Abstract
Colorectal cancer (CRC) is the third most commonly diagnosed cancer in males and second in females worldwide. Unfortunately 40–50% of patients already have metastatic disease at presentation when prognosis is poor with a 5-year survival of <10%. Reactive oxygen species (ROS) have been proposed to play a crucial role in tumor metastasis. We now show that higher levels of ROS accumulation are found in a colorectal cancer-derived metastatic cell line (SW620) compared with a cell line (SW480) derived from the primary lesion from the same patient. In addition, ROS accumulation can affect both the migratory and invasive capacity of SW480 and SW620 cells. To explore the molecular mechanism underlying ROS-induced migration and invasion in CRC, we have compared protein expression patterns between SW480 and SW620 cells using a two-dimensional electrophoresis-based proteomics strategy. A total of 63 altered proteins were identified from tandem MS analysis. Cluster analysis revealed dysregulated expression of multiple redox regulative or ROS responsive proteins, implicating their functional roles in colorectal cancer metastasis. Molecular and pathological validation demonstrated that altered expression of PGAM1, GRB2, DJ-1, ITGB3, SOD-1, and STMN1 was closely correlated with the metastatic potential of CRC. Functional studies showed that ROS markedly up-regulated expression of ITGB3, which in turn promoted an aggressive phenotype in SW480 cells, with concomitant up-regulated expression of STMN1. In contrast, knockdown of ITGB3 expression could mitigate the migratory and invasive potential of SW620 or H2O2-treated SW480 cells, accompanied by down-regulated expression of STMN1. The function of ITGB3 was dependant on the surface expression of integrin αvβ3 heterodimer. Furthermore, STMN1 expression and the PI3K-Akt-mTOR pathway were found to be involved in ROS-induced and ITGB3-mediated migration and invasion of colorectal cancer cells. Taken together, these studies suggest that ITGB3 plays an important role in ROS-induced migration and invasion in CRC.
Colorectal cancer (CRC)1 is a major cause of cancer-related deaths worldwide. Although around 50% of CRC patients can be cured by surgery and multimodal treatment before tumor cell dissemination, 40–50% of patients have metastatic disease at presentation and of these, 90% die within 5 years of diagnosis (1–4). Early diagnosis of individuals at high risk of metastasis is hampered by the lack of understanding of the molecular mechanisms involved.
Reactive oxygen species (ROS), such as superoxide anion (O2−), hydrogen peroxide (H2O2), and hydroxyl radicals (OH−), are generated endogenously by all aerobic organisms. These ROS appear to be involved in the regulation of various physiological pathways, including signal transduction, apoptosis, and differentiation (5, 6). Recently, emerging evidence has suggested the involvement of ROS and the aberrant activation of redox-sensitive signaling pathways in tumor invasion and migration (7, 8). Some ROS-regulated proteins play key roles in epithelial-mesenchymal transition and tumor metastasis, including E-cadherin, integrins, and matrix metalloproteinases (8–10).
Binding of integrins to binding partners on the extracellular matrix (ECM) results in activation of diverse intracellular signaling pathways, which are implicated in cell survival, proliferation, and migration (11, 12). In vitro and in vivo experiments have demonstrated that abnormal integrin function correlates with tumor invasion through binding to ECM, which in turn regulates intracellular signaling pathways associated with tumor progression (12–15). Most integrins interact with cytoplasmic signaling pathways such as PI3K and MAPK by activation of focal adhesion kinase and Src family kinases at the attachment sites. Some members of the Ras and Rho families of small GTPases are essential as signaling intermediates in this process (11–15).
Integrins are heterodimeric cell surface glycoproteins consisting of α and β subunits (11–15). There are two members in the β3 integrin family, αIIbβ3 and αvβ3. αIIbβ3 is a receptor expressed mainly on the surface of platelets and megakaryocytes, whereas αvβ3 is expressed on the surface of endothelial cells, smooth muscle cells, monocytes, and platelets (16). The expression of β3 integrin is mainly associated with tumor metastasis (16) and has been reported to increase the metastatic potential of melanoma, breast cancer, and lymphoma cells, but interestingly has been found to have the opposite effect in ovarian cancer (17–20).
STMN1, a ubiquitous cytosolic phosphoprotein, binds tubulin to control microtubule polymerization (21). It mainly regulates cell cycle progression through mitosis, and has also been reported to be involved in carcinogenesis, cell proliferation, tumor metastasis, and chemoresistance (22–25). Overexpression of STMN1 has been reported in many human malignancies, including leukemia, osteosarcoma, as well as breast, ovarian, liver, lung, and colon cancers (22–25). In addition, dysregulated expression of STMN1 has been found to be associated with the activation of intracellular pathways such as MAPK and PI3K, which can be triggered by integrin engagement in response to numerous intra- or extracellular stimuli such as ROS (8–15, 26–29).
Two-dimensional gel electrophoresis (2-DE) based proteomics has been shown to be a powerful tool to rapidly profile differentially expressed proteins associated with a number of diseases (30–32). The paired colorectal cancer cell lines SW480 (derived from a surgical specimen of a primary colon adenocarcinoma) and SW620 (derived from a lymph node metastasis of the same patient), have been widely used in biochemical, immunological, genetic, and proteomics studies for screening disease markers associated with colon cancer progression (33–40) notwithstanding some controversy regarding the metastatic potential of these cells (41). In the current study, these cell lines were employed as a model in a 2DE-MS-based proteomics approach to investigate changed expression in ROS signaling, with an aim to better understanding the molecular mechanisms underlying ROS-induced colon cancer progression.
MATERIALS AND METHODS
Cell Lines, Transfections, and Treatments
SW480 and SW620 cell lines were purchased from American Type Culture Collection (ATCC, Rockville, MD). Cells were maintained in Dulbecco's Modified Eagle's Medium (DMEM, Invitrogen) containing 10% fetal calf serum (Hyclone, Logan, UT), penicillin (107 U/L) and streptomycin (10 mg/L) at 37 °C in a humidified chamber containing 5% CO2. For ROS treatment, the media were changed daily whereas SW480 cells were incubated for 4 days in the presence of 400 μmol/L H2O2.. In some experiments, cells were pretreated with 5 mmol/L N-acetylcysteine (NAC) for 30 min before addition of H2O2.
siRNA-ITGB3, corresponding to human ITGB3 at nucleotides 701–721 (5′-CCGCTTCAATGAGGAAGTGAA-3′), and siRNA-STMN1 corresponding to human STMN1 at nucleotides 254–272 (5′-AGGCAAUAGAAGAGAACAAdtdt-3′) were synthesized following the published literature (42, 43). The scrambled sequence, with no homology to any human gene, was used as negative control (NC). For siRNA treatment, both SW480 (in the presence of 400 μmol/L H2O2) and SW620 cells were transfected with siRNA-ITGB3 or the NC siRNA using lipofectamine RNAiMAX (Invitrogen) according to the manufacturer's instructions. Cells treated with lipofectamine RNAiMAX alone were used as a mock control. For motility and proliferation assays, cells were transfected with siRNA for 24 h before use. For expression analyses, cells were harvested at 48 h post-siRNA transfection.
ITGB3-plasmid encoding full-length human ITGB3 was a kind gift from Professor Erik H. J. Danen (Leiden University, The Netherlands) (44). For ITGB3 expression, the ITGB3-plasmid and control empty vector were separately transfected into SW480 cells using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions and the stable transfectants were selected in the presence of 0.8 mg/ml G418 (Invitrogen).
Clinical Specimens
All colorectal carcinomas and corresponding adjacent normal tissues were obtained from patients who underwent surgical resections at West China Hospital or Sichuan Provincial People's Hospital (Chengdu, China). All paired samples were immediately frozen in liquid nitrogen. Ethics approval was obtained from the Institutional Ethics Committee of Sichuan University. Tumor differentiation was characterized according to WHO classification, whereas the surgical pathologic stage was analyzed according to the TNM classification system of the International Union against Cancer (45). Clinico-pathological information for these patients is summarized in Table I.
Table I. Clinico-pathologic parameters of all patients.
| Clinico-pathologic features | Number | % |
|---|---|---|
| Gender | 30 | |
| Male | 16 | 53.3% |
| Female | 14 | 46.7% |
| Age | 60.5 ± 13.2 (35–84)a | |
| ≤60 | 19 | 63.3% |
| >60 | 11 | 36.7% |
| Histodifferentiation grading | ||
| Well differentiated | 10 | 33.3% |
| Moderately differentiated | 16 | 53.3% |
| Poorly differentiated | 4 | 13.4% |
| T stage | ||
| T1 | 3 | 10% |
| T2 | 2 | 6.7% |
| T3 | 17 | 56.7% |
| T4 | 8 | 26.6% |
| N (lymph nod metastasis) | ||
| N0 | 15 | 50% |
| N1 N2 | 15 | 50% |
| Surgical pathologic stage | ||
| I | 3 | 10% |
| II | 9 | 30% |
| III | 11 | 36.7% |
| IV | 7 | 23.3% |
a Mean age in years.
Reactive Oxygen Species (ROS) Measurement
Intracellular ROS were detected by staining cells with 2′,7′-dichlorofluorescein diacetate (DCFH-DA) (Genmed Scientifics Inc., Burlington, MA, USA) according to the manufacturer's instructions. The DCFH-DA signal was observed using an inverted fluorescent microscope or measured with a Molecular Devices SPECTRAMAX M5 fluorimeter (490 nm excitation and 530 nm emission).
