Abstract
Antibody-based microarrays are a rapidly evolving affinity-proteomic methodology that recently has shown great promise in clinical applications. The resolution of these proteomic analyses is, however, directly related to the number of data-points, i.e. antibodies, included on the array. Currently, this is a key bottleneck because of limited availability of numerous highly characterized antibodies. Here, we present a conceptually new method, denoted global proteome survey, opening up the possibility to probe any proteome in a species-independent manner while still using a limited set of antibodies. We use context-independent-motif-specific antibodies directed against short amino acid motifs, where each motif is present in up to a few hundred different proteins. First, the digested proteome is exposed to these antibodies, whereby motif-containing peptides are enriched, which then are detected and identified by mass spectrometry. In this study, we profiled extracts from human colon tissue, yeast cells lysate, and mouse liver tissue to demonstrate proof-of-concept.
Delineating the composition of the human proteome(s) will be instrumental in advancing our fundamental knowledge about the underlying biology in health and disease (1–3). To generate detailed protein expression profiles, or atlases of complex proteomes, the development of rapid and highly multiplexed proteomic research tools will be crucial. For in-depth characterization of biological systems, classical separation techniques, such as liquid chromatography and two-dimensional gels, linked with single or tandem MS has been established as a powerful hypothesis-generating tool (2, 4, 5). Despite the fact that MS-based set-ups have constituted the cornerstone in many proteomic profiling endeavors, the impact of this work in terms of, for example, defining disease-associated biomarkers, have so far been limited, mainly because of technical shortcomings (5, 6).
To meet methodological issues, such as sensitivity, dynamic range, resolution, and reproducibility, affinity-proteomic based on antibody microarrays has in recent years become an established proteomic technology for differential protein expression profiling of complex proteomes (7–9). To date, the technology has been applied in several clinical applications, outlining the potential for example in disease diagnostics and patient stratification (10–12), setting a new standard for array-based proteomics (7, 13, 14). The resolution of an antibody microarray data set correlate directly to the number of antibodies included on the array and the range of their specificities, which tends to be a key bottleneck (13, 15). Even though current antibody microarray formats have been shown to be capable of discriminating among various proteomes (10–12, 16), an improved analytical resolution will be crucial for pin-pointing candidate biomarkers that can predict, for example, disease progression and response to treatment, with high specificity and sensitivity (7).
To advance further and set a novel standard for proteomics, the most attractive features of affinity proteomics could be combined with those of MS-based proteomics. This has previously been attempted, where antibodies have initially been used to enrich specific proteins (17–20) or peptides (21–24) followed by subsequent MS analysis. However, all of these platforms mainly rely on the conventional approach of using one binder per unique peptide and protein, creating major logistical issues when scaling up for global profiling efforts. In addition, the use of polyclonal antibodies as capturing agents (21) has inherent disadvantages because they are derived from a nonrenewable resource.
Here, we describe a conceptually novel method, denoted global proteome survey (GPS)1, with inherent capability of probing any proteome in a discovery mode, in a species independent manner, while still using a limited number of antibodies. As the affinity reagent, we used renewable human recombinant scFv antibodies specific for short amino acid sequences or motifs, each motif being present in up to a few hundred proteins. Next, the biological sample is digested, exposed to these antibodies, whereby motif-containing peptides are specifically captured, enriched, and subsequently detected and identified (and quantified) using single- or tandem-MS. In this manner, 100 of these context independent motif specific (CIMS) antibodies, would theoretically cover almost 50% of the nonredundant human proteome (25), a concept supported by a recent in-silico study of the human proteome (26).
EXPERIMENTAL PROCEDURES
Design of Selection Peptide Motifs
We defined the tandem MS (MS/MS) addressable cohort of the human proteome, release 56.1 of UniProtKB/Swiss-Prot, after in silico trypsination, setting a molecular weight cutoff in the range of 500 to 3500 Da using SignPept, an in-house developed software. Using criteria defined to exclude cysteines and methionines, but tailored to contain a C-terminal arginine or lysine, we designed 27 four or six amino acids long (27, 28) peptide selection motifs to be present in a few to several hundred (<700) of these human peptides using MS-pattern and/or SignPept (Dr. F. Levander, Department. of Immunotechnology, Lund University, Sweden). The theoretical cleavage specificities of trypsin have been built into the SignPept software, and mimics the rules employed by PeptideCutter (http://expasy.org/tools/peptidecutter). Further, the effect of motif wobbling on the frequency of motif occurrence was investigated using SignPept. The synthesized peptides were composed of an N-terminal biotin, a SGSG-linker, followed by the C-terminal located selection motif (Innovagen, Lund, Sweden).
Selection of CIMS antibodies
Human recombinant scFv antibodies were selected from the phage display library, n-CoDeR (29). Three consecutive rounds of selection were performed, using biotinylated peptide motifs as antigens. In selection round one, about 1013 colony-forming units of phage were mixed with 50 nm antigen in a total volume of 3 ml. The selection buffer was phosphate-buffered saline (PBS) containing 3% (w/v) bovine serum albumin (BSA), 0.05% (v/v) Tween-20, and 0.02% (w/v) sodium azide. The antigen and phage mixture was incubated for ∼16 h at room temperature. Biotinylated peptides were captured on ∼108 streptavidin-conjugated magnetic beads (Dynabeads M-280, Dynal, Oslo, Norway) during a 30 min incubation. Before use, Dynabeads were blocked with 5% (w/v) BSA in selection buffer. Following peptide capture, beads were washed a total of nine times, using a Magnetic Particle Concentrator (Dynal, Oslo, Norway), three times with selection buffer, three times with PBS containing 0.05% (v/v) Tween-20 and three times with PBS. Captured phages were then eluted by addition of 400 μl of a 1 mg/ml trypsin solution for 30 min, after which trypsin was inactivated by addition of 40 μl of a 2 mg/ml aprotinin solution. All incubations were performed with gentle end-over-end rotation. Log phase Escherichia coli was infected with the eluted phage pool and a new, amplified phage pool was produced essentially as described by Engberg et al. (30), using E. coli strain HB101F′ (constructed from HB101, Invitrogen, Carlsbad, CA) and 20-fold excess of helper phage R408 (Stratagene, La Jolla, CA).
In selection round two, about 1011 colony-forming units of amplified phage were mixed with 20 nm antigen in a total volume of 1 ml and ∼3 × 107 streptavidin-conjugated magnetic beads were used to capture biotinylated peptide motifs. Bound phage were eluted by addition of 400 μl of 10 mm glycin-HCl, pH 2.2 for 30 min. A few μl of 1 m Tris-HCl, pH 9.0, was then added to neutralize the acid. The eluted phage pool was not amplified, but used directly in the third selection round. Thus, in selection round three, peptides were preloaded on avidin-coated wells of a microtiter plate, with 8 wells each coated with 0.5 μg avidin and loaded with 10 pmol peptide. Wells were then blocked with 5% (w/v) BSA in selection buffer. About 106 eluted phages from round two were diluted to 800 μl in selection buffer and then added to peptide-loaded wells, 100 μl per well. The plate was incubated for ∼16 h at room temperature with gentle agitation. Wells were washed three times with selection buffer, three times with PBS containing 0.05% (v/v) Tween-20, and three times with PBS. Captured phages were eluted using trypsin, 100 μl per well, as described above.
To counteract selection of irrelevant (nonspecific) phages, each selection round was stringently preceded by a preselection, designed to eliminate phage clones of certain antigen specificities. The starting phage stocks of selection rounds one and two were preselected against irrelevant biotinylated peptide motifs followed by capture on streptavidin-conjugated magnetic beads. The phage stock used in round three was preselected against avidin coated on a microtiter plate. Enrichment of irrelevant phages was also counteracted by addition of irrelevant nonbiotinylated peptide motifs as competitors to the phage and antigen mixture.
Screening of CIMS Antibodies
A log phase culture of E. coli strain HB101F′ was infected with the eluted phage pool from selection round three and phagemid DNA was amplified and isolated essentially as described before (30). Phage-specific DNA was eliminated by EagI digestion and re-ligated material was transformed into chemically competent E. coli strain TOP10 (Invitrogen). Individual colonies were obtained by spreading transformed bacteria on 22 × 22 cm LA plates containing 1% (w/v) glucose and 100 μg/ml ampicillin. Each colony carries a plasmid encoding a specific scFv clone with two general C-terminal affinity tags, one c-Myc tag and one 6xHis tag. Colony picking, expression of soluble scFv, and a primary ELISA screening were performed in 384-well format, using an integrated robotic workstations (31). First, colonies were transferred from LA plates to master plates containing Luria broth, 1% (w/v) glucose, and 100 μg/ml ampicillin. Master plates were then replicated to expression plates containing the medium from above, but without glucose. Expression of scFv was induced during bacterial growth by addition of isopropyl β-d-thiogalactoside (IPTG) to a concentration of 0.4 mm. Finally, expression supernatants were screened by ELISA against biotinylated peptides loaded on streptavidin. Detection was performed using an horseradish peroxidase (HRP)-conjugated anti-His antibody (R&D Systems, Minneapolis, MN) and the SuperSignal ELISA Pico Chemiluminescent substrate (Pierce, Rockford, IL).
Bacterial clones, identified as “actives” in the primary ELISA screening, were cherry picked into 96-well micro-titer plate-format, the scFv were re-expressed and clones retested on the ELISA system (31). The scFv-encoding gene of confirmed actives was sequenced to identify unique hit clones (31).
Production of CIMS Antibodies
All scFv antibodies were produced in 100 ml E. coli cultures and purified using affinity chromatography on nickel-nitrilotriacetic acid (Ni2+-NTA) agarose (Qiagen, Hilden, Germany). Bound molecules were eluted with 250 mm imidazole, dialyzed against PBS (pH 7.4) for 72 h and then stored at +4 °C until further use. The protein concentration was determined by measuring the absorbance at 280 nm using a Nanodrop-1000. The integrity and purity of the scFv antibodies was evaluated by running Protein 80 chips on Agilent Bioanalyzer (Agilent, Waldbronn, Germany). Because of logistical liquid chromatography tandem MS (LC-MS/MS) limitations, 14 of 91 selected scFv antibodies, directed against eight motifs, were first selected for protein microarray characterization. Seven of these 14 antibodies were then included in the subsequent LC-MS/MS analysis.
