A role for the adenovirus inducible E2F transcription factor in a proliferation dependent signal transduction pathway. EMBO J. 9, 2179–2184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Jorissen R. N., Gibbs P., Christie M., Prakash S., Lipton L., Desai J., Kerr D., Aaltonen L. A., Arango D., Kruhoffer M., Orntoft T. F., Andersen C. L., Gruidl M., Kamath V. P., Eschrich S., Yeatman T. J., Sieber O. M. (2009) Metastasis-associated gene expression changes predict poor outcomes in patients with Dukes stage B and C colorectal cancer. Clin. Cancer Res. 15, 7642–7651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sancar A., Lindsey-Boltz L. A., Unsal-Kacmaz K., Linn S. (2004) Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annu. Rev. Biochem. 73, 39–85 [DOI] [PubMed] [Google Scholar]
- 34. Giacinti C., Giordano A. (2006) RB and cell cycle progression. Oncogene 25, 5220–5227 [DOI] [PubMed] [Google Scholar]
- 35. Doxsey S., McCollum D., Theurkauf W. (2005) Centrosomes in cellular regulation. Annu. Rev. Cell Dev. Biol. 21, 411–434 [DOI] [PubMed] [Google Scholar]
- 36. Carrera A. C., Alvarado-Kristensson M. (2009) SADB kinases license centrosome replication. Cell. Cycle 8, 4005–4006 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.