2-DE and Image Analysis
Cells (3 × 107) were lysed in 1 ml lysis buffer (7 m urea, 2 m thiourea, 4% 3-[(3-cholamidopropyl)dimethylammonio]propanesulfonate, 100 mm dithiotreitol, 0.2% pH 3–10 ampholyte, BioRad) containing protease inhibitor mixture 8340 (Sigma). Samples were kept on ice and sonicated in six cycles of 10 s, each consist of 5 s sonication, followed by a 10 s break. After centrifugation at 14,000 rpm for 1 h at 4 °C, the supernatant was collected and the protein concentrations determined using the DC Protein Assay Kit (Bio-Rad). Protein samples (2 mg) were applied to immobilized pH gradient strip (17 cm, pH 3–10 NL, Bio-Rad) using a passive rehydration method. After 12–16 h of rehydration, the strips were transferred to an isoelectric focusing (IEF) Cell (Bio-Rad). IEF was performed as follows: 250 V for 30 min, linear; 1000 V for 1 h, rapid; linear ramping to 10,000 V for 5 h, and finally 10,000 V for 6 h. Once IEF was completed, the strips were equilibrated in equilibration buffer (25 mm Tris-HCl, pH 8.8, 6 m urea, 20% glycerol, 2% SDS, and 130 mm dithiotreitol) for 15 min, followed by the same buffer containing 200 mm iodoacetamide instead of dithiotreitol for another 15 min. The second dimension was performed using 12% SDS-PAGE at 30 mA constant current per gel. The gels were stained using CBB R-250 (Merck, Germany) and scanned with a Bio-Rad GS-800 scanner. Triplicate samples were analyzed at each time point of treatment to ensure the reproducibility of analyses. The maps were analyzed by PDQuest software Version 6.1 (Bio-Rad). Each gel spot was normalized as a percentage of the total quantity of all spots in that gel and evaluated in terms of O.D. Only those spots that changed consistently and significantly (>2.0-fold) were selected for MS analysis.
In-gel Digestion
In-gel protein digestion was carried out using mass spectrometry grade trypsin (Trypsin Gold. Promega, Madison, WI) according to the manufacturer's instructions. Briefly, spots were cut out of the gel (1–2 mm diameter) using a razor blade, and destained twice with 100 mm NH4HCO3/50% acetonitrile (ACN) at 37 °C for 45 min. After dehydration and drying, the gels were preincubated in 10–20 μl trypsin solution for 1 h. Sufficient digestion buffer (40 mm NH4HCO3/10% ACN) to cover the gels was then added followed by overnight incubation at 37 °C. Peptide digests were extracted using MiliQ water, followed twice by extraction with 50% ACN/5% trifluoroacetic acid for 1 h each time. The combined extracts were dried in a vacuum concentrator at room temperature. The samples were then subjected to mass spectrometric analysis.
Electrospray Ionization-Quadrupole-Time-of-Flight (ESI-Q-TOF)
Mass spectra were acquired using a Q-TOF mass spectrometer (Micromass, Manchester, UK) fitted with an ESI source (Waters, Milford, MA). Tryptic digests were dissolved in 18 μl 50% ACN. Tandem MS (MS/MS) was performed in a data-dependent mode in which the top ten most abundant ions for each MS scan were selected for MS/MS analysis. The MS/MS data were acquired and processed using MassLynx V4.1 software (Micromass) and Mascot from Matrix Science in June 2009 was used to search the database using the following parameters: Database, SwissProt 57.3/NCBI(468851 sequences); taxonomy, homo species (20401 sequences); enzyme, trypsin; and allowance of one missed cleavage. Carbamidomethylation was selected as a fixed modification and oxidation/phosphorylation was allowed to be variable. The peptide and fragment mass tolerances were set at 1 and 0.2 Da, respectively. The data format was selected as Micromass PKL and the instrument was selected as ESI-Q-TOF. Proteins with probability based MOWSE scores exceeding their threshold (p < 0.05) were considered to be positively identified. If proteins were identified by a single peptide, the spectrum was validated manually. For a protein to be accepted, the assignment had to be based on four or more y- or b-series ions.
Western Blot
Proteins were extracted in radioimmunoprecipitation assay buffer (50 mm Tris-base, 1.0 mm EDTA, 150 mm NaCl, 0.1% SDS, 1% TritonX-100, 1% Sodium deoxycholate, 1 mm phenylmethylsulfonyl fluoride) and quantified using the DC protein assay kit (Bio-Rad, USA). Samples were separated on 8% or 12% SDS-PAGE and transferred to polyvinylidene difluoride membranes (Amersham Biosciences). The membranes were blocked overnight with phosphate-buffered saline (PBS) containing 0.1% Tween 20 in 5% skimmed milk at 4 °C, and subsequently probed using the primary antibodies: goat-anti-PGAM1 (diluted 1:1000, Abcam, Cambridge, UK), mouse-anti-DJ-1 (diluted 1:1000, Santa Cruz, Santa Cruz, CA), mouse-anti-GRB2 (diluted 1:300, Santa Cruz), mouse-anti-ITGB3 (diluted 1:300, Santa Cruz), mouse-anti-SOD-1 (diluted 1:1000, Santa Cruz), mouse-anti-STMN1 (diluted 1:1000, Santa Cruz), rabbit-anti-Akt (diluted 1:1000, Cell Signaling, Danvers, MA), mouse-anti-Phospho-Akt (S473) (diluted 1:1000, Cell Signaling), rabbit-anti-mTOR (diluted 1:1000, Cell Signaling), rabbit-anti-Phospho-mTOR (S2448) (diluted 1:500, Cell Signaling), rabbit-anti-p38 MAPK (diluted 1:1000, Cell Signaling) and rabbit-anti- Phospho-p38 MAPK (Thr180/Tyr182) (diluted 1:1000, Cell Signaling). Blots were incubated with the respective primary antibodies for 2 h at room temperature and washed 3 times in Tris-buffered saline with Tween 20. Subsequently, the blots were incubated with secondary antibody conjugated to Horseradish Peroxidase for 2 h at room temperature. Target proteins were detected by enhanced chemiluminescence reagents (Amersham Biosciences, Piscataway, NJ). β-actin was used as an internal control.
Immunohistochemistrical Analysis
Tissues were formalin-fixed and paraffin-embedded, and sections were consecutively cut (3–4 μm thickness) for immunohistochemistry analysis using a Dako EnVision System (Dako Cytomation GmbH, Hamburg, Germany) according to the manufacturer's instructions. Briefly, the paraffin-sections were dewaxed, rehydrated, and incubated in 3% H2O2 for 10 min in dark at room temperature to quench the endogenous peroxidase activity. Antigen retrieval was performed in citrate buffer (pH 6.0) using the autoclave sterilizer method. Subsequently, the sections were blocked with normal rabbit or goat serum diluted in PBS (pH 7.4) for 20 min at 37 °C, followed by an incubation at 4 °C overnight with the primary antibodies: anti-PGAM1 (diluted 1:150, Abcam, Cambridge, UK), mouse-anti-DJ-1 (diluted 1:150, Santa Cruz), mouse-anti-GRB2 (diluted 1:25, Santa Cruz), mouse-anti-ITGB3 (diluted 1:50, Santa Cruz), mouse-anti-SOD-1 (diluted 1:100, Santa Cruz) and mouse-anti-STMN1 (diluted 1:150, Santa Cruz). After rinsing in fresh PBS for 15 min, slides were incubated with horseradish peroxidase-linked rabbit anti-goat or anti-mouse antibodies at 37 °C for 40 min, followed by reaction with 3,3′-diaminobenzidine substrate solution (Dako Cytomation) and counterstaining with Mayer's hematoxylin. The immunohistochemical staining was assessed and scored by calculating the percentage of positive CRC cells and the immunostaining intensity as described previously (46).
Indirect Immunofluorescence
For immunofluorescence, cells were fixed with 4% paraformaldehyde in PBS for 30 min. After being washed twice with ice-cold PBS, fixed cells were blocked with PBS containing 5% bovine serum albumin at room temperature for 30 min, after incubation with anti-ITGB3 (diluted 1:50, Santa Cruz) or LM609(diluted 1:100, anti-αVβ3 integrin, Millipore, Billerica, MA) antibody at 4 °C overnight. The cells were washed twice with PBS and treated with fluorescein-conjugated secondary antibodies (1:100 dilution) in PBS for 1h at room temperature, followed by Hoechst staining of the cell nucleus (blue). Images were acquired using a Zeiss Imager. Z1 fluorescence microscope equipped with an AxioCam MRc5 digital CCD camera (Carl Zeiss Microimaging, Jena, Germany).
In Vitro Motility and Proliferation
Cell migration assays were performed in 24-well transwell chambers, (Corning, Corning, NY). Cells in serum-free medium (2.5 × 104 cells per well) were added to the upper chamber. After 24 h, the number of cells that migrated through the membrane to the lower chamber was counted. For invasion assays, matrigel (1:3, BD) was added to the transwell membrane chambers, incubated for 4 h, and cells were seeded. Cells, which had migrated to the lower chamber, were counted after 72 h. For blocking experiments, cells were added to the upper chambers in the presence of LM609 (10 μg/ml) or control mouse IgG (10 μg/ml), in addition, filter undersides were precoated with fibrinogen (20 μg/ml) in PBS and blocked with 2.5% bovine serum albumin in 0.2% Tween 20/PBS in migration assay. Migration and invasion assays were performed in triplicate for each cell line tested.
For the scratch wound healing assay, wounds were created in confluent cells by scraping the cell surface using a sterile pipet tip, and then washed with medium to remove free-floating cells and debris. Wound healing within the scraped line was documented daily over 72 h, and repeated in duplicate at least three times. For evaluation of cell growth in soft agar, cells (3000 cells/well) were resuspended in DMEM containing 10% FBS with 0.35% agarose and layered on top of 0.5% agarose in DMEM on 6-well plates. Cultures were maintained for 14 days and plates were stained with 0.3 ml of 0.005% Crystal Violet for more than 1 h. Colonies (larger than 200 μm) were then counted using a dissecting microscope. Each experiment was done in triplicate.
Statistics
All quantitative data were recorded as mean ± S.D. Comparisons between two groups were performed by Student's t test. Differences among multiple groups were assessed by one-way ANOVA analysis. Relevance analysis of ordinal data was performed by cross χ2 test. Statistical significance was defined as p < 0.05.