Affinity Measurements of CIMS-binders
A competitive inhibition affinity assay was designed, based on fluorescent labeled synthetic peptides, CIMS antibody functionalized magnetic beads and the KingFisher Flex system (Thermo Fisher Scientific), an automated magnetic particle processing. Measurements were performed for the CIMS-1-A05, CIMS-1-B03, CIMS-15-A06, CIMS-17-E02, CIMS-32–3A-G03, CIMS-33–3D-F06, and CIMS-34–3A-D10. Briefly, the antibody conjugated beads were prewashed with 0.03% (w/v) 3-[(3-cholamidopropyl)dimethylammonio]propanesulfonate (CHAPS) in PBS and then transferred into a 96-well plate prefilled with 3 μl of a CIMS-scFv bead solution. A plate containing 100 nm (CIMS-17-E02) or 200 nm (all other CIMS antibodies) fluorescent N-terminally labeled (tetramethylrhodamine) synthetic selection peptide (80% purity) (Thermo Biopolymers, Germany) and various amounts of an identical non-tetramethylrhodamine labeled synthetic peptide (Thermo Biopolymers) was prepared. Eight different concentrations were used and the range of the nonlabeled competitive peptide was, for the majority of the measurements, between 0 and 5000 nm. The plates were incubated for 1 h, with mixing for 40 s (medium speed) every 3.5 min. The magnetic beads were then transferred to the elution plate containing 45 μl of 5% acetic acid and incubated for 2 min. Finally, the eluates and the incubation solutions were transferred into 384-well black, low-volume plates (Corning, Corning, NY) and the fluorescent intensities were determined using the FLUOstar Omega microplate reader (BMG Labtech, Durham, NC). The values were corrected for any background signal intensity using 0.03% (w/v) CHAPS in PBS as cut-off value. In addition, a control, un-conjugated magnetic beads were run in parallel for all conditions and peptides. A control bead background intensity was determined and subtracted from the intensity values obtained for all samples. The dissociation constant (KD) was calculated using Scatchard plots.
Protein Microarray Screening
The reactivity patterns of purified CIMS antibodies against intact human proteins was evaluated by protein array screening using arrays comprising 37,200 human fetal brain proteins, generated from the hEx1 cDNA expression library (32, 33) (ImaGenes, Germany). The polyvinylidene difluoride arrays were soaked in 95% (v/v) ethanol, rinsed in deionized water, and washed clean of residual bacterial colonies with 20 mm Tris, pH 7.4, 500 mm NaCl, 0.05% (v/v) Tween-20 (TBST), with 0.5% (v/v) Triton X-100. For CIMS scFv profiling, the protein arrays were blocked in 2% (w/v) nonfat, dry milk powder in 20 mm Tris HCl pH 7.4, 150 mm NaCl (TBS) for 2 h, then washed twice in TBST and subsequently incubated with scFv at a concentration of 1 μg/ml in TBS. The protein arrays were then washed in TBST three times for 10 min each and subsequently incubated with mouse anti-c-myc antibody (9E10) (Santa Cruz, Santa Cruz, CA) at a concentration of 0.2 μg/ml. This secondary anti-c-myc antibody is specific for the myc affinity-tag carried by all CIMS scFv's. The tertiary labeled antibody used was goat anti-mouse alkaline phosphatase antibody (Sigma). The arrays were illuminated with long-wave UV light and the images were taken using a high resolution CCD detection system (Fuji). Image analysis was performed with VisualGrid (GPC Biotech, Martinsreid, Germany). The EMBOSS Pairwise Alignment software, using the needle algorithm, was used to map potential epitopes on the bound proteins based on the original selection peptide motifs as well as the experimentally refined binding motifs, as delineated from the captured peptides identified by LC-MS/MS.
Preparation of Trypsin-digested Mouse Liver Proteomes
Protein was extracted from Mus musculus liver. A 1.02 g liver was minced in 0.5 mm Tris-HCl buffer, pH 7.4, containing 0.25 m sucrose and 0.75 mm magnesium chloride. The mixture was first sonicated to release proteins and then centrifuged to remove nonsoluble material, 600 × g for 15 min followed by 25000 × g for 100 min. Protein in the supernatant was precipitated by adding ammonium sulfate to 72% saturation, centrifuged at 25000 × g for 100 min and resuspended in 2.4 ml 25 mm ammonium carbonate. Once more the mixture was sonicated and centrifuged, 25,000 × g for 40 min. The supernatant was filtered through a 0.22 μm filter and then desalted on a PD-10 column (Amersham Biosciences, Uppsala, Sweden). Protein was eluted in 25 mm ammonium carbonate and fractions containing most of the protein, identified from A280 measurement, were pooled (∼4 ml). All steps of the extraction process were performed either on ice or at 4 °C. The protein concentration of the extracted protein pool was estimated to 5.6 mg/ml using the BCA Protein Assay (Pierce, Rockford, IL) using BSA as a standard.
Extracted protein (8.8 mg, 1.6 ml) was reduced, alkylated and trypsin digested. First, dithiothreitol and SDS were added to 16 mm and 0.2% (w/v), respectively, and the sample was reduced for 30 min at 50 °C. Then iodoacetamide was added to 27 mm and the sample was alkylated for 30 min at 37 °C. Excess reagents were removed using a PD-10 column and protein was eluted in PBS. Fractions containing most of the protein were pooled (∼3 ml). Finally, 37 μl of a 10 mg/ml trypsin solution was added, the sample was digested for 20 h at 37 °C and then stored at −80 °C.
Preparation of Trypsin-digested Human Colon Tissue Proteomes
Protein was extracted from human colon tissue from colon cancer patients kindly supplied by Prof. X-F Sun (Department of Oncology, University of Linköping, Linköping, Sweden), and stored at −80 °C until use. The sample collection was approved by the Regional Ethical Committee. Tissue pieces (about 100 mg/sample) were homogenized in Teflon containers, precooled in liquid nitrogen, by fixating the bomb in a shaker for 2 × 30 s with quick cooling in liquid nitrogen in between the two shaking rounds. The homogenized tissue powder was then collected in lysis buffer containing 8 m urea, 30 mm Tris, 5 mm magnesium acetate and 4% (w/v) CHAPS (pH 8.5) was added (1.5 mg tissue/30 μl buffer). The tubes were briefly vortexed and then incubated on ice for 30 min with brief vortex of the sample every 5 min. The sample was then centrifuged at 13,000 rpm, and the supernatant transferred to new tubes followed by a second centrifugation. The buffer was exchanged to 0.3 m HEPES, 1 m Urea using Zeba desalting spin columns (Pierce) before the protein concentration was determined using Total Protein Kit, Micro Lowry (Sigma). Finally, the sample was aliquoted and stored at −80 °C until further use.
Thawed protein extracts were reduced, alkylated, and trypsin digested. First, SDS and tris(2-carboxyethyl) phosphine hydrochloride (Thermo Scientific) were added to 0.1% (w/v) and 5 mm, respectively, and the sample was reduced for 30 min at 37 °C. The samples were cooled down to room temperature before iodoacetamide was added to 40 mm and the sample was alkylated for 30 min at room temperature. Next, sequencing-grade modified trypsin (Promega, Madison, Wisconsin) was added at 20 μg per mg of protein for 16 h at 37 °C. To ensure complete digestion, a second round of trypsin (10 μg per mg protein) was added and the tubes were incubated for an additional 3 h at 37 °C. Finally, the samples were aliquoted and stored at −80 °C until further use.
Preparation of Trypsin-digested Yeast Proteomes
Colonies of a Saccharomyces cerevisiae, (strain W303–1A point mutated wild type with mutations leu2–3, 112 ura3–1, trp1–1, his3–11/15, ade2–1, can1–100, GAL SUC2 mal0, and genotype MATa) were grafted to a 500 ml preculture flask and grown at 30 °C on a rotary shaker at 200 rpm in synthetic yeast nitrogen medium containing ammonium sulfate (50 g/l) and supplemented with 2% (w/v) glucose and necessary amino acids and nucleotides to a final concentration of 120 μg/ml each. A liquid culture with 0.05% (w/v) glucose and 3% (v/v) ethanol were inoculated with 1% (v/v) from the overnight preculture. The cultures were grown in 30 °C on a rotary shaker at 200 rpm and harvested in log phase, after about 4 generations, at OD 0.5. Cell density was estimated at 600 nm. The cultures were centrifuged at 4 °C (at 5000 rpm for 10 min. The pellets were dissolved in 20 ml ice-cold Milli-Q water and centrifuged at 3000 rpm for 3 min. The pellets were again resuspended in 1 ml Milli-Q water and finally collected by a centrifugation using 13,200 rpm for 1 min at 4 °C. Samples were quick-frozen in liquid nitrogen and stored at −80 °C.
Pellets were thawed on ice, resuspended in 800 μl ice-cold water and transferred to tubes containing 0.7g acid washed glass beads (diameter 0.55 mm). Samples were vortexed 4 × 60 s, with 60 s intervals on ice, at 4 °C. To each sample, 100 μl of a solution containing 3% (w/v) SDS, 140 mm Tris-HCl, 110 mm TrisBase, and 600 Mm dithiothreitol, was added and then incubated for 5 min at 85 °C. The samples were allowed to cool on ice for a short time before adding a solution containing 1.5 mm Tris-HCl and TrisBase, 1 m MgCl2, DNAse1, and RNase (A), followed by incubation for 30 min on ice. Cell material was removed by centrifugation at 4 °C, 13,200 rpm for 15 min, and the supernatant stored at −80 °C.
The buffer was exchanged to 0.15 m HEPES, 0.5 m Urea using Zeba desalting spin columns (Pierce) before the protein concentration was determined using Total Protein Kit, Micro Lowry (Sigma). Finally, the sample was aliquoted and stored at −80 °C until further use. Thawed protein extracts were reduced, alkylated and trypsin digested. First, SDS and Tris(2-Carboxyethyl) phosphine Hydrochloride (Thermo Scientific, Rockford, IL, USA) were added to 0.05% (w/v) and 5 mm, respectively, and the sample was reduced for 30 min at 37 °C. The samples were cooled down to room temperature before iodoacetamide was added to 10 mm and the sample was alkylated for 30 min at room temperature. Next, sequencing-grade modified trypsin (Promega) was added at 20 μg per mg of protein for 16 h at 37 °C. To ensure complete digestion, a second round of trypsin (10 μg per mg protein) was added and the tubes were incubated for an additional 3 h at 37 °C. Finally, the digested samples were aliquoted and stored at −80 °C until further use.
Fabrication of CIMS Antibody Functionalized Micro-columns
Purified CIMS antibodies were individually coupled to POROS 20 AL bead (Applied Biosystems, Foster City, CA) using standard protocols. Briefly, 500 μg scFv was concentrated to 200 μl and the buffer exchanged to 25 mm sodium phosphate pH 7.2 containing 0.5 m Na2SO4. After addition of 20 μl 200 mg/ml sodium cyanoborohydride solution, the antibodies were coupled to 30 mg beads. The coupling was performed at room temperature under salting out conditions (stepwise increase of 0.1 m sodium sulfate from 0.5 to 1.1 m every 30 min) by adding a 25 mm sodium phosphate buffer, pH 7.2, containing 1.5 m Na2SO4 and 200 (or 100) mg/ml sodium cyanoborohydride. The final concentration of sodium cyanoborohydride was 45 (or 26) mg/ml. Next, the mixture was incubated over night at room temperature with gentle shaking. Beads were then separated from the reaction buffer by centrifugation. To quench residual aldehyde functionality, beads were resuspended in 150 μl (or 200 μl) 0.2 m Tris-HCl buffer, pH 7.4, containing 250 mg/ml (or 200 mg/ml) sodium cyanoborohydride and incubated for 2 h at room temperature. Beads were then washed three times in PBS and finally re-suspended in 400 μl PBS and stored at 4 °C until use. Micro-columns were fabricated by packing about 10 μl of bead slurries in GEloader tips (Eppendorf) to generate 20 to 30 mm long columns.
Fabrication of CIMS Antibody Functionalized Magnetic Beads
Purified scFvs were individually coupled to magnetic beads using manufacture provided protocols with some modifications. Briefly, 300 μl (∼ 9 mg) of beads, M-270 carboxylic acid-activated (Invitrogen Dynal, Oslo, Norway), were first washed and incubated twice with 300 μl 25 mm 2-(N-morpholino)ethanesulfonic acid (MES) (pH 6) and slow mixing for 10 min. N-Ethyl-N′-(3-dimethylaminopropyl)carbodiimide (Sigma) was freshly dissolved in cold 25 mm MES, pH 6 to a concentration of 25 mg/ml. Similarly, a 47 mg/ml solution of Sulfo-NHS (Thermo Scientific) solution was prepared in 25 mm MES, pH 6. 150 μl of both the N-Ethyl-N′-(3-dimethylaminopropyl)carbodiimide-solution and Sulfo-NHS solution were added to the beads and incubated for 30 min at room temperature with slow mixing. The beads were washed once with 25 mm MES and PBS, respectively. Then, 180–250 μg purified scFv was added to the beads and incubated for 45 min under slow mixing at room temperature. To block unreacted surface, the beads were washed with 2 × 300 μl 50 mm Tris buffer, pH 7.4 and incubated for 15 min at room temperature. Finally, the beads were washed four times with PBS containing 0.005% (v/v) Tween-20, transferred to new tubes and resuspended in 300 μl PBS/0.005% (v/v) Tween-20 and stored at 4 °C until use.
Analysis of Synthetic Peptide Mixtures
The individual columns were equilibrated with 50 μl 5% (v/v) HAc followed by 50 μl PBS (pH 7.4). Next, a synthetic peptide mixture consisting of ten different selection peptides, denoted peptide mix-1 (M-1, M-13, M-14, M-16, M-17, M-29, M-30, M-32, M-33, and M-34) or mix-2 (M-13, M-14, M-16, M-17, M-29, M-30, M-32, M-33, M-34, and M-35), was applied. Peptide mix-1 was applied to columns functionalized with either CIMS-1-A05 or CIMS-1-B03, whereas peptide mix-2 was applied to either CIMS-17-E02, CIMS-M32–3A-G03, CIMS-33–3D-F06, or CIMS-34–3A-D10 functionalized columns. A two-step washing procedure was performed by adding 15 μl and 10 μl PBS (pH 7.4), respectively, subsequently any captured peptides were eluted by adding 5 μl 5% (v/v) HAc. Before mass spectrometry analysis, a sample cleanup was performed to remove any contaminating bead particles using small pieces of a C18-filter (3 m Empore) (3 m Center, St. Paul, MN) packed into a GEloader tip (Eppendorf) as previously described (34). The filter was activated by adding 10 μl 50% (v/v) acetonitrile followed by a wash with 10 μl 0,1% (v/v) trifluoroacetic acid (TFA). The sample was then added and rinsed using 20 μl 0.1% TFA. Bound peptides were eluted by adding 5 μl 70% (v/v) acetonitrile, 0.1% (v/v) TFA. Mass spectrometry was performed using a matrix-assisted laser desorption ionization/time of flight (MALDI-TOF) micro MX (Waters, Milford, MA) instrument. Standard 96-well stainless steel MALDI target plate (Waters) was used, were 0.5 μl matrix solution (2.5 mg/ml α-cyano-4-hydroxycinnamic acid in 50% (v/v) acetonitrile, 0.05% (v/v) trifluoroacetic acid) was first spotted followed by 0.5 μl sample. Mass scanning was performed between 800 and 3500 Da and each spectrum represented an average of up to 400 laser shots.
GPS of Complex Proteomes Using Micro-column Setup
Crude trypsin digested mouse liver extracts or human colon tissue extracts were applied to CIMS antibody functionalized micro-columns. For the mouse liver extracts CIMS-1-A05, CIMS-1-B03, CIMS-17-E02, and CIMS-33–3D-F06 were tested and for the colon samples CIMS-1-A05, CIMS-1-B03, CIMS-15-A06, CIMS-17-E02, CIMS-32–3A-G03, CIMS-33–3D-F06, and CIMS-34–3A-D10 were tested. Before sample application, the columns were washed with 2 × 30 μl 5% (v/v) HAc and equilibrated with 2 × 30 μl PBS.
In the case of crude trypsin digested mouse liver extracts, 10 μl of a 2-fold dilution in PBS was applied (about 20 μg). Columns were washed with 2 × 15 μl PBS and captured peptides were eluted with 7 μl 5% (v/v) HAc. The eluted peptides were desalted using a reversed-phase material, POROS 20 R2 media (Applied Biosystems). The R2 beads were packed into micro columns of ∼5 mm length in the same way as described for the affinity columns. R2 columns were washed with 30 μl 75% (v/v) acetonitrile, 1% (v/v) TFA and equilibrated with 2 × 15 μl 0.1% (v/v) TFA. Samples from the affinity columns (∼7 μl) were added and R2 columns were washed with 2 × 15 μl 0.1% (v/v) TFA. Finally peptides were eluted with 3 μl 75% (v/v) acetonitrile, 0.1% (v/v) TFA directly onto a MALDI target. MALDI-TOF-TOF analysis was done using a 4700 Proteomics Analyzer (Applied Biosystems, Framingham, MA). 0.5 μl per well of 5 mg/ml solution of α-cyano-4-hydroxycinnamic acid in 75% (v/v) acetonitrile, 1% (v/v) TFA was used as matrix solution. Database searches were performed using Mascot Distiller (v 2.3.1.0) (Matrix Science, London, UK) against the Mus musculus (Swiss-Prot 57.12, 23692 sequences for Mus musculus (house mouse) as taxonomy) with the following parameters: peptide mass tolerance ±0.3 Da; fragment mass tolerance: ±0.3 Da; enzyme: trypsin; missed cleavages: 1; fixed modification: carbamidomethyl (C); variable modification: methionine oxidation. In addition a TrypChymo search with two and three missed cleavages were performed. Only peptides that passed the Mascot threshold set for positive MS/MS identification (p < 0.05) were considered as valid hits. All generated MS and MS/MS data for the mouse liver set were then uploaded and stored in the The Proteios Software Environment (ProSE) (35) and deposited in the public PRoteomics IDEntifications database (PRIDE) (36) (accession numbers 13401–13512).
In the case of crude trypsin digested colon extracts, 10 μl of a 2 μg/μl extract (diluted in PBS) was added. A two step washing procedure was performed by adding 15 μl and 10 μl PBS (pH 7.4), respectively and any captured peptides were eluted by adding 5 μl 5% (v/v) HAc. Next, the eluted peptides were cleaned-up using regular C18-filter (3 m Empore) as described above for the synthetic peptide mixtures. The captured human colon peptides were analyzed using a Micromass electrospray ionization TOF (ESI-QTOF) Ultima API (Waters) coupled to a CapLC HPLC. Before injection on the ESI-QTOF Ultima API, all samples were almost completely dried down by speedvac and then redissolved in 8 μl 0.1% (v/v) formic acid. The auto-sampler injected 6 μl of sample, and the peptides were trapped on a precolumn (C18, 300 μm×5 mm, 5 μm, 100 Å, LC-Packings) and separated on a reverse-phase analytical column (Atlantis, C18, 75 μm ×150 mm, 3 μm, 100 Å, Waters). The flow rate was 200 nl/min. Solvent A consisted of 2% (v/v) acetonitrile and 98% water with 0.1% (v/v) formic acid. Solvent B consisted of 90% (v/v) acetonitrile, 10% water, and 0.1% (v/v) formic acid. The total runtime was 90 min starting with a 5 min wash of the peptides. Mass spectra were acquired from m/z 400–1600 for 1.9 s followed by three data-dependent MS/MS scans from m/z 50–1800 for 1 s each. The collision energy used to perform MS/MS was automatically varied according to the mass and charge state of the eluting peptide. Only spectra from ions with charge state 2 and 3 were acquired for MS/MS analysis. Mascot Distiller (version 2.2.1.0) was used to process the generated raw-data files into mzData file format. All generated MS and MS/MS data for the colon set were then uploaded and stored in the The Proteios Software Environment (ProSE) (35) and deposited in PRIDE (36) (accession numbers 13401–13512). The Proteios platform was used for performing automated database searches in both Mascot and X! Tandem against the human International Protein Index (IPI) protein version 3.60 database (consisting of forward and random sequences with the same amino acid distribution and size resulting in a total of 160824 sequences for the combined database) with the following parameters: enzyme: trypsin; missed cleavages: 1; fixed modification: carbamidomethyl (C); variable modification: methionine oxidation. A peptide mass tolerance of 100 ppm and fragment mass tolerance of 0.1 Da was used. This was followed by automated combination of the search results from the two different search engines with predetermined false discovery rate (FDR of <0.05 was used). No significantly scored (MS/MS) background binding peptides were detected for blank columns, containing no antibody.