RESULTS
ROS Promotes the Migration and Invasion of CRC Cells
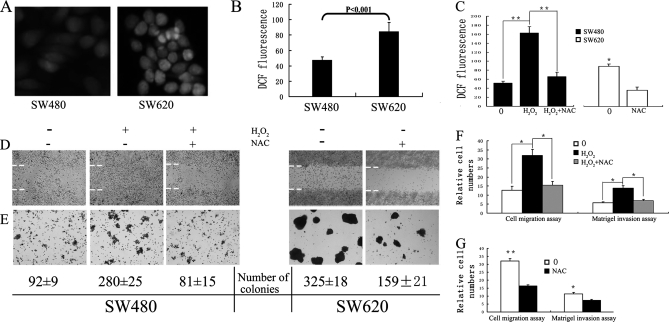
Recent studies have revealed that aberrant generation of cellular ROS is associated with tumor metastasis (7–10), and that accumulation of cellular ROS induced by H2O2 can promote a malignant phenotype in certain cancer cells, such as prostate cancer cells and hepatocellular carcinoma cells (7). In an initial study, levels of intracellular ROS, detected using cell-permeable 2′,7′-dichlorofluorescin diacetate as a probe, were found to be twofold higher in the SW620 cells derived from a CRC lymph node metastasis compared with the corresponding SW480 cell line derived from the primary tumor. (Fig. 1A and 1B).
Fig. 1.
ROS promotes the proliferation and migration of SW480 cells. A, Representative fluorescence images showing intracellular ROS levels of SW480 and SW620 detected by staining cells with 2′,7′-dichlorofluorescein diacetate. B, Intracellular ROS in SW480 and SW620 cells measured using a Molecular Devices SPECTRAMAX M5 fluorimeter (490 nm excitation and 530 nm emission). The x axis shows the average value of three independent experiments (each experiment had at least three triplicates). C, Intracellular ROS in SW480 and SW620 treated with or without H2O2 or/and NAC were measured using a Molecular Devices SPECTRAMAX M5 fluorimeter. D, A confluent monolayer was wounded with a sterile pipet tip and SW480 and SW620 cells, with or without H2O2 or/and NAC treatment, were allowed to migrate for 72 h before analysis. Dashed lines indicate the original wound boundaries. E, Representative photographs of soft agar colony formation 14 days after culture of cells, with mean colony counts from three independent experiments indicated below. F and G, The number of migrated and invaded SW480 (F) or SW620 (G) cells from five random fields were counted and presented with relative cell numbers. Migration proceeded for 24 h, while invasion was measured after 72 h. 0, untreated cells; H2O2, H2O2-treated cells; NAC, NAC treated cells. (*) p < 0.05; (**) p < 0.01. All data were from at least three independent experiments and are shown as mean ± S.D.
To evaluate the potential significance of ROS in colon cancer metastasis, SW480 cells were continuously treated with 400 μmol/L H2O2 (as ROS source) for 4 days in the presence or absence of NAC (Figs. 1C–1F). The migratory and invasive capacity of SW480 cells treated with or without H2O2 was measured by various methods including a wound healing (migration) assay (Figs. 1D and 1F), soft agar colony formation assay (Fig. 1E) and matrigel invasion (Fig. 1F). In the wound healing assay, the number of H2O2-treated SW480 cells migrating into the wound area was much higher than the control (Fig. 1D). Consistent with this observation, H2O2 produced about threefold more colonies in soft agar (Fig. 1E), induced SW480 cell migration by approximate threefold (Fig. 1F) and increased the invasion potential as demonstrated by martigel invasion by more than twofold (Fig. 1F). These effects were inhibited after treatment of cells with the antioxidant NAC (Figs. 1D–1F) which decreased the intracellular ROS level in both H2O2-treated SW480 and SW620 cells (Fig. 1C). Furthermore, treatment with NAC also reduced the proliferative, migratory and invasive capacity of SW620 cells (p < 0.01) (Figs. 1D–1E and 1G). These results demonstrate the functional potential of ROS in promoting cancer cell invasion and migratory potential.
Proteomic Analysis of SW480 and SW620 Protein Expression
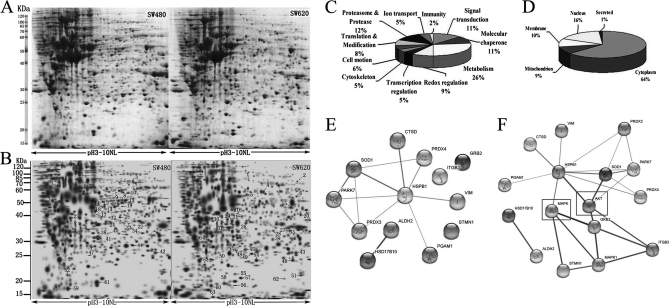
To explore the molecular mechanisms underlying ROS-induced migration and invasion of colorectal cancer cells, 2-DE based proteomics was used to profile differentially expressed proteins in SW480 and SW620 cells. Representative 2-DE maps are shown in Fig. 2A. Approximately 1200–1300 protein spots were detected by CBB R-250 staining in a single 2-DE gel. Each protein spot was normalized as a percentage of the total intensity of all spots in the gel. By comparing 2-DE patterns, differentially expressed proteins were defined as statistically meaningful (p < 0.05) based on both of the following two criteria: 1) intensity alterations of >2.0-fold and 2) observed in at least three individual experiments. Using these criteria, a total of 63 spots were identified as being differentially expressed. Of these, 28 proteins were down-regulated whereas 35 proteins were up-regulated in SW620 compared with SW480 cells (Figs. 2A and 2B).
Fig. 2.
Proteomic analysis of SW480 and SW620 protein expression. A, Representative two-dimensional gel images of human colorectal cancer cell lines SW480 and SW620. Total protein extracts were separated on pH 3–10 nonlinear immobilized pH gradient strips in the first dimension followed by 12% SDS-PAGE in the second dimension and visualized by CBB staining. B, Reference maps were generated from Fig 2A using PDQuest software. Sixty-three differentially expressed spots (28 up-regulated in SW480, 35 up-regulated in SW620) were identified (as numbered). Details for each numbered spot are reported in Table II. C, Sixty-three identified proteins were classified into 11 groups. These included metabolism (26%), redox regulation (9%), cell motion (6%), signal transduction (11%), and cell cytoskeleton (5%). D, These proteins were found to be located in the cytoplasm (64%), nucleus (16%), mitochondrion (9%), cell membrane (10%), or secreted (1%). (E and F) In silico protein interaction analysis. Regulated proteins involved in redox regulation or ROS signaling were analyzed for protein interactions using the web based software tool STRING, without (E) or with (F) addition of MAPK and AKT, which are critical kinases in ROS signaling.
Protein Identification, Validation, and Bioinformatics Analysis
Sixty-three spots with differential expression levels were subjected to MS/MS analysis. The MS/MS data were queried using the search algorithm MASCOT against the Expasy protein sequence database. Proteins were identified based on a number of criteria including pI, MW, the number of matched-peptides, and MOWSE score (Table II). The identified proteins were divided into various groups based on their biological functions and subcellular localization (Figs. 2C and 2D). This implicated roles in metabolism (26%), redox regulation (9%), cell motion (6%), signal transduction (11%), and cell cytoskeleton. The proteins were found to be located in the cytoplasm (64%), nucleus (16%), mitochondrion (9%), cell membrane (10%), or secreted (1%). For a macroscopic presentation, cluster maps and protein interaction and function networks were generated using Cluster or the KEGG-based software tool Cytoscape respectively (supplementary Fig. S1A and B). Fourteen proteins, accounting for 23% of the proteins identified, were found to be associated with redox regulation or ROS signaling (Table III). The intrinsic interaction of these proteins was analyzed using the web-based tool STRING. The redox regulative proteins and ROS responsive proteins were grouped in different clusters (Fig. 2E). Interestingly, these proteins could be linked together when MAPK and AKT, two critical kinases in ROS signaling, were added into the analysis (Fig. 2F).
Table II. Protein spots were identified by ESI-Q-TOF.