GPS Experiments of Complex Proteomes Using Magnetic Bead Setup
Crude trypsin digested human colon extracts or yeast extracts were applied to CIMS antibody functionalized magnetic beads. For each GPS-experiment experiment 25–30 μl (the latter for yeast experiments) of a conjugated CIMS-scFv bead solution was used. The conjugated beads were prewashed with 200 μl PBS. A tryptic digest of 20 μg was defrosted and diluted (using PBS) into a final volume of 20 μl. Incubation time was 15 min with gentle mixing. The tubes were then placed on a magnet and the supernatant removed. Two washes with PBS (53 μl and 45 μl) were performed and the beads transferred to new tubes in between each washing step. Finally the beads were incubated with 7.5 μl of a 5% (v/v) acetic acid solution for 1 min to elute the captured peptides. The eluate was then ready for mass spectrometry analysis and no further cleanup needed. The captured peptides were analyzed using a Micromass ESI-QTOF Ultima API (Waters) coupled to a CapLC HPLC (as described above) and a ESI-LTQ-Orbitrap (Thermo Electron, Bremen, Germany) coupled to an Eksigent two-dimensional nano HPLC (Eksigent technologies, Dublin, CA). For the colon samples replicate capture experiments were done for each of the instrumentation configurations (i.e. in total four capture experiments (two capture and LC-MS/MS runs per instrumentation) however the digests were not from the same pieces of tissue and thereby data not directly comparable among the instruments). In the case of yeast samples replicate capture experiments were done and captured peptides only analyzed with the LTQ-Orbitrap. The auto-sampler for the LTQ-Orbitrap injected 6 μl of sample, and the peptides were trapped on a precolumn (Zorbax 300SB-C18 5 × 0.3 mm, 5 μm, Agilent) and separated on a reversed-phase analytical column (Zorbax 300SB-C18 150 × 0.75 mm, 3.5 μm, Agilent). The flow rate was 400 nl/min. Solvent A consisted of 0.1% (v/v) formic acid in water and solvent B of 0.1% (v/v) formic acid in acetonitrile. The total runtime was 90 min starting with a 15 min wash of the peptides. The LTQ-Orbitrap was operated in the data-dependent mode to automatically switch between Orbitrap-MS and LTQ-MS/MS acquisition. Survey full scan MS spectra (from m/z 400 to 2000) were acquired in the Orbitrap with a resolution of 60000 at m/z 400 using the lock mass option for internal calibration. The seven most intense ions with charge state 2 and up were sequentially isolated for CID-fragmentation in the LTQ with a normalized collision energy of 35%. The resulting fragment ions were recorded in the LTQ. In contrast to the QTOF set-up, significantly scoring (MS/MS) background binding peptides were detected for blank beads (no coupled antibody) for the Orbitrap set-up (supplemental Table S6). Based on the relatively low number of background binding peptides, the colon data was left unfiltered. For the yeast data, any background binding peptides identified in both the blank runs (supplemental Table S6) and in any of the CIMS-binder runs were removed and not included in the analysis.
GPS Multiplex Experiments of Complex Proteomes Using Magnetic Bead Setup
Briefly 30 μl solution of the CIMS-17-E02 and CIMS-33–3D-F06 conjugated beads were mixed and then prewashed with 200 μl PBS + 100 μl PBS. A tryptic digest (40 μg was defrosted and then diluted using PBS) into a final volume of 40 μl (human colon digest) or 55 μl (yeast) and then incubated with the beads for 15 min with gentle mixing. The tubes were then placed on a magnet and the supernatant removed. Two washes with PBS were performed using 100 and 90 μl, and the beads were transferred to new tubes in between each wash step. Finally the beads were incubated with 7.5 μl of a 5% (v/v) acetic acid solution for 1 min. The eluate was then ready for mass spectrometry analysis and no further cleanup needed. The captured peptides were analyzed using an ESI-LTQ-Orbitrap (Thermo Electron, Bremen, Germany) coupled to an Eksigent 2D nano HPLC (Eksigent technologies, Dublin, CA). The auto-sampler injected 6 μl of sample, and the peptides were trapped on a precolumn (Zorbax 300SB-C18 5 × 0.3 mm, 5 μm, Agilent, Santa Clara, CA) and separated on a reversed-phase analytical column (Zorbax 300SB-C18 150 × 0.75 mm, 3.5 μm, Agilent). The flow rate was 400 nl/min. Solvent A consisted of 0.1% (v/v) formic acid in water and solvent B of 0.1% (v/v) formic acid in acetonitrile. The total runtime was 90 min starting with a 15 min wash of the peptides. The LTQ-Orbitrap was operated in the data-dependent mode to automatically switch between Orbitrap-MS and LTQ-MS/MS acquisition. Survey full scan MS spectra (from m/z 400 to 2000) were acquired in the Orbitrap with a resolution of 60,000 at m/z 400 using the lock mass option for internal calibration. The seven most intense ions with charge state 2 and up were sequentially isolated for CID-fragmentation in the LTQ with a normalized collision energy of 35%. The resulting fragment ions were recorded in the LTQ.
Database Searches for all Magnetic Bead Experiments
All generated raw-data files were processed and converted into mzData format (Mascot Distiller (version 2.2.1.0) used for files generated by the ESI-QTOF instrument, and Proteome Discovery (version 1.0) for files generated by the LTQ-Orbitrap). All generated MS and MS/MS data from all magnetic bead experiments (colon sets and yeast set) were then uploaded into separate projects, stored in ProSE (35), and deposited in PRIDE (36) (accession numbers 13401–13512). The Proteios platform was used for performing automated database searches in both Mascot and X! Tandem against the human IPI protein database (version 3.60 and against a random generated decoy IPI version 3.60 with same amino acid distribution) for the colon samples and the Saccharomyces cerevisae Swiss-Prot (13-Oct 2009 and against a random generated decoy version with same amino acid distribution) for the yeast samples. In both cases with the following parameters: enzyme: trypsin; missed cleavages: 1; fixed modification: carbamidomethyl (C); variable modification: methionine oxidation. A peptide mass tolerance of 100 ppm and fragment mass tolerance of 0.1 Da was used for analysis done with the ESI-QTOF whereas for analysis done on the LTQ-Orbitrap a peptide mass tolerance of 3 ppm and fragment mass tolerance of 0.5 Da was used. This was followed by automated combination of the search results from the two different search engines with predetermined false discovery rates. A FDR of 0.05 was used for the human colon sets (QTOF and the LTQ-Orbitrap), whereas a FDR of 0.01 was used for the yeast set.
Label-free Quantitative GPS LC-MS/MS Experiments of Complex Proteomes
To address the reproducibility of the capture step, in terms of both peptide identification and quantification, a set of in total 18 capture experiments were performed. 35 μl of the conjugated CIMS-33–3D-F06 bead solution was used for each capture experiment and exposed to either a tryptic digest, from either colon or yeast, in a final volume of 30 μl (diluted with PBS). Incubation time was 15 min with gentle mixing followed by two washes with PBS (65 μl and 50 μl) and the beads were transferred to new tubes for each washing step. Finally, the beads were incubated with 8.5 μl of a 5% (v/v) acetic acid solution for 2 min. The eluate was then ready for mass spectrometry analysis and no further cleanup needed. For all samples, the ESI-LTQ-Orbitrap XL (Thermo Electron, Bremen, Germany) was used as previously described with some minor modifications. Briefly, the total runtime was changed to 70 min starting with a 5 min wash of the peptides. Further, the flow rate was 350 nl/min with solvent A, containing of 0.1% (v/v) formic acid in water and solvent B of 0.1% (v/v) formic acid in acetonitrile. For the first three captures (20 μg digest of both colon and yeast samples), the LTQ-Orbitrap was operated in a data-dependent acquisition (Top-7) manner to automatically switch between Orbitrap-MS and LTQ-MS/MS acquisition. Inclusion lists were made based on all the identified peptides from the first three capture experiments. Six new independent capture experiments were then performed (three using 20 μg digest and three with 5 μg digest) on both the colon and yeast tryptic digested proteomes. The eluates from these six captures were then analyzed in the same way as previously described with the exception that inclusion lists now were used in combination with the option that if no ions on the list were present the four most intense ions with charge state 2 and up could sequentially be isolated for CID-fragmentation. All generated raw-data files were processed and converted into mzData format using Proteome Discovery (version 1.0). The Proteios platform was used for automated database searches in both Mascot and X! Tandem. The same search parameters, as previously described, were used with the only exception that the human samples were searched against the human IPI protein database (version 3.71) (consisting of forward and random sequences with the same amino acid distribution and size, resulting in a total of 173490 sequences for the combined database). The automated combination of results for either the colon and yeast data, using the different search engines (with a predetermined false discovery rate of 0.01), was used. Data has been deposited in PRIDE (36) (accession numbers 16449–16469). For assessing the quantitative reproducibility, the Progenesis-LC-MS software (v 2.5) with default settings was used. The raw data files were converted to mzXML using the ProteoWizard software package before using the Progenesis-LC-MS software. The built-in peptide feature finding tool and Mascot search tool (p value 0.05) were used (no X! Tandem) and the generated raw abundance values were extracted and used for downstream quantitative analysis i.e. calculating the coefficient of variation (CV) and ratios for the replicate independent capture experiments.
Redefining of Motives
In all cases, the MS/MS identified peptides for each CIMS antibody in all experiments (mouse liver, human colon and yeast) were used to manually re-annotate and redefine the peptide binding motifs. The tool weblogo (37) was used to generate the motif figures for each binders enriched sequence motifs.
Structural Analysis of CIMS antibodies
Three-dimensional structural homology models were generated for selected CIMS antibodies using Web Antibody Modeling server (http://antibody.bath.ac.uk). The model structures were visualized using Pymol (Delano Scientific LCC).
Bioinformatic Analysis
High level GO terms that represent the major biological processes and compartments mapped for all peptides identified in the human colon sample and from the S. cerevisiae extracts were generated by using the Generic Gene Ontology (GO) TERM MAPPER tool at http://go.princeton.edu/cgi-bin/GOTermMapper.
The compiled set of identified peptides for the different samples were checked for presence in the PeptideAtlas and the following builds: Human Jan-2010 Build with 59921 distinct peptides and 4553414 peptide spectrum matches. Yeast May-2009 build with 58719 distinct peptides and 2697580 peptide spectrum matches. Mouse build with 17853 distinct peptides and 576448 peptide spectrum matches.
RESULTS
We present proof-of-concept for a conceptually new method opening up the possibility to probe proteomes in a species independent manner, while still using a limited set of antibodies. An overall workflow outlining the steps in the design and development of the GPS methodology is schematically shown in supplemental Fig. S1.
Design of Selection Peptide Motifs
In silico trypsin digestion of the human proteome, composed of 20,325 nonredundant proteins (release 56.1 of UniProtKB/Swiss-Prot) generated 1,193,062 peptides, of which 58.5% represents the mass fraction ranging from 500 to 3500 Da that theoretically could be detected using a high-performing mass spectrometer. To target a general subset of this collection of addressable peptides, we designed 27 selection peptide motifs, consisting of either 4 or 6 amino acids (27, 28), without imposing any restrictions on the original wild-type proteins (e.g. abundance and functional classification) Table I and supplemental Table S1). The motifs were selected to contain a C-terminal lysine or arginine to mimic tryptic peptides, whereas residues sensitive for oxidation or dimerization, such as methionine and cysteine, were excluded. If one requires a perfect match for identification, the results showed that each motif was present in 0 to 670 proteins, representing a total coverage of about 2144 proteins (supplemental Table S1). However, allowing a single residue to vary in each position of the motif, except in the C terminus, significantly increased the theoretical coverage. This is illustrated by a 34.3 times increase, from 124 to 4259 hits, for eight of the selected model motifs (Table I).