| Spot no. | Accession no. | Protein name | Gene name | Theoretical Mr | Theoretical pI | No. of peptide | Coverage % | Score |
|---|---|---|---|---|---|---|---|---|
| 1 | P05106 | Integrin β-3 | ITGB3 | 84389 | 4.97 | 3 | 4 | 120 |
| 2 | Q96AE4 | Far upstream element-binding protein | FUBP1 | 67560 | 7.18 | 8 | 14 | 95 |
| 3 | P13647 | Keratin, type II cytoskeletal 5 | KRT5 | 62378 | 7.58 | 17 | 24 | 446 |
| 4 | P78371 | T-complex protein 1 subunit β | CCT2 | 57357 | 6.02 | 7 | 19 | 238 |
| 5 | P05091 | Aldehyde dehydrogenase, mitochondrial | ALDH2 | 56381 | 6.63 | 10 | 29 | 365 |
| 6 | Q6ICS0 | ANXA11 protein | ANXA11 | 54336 | 6.91 | 3 | 9 | 135 |
| 7 | P08670 | Vimentin | VIM | 53520 | 5.06 | 5 | 12 | 155 |
| 8 | P20073 | Annexin A7 | ANXA7 | 52739 | 5.52 | 2 | 5 | 113 |
| 9 | Q6IAT1 | GDI2 protein | GDI2 | 50663 | 6.11 | 16 | 30 | 401 |
| 10 | Q9Y265 | RuvB-like 1 | RUVBL1 | 50228 | 6.02 | 25 | 26 | 178 |
| 11 | P26641 | Elongation factor 1-γ | EEF1G | 49987 | 6.27 | 6 | 8 | 103 |
| 12 | O75439 | Mitochondrial-processing peptidase subunit β | PMPCB | 49487 | 5.77 | 2 | 8 | 59 |
| 13 | P31943 | Heterogeneous nuclear ribonucleoprotein H | HNRNPH1 | 49229 | 5.89 | 4 | 12 | 98 |
| 14 | P35998 | 26S protease regulatory subunit 7 | PSMC2 | 48502 | 5.72 | 10 | 28 | 279 |
| 15 | P22234 | Multifunctional protein ADE2 | PAICS | 46948 | 7.09 | 5 | 18 | 152 |
| 16 | P07339 | Cathepsin D | CTSD | 44552 | 6.10 | 2 | 7 | 45 |
| 17 | Q53XU2 | Proteasome (Prosome, macropain) 26S subunit, non-ATPase, 13 | PSMD13 | 42918 | 5.53 | 17 | 48 | 448 |
| 18 | Q9Y570 | Protein phosphatase methylesterase 1 | PPME1 | 42184 | 5.67 | 6 | 7 | 175 |
| 19 | Q15019 | Septin-2 | SEPT2 | 41487 | 6.15 | 4 | 24 | 184 |
| 20 | Q9Y617 | Phosphoserine aminotransferase | PSAT1 | 40422 | 7.56 | 7 | 32 | 174 |
| 21 | Q53GF8 | Isocitrate dehydrogenase 3 (NAD+) α variant | No gene name | 39619 | 6.46 | 5 | 18 | 295 |
| 22 | P07355 | Annexin A2 | ANXA2 | 38472 | 7.56 | 43 | 51 | 838 |
| 23 | P37837 | Transaldolase | TALDO1 | 37540 | 6.36 | 2 | 6 | 89 |
| 24 | P05198 | Eukaryotic translation initiation factor 2 subunit 1 | EIF2S1 | 35980 | 5.02 | 9 | 40 | 245 |
| 25 | Q15293 | Reticulocalbin-1 | RCN1 | 35867 | 4.68 | 9 | 27 | 198 |
| 26 | P09525 | Annexin A4 | ANXA4 | 35751 | 5.85 | 6 | 27 | 348 |
| 27 | P60891 | Ribose-phosphate pyrophosphokinase 1 | PRPS1 | 34703 | 6.56 | 7 | 28 | 241 |
| 28 | P11908 | Ribose-phosphate pyrophosphokinase 2 | PRPS2 | 34637 | 6.17 | 10 | 23 | 146 |
| 29 | P05388 | 60S acidic ribosomal protein P0 | RPLP0 | 34273 | 5.72 | 24 | 38 | 586 |
| 30 | P52907 | F-actin-capping protein subunit α-1 | CAPZA1 | 32791 | 5.45 | 7 | 43 | 240 |
| 31 | Q15181 | Inorganic pyrophosphatase | PPA1 | 32660 | 5.54 | 8 | 23 | 149 |
| 32 | P00491 | Purine nucleoside phosphorylase | NP | 32117 | 6.45 | 19 | 44 | 226 |
| 33 | P45880 | Voltage-dependent anion-selective channel protein 2 | VDAC2 | 31566 | 7.50 | 4 | 11 | 98 |
| 34 | P10768 | S-formylglutathione hydrolase | ESD | 31462 | 6.54 | 7 | 20 | 124 |
| 35 | Q13162 | Peroxiredoxin-4 | PRDX4 | 30539 | 5.86 | 13 | 51 | 324 |
| 36 | P27707 | Deoxycytidine kinase | DCK | 30518 | 5.14 | 4 | 15 | 59 |
| 37 | Q14847 | LIM and SH3 domain protein 1 | LASP1 | 29717 | 6.61 | 2 | 9 | 61 |
| 38 | P61289 | Proteasome activator complex subunit 3 | PSME3 | 29506 | 5.69 | 5 | 17 | 145 |
| 39 | P18669 | Phosphoglycerate mutase 1 | PGAM1 | 28672 | 6.75 | 18 | 63 | 388 |
| 40 | P25788 | Proteasome subunit α type-3 | PSMA3 | 28302 | 5.19 | 8 | 29 | 279 |
| 41 | P78417 | Glutathione S-transferase ω-1 | GSTO1 | 27565 | 6.24 | 5 | 26 | 141 |
| 42 | Q99714 | 3-hydroxyacyl-CoA dehydrogenase type-2 | HSD17B10 | 26791 | 7.87 | 21 | 69 | 566 |
| 43 | Q15056 | Eukaryotic translation initiation factor 4H | EIF4H | 27253 | 6.92 | 5 | 40 | 274 |
| 44 | P60174 | Triosephosphate isomerase | TPIS | 26538 | 6.51 | 15 | 52 | 456 |
| 45 | P62993 | Growth factor receptor-bound protein 2 | GRB2 | 25206 | 5.89 | 2 | 22 | 136 |
| 46 | Q9BZK3 | Putative nascent polypeptide-associated complex subunit α-like protein | NACAP1 | 23306 | 4.53 | 8 | 13 | 108 |
| 47 | P04792 | Heat shock protein β-1 | HSPB1 | 22782 | 5.98 | 15 | 41 | 304 |
| 48 | Q5W0H4 | Tumor protein, translationally controlled 1 | TPT1 | 21525 | 5.34 | 20 | 74 | 470 |
| 49 | P30048 | Thioredoxin-dependent peroxide reductase | PRDX3 | 21468 | 5.77 | 9 | 55 | 356 |
| 50 | Q9BY32 | Inosine triphosphate pyrophosphatase | ITPA | 21445 | 5.50 | 4 | 22 | 79 |
| 51 | P30086 | Phosphatidylethanolamine-binding protein 1 | PEBP1 | 20925 | 7.43 | 2 | 18 | 65 |
| 52 | Q13185 | Chromobox protein homolog 3 | CBX3 | 20811 | 5.23 | 4 | 36 | 136 |
| 53 | Q04760 | Lactoylglutathione lyase | GLO1 | 20646 | 5.12 | 7 | 29 | 73 |
| 54 | Q99497 | DJ-1 | PARK7 | 19891 | 6.33 | 23 | 48 | 283 |
| 55 | P33316 | 5′-triphosphate nucleotidohydrolase, mitochondrial | DUT | 19345 | 7.96 | 14 | 61 | 257 |
| 56 | Q96CE4 | Stathmin | STMN1 | 17336 | 5.76 | 18 | 36 | 271 |
| 57 | P15531 | Nucleoside diphosphate kinase A | NME1 | 17148 | 5.83 | 26 | 51 | 402 |
| 58 | Q9BPX5 | Actin-related protein 2/3 complex subunit 5-like protein | ARPC5L | 16941 | 6.15 | 2 | 16 | 101 |
| 59 | P63241 | Eukaryotic translation initiation factor 5A-1 | EIF5A | 16701 | 5.08 | 22 | 46 | 488 |
| 60 | Q53S41 | Putative uncharacterized protein YWHAQ | YWHAQ | 16583 | 4.30 | 4 | 26 | 45 |
| 61 | P00441 | Superoxide dismutase [Cu-Zn] | SOD1 | 15935 | 5.70 | 12 | 62 | 205 |
| 62 | Q14019 | Coactosin-like protein 50 | COTL1 | 15813 | 5.55 | 3 | 17 | 84 |
| 63 | O75347 | Tubulin-specific chaperone A | TBCA | 12723 | 5.25 | 12 | 27 | 219 |
Table III. Proteins identified to be involved in Redox process.
| Spot no. | Accession number | Protein name | Average ratio | Subcellular location b | Main function | Redox process |
|---|---|---|---|---|---|---|
| 1 | P05106 | Integrin β-3 | 2.235 | Membrane | Signal transduction | Ros response |
| 5 | P05091 | Aldehyde dehydrogenase, mitochondrial | 2.349 | Mitochondrion | Alcohol metabolism | Redox regulation |
| 7 | P08670 | Vimentin | 0.343 | Cytoplasm | Cell motion | Ros response |
| 16 | P07339 | Cathepsin D | 2.372 | Lysosome | Protease | Redox regulation |
| 21 | Q53GF8 | Isocitrate dehydrogenase 3 (NAD+) α variant | 2.945 | Mitochondrion | Oxidation Reduction | Redox regulation |
| 35 | Q13162 | Peroxiredoxin-4 | 2.178 | Cytoplasm | Redox regulation | Redox regulation |
| 39 | P18669 | Phosphoglycerate mutase 1 | 6.364 | Cytoplasm | Metabolism | Redox regulation |
| 43 | Q99714 | 3-hydroxyacyl-CoA dehydrogenase type-2 | 2.825 | Mitochondrion | Lipid metabolic | Redox regulation |
| 45 | P62993 | Growth factor receptor-bound protein 2 | 2.117 | Cytoplasm | Signal transduction | Ros response |
| 47 | P04792 | Heat shock protein β-1 | 0.202 | Cytoplasm | Molecular chaperon | Redox regulation |
| 49 | P30048 | Thioredoxin-dependent peroxide reductase | 3.245 | Mitochondrion | Redox regulation | Redox regulation |
| 54 | Q99497 | Protein DJ-1 | 2.446 | Nucleus. Cytoplasm | Signal transduction | Redox regulation |
| 56 | Q96CE4 | Stathmin | 3.534 | Cytoplasm | Cell motion | Ros response |
| 61 | P00441 | Superoxide dismutase | 0.428 | Cytoplasm | Antioxidant | Redox regulation |
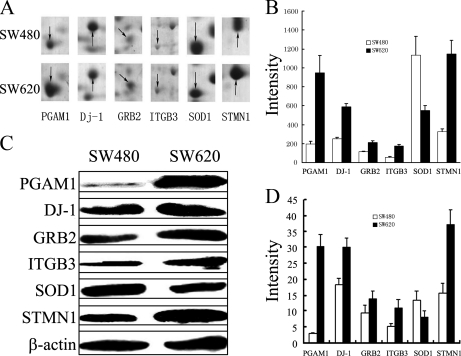
Six proteins (PGAM1, GRB2, DJ-1, ITGB3, SOD-1, and STMN1), which have a well documented involvement in tumor progression and are closely associated with ROS-induced cancer metastasis in a number of reports (8–15, 22–23), were chosen for further validation by image analysis (Figs. 3A and 3B) and Western blot analysis (Figs. 3C and 3D). Consistent with the observations from 2-DE analysis, expression of PGAM1, GRB2, DJ-1, ITGB3, STMN1 were up-regulated, whereas SOD1 was down-regulated in SW620, compared with SW480 cells.
Fig. 3.
The differential expression of PGAM1, GRB2, DJ-1, ITGB3, SOD1 and STMN1. A, Enlargement of selected regions in Fig. 2A. Spots selected are PGAM 1 (Spot 39), DJ-1(Spot 54), GRB2 (Spot 45), ITGB3 (Spot 1), SOD1 (Spot 61), and STMN1 (Spot 56). B, Relative expression of PGAM1, GRB2, DJ-1, ITGB3, SOD1, and STMN1. Each gel spot was quantified using PDQuest 2-DE software. C, The relative expression of PGAM1, GRB2, DJ-1, ITGB3, SOD1, and STMN1 in SW480 and SW620 cells was further validated by Western blot analysis. β-actin was used as a loading control. D, Bands were analyzed by densitometry using Quality-One software (Bio-Rad laboratories, Richmond, CA). The x axis shows the average intensity from four individual experiments. Data is presented as mean ± S.D.
Immunostaining for ROS-related Proteins in Colorectal Carcinoma
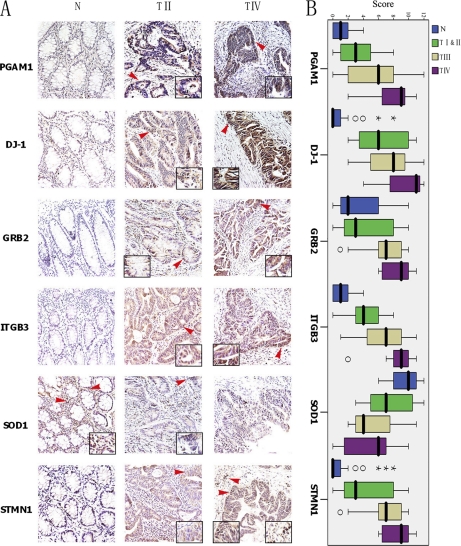
To examine the clinical significance of the aforementioned ROS-related proteins in the progression of colorectal carcinoma, immunohistochemical analysis was performed using 30 matched pairs of paraffin-embedded CRC tissues (the clinico-pathological parameters for the patients are summarized in Table I). Representative data is shown in Fig. 4A. Immunostaining of PGAM1, GRB2, DJ-1, ITGB3, and STMN1 was significantly stronger in cancer tissues than the matched normal colon mucosae (p < 0.001). By contrast, compared with corresponding normal tissues, SOD-1 was down-regulated in cancer tissues (p < 0.001). In addition, to determine whether expression of the ROS-related proteins within these lesions correlated with the surgical pathologic stage of colorectal carcinoma progression, the relative staining intensity for each case was scored and analyzed using SPSS software. The results showed that the immunoreactivity scores showed significant correlation with surgical-pathologic stage (Fig. 4B). Expression of PGAM1, GRB2, DJ-1, ITGB3, and STMN1 was increased in high-grade (Stage III and IV) tumors compared with low-grade (Stage I and II) tumors, whereas SOD-1 expression reduced with increasing grade (p < 0.05, Table 4 and Fig. 4B). Moreover, stronger immunoreactivity of PGAM1, ITGB3, and STMN1 was shown to be more likely present in stage IV (p < 0.05, χ2 test). Taken together, these results suggest that expression of a panel of ROS-related proteins could be a valuable indicator for stage progression and metastatic potential of CRC, especially PGAM1, ITGB3, and STMN1.
Fig. 4.
The relevance between the altered protein expression and tumor stage in CRC. A, Representative immunostaining of PGAM1, DJ-1, GRB2, ITGB3, SOD-1, and STMN1. N, normal colonic mucosa; TII, Stage II; and TIV, Stage IV. Positive staining is indicated by red arrows. B, SPPS immunohistochemical scores for PGAM1, DJ-1, GRB2, ITGB3, SOD-1, and STMN1 in normal colonic mucosa, stage I and II, stage III, and stage IV CRC. The average immunoreactivity scores of PGAM1, GRB2, DJ-1, ITGB3, STMN1 were significantly higher in cancer tissues than in normal tissues (One-way ANOVA analysis, p < 0.001). In contrast, SOD1 was markedly lower in cancer tissues than in normal tissues. In addition, the average immunoreactivity scores of the six ROS-related proteins was significantly correlated with surgical-pathological stage (p < 0.05).
Table IV. Relationship between clinico-pathologic stage of CRC and the six metastasis-associated proteins immunoreactivity.
| Normal |
Stage I and II |
Stage III |
Stage IV |
|||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| – | + | ++ | +++ | Score | – | + | ++ | +++ | Score | – | + | ++ | +++ | Score | – | + | ++ | +++ | Score | |
| PGMA1 | 15 | 15 | 1.00 ± 1.20 | 3 | 6 | 3 | 3.25 ± 2.93 | 2 | 3 | 4 | 2 | 5.36 ± 4.08 | 1 | 2 | 4 | 7.71 ± 3.03 | ||||
| Dj-1 | 18 | 10 | 2 | 1.13 ± 1.98 | 3 | 5 | 4 | 6.58 ± 3.26 | 3 | 3 | 5 | 7.36 ± 3.29 | 3 | 4 | 9.29 ± 3.04 | |||||
| GRB2 | 3 | 16 | 9 | 2 | 4.01 ± 3.17 | 1 | 6 | 2 | 3 | 4.58 ± 3.50 | 2 | 4 | 5 | 6.55 ± 2.88 | 3 | 4 | 8.43 ± 2.07 | |||
| ITGB3 | 15 | 15 | 1.23 ± 1.45 | 1 | 7 | 4 | 4.17 ± 2.25 | 3 | 4 | 4 | 6.45 ± 3.30 | 1 | 1 | 5 | 8.29 ± 3.04 | |||||
| SOD1 | 9 | 21 | 9.43 ± 1.89 | 3 | 5 | 4 | 7.42 ± 3.12 | 6 | 3 | 2 | 5.36 ± 3.23 | 2 | 1 | 3 | 1 | 4.71 ± 3.86 | ||||
| STMN1 | 20 | 7 | 3 | 1.17 ± 2.23 | 1 | 7 | 2 | 2 | 4.25 ± 3.25 | 2 | 3 | 2 | 4 | 5.27 ± 3.80 | 1 | 1 | 5 | 7.86 ± 3.39 | ||
ITGB3 Depletion Attenuates ROS-induced Migration and Invasion of SW480 Cells
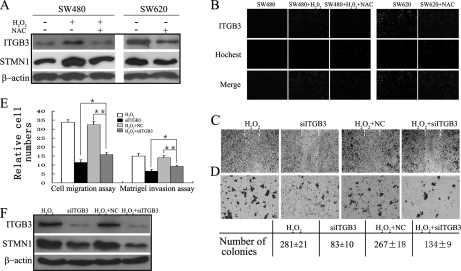
Western blot analysis showed that treatment of SW480 cells with H2O2 resulted in significant up-regulated expression of ITGB3 and STMN1 (Fig. 5A), in agreement with our observations from 2-DE analysis (Fig. 3A) and subsequent Western blot validation (Fig. 3C), in which expression of both proteins was higher in SW620 than in SW480. NAC could block expression of ITGB3 and STMN1 in H2O2-treated SW480 cells and down-regulate their expression in SW620 cells (Fig. 5A), which we have shown to have elevated intracellular ROS (Figs. 1A, 1B).
Fig. 5.
ITGB3 depletion attenuates ROS-induced migration and invasion in SW480 cells. A, Western blot analysis of SW480 and SW620 cells in the presence (+) or absence (-) of H2O2 and NAC. Blots were probed with anti ITGB3 and STMN1 antibodies (see Materials and Methods). Actin was used as a loading control. B, Immunofluorescent detection of ITGB3 on the surface of SW480 and SW620 treated with or without H2O2 or/and NAC. (C–F) For the following experiments H2O2-treated SW480 cells were transfected with siRNA-ITGB3 (H2O2+siITGB3) or a negative control siRNA (NC+siITGB3). Cells treated with lipofectamine RNAiMAX alone were used as mock control (H2O2) and SW480 cells transfected with siRNA-ITGB3 were used as a function control (siITGB3). C, Wound healing assay. Photographs were taken at 72 h postwounding. Dashed lines indicated the original wound boundaries. D, Soft agar colony formation. Mean colony counts from three independent experiments at shown below. ITGB3 knockdown prevented colony formation in H2O2-treated SW480 cells. Photographs were taken after 2 weeks. E, Cell migration and matrigel invasion assays. Migration proceeded for 24 h, whereas invasion was analyzed after 72 h. (*) p < 0.05; (**) p < 0.01. All data were from at least three independent experiments and are shown as mean ± S.D. F, Western blot analysis showing expression of ITGB3 and STMN1 in ITGB3-siRNA-transfected, NC-transfected, and mock control cells.
Previous studies revealed involvement of integrins in malignant progression and tumor invasion both in vitro and in vivo (12–15). Many of these integrins are downstream effectors in ROS-triggered cancer metastasis (8). Therefore, it was of particular interest to examine the functional role of ITGB3 in ROS-induced migration and invasion. Integrins are heterodimeric cell surface glycoproteins, therefore immunofluorescence analysis was performed to show that ITGB3 was displayed the cell surface (Fig. 5B). Consistent with the observation in Western analysis (Fig. 5A), H2O2 could induce the expression of ITGB3 on the surface of SW480, whereas NAC block it in both SW480 and SW620 cells.
Furthermore, H2O2-treated SW480 cells were transfected with siRNA-ITGB3 (siITGB3) or a negative control siRNA (NC). Intriguingly, ITGB3 knockdown markedly attenuated the H2O2-induced migration of SW480 cells in a wound healing assay (Fig. 5C) and decreased the number of colonies in soft agar (Fig. 5D). In addition, the migratory capacity of siITGB3 cells was also reduced by 55%, whereas the invasiveness was reduced by 35% (Fig. 5E). Notably, knockdown of ITGB3 expression significantly mitigated H2O2-induced over-expression of STMN1 in SW480 cells (Fig. 5F). These data suggest that ITGB3 plays an important role in ROS-induced invasion and migratory potential of SW480, and that STMN1 is possibly involved in this process.
Interestingly, the proliferative, migratory and invasive capacity of siITGB3 SW480 cells was lower than siITGB3 H2O2-treated cells (p < 0.05) (Figs. 5C–5E). This suggests that other molecules beside ITGB3 may be also involved in ROS-induced invasion and migration.