Table I. Examples of selection peptide motifs. Eight of 27 motifs are shown (see supplementary Table 1 online).
| Motif | Sequence | No. of motif-carrying nonredundant proteinsa |
||
|---|---|---|---|---|
| Perfect match | Perfect match and 500–3500 | Wobbling and 500–3500b | ||
| Da limit | Da limit | |||
| M-1 | EDFR | 126 | 118 | 3959 |
| M14 | DFAEDK | 0 | 0 | 10 |
| M-15 | LTEFAK | 2 | 2 | 34 |
| M-16 | TEEQLK | 4 | 4 | 116 |
| M-17 | SSAYSR | 0 | 0 | 45 |
| M-32 | QEASFK | 0 | 0 | 18 |
| M-33 | LSADHR | 0 | 0 | 31 |
| M-34 | SEAHLR | 0 | 0 | 46 |
| All 27 motifs | 2393 | 2144 | - | |
a The number of non-redundant proteins covered by motif-carrying peptides. The nonredundant human proteome (relase 56.1 of UniProt/Swiss-Prot), trypsin digested in silico, was used as peptide database.
b A single amino acid residue was allowed to vary in any position of the motif, except for in the C-terminus.
Selection of CIMS Antibodies
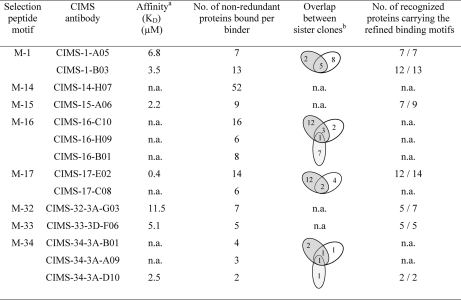
Human recombinant scFv antibodies were selected from a large phage display library (29), using short synthetic peptides, composed of an N-terminal biotin, a SGSG-linker, followed by the C-terminal located peptide selection motif bound to streptavidin conjugated magnetic beads. In total, 91 nonredundant scFv's were selected of which 14 binders that were directed against 8 motifs were further characterized in this proof-of-concept study (Table II). Notably, the selection step was designed to generate motif specific binders, but all four or six residues of the binding motifs were not required as fixed residues for recognition. This enabled us to select a set of CIMS antibodies, denoted sister clones, against each selection peptide, displaying similar but distinct binding patterns, increasing the repertoire of binders even further, while still using a limited set of selection motifs. Further, the dissociation constant (KD) was determined for the antibodies, and representative results are shown in Table II (supplemental Fig. S2). The antibodies were found to display KD values in the range of 0.4 to 11.5 μm. Hence, the antibodies displayed affinities as could be expected for anti-peptide antibodies directly selected from scFv phage-display libraries and not subjected to affinity maturation.
Table II. Binding patterns of 14 CIMS antibodies against full-length semidenatured human proteins as determined by protein array screening.
a Determined against selection peptide.
b Sister clones are defined as CIMS antibodies selected against the same selection peptide motif, displaying similar but distinct binding patterns. n.a., not available.
Protein Array Characterization of Selected CIMS Antibodies
First, we investigated the binding patterns of the 14 CIMS antibodies by protein array screening, based on 37, 200 redundant human recombinant brain proteins (32) (Table II and supplemental Table S2). The 14 CIMS antibodies, including six sister clones, were found to display distinct binding patterns. In more detail, the CIMS antibodies were found to bind between 2 and 52 nonredundant proteins per binder. In total, the 14 CIMS antibodies bound to 152 proteins, corresponding to 113 unique protein identities. Next, we compiled all the LC-MS/MS data to map the experimentally refined and validated peptide binding motifs (available for seven of the CIMS antibodies) (see below, and Fig. 2 and supplemental Fig. S3) onto the recognized brain proteins. The data showed that the motifs were present in 71–100% of the recognized proteins (Table II), demonstrating that CIMS antibodies displaying distinct reactivity patterns could be generated. Further experiments will be required to elucidate the frequency of motif-carrying brain proteins that were not recognized, as the sequence for many of the arrayed proteins has not been determined.
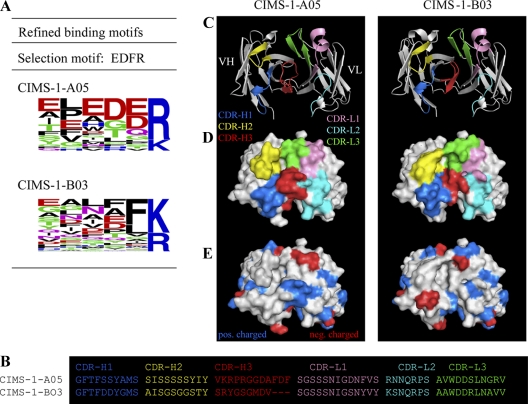
Fig. 2.
Structural analysis of two CIMS antibodies, denoted sister clones, and selected against the same selection motif. A,Experimentally refined binding motif amino acid sequences as compiled from all MS/MS detected captured peptides, and the frequency of each individual residue in the last six C-terminal positions is indicated. The two sister clones were selected against the same original selection peptide motif, M-1 EDFR. B, Comparison of the amino acid sequence of the complementarity determining regions (CDRs) of the two sister clones. C, Three-dimensional homology models, shown as a ribbon structure (top view) and with color coded CDRs of the two sister clones. D, Space filling models of the three-dimensional structures shown in C. E, Space filling models of the structures shown in C, onto which the electrostatics have been mapped.
Analysis of Peptide Mixtures Using CIMS Antibodies
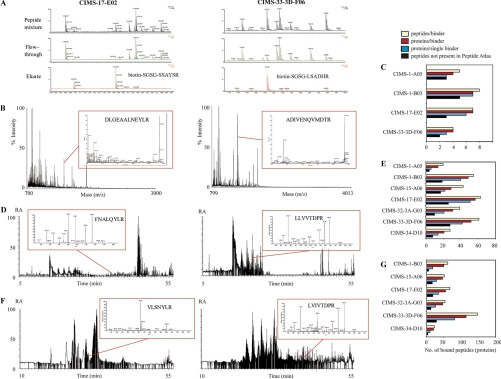
Next, we tested the principle of the GPS method by applying a mixture of 10 synthetic peptides, harboring the selection motifs, to six micro-columns containing different CIMS antibodies covalently coupled to POROS-AL beads. The micro-columns were interfaced with a MALDI-TOF mass spectrometer allowing a rapid read-out (Fig. 1A and supplemental Fig. S4). In all cases, the CIMS antibodies were found to bind and enrich the corresponding selection peptide, whereas the remaining nine irrelevant peptides were found predominantly in the flow-through.
Fig. 1.
First prototype GPS set-up targeting synthetic peptide mixtures or tryptic digests of complex proteomes. A, MALDI-TOF MS analysis of a mixture of 10 synthetic selection peptide motifs (mix-1 and 2), applied to micro-columns containing immobilized CIMS-17-E02 or CIMS-33–3D-F06 antibodies. B, MALDI-MS/MS analysis of tryptic digests from crude mouse liver homogenates applied to micro-columns containing immobilized CIMS-17-E02 or CIMS-33–3D-F06 antibodies. C, Statistics of the number of bound mouse peptides/proteins per CIMS antibody, as determined in B, and illustrated for four different CIMS antibodies. D, LC-MS/MS analysis of tryptic digests from crude human colon tissue applied to micro-columns or beads containing immobilized CIMS-17-E02 or CIMS-33–3D-F06 antibodies. E, Statistics of the number of bound human peptides/proteins per CIMS antibody, as determined in D, and illustrated for seven different CIMS antibodies. F, LC-MS/MS analysis of tryptic digests from crude yeast cell lysates applied to beads containing immobilized CIMS-17-E02 or CIMS-33–3D-F06 antibodies. G, Statistics of the number of bound yeast peptides/proteins per CIMS antibody, as determined in F, and illustrated for six different CIMS antibodies.
Analysis of Complex, Cross-species Proteomes Using CIMS Antibodies
To validate the GPS method and to demonstrate cross-species capability, we analyzed tryptic digests from crude mouse liver homogenates, human liver tissue extracts, and yeast (S. Cerevisiae) cell lysates (Fig. 1B–1G and supplemental Tables S3 to S5). The digested samples were analyzed on micro-columns (mouse and human species) or magnetic beads (human and yeast species), functionalized with single CIMS antibodies (in total four to seven different CIMS binders were used), and directly interfaced with tandem MS-based set-ups, including MALDI-TOF/TOF (mouse), ESI-QTOF (human), and ESI-LTQ-Orbitrap (human, yeast). We could demonstrate that it was possible to capture, enrich and identify distinct subpopulations of peptides from mouse liver (Fig. 1B and 1C), human colon (Fig. 1C and 1E), and yeast (Fig. 1E and 1F). The overlap among different binders was very low in all three proteomes (supplemental Tables S3 to S5). The fact that significantly more proteins were identified in the yeast and human colon samples compared with mouse liver samples reflected the progression of the project, e.g. use of more sophisticated MS/MS set-ups and streamlined procedures (supplemental Fig. S1). In addition the background binding was found to be relatively low (supplemental Table S6). In the case of mouse liver, four CIMS antibodies were found to bind between four and eight nonredundant peptides each, corresponding to 21 unique mouse proteins (Fig. 1C). Notably, 54% of these mouse peptides have not previously been reported in MS/MS experiments (38). For human colon tissue, seven CIMS antibodies were found to bind between 20 and 63 nonredundant peptides per binder (Fig. 1E). Of the 257 different and identified peptides, 110 (42%) have previously not been reported in the PeptideAtlas. In total, the seven CIMS antibodies recognized 217 different human proteins. In the case of yeast cell lysates, the data showed that six CIMS antibodies bound between 23 and 148 nonredundant peptides per binder, corresponding to 251 different proteins (Fig. 1G). Of note, of the 349 identified peptides, 87 (23%) have previously not been reported in the PeptideAtlas. Taken together, the GPS method could be used to probe crude complex proteomes in a species independent manner yielding a broad coverage.
Analysis of the Specificity of CIMS Antibodies
The anticipated peptide motif specificity was refined and validated by compiling all the experimental LC-MS/MS data (supplemental Tables S3 to S5) and reviewing the motifs bound by the seven CIMS antibodies tested (Fig. 2A and supplemental Fig. S3). The CIMS antibodies were found to specifically enrich peptides containing the selection motif and a narrow set of motif-like sequences in a clone dependent manner. Further, the sequence analysis showed that a few amino acid positions appeared to be more essential in defining these linear epitopes. Although physico-chemical requirements, such as charge, size, and polarity, were placed on these anchor positions, the others were found to vary more freely. The three most C-terminal residues frequently appeared to play a central role in defining the epitopes, whether a four or a six amino acids selection motif had been used during the selection step.