Overexpression of ITGB3 Promotes an Aggressive Phenotype in Colorectal Cancer Cells
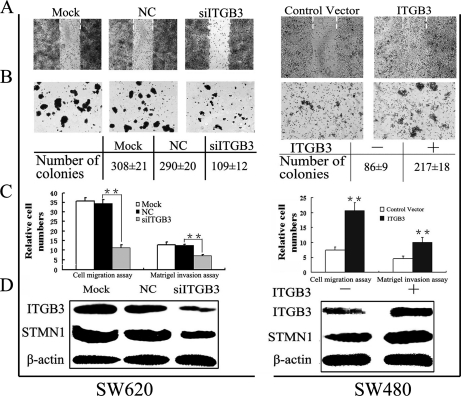
To further determine the functional roles of ITGB3 in migration and invasion of colorectal cancer cells independent of H2O2-mediated ROS intervention, we examined the biological effects of stable expression of ITGB3 in SW480 cells or siITGB3-mediated knockdown of ITGB3 expression in SW620 cells. Suppression of ITGB3 expression in SW620 caused a significant decrease in cell migration (to 30%) and invasion (to 55%), whereas ITGB3 overexpression in SW480 markedly enhanced cell migration (to 270%) and increased the invasive ability up to 220%, as evidenced by both the wound healing and soft agar assays (Figs. 6A–6C). Interestingly, Western blot analysis showed that expression of STMN1 directly correlated with that of ITGB3 (Fig. 6D).
Fig. 6.
ITGB3 promotes an aggressive cancer phenotype in colorectal cancer cells. siRNA-ITGB3 (siITGB3) and the negative control siRNA (NC) were transfected into highly metastatic SW620 cells. Cells treated with lipofectamine RNAiMAX alone were used as a control (Mock). Meanwhile, ITGB3 was consistently overexpressed in SW480 cells, whereas the empty vector was used as control. A, Wound healing model. Dashed lines indicated the original wound boundaries. Data were analyses at 72 h postwounding. B, Soft agar colony formation. Data were analyzed after 2 weeks. Mean colony counts at the bottom were from three independent experiments. C, Quantitative analysis of cell migration and matrigel invasion assays. Migration was proceed for 24 h, whereas invasion was 72 h. (*) p < 0.05; (**) p < 0.01. All data were from at least three independent experiments and shown as mean ± S.D. D, Western blot analysis of ITGB3 and STMN1 expression.
These studies demonstrate that ITGB3 can positively regulate the migration and invasion of colorectal cancer cells in these models, and in this context STMN1 might be one of the downstream effectors in ITGB3-mediated ROS signaling.
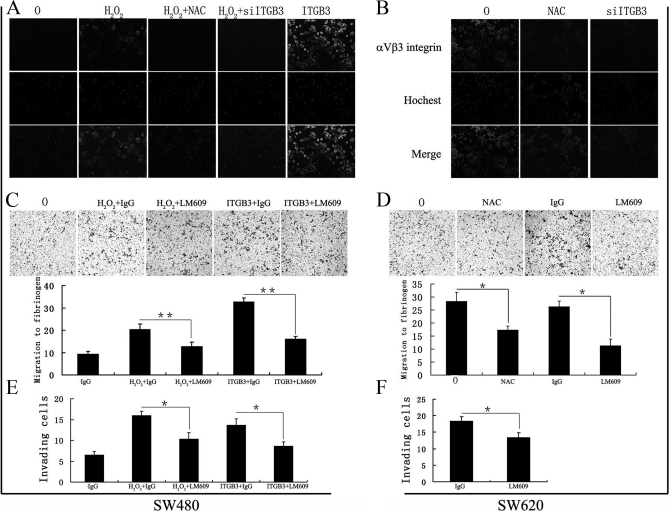
αvβ3 Expression is Necessary in ROS-ITGB3-induced Migration and Invasion of Colorectal Cancer Cells
The αvβ3-integrin, which has been reported to have a role in angiogenesis and metastasis (14–16), is consistently overexpressed in epithelial tumors (47) and, in line with this, SW480 colorectal cancer cells also express low levels of the αvβ3 complex on their cell surface (48). To determine whether ITGB3-mediated ROS signaling was induced by heterodimer αvβ3, we firstly examined αvβ3 expression and location by immunofluorescence. As shown in Fig. 7, H2O2 and ITGB3 increased integrin αvβ3 expression on the surface of SW480, but these effects were inhibited after treatment of cells with NAC or siITGB3 (Fig. 7A) which also reduced expression of αvβ3 on the surface of SW620 cells (Fig. 7B). To further verify the biological role of integrin αvβ3 in ROS-induced and ITGB3-mediated migration and invasion of colorectal cancer cells, we examined the migration toward fibrinogen, which is recognized only by αvβ3 on most tumor cells (49–51), and investigated the invasion capacity of H2O2-treated and ITGB3-overexpressing SW480 cells, as well as SW620 cells in the presence of an anti-integrin αvβ3 (LM609) or control (mouse IgG) antibody. LM609 has been shown to block adhesion of a human melanoma cell line (M21) to vitronectin, fibrinogen, and von Willebrand factor, as well as to a synthetic RGD containing peptide (52). The results showed that LM609 inhibited migration to fibrinogen in H2O2 or ITGB3 treated SW480 (Fig. 7C) and SW620 cells (Fig. 7D), and that NAC could also inhibit migration of SW620 cells to fibrinogen (Fig. 7D) (p < 0.01), which demonstrated the role of αvβ3 in this function. In addition, LM609 blocked the increased invasive ability of SW480 cells induced by H2O2 or ITGB3 (Fig. 7E) and decreased the invasiveness of SW620 (to 60%) (Fig. 7F). These results suggest that ROS-ITGB3-induced migration and invasion involve the heterodimer complex αvβ3.
Fig. 7.
αvβ3 expression is necessary in ROS-ITGB3-induced migration and invasion of colorectal cancer cells. (A) and (B) immunofluorescent detection of αvβ3 expression on surface of SW480 (A) and SW620 (B) cells treated with or without H2O2, ITGB3, NAC, and/or siITGB3. C, Migration of H2O2-treated and ITGB3-overexpression SW480 toward fibrinogen in the presence of integrin αvβ3 antibody (LM609, 10 μg/ml) or control antibody (mouse IgG, 10 μg/ml). D, Migration of SW620 cells to fibrinogen in the presence of NAC or LM609. E, LM609 blocked the increased invasive ability of SW480 cells induced by H2O2 or ITGB3. F, LM609 decreased the invasive ability of SW620. Migration was allowed to proceed for 24 h, whereas invasion was 72 h. 0, untreated cells; H2O2, H2O2-treated cells; NAC, NAC treated cells. (*) p < 0.05; (**) p < 0.01. All data were from at least three independent experiments and shown as mean ± S.D.
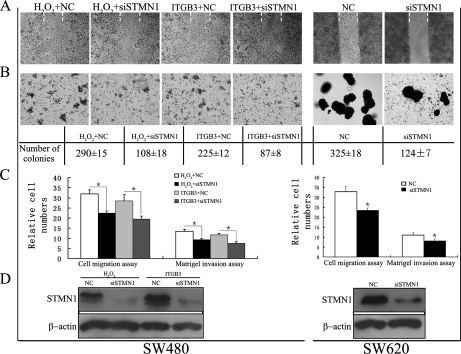
STMN1 Expression is Important in ROS-ITGB3-induced Migration and Invasion of Colorectal Cancer Cells
To verify whether STMN1 expression was involved in ROS-induced and ITGB3-mediated migration and invasion of colorectal cancer cells, STMN1 expression was knocked down in H2O2-treated and ITGB3-overexpressing SW480 as well as SW620 cells (Fig. 8D). Suppression of STMN1 reduced cell migration in the wound healing assay (Fig. 8A) and colony number in soft agar (Fig. 8B) induced by H2O2 or ITGB3. Additionally, it partially counteracted H2O2- or ITGB3-imposed cell migration and invasion in SW480 cells (Fig. 8C) (p < 0.05). Knockdown of STMN1 expression in SW620 cells significantly decreased the number of colonies in soft agar (Fig. 8B), and the migratory and invasive capacity of siSTMN1 SW620 cells were also reduced by about 30 and 25%, respectively (Fig. 8C). These results demonstrate that STMN1 is involved in ROS-ITGB3-induced proliferation and plays an important role in ROS-induced and ITGB3-mediated migration and invasion of colorectal cancer cells.
Fig. 8.
STMN1 expression is important in ROS-ITGB3-induced migration and invasion of colorectal cancer cells. siRNA-STMN1 (siSTMN1) and the negative control siRNA (NC) were transfected into H2O2-treated and ITGB3-overexpression SW480 cells as well as SW620 cells. A, Wound healing model. Dashed lines indicated the original wound boundaries. Data were analyses at 72 h postwounding. B, Soft agar colony formation. Data were analyzed after 2 weeks. Mean colony counts at the bottom were from three independent experiments. C, Quantitative analysis of cell migration and matrigel invasion assays. Migration was proceed for 24 h, whereas invasion was 72 h. (*) p < 0.05; (**) p < 0.01. All data were from at least three independent experiments and shown as mean ± S.D. D, The expression of STMN1 in cells transfected with or without siSTMN1.