Moreover, primary (Fig. 2B) and tertiary (Fig. 2B–2E) structural analyses outlined that sister clones that had been selected against the same original peptide motif, but showed distinct binding patterns, also displayed key structural differences in their antigen binding-sites. Using two sister clones, CIMS-1-A05 and CIMS-1-B03, as model example, the main structural differences were manifested by a longer CDR-H3 and a more protruding CDR-H2 in CIMS-1-A05, creating a more cleft-like structure. In addition, CDR-H1 and CDR-L3 were also differently positioned, resulting in a more flattened binding site with a central cavity in CIMS-1-B03, as well as a different composition and distribution of charged residues.
Assay Characterization-multiplexing, Reproducibility, Quantification, and Dynamic Range
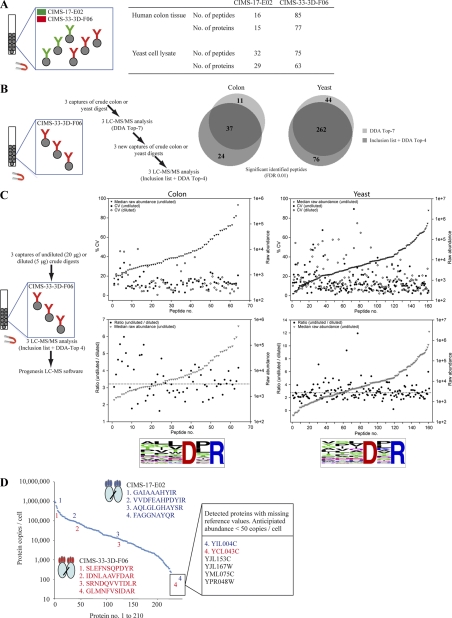
To outline the potential for increased throughput, we demonstrated that the set-up could be multiplexed by using several CIMS antibodies at the same time (Fig. 3A). In these proof-of-concept testes, we simultaneously profiled peptides captured by a mixture of two CIMS antibodies targeting human colon tissue and yeast cell lysates, respectively. In the case of human colon, the two CIMS antibodies captured 16 and 85 nonredundant peptides, respectively corresponding to in total 91 nonredundant human proteins. For yeast cell lysates, the same two CIMS antibodies captured 32 and 75 nonredundant peptides, respectively representing nonredundant 89 yeast proteins. The reproducibility of this two-plexed GPS set-up, including both the affinity capture and the LC-MS/MS step, was evaluated after six replicate captures, targeting colon tissue digest (supplemental Fig. S5). A total of 114 peptides were identified with significant scores (fixed or variable modified peptides were excluded), of which 21.1% were present in all six runs, 9.6% in five runs, 8.8% in four runs, 12.3% in three runs, 14.0% in two runs, and 34.2% in individual runs.
Fig. 3.
Characterization of the GPS set-up. A, Multiplexing of GPS, as illustrated for two CIMS antibodies, CIMS-17-E02 and CIMS-33–3D-F06, which were mixed and subsequently exposed to tryptic digests from human colon tissue or yeast cell lysates, respectively and then analyzed by LC-MS/MS. The statistics of the number of bound human or yeast peptides or proteins per CIMS antibody are shown. B, Peptide identification reproducibility of the complete GPS-setup, including both affinity capture and LC-MS/MS as illustrated for CIMS-33–3D-F06 exposed to human tryptic colon digest or yeast cell lysates. All peptides identified with fixed or variable modifications were excluded. C, Reproducibility of the entire GPS setup for label-free quantitative LC-MS/MS, as illustrated for CIMS-33–3D-F06 exposed to different amounts of human tryptic colon digests or yeast cell lysates. Raw abundance values (nonnormalized area under the peak values generated by Progenesis LC-MS software) are plotted. Data plotted were limited to peptides containing the experimentally refined binding motif (the frequency of the last six C-terminal amino acids is indicated for the peptides plotted). D, Dynamic range of the GPS set-up, illustrated for six CIMS antibodies immobilized on magnetic beads and exposed to tryptic digest of yeast cell lysates. The identified peptides (proteins) were plotted against the á priori known protein abundance levels of all individual yeast proteins (39). Among the detected peptides, four are indicated for two CIMS antibodies each, CIMS-17-E02 and CIMS-33–3D-F06, demonstrating the dynamic coverage. Eight detected peptides for which reference values are missing (39) (not plotted) and for which the abundance is anticipated to be <50 protein copies/cell are shown.
The reproducibility for a 1-plexed GPS set-up, including both the affinity capture and the LC-MS/MS step, is illustrated in Fig. 3B (supplemental Tables S7 and S8) for one antibody, targeting either colon or yeast digest. In the case of colon, the data showed that 37 peptides (51%) was reproducibly identified, whereas 11 (15.2%) or 24 (33.5%) peptides were only significantly scored in one of the two data acquisition approaches. In the case of yeast, the results showed that 262 peptides (68.5%) was reproducibly identified, whereas only 44 (11.5%) or 76 (19.9%) peptides were identified in one of the two data acquisition approaches. Hence, the results demonstrated an adequate reproducibility for the entire assay, i.e. including all steps of the GPS set-up.
Furthermore, we also demonstrated that quantitative information could be retrieved in a reproducible manner by running a label-free quantitative GPS LC-MS set-up (Fig. 3C and supplemental Table S9). In these experiments, we profiled the same tryptic digest from either colon or yeast at two concentrations (5 versus 20 μg), using one CIMS antibody. In total, 64 colon and 161 yeast peptides, present at a dynamic range of about four orders of magnitude, were monitored in three replicate runs. Notably, a median CV value of 11.1% to 12.7% was obtained for the colon peptides and 11.0% to 13.7% for the yeast peptides, further highlighting an adequate reproducibility of the entire GPS set-up. Furthermore, a 3.2 (colon) and 2.7 (yeast) times higher median signal intensities (the theoretical ratio was 4.0) were obtained for all the peptides monitored in the highest concentrated sample using nonnormalized data, indicating the quantitative capability of the set-up.
To evaluate the dynamic range of the GPS method, we matched the yeast peptides detected by the six CIMS antibodies (Fig. 1g and supplemental Table 5) with the known yeast protein abundance levels (39) (Fig. 3d and supplemental Table 10). The data showed that the GPS method detected yeast protein covering the full dynamic range, from high-abundant species (1 × 106 copies/cell) to low-abundant proteins (∼2 × 102 copies/cell). In addition, a set of 45 additional peptides were detected that could not be correlated as the reference values were missing (supplemental Table S10). Notably, 6 of these peptides were anticipated to be present only at very low levels (< 50 protein copies/cell) (39), further highlighting the capability of the GPS method to probe a wide dynamic range.
Biological Relevance of Captured Protein
To assess the biological relevance of the proteins identified, we analyzed the functionality and localization of the 217 colon proteins (supplemental Table S4) and 251 yeast proteins (supplemental Table S5). In the case of colon proteins, the three most represented functional groups were, (1) cellular processes, (2) metabolic processes, and (3) regulation of biological processes (supplemental Fig. S6a). The identified proteins were mainly located intracellular (89%), in the plasma membrane (34%), nucleus (26%), cytosol (16%) and/or in various organelles, outlining the broad coverage obtained (supplemental Fig. S6b). Notably, at least 16 of these proteins have previously been found to be up- or down-regulated in human colon cancer (supplemental Fig. S6c), underlining that biologically relevant proteins were detected using GPS.
In the case of yeast cell lysates, the top functional groups represented were (1) translation, (2) transportation, and (3) cellular amino acid and derivative metabolic process, reflecting the yeast cells were in log-phase (supplemental Fig. S6d). The identified proteins were broadly distributed among different compartments of the yeast cell, including e.g. the membrane (9–17%), nucleus (27%), ribosome (16%), and endoplasmic reticulum (7%) (supplemental Fig. S6e), outlining the coverage of the GPS method.
DISCUSSION
Despite recent achievements within the field of affinity proteomics (7), two major bottlenecks remain before antibody microarray-based analysis can be performed in a more global-scale and/or in discovery mode. Today, antibodies of known specificities are arrayed, meaning that we have preselected what protein families that will be targeted, thereby excluding the possibility to discover novel proteins. Furthermore, the number of readily available antibodies, designed for microarray applications (7), and directed against proteins of the human proteome is limited. Both of these limitations could be resolved by designing motif-specific antibodies, where each motif is present in several different proteins, including also proteins not yet annotated.
In this study, we have demonstrated proof-of-concept for a method based on such motif specific antibodies, denoted CIMS-antibodies, combining the best features of affinity proteomics and mass spectrometry. Consequently, by using a probe source based on recombinant CIMS-antibodies, the above limitations have been reduced, because (1) antibodies with novel motif specificities could be designed, selected and validated, and (2) a limited number of antibodies could be used to target a large set of proteins both in a discovery mode as well as in a species independent manner. In addition, the issue of antibody availability/renewability, hampering previous platforms based on affinity proteomics and MS (21–24) has been resolved using a recombinant antibody library as probe source. Based on experimental LC-MS/MS data, the CIMS antibodies were found to specifically recognizing a linear epitope composed of two to four conserved residues, whereas the identity of the neighboring residues was less crucial (Fig. 2A, supplemental Fig. S3, and supplemental Tables S3–S5). Similar observations have been made by Choulier and coworkers, indicating that antibodies recognized linear epitopes with a length of about six amino acids (27, 28). Changes among “allowed” residues in the nonessential positions do not abolish binding, but might influence the affinity of antibody-peptide interaction. It should be noted that the selections in this study were not designed to optimize the affinity, although anti-protein scFv antibodies selected from the present antibody library, using standard protocols, frequently display nm affinities (29). The anti-peptide CIMS antibodies were found to display affinities (KD values) in the μm range (Table I and supplemental Fig. 2) and these affinities were still sufficient to perform adequately in the GPS approach. A higher affinity could be generated by adopting more stringent selection criteria or subjecting the present CIMS antibodies to affinity maturation.