Akt-mTOR and p38 MAPK Signaling Pathways are Involved in ROS-ITGB3-induced Migration and Invasion of Colorectal Cancer Cells
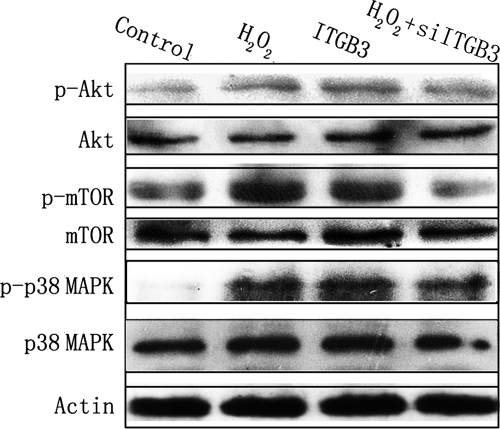
Both ROS and integrins can induce activation of PI3K and MAPK pathways, and both these pathways have been implicated in tumor metastasis (8–15). Additionally, recent studies have demonstrated that STMN1 is the direct target of both p38 MAPK and PI3K-Akt-mTOR pathways (26–29). Therefore, the crosstalk between these two pathways and ITGB3-mediated ROS signaling was investigated. The phosphorylation status of Akt, mTOR and p38 MAPK were measured by Western blot analysis in SW480 cells treated with H2O2, ITGB3, or a combination of H2O2 and siITGB3, respectively. As shown in Fig. 9, H2O2 treatment or ITGB3 overexpression stimulated the phosphorylation of Akt (S473), mTOR (S2448) and p38 MAPK (Thr180/Tyr182) in SW480 cells. In addition, knockdown of ITGB3 expression in H2O2-treated SW480 cells inhibited phosphorylation of both Akt and mTOR, whereas it slightly attenuated phosphorylation of p38 MAPK. These studies show that ITGB3 is a key mediator in ROS-induced activation of PI3K-Akt-mTOR pathway, but whereas ITGB3 plays a role in the activation of ROS-induced p38 MAPK pathway, other mediators may also be involved.
Fig. 9.
Akt-mTOR and p38 MAPK signaling pathways are involved in ROS-ITGB3-induced migration and invasion of colorectal cancer cells. The phosphorylation status of Akt, mTOR and p38 MAPK in SW480 cells treated with H2O2, ITGB3, or a combination of H2O2 and siITGB3 was measured by Western blot analysis. Details of antibodies used are given in Materials and Methods. Actin was used as a loading control.
DISCUSSION
Metastasis (the spread of cancer cells from a primary site to distant organs) consists of a series of interdependent, sequential biological processes including invasion of the tissue surrounding the primary tumor, intravasating either the lymphatics or bloodstream, surviving and eventually arresting in the circulation, extravasating into a tissue and growing at the new site (53, 54). Complex and redundant pathways are involved in this process (53). In this study, we found that a higher level of ROS was accumulated in SW620, compared with SW480. Treatment with H2O2 (as ROS source) could promote the migration and invasion of SW480 cells, whereas NAC could inhibit the effect both in H2O2-treated SW480 and SW620 cells (Fig. 1). The intracellular redox state is a key determinant of cell fate: excessive production of ROS usually results in cytotoxic effects and may lead to apoptotic cell death, whereas a certain level of ROS can act as a second-messenger for regulation of diverse cellular processes such as cell survival, proliferation and differentiation (5–7). ROS were recently proposed to be involved in tumor metastasis (7, 8). Chronic and sustained generation of ROS can activate epithelial mesenchymal transition- and metastasis-related genes, including E-cadherin, integrins and matrix metalloproteinasess which exert their effects through several important signal-transduction pathways including MAPK, PI3K, Rho-GTPase, and the Smads cascade (8–10).
In this study, 2DE-based proteomics in combination with bioinformatics analyses revealed dysregulation of a cluster of ROS-related proteins, including HSPB1, ITGB3, GRB2, DJ-1, SOD-1, PGAM1, and STMN1(Figs. 2 and 3, Table II). Association of these proteins with cancer metastasis has been proposed in diverse reports. Thus, in line with this study, Zhao and colleagues found that the expression of HSP27 (HSPB1) in SW620 was higher than SW480 and HSP27 overexpression played an important role in metastasis and progression of CRC (38). Overexpression of ITGB3 promoted tumor metastasis (16), and antagonizing ITGB3 or its heterodimers (combination with integrin αΙΙb or αv) lead to decreased colon cancer metastasis in mice (55). Both DJ-1 and SOD-1 are involved in antioxidative stress reactions and play important roles in the progression of certain cancers (56, 57). DJ-1 was originally identified as a putative oncogene (58) and has been shown to be a negative regulator of the tumor suppressor PTEN (59). Pardo et al. discovered DJ-1 as a potential serum biomarker for uveal malignant melanoma (60). SOD-1 has been shown to exert variable roles in cancer cells: overexpression of SOD-1 inhibited breast cancer cell growth and invasion (61), whereas in choline tetrathiomolybdate (ATN-224, a copper binding compound that inhibits SOD-1) treated mice, a rapid decrease in SOD-1 activity in blood cells was observed to correlate with inhibition of angiogenesis (62). Dysregulation of STMN1, GRB2, and PGAM1 has been linked with colon cancer metastasis (63–65). In addition, ITGB3 (8–10), GRB2 (8–10), DJ-1(56), SOD-1(57), PGAM1 (66), and STMN1 (67) are all involved in redox regulation or are regulated by ROS. Taken together, these previous studies suggest the six ROS-related proteins may be associated with CRC progression, which was in good agreement with our current molecular and pathological validation (Fig. 4).
Notably, our results have demonstrated that up-regulated expression of ITGB3 can be triggered by extracellular ROS stimuli (Figs. 5A and 5B). Binding of integrins to ligands on the extracellular matrix (ECM) could transmit both mechanical and chemical signals to induce cell cytoskeleton remodeling during adhesion and migration, thus the activity and expression of integrins can be influenced by a variety of factors including ROS (8, 13). Integrins can contribute directly to the control and progress of metastatic dissemination, and aberrant expression and distribution of integrins has been observed in each step of the metastatic cascade (12–15). For example, the epithelial mesenchymal transition of colon carcinoma is coincident with an increase in αvβ6 integrin expression (68), whereas the up-regulated expression of αvβ3 in melanomas facilitates penetration of the basement membrane and invasion into the underlying stroma (69). Using functional ITGB3 analysis, we found that silencing ITGB3 expression could abolish ROS-induced migration and invasive ability of SW480 cells (Figs. 5C–5F). Correlatively, either knockdown of ITGB3 in SW620 cells (high ROS level) or overexpression of ITGB3 in SW480 cells (low ROS level) significantly altered migratory and invasive potential of the two cell lines respectively (Fig. 6). Our data thus provides new evidence that ITGB3 is a key effector in ROS-induced cell migration and invasion.
ITGB3 can combine with integrin αIIb or αv to form heterodimers, αIIbβ3 and αvβ3 function as key surface adhesion and cell signaling receptors (11–18). The αvβ3 heterodimer has been implicated in the malignant behavior of various tumor types, including melanoma, glioma, breast, and ovarian cancer (16, 18, 49). Integrin αvβ3 expression in colon carcinoma was correlated with survival, with increased expression also correlating with the presence of liver metastases (70), whereas activation of the signaling function of αvβ3 in the human intestinal carcinoma Caco-2 cells led to substantial stimulation of anoikis (71). In this study, we found that ROS accumulation could affect the expression of integrin αvβ3 heterodimer on the surface of SW480 and SW620 (Figs. 7A and 7B). In addition, the αvβ3-inhibiting antibody LM609 could inhibit H2O2- or ITGB3-stimulated migration to fibrinogen and invasion in SW480 and SW620 cells (Figs. 7C–7F). These results reveal that αvβ3 integrin participates in ROS-ITGB3-induced migration and invasion of colorectal cancer cells.
Integrins can influence cell migration and invasion by directly mediating adhesion to the extracellular matrix or regulating intracellular signaling pathways that control cytoskeletal organization, force dependent effects and survival (11–15). Integrin signaling initiates from recruitment of focal adhesion kinase and induction of its phosphorylation through β subunits of integrins, which results in recruitment of adaptor proteins, and activation of PI3K and MAPK signaling, which have been demonstrated to play crucial roles in cell migration and invasion (11–15). Events downstream of ROS signaling also involve the PI3K and MAPK pathways, thus involvement of ITGB3 in ROS-induced activation of these two pathways is possible (8–15). In the current study, we demonstrate that accumulation of either ROS or ITGB3 can induce activation of both PI3K and p38 MAPK pathways (Fig. 9). We also show that ROS-induced activation of the PI3K pathway can be attenuated by specific siRNA targeting ITGB3, leading to the notion that ITGB3 is not only a direct effector but also a mediator of ROS signaling. However, there remain questions to answer as to whether other signaling pathways are also involved in ROS-ITGB3-induced colorectal cancer cell migration and invasion.
STMN1 is a ubiquitous cytosolic phosphoprotein that regulates microtubule expression during the assembly of the mitotic spindle, cell proliferation, differentiation, and cell motility (21). The STMN1 gene is expressed at a high level in many human neoplasms (22–25). Recent studies have implicated STMN1 in colorectal cancer progression (63, 67). In this study, we have shown that exogenous overexpression of ITBG3 augmented the intracellular level of STMN1 in SW480 cells, whereas silencing ITGB3 significantly reduced ROS-induced STMN1 accumulation (Figs. 5F and 6D). Further more, knockdown of STMN1 expression also reduced the proliferative, migratory, and invasive capacity of H2O2- or ITGB3-treated SW480 cells and SW620 cells. These results suggest that STMN1 is a downstream effector in ITGB3-mediated ROS signaling (Fig. 8).
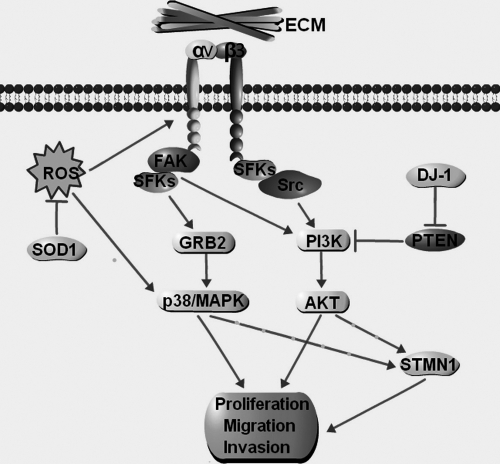
Based on the results obtained, we propose a scheme to illustrate the functional role of ITGB3 in ROS-induced migration and invasion of colorectal cancer cells (Fig. 10). The accumulation of cellular ROS up-regulates expression of ITGB3, which may increase the migration and invasion potential of colorectal cancer cells, either directly or indirectly, by activating downstream targets such as STMN1 through PI3K and MAPK signaling pathways. Association of the additional ROS-related proteins in the proposed signaling cascade (Fig. 10) has been supported by other studies. Thus, SOD-1 has antioxidant effect which could reduce the accumulation of ROS (57). GRB2 is an important adaptor protein in integrin signaling (11–15). DJ-1 is a suppressor of PTEN which antagonizes the PI3K pathway (72). In addition, Saal et al. found that over-expression of PTEN could down-regulate the expression of STMN1 in breast cancer (27).