The first generation of selection motifs was designed (Table I and supplemental Table S1) mainly by considering the frequency of which the motifs occurred in the human peptidome. Despite this, the motifs were found to be highly applicable, even across species, allowing us to select binders targeting a wide range of proteins with heterogeneous origin (regarding e.g. specie and location) displaying a broad range of physico-chemical properties. In this context, it should be noted that the concept of designing unique motifs allowing a limited set of binders to cover a significant part of the human proteome (25) was supported by a recent in-silico study (26). Notably, the GPS method also detected several peptides previously not reported in the PeptideAtlas for all three species targeted (Fig. 1 and supplemental Tables S3–S5). Hence, the GPS methodology could provide a complementary approach to existing proteomic technologies, in the end further increasing the tentative coverage. Moreover, the GPS method was capable of targeting peptides (proteins) across the entire known dynamic range (at least four orders of magnitude) in yeast, and even targeting protein species anticipated to be very rare (<50 copies/cell) (Fig. 3D) (39). This shows that even low-abundant proteins could be detected in discovery mode using affinity proteomics. In future efforts, the design of the motifs (and choice of digesting enzyme) could be refined to tailor the method toward certain, preferred targets, e.g. low-abundant proteins and/or certain groups of functional proteins. Depending on the design of the motifs combined with how the antibodies were selected, the GPS set-up could thus also be customized toward either assays targeting preselected proteins/groups of proteins or protein profiling aiming for a more broad coverage. The former set-up could also be combined with a multiple reaction monitoring-based approach (40). Additional work will be required to explore the use of the GPS methodology for studying different protein variants (e.g. post-translational modifications and isoforms). By multiplexing the method (Fig. 3), the number of runs required to perform these tasks could be anticipated to be moderate. Future work will also be required to investigate how many antibodies that could be mixed in a single capture step, whereas maintaining overall assay performance. In addition, the GPS methodology is compatible with different forms of automation (41), in the end providing economically advantageous set-ups compatible with high-throughput.
The feasibility of the GPS approach could potentially be monitored by evaluating the overlap between the experimentally observed proteins (supplemental Tables S3–S5 and S7–S8) and the trypsin digested predictions in Table I. The predictions in Table I were performed assuming a four or six fixed amino acid motif, with the added possibility of allowing wobbling in one position (except for the C-terminal residue). However, the experimental data, showed that the CIMS antibodies recognized motifs characterized by a few key residues, i.e. a shorter motif than anticipated, reflecting the selection criteria adopted, thereby impairing the evaluation. Consequently, we will address this issue in future studies, where we will design the second generation of motifs based on the present knowledge and in combination with a range of selection criteria.
The experimental observation that the CIMS antibodies recognized shorter motifs, could, combined with the ability to detect low abundance proteins, potentially result in more tentative peptides to be identified by each antibody than anticipated (Table I). It is, however, difficult to evaluate the false negative rate by comparing the results for the refined motifs generated from the experimental data in an in-silico trypsin prediction versus the number of experimentally identified targets. The reason being that the number of actually MS/MS detected and identified peptides, used as read-out, is dependent on many interchelating factors, such as MS/MS performance and antibody affinity. But depending on the selection criteria imposed, CIMS antibodies displaying binding patterns more stringent e.g. six amino acids motif could be selected. This would not only allow us to adequately evaluate the false binding rate, but also to generate CIMS antibodies displaying the kind of binding patterns required by the GPS-based application at hand. It was interesting to note that the CIMS antibodies displayed reactivity against intact, semidenatured, arrayed proteins (protein arrays) and not only digested peptides in solution. However, the protein array experiments should only be seen as a complementary method to control the reactivity of the CIMS antibodies. Additional experiments will be required to delineate the potential use of CIMS antibodies against also motif-carrying intact proteins.
Using the GPS methodology, the identification of a protein will mainly be based on a single peptide MS/MS hit, placing high demands on the technology to minimize any false positives (42). Here, we approached the identification by using a combination of Mascot, X!Tandem, and FDR, as provided by the Proteios software environment, for defining positive hits with high confidence level (35). To handle this key issue, several approaches can be envisioned to increase the confidence level even further. By taking advantage of the á priori information about the amino acid sequence of the binding motifs, a filter function could be devised to identify tentative false positive hits while improving true positive (42). Using instrumentation only with very high parent ion mass (>5ppm) accuracy and resolution enables the elemental composition of peptide to be determined, which greatly improves the effectiveness of database searching. Furthermore, by collecting alternating scans of collisionally activated dissociation and electron transfer dissociation for peptide fragmentation generates two independent, orthogonal MS/MS spectra for database searching. Moreover, chemical tagging of the amino acids, including for example isobaric tags for relative and absolute quantification and stable isotope labeling by amino acids in cell culture, would also be beneficial providing improved ionization and would provide a means for quantitative and multiplexed analysis (43). The combination of high mass accuracy, dual fragmentation type data and chemical modification would increase the database search confidence levels by an order or two of magnitude. In comparison, by using stable isotope labeling by amino acids in cell culturein combination with one-dimensional gel electrophoresis and isoelectric focusing of peptides followed by high resolution analysis on a LTQ-Orbitrap, Graumann and coworkers succeeded in profiling mouse embryonic stem cells to a depth of 5111 proteins (44). Hence, by using advanced MS/MS set-ups, the principle underlying GPS could be demonstrated, whereas the initial applications in e.g. yeast also demonstrated sensitivity and dynamic range.
However, backtracking (single) degenerate peptides, present in two or more proteins (database entries) will be challenging, not only for our GPS approach, but for any MS-based approach. In regular profiling efforts, Occam's razor (get the simplest list of proteins sufficient to explain the observed peptides assigned to MS/MS spectra in the data set) is often used (not necessarily optimal). In the GPS approach, it will be essential to design adequate (unique) motifs and select high-performing antibodies and/or to use a set of CIMS antibodies targeting two or more peptides per protein (multicoverage) to minimize this issue. In this context, it is of interest to note that Planatscher et al. have showed that about only 2000 solutions (motifs) will be required to cover the entire human proteome and to provide multicoverage for around 13,800 proteins (26).
The repeatability for the entire GPS set-up, i.e. including both the affinity capture and subsequent LC-MS/MS step, was found to be in the 50–69% range at a false discovery rate of 0.01 (Fig. 3B). To the best of our knowledge, one could routinely expect to achieve a 35–50% peptide overlap reproducibility among technical LS-MS/MS runs in a discovery mode (45), and without adding/including a sample fractionation strategy (e.g. strong cation exchange), which most likely would cause the reproducibility to drop even further. Furthermore, the reproducibility of the label-free quantification was found to display CV values in the range of 11–14% (Fig. 3C). This is comparable to other existing parallel methods, such as the golden-standard MRM, where CV values around 10–15% have routinely been achieved (40). Notably, the GPS measurements of the peptides were performed in the entire range of the mass spectrometry detector (roughly four orders of magnitude). As could be expected, a higher variance was observed for the low-abundance peptides (Fig. 3C). Further, looking at the experimental ratios of the levels between the dilutions (2.7 and 3.2), the values were lower than the theoretically expected ratio (4.0). This discrepancy could be explained by the fact that we reported nonnormalized data. Although additional experiments will be required to delineate feature of label-free quantitative capability in more detail, we anticipate the method to compare well with traditional quantitative approaches. In all, the GPS methodology displayed adequate performance (e.g. reproducibility), supporting the applicability of the methodology.
Taken together, we have demonstrated proof-of-principle for that a limited number of motif specific recombinant antibodies could be used to profile crude, digested proteomes when combined with a mass spectrometry-based read-out in a reproducible manner. The GPS platform was shown to provide a broad and novel coverage, and to have the potential to reach deep into a proteome in a species independent manner. Furthermore, the methodology has potential for both discovery projects (e.g. unbiased biomarker discovery) as well as for focused profiling efforts (e.g. directed pathway analysis) targeting complex proteomes.
Acknowledgments
We thank Dr. Fredrik Levander for assistance with the Proteios Software Environment, Kristofer Wårell for running the SignPept analysis, Mats Mågård and Dr. Karin M. Hansson for assistance with LC-MS/MS instruments, Liselotte Andersson for assistance with the MALDI-TOF instrument, Nicolai Bache for MALDI-TOF-TOF analysis, and Björn Hambe for developing the affinity columns.
Footnotes
* This project was supported by grants from Swedish National Research Council (VR-NT), the Foundation for Strategic Research (SSF) (Strategic Center for Translational Cancer Research-CREATE Health), and Vinnova.
 This article contains supplemental material.
This article contains supplemental material.
1 The abbreviations used are:
- GPS
- global proteome survey
- CIMS
- context independent motif specific
- FDR
- false discovery rate
- LTQ
- linear ion trap
- PBS
- phosphate-buffered saline
- BSA
- bovine serum albumin
- IPI
- International Protein Index.
REFERENCES
- 1. Cho W. C. (2007) Contribution of oncoproteomics to cancer biomarker discovery. Mol. Cancer 6, 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Hanash S. (2003) Disease proteomics. Nature 422, 226–232 [DOI] [PubMed] [Google Scholar]
- 3. Hu S., Loo J. A., Wong D. T. (2006) Human body fluid proteome analysis. Proteomics 6, 6326–6353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bengtsson S., Krogh M., Szigyarto C. A., Uhlen M., Schedvins K., Silfverswärd C., Linder S., Auer G., Alaiya A., James P. (2007) Large-scale proteomics analysis of human ovarian cancer for biomarkers. J. Proteome Res. 6, 1440–1450 [DOI] [PubMed] [Google Scholar]
- 5. Cravatt B. F., Simon G. M., Yates J. R., 3rd (2007) The biological impact of mass-spectrometry-based proteomics. Nature 450, 991–1000 [DOI] [PubMed] [Google Scholar]
- 6. Listgarten J., Emili A. (2005) Practical proteomic biomarker discovery: taking a step back to leap forward. Drug Discov. Today 10, 1697–1702 [DOI] [PubMed] [Google Scholar]
- 7. Borrebaeck C. A. K., Wingren C. (2007) High-throughput proteomics using antibody microarrays: an update. Expert Rev. Mol. Diagnos. 7, 673–686 [DOI] [PubMed] [Google Scholar]
- 8. Kusnezow W., Banzon V., Schroder C., Schaal R., Hoheisel J. D., Ruffer S., Luft P., Duschl A., Syagailo Y. V. (2007) Antibody microarray-based profiling of complex specimens: systematic evaluation of labeling strategies. Proteomics 7, 1786–1799 [DOI] [PubMed] [Google Scholar]
- 9. Wingren C., Ingvarsson J., Dexlin L., Szul D., Borrebaeck C. A. K. (2007) Design of recombinant antibody microarrays for complex proteome analysis: choice of sample labeling-tag and solid support. Proteomics 7, 3055–3065 [DOI] [PubMed] [Google Scholar]
- 10. Carlsson A., Wingren C., Ingvarsson J., Ellmark P., Baldertorp B., Fernö M., Olsson H., Borrebaeck C. A. K. (2008) Serum proteome profiling of metastatic breast cancer using recombinant antibody microarrays. Eur. J. Cancer 44, 472–480 [DOI] [PubMed] [Google Scholar]
- 11. Ingvarsson J., Wingren C., Carlsson A., Ellmark P., Wahren B., Engström G., Harmenberg U., Krogh M., Peterson C., Borrebaeck C. A. K. (2008) Detection of pancreatic cancer using antibody microarray-based serum protein profiling. Proteomics 8, 2211–2219 [DOI] [PubMed] [Google Scholar]
- 12. Sanchez-Carbayo M. (2006) Antibody arrays: technical considerations and clinical applications in cancer. Clin. Chem. 52, 1651–1659 [DOI] [PubMed] [Google Scholar]
- 13. Borrebaeck C. A. K. (2006) Antibody microarray-based oncoproteomics. Expert Opin. Biol. Ther. 6, 833–838 [DOI] [PubMed] [Google Scholar]
- 14. Kingsmore S. F. (2006) Multiplexed protein measurement: technologies and applications of protein and antibody arrays. Nat. Rev. Drug Discov. 5, 310–320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wingren C., Ingvarsson J., Lindstedt M., Borrebaeck C. A. K. (2003) Recombinant antibody microarrays–a viable option?. Nat. Biotechnol. 21, 223. [DOI] [PubMed] [Google Scholar]
- 16. Shafer M. W., Mangold L., Partin A. W., Haab B. B. (2007) Antibody array profiling reveals serum TSP-1 as a marker to distinguish benign from malignant prostatic disease. Prostate 67, 255–267 [DOI] [PubMed] [Google Scholar]
- 17. Hurst G. B., Buchanan M. V., Foote L. J., Kennel S. J. (1999) Analysis for TNF-alpha using solid-phase affinity capture with radiolabel and MALDI-MS detection. Anal. Chem. 71, 4727–4733 [DOI] [PubMed] [Google Scholar]
- 18. Kiernan U. A., Tubbs K. A., Nedelkov D., Niederkofler E. E., Nelson R. W. (2002) Comparative phenotypic analyses of human plasma and urinary retinol binding protein using mass spectrometric immunoassay. Biochem. Biophys. Res. Commun. 297, 401–405 [DOI] [PubMed] [Google Scholar]
- 19. Lacey J. M., Bergen H. R., Magera M. J., Naylor S., O'Brien J. F. (2001) Rapid determination of transferrin isoforms by immunoaffinity liquid chromatography and electrospray mass spectrometry. Clin. Chem. 47, 513–518 [PubMed] [Google Scholar]
- 20. Niederkofler E. E., Tubbs K. A., Gruber K., Nedelkov D., Kiernan U. A., Williams P., Nelson R. W. (2001) Determination of beta-2 microglobulin levels in plasma using a high-throughput mass spectrometric immunoassay system. Anal. Chem. 73, 3294–3299 [DOI] [PubMed] [Google Scholar]
- 21. Anderson N. L., Anderson N. G., Haines L. R., Hardie D. B., Olafson R. W., Pearson T. W. (2004) Mass spectrometric quantitation of peptides and proteins using Stable Isotope Standards and Capture by Anti-Peptide Antibodies (SISCAPA). J. Proteome Res. 3, 235–244 [DOI] [PubMed] [Google Scholar]
- 22. Lisek C. A., Bailey J. E., Benson L. M., Yaksh T. L., Jardine I. (1989) Quantitation of endogenous substance P by on-line microcolumn liquid chromatography/continuous-flow fast-atom bombardment mass spectrometry. Rapid Commun. Mass Spectrom. 3, 43–46 [DOI] [PubMed] [Google Scholar]
- 23. Scrivener E., Barry R., Platt A., Calvert R., Masih G., Hextall P., Soloviev M., Terrett J. (2003) Peptidomics: A new approach to affinity protein microarrays. Proteomics 3, 122–128 [DOI] [PubMed] [Google Scholar]
- 24. Stemmann O., Zou H., Gerber S. A., Gygi S. P., Kirschner M. W. (2001) Dual inhibition of sister chromatid separation at metaphase. Cell 107, 715–726 [DOI] [PubMed] [Google Scholar]
- 25. Wingren C., James P., Borrebaeck C. A. K. (2009) Strategy for surveying the proteome using affinity proteomics and mass spectrometry. Proteomics 9, 1511–1517 [DOI] [PubMed] [Google Scholar]
- 26. Planatscher H., Supper J., Poetz O., Stoll D., Joos T., Templin M. F., Zell A. (2010) Optimal selection of epitopes for TXP-immunoaffinity mass spectrometry. Algorithms Mol. Biol. 5, 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Choulier L., Laune D., Orfanoudakis G., Wlad H., Janson J., Granier C., Altschuh D. (2001) Delineation of a linear epitope by multiple peptide synthesis and phage display. J. Immunol. Methods 249, 253–264 [DOI] [PubMed] [Google Scholar]
- 28. Choulier L., Orfanoudakis G., Robinson P., Laune D., Ben Khalifa M., Granier C., Weiss E., Altschuh D. (2002) Comparative properties of two peptide-antibody interactions as deduced from epitope delineation. J. Immunol. Methods 259, 77–86 [DOI] [PubMed] [Google Scholar]
- 29. Söderlind E., Strandberg L., Jirholt P., Kobayashi N., Alexeiva V., Aberg A. M., Nilsson A., Jansson B., Ohlin M., Wingren C., Danielsson L., Carlsson R., Borrebaeck C. A. K. (2000) Recombining germline-derived CDR sequences for creating diverse single-framework antibody libraries. Nat. Biotechnol. 18, 852–856 [DOI] [PubMed] [Google Scholar]
- 30. Engberg J., Andersen P. S., Nielsen L. K., Dziegiel M., Johansen L. K., Albrechtsen B. (1996) Phage-display libraries of murine and human antibody Fab fragments. Mol. Biotechnol. 6, 287–310 [DOI] [PubMed] [Google Scholar]
- 31. Hallborn J., Carlsson R. (2002) Automated screening procedure for high-throughput generation of antibody fragments. BioTechniques Suppl. 33, 530–537 [PubMed] [Google Scholar]
- 32. Büssow K., Nordhoff E., Lübbert C., Lehrach H., Walter G. (2000) A human cDNA library for high-throughput protein expression screening. Genomics 65, 1–8 [DOI] [PubMed] [Google Scholar]
- 33. Büssow K., Cahill D., Nietfeld W., Bancroft D., Scherzinger E., Lehrach H., Walter G. (1998) A method for global protein expression and antibody screening on high-density filters of an arrayed cDNA library. Nucleic Acids Res. 26, 5007–5008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Rappsilber J., Ishihama Y., Mann M. (2003) Stop and go extraction tips for matrix-assisted laser desorption/ionization, nanoelectrospray, and LC/MS sample pretreatment in proteomics. Anal. Chem. 75, 663–670 [DOI] [PubMed] [Google Scholar]
- 35. Häkkinen J., Vincic G., Månsson O., Wårell K., Levander F. (2009) The proteios software environment: an extensible multiuser platform for management and analysis of proteomics data. J. Proteome Res. 8, 3037–3043 [DOI] [PubMed] [Google Scholar]
- 36. Vizcaino J. A., Côte R., Reisinger F., Foster J. M., Mueller M., Rameseder J., Hermjakob H., Martens L. (2009) A guide to the Proteomics Identifications Database proteomics data repository. Proteomics 9, 4276–4283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Crooks G. E., Hon G., Chandonia J. M., Brenner S. E. (2004) WebLogo: a sequence logo generator. Genome Res. 14, 1188–1190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Deutsch E. W., Lam H., Aebersold R. (2008) PeptideAtlas: a resource for target selection for emerging targeted proteomics workflows. EMBO Rep. 9, 429–434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ghaemmaghami S., Huh W. K., Bower K., Howson R. W., Belle A., Dephoure N., O'Shea E. K., Weissman J. S. (2003) Global analysis of protein expression in yeast. Nature 425, 737–741 [DOI] [PubMed] [Google Scholar]
- 40. Addona T. A., Abbatiello S. E., Schilling B., Skates S. J., Mani D. R., Bunk D. M., Spiegelman C. H., Zimmerman L. J., Ham A. J., Keshishian H., Hall S. C., Allen S., Blackman R. K., Borchers C. H., Buck C., Cardasis H. L., Cusack M. P., Dodder N. G., Gibson B. W., Held J. M., Hiltke T., Jackson A., Johansen E. B., Kinsinger C. R., Li J., Mesri M., Neubert T. A., Niles R. K., Pulsipher T. C., Ransohoff D., Rodriguez H., Rudnick P. A., Smith D., Tabb D. L., Tegeler T. J., Variyath A. M., Vega-Montoto L. J., Wahlander A., Waldemarson S., Wang M., Whiteaker J. R., Zhao L., Anderson N. L., Fisher S. J., Liebler D. C., Paulovich A. G., Regnier F. E., Tempst P., Carr S. A. (2009) Multi-site assessment of the precision and reproducibility of multiple reaction monitoring-based measurements of proteins in plasma. Nat. Biotechnol. 27, 633–641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Whiteaker J. R., Zhao L., Anderson L., Paulovich A. G. (2010) An automated and multiplexed method for high throughput peptide immunoaffinity enrichment and multiple reaction monitoring mass spectrometry-based quantification of protein biomarkers. Mol. Cell. Proteomics 9, 184–196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Helsens K., Timmerman E., Vandekerckhove J., Gevaert K., Martens L. (2008) Peptizer: A tool for assessing false positive peptide identifications and manually validating selected results. Mol. Cell. Proteomics 7, 2364–2372 [DOI] [PubMed] [Google Scholar]
- 43. Chen X., Sun L., Yu Y., Xue Y., Yang P. (2007) Amino acid-coded tagging approaches in quantitative proteomics. Expert Rev. Proteomics 4, 25–37 [DOI] [PubMed] [Google Scholar]
- 44. Graumann J., Hubner N. C., Kim J. B., Ko K., Moser M., Kumar C., Cox J., Schöler H., Mann M. (2008) Stable isotope labeling by amino acids in cell culture (SILAC) and proteome quantitation of mouse embryonic stem cells to a depth of 5,111 proteins. Mol. Cell. Proteomics 7, 672–683 [DOI] [PubMed] [Google Scholar]
- 45. Tabb D. L., Vega-Montoto L., Rudnick P. A., Variyath A. M., Ham A. J., Bunk D. M., Kilpatrick L. E., Billheimer D. D., Blackman R. K., Cardasis H. L., Carr S. A., Clauser K. R., Jaffe J. D., Kowalski K. A., Neubert T. A., Regnier F. E., Schilling B., Tegeler T. J., Wang M., Wang P., Whiteaker J. R., Zimmerman L. J., Fisher S. J., Gibson B. W., Kinsinger C. R., Mesri M., Rodriguez H., Stein S. E., Tempst P., Paulovich A. G., Liebler D. C., Spiegelman C. (2010) Repeatability and reproducibility in proteomic identifications by liquid chromatography-tandem mass spectrometry. J. Proteome Res. 9, 761–776 [DOI] [PMC free article] [PubMed] [Google Scholar]