Fig. 10.
Schematic illustrating the potential role of integrin β3 (ITGB3) in ROS-induced migration and invasion of colorectal cancer cells. The accumulation of ROS could up-regulate the expression integrin β3, which may alter the expression of the αvβ3 integrin heterodimers. This change may increase the activity of proliferation, migration, and invasion of colorectal cancer cells directly or by activating the downstream targets such as STMN1 through PI3K or MAPK pathways. Integrin signaling is predominantly initiated through the recruitment of focal adhesion complexes, which are composed of integrins, Src-family kinases (SFKs), and focal adhesion kinase. SOD-1 is an antioxidase that can reduce the accumulation of ROS. GRB2 is an important adaptor protein in integrin signaling. DJ-1 is a suppressor of PTEN, which antagonizes PI3K-Akt pathway. ECM, extracellular matrix.
In conclusion, we have demonstrated that ITGB3 or the ITGB3-STMN1 axis play important roles in ROS-induced migration and invasion of colorectal cancer cells. Although the use of cell lines as model systems to study colon cancer metastasis may not represent the disease in its entirety, information provided in this study will assist an improved understanding of the molecular mechanisms underlying ROS-triggered colorectal tumorigenesis, and underscores the clinical potential of ITGB3 or the ITGB3-STMN1 axis for the early detection or therapeutic treatment of colon cancer metastasis.
Footnotes
* This work was supported by grants from the National 973 Basic Research Program of China (2011CB910703′2010CB529900), and Chinese NSFC (81072022′ 81172173′ 30872569), ECN was supported, in part, by NHMRC project grant 603130.
 This article contains supplemental Fig. S1.
This article contains supplemental Fig. S1.
1 The abbreviations used are:
- CRC
- Colorectal cancer
- 2-DE
- 2-dimensional polyacrylamide gel electrophoresis
- ESI-Q-TOF
- Electrospray ionization quadrupole time-of-flight
- ROS
- Reactive oxygen species
- ITGB3
- Integrin β3
- STMN1
- Stathmin 1
- PGAM1
- Phosphoglycerate mutase 1
- GRB2
- Growth factor receptor-bound protein 2
- SOD-1
- Superoxide dismutase 1
- PTEN
- Phosphatase and tensin homolog
- NAC
- N-acetylcystein
- ACN
- acetonitrile
- MS/MS
- tandem MS
- PBS
- phosphate-buffered saline
- MOWSE
- molecular weight search.
REFERENCES
- 1. Saif M. W. (2006) Targeted agents for adjuvant therapy of colon cancer. Clin. Colorectal Canc. 6, 46–51 [DOI] [PubMed] [Google Scholar]
- 2. Etzioni R., Urban N., Ramsey S., McIntosh M., Schwartz S., Reid B., Radich J., Anderson G., Hartwell L. (2003) The case for early detection. Nat. Rev. Cancer 3, 243–252 [DOI] [PubMed] [Google Scholar]
- 3. Chambers A. F., Groom A. C., MacDonald I. C. (2002) Dissemination and growth of cancer cells in metastatic sites. Nat. Rev. Cancer 2, 563–572 [DOI] [PubMed] [Google Scholar]
- 4. Davies R. J., Miller R., Coleman N. (2005) Colorectal cancer screening: prospects for molecular stool analysis. Nat. Rev. Cancer 5, 199–209 [DOI] [PubMed] [Google Scholar]
- 5. Poli G., Leonarduzzi G., Biasi F., Chiarpotto E. (2004) Oxidative stress and cell signalling. Curr. Med. Chem. 11, 1163–1182 [DOI] [PubMed] [Google Scholar]
- 6. Chandel N. S., Budinger G. R. S. (2007) The cellular basis for diverse responses to oxygen. Free Radic. Biol. Med. 42, 165–174 [DOI] [PubMed] [Google Scholar]
- 7. López-Lázaro M. (2007) Dual role of hydrogen peroxide in cancer: Possible relevance to cancer chemoprevention and therapy. Cancer Lett. 252, 1–8 [DOI] [PubMed] [Google Scholar]
- 8. Wu W. S. (2006) The signaling mechanism of ROS in tumor progression. Cancer Metast. Rev. 25, 695–705 [DOI] [PubMed] [Google Scholar]
- 9. Koshikawa N., Hayashi J., Nakagawara A., Takenaga K. (2009) Reactive oxygen species-generating mitochondrial DNA mutation up-regulates hypoxia-inducible factor-1 alpha gene transcription via phosphatidylinositol 3-kinase-Akt/protein kinase C/histone deacetylase pathway. J. Biol. Chem. 284, 33185–33194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wu W. S., Wu J. R., Hu C. T. (2008) Signal cross talks for sustained MAPK activation and cell migration: the potential role of reactive oxygen species. Cancer Metast. Rev. 27, 303–314 [DOI] [PubMed] [Google Scholar]
- 11. Giancotti F. G., Ruoslahti E. (1999) Transduction - Integrin signaling. Science 285, 1028–1032 [DOI] [PubMed] [Google Scholar]
- 12. Huveneers S., Truong H., Danen E. H. J. (2007) Integrins: Signaling, disease, and therapy, Int. J. Radiat. Biol. 83, 743–751 [DOI] [PubMed] [Google Scholar]
- 13. Hood J. D., Cheresh D. A. (2002) Role of integrins in cell invasion and migration. Nat. Rev. Cancer 2, 91–100 [DOI] [PubMed] [Google Scholar]
- 14. Avraamides C. J., Garmy-Susini B., Varner J. A. (2008) Integrins in angiogenesis and lymphangiogenesis. Nat. Rev. Cancer 8, 604–617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Guo W., Giancotti F. G. (2004) Integrin signalling during tumour progression. Nat. Rev. Mol. Cell Biol. 5, 816–826 [DOI] [PubMed] [Google Scholar]
- 16. Switala-Jelen K., Dabrowska K., Opolski A., Lipinska L., Nowaczyk M., Gorski A. (2004) The biological functions of beta 3 integrins. Folia Biol-Prague 50, 143–152 [DOI] [PubMed] [Google Scholar]
- 17. Galliher A. J., Schiemann W. P. (2006). Beta(3) Integrin and Src facilitate transforming growth factor-beta mediated induction of epithelial-mesenchymal transition in mammary epithelial cells. Breast Cancer Res. 8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chen J., Zhang J., Zhao Y., Li J., Fu M. (2009) Integrin beta 3 down-regulates invasive features of ovarian cancer cells in SKOV3 cell subclones. J. Cancer Res. Clin. Oncol. 135, 909–917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hieken T. J., Farolan M., Ronan S. G., Shilkaitis A., Wild L., Das Gupta T. K. (1996) beta 3 integrin expression in melanoma predicts subsequent metastasis. J. Surg. Res. 63, 169–173 [DOI] [PubMed] [Google Scholar]
- 20. Vacca A., Ria R., Presta M., Ribatti D., Iurlaro M., Merchionne F., Tanghetti E., Dammacco F. (2001) alpha(v)beta(3) integrin engagement modulates cell adhesion, proliferation, and protease secretion in human lymphoid tumor cells. Exp. Hematol. 29, 993–1003 [DOI] [PubMed] [Google Scholar]
- 21. Rubin C. I., Atweh G. F. (2004) The role of stathmin in the regulation of the cell cycle. J. Cell Biochem. 93, 242–250 [DOI] [PubMed] [Google Scholar]
- 22. Mistry S. J., Atweh G. F. (2002) Role of stathmin in the regulation of the mitotic spindle: Potential applications in cancer therapy. Mt. Sinai J. Med. 69, 299–304 [PubMed] [Google Scholar]
- 23. Rana S., Maples P. B., Senzer N., Nemunaitis J. (2008) Stathmin 1: a novel therapeutic target for anticancer activity. Expert Rev. Anticancer Ther. 8, 1461–1470 [DOI] [PubMed] [Google Scholar]
- 24. McGrogan B. T., Gilmartin B., Camey D. N., McCann A. (2008) Taxanes, microtubules and chemoresistant breast cancer. Biochim. Biophys. Acta 1785, 96–132 [DOI] [PubMed] [Google Scholar]
- 25. Sherbet G. V., Cajone F. (2005) Stathmin in cell proliferation and cancer progression. Cancer Genomics Proteomics July 2, 227–237 [PubMed] [Google Scholar]
- 26. Beretta L., Dubois M. F., Sobel A., Bensaude O. (1995) Stathmin is a major substrate for mitogen-activated protein-kinase during heat-shock and chemical stress in HeLa cells. Eur. J. Biochem. 227, 388–395 [DOI] [PubMed] [Google Scholar]
- 27. Saal L. H., Johansson P., Holm K., Gruvberger-Saal S. K., She Q. B., Maurer M., Koujak S., Ferrando A. A., Malmström P., Memeo L., Isola J., Bendahl P. O., Rosen N., Hibshoosh H., Ringnér M., Borg A., Parsons R. (2007) Poor prognosis in carcinoma is associated with a gene expression signature of aberrant PTEN tumor suppressor pathway activity. Proc. Natl. Acad. Sci. U.S.A. 104, 7564–7569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mizumura K., Takeda K., Hashimoto S., Horie T., Ichijo H. (2006) Identification of Op18/stathmin as a potential target of ASK1-p38 MAP kinase cascade, J. Cell Physiol. 206, 363–370 [DOI] [PubMed] [Google Scholar]
- 29. Ogino S., Nosho K., Baba Y., Kure S., Shima K., Irahara N., Toyoda S., Chen L., Kirkner G. J., Wolpin B. M., Chan A. T., Giovannucci E. L., Fuchs C. S. (2009) A Cohort Study of STMN1 Expression in Colorectal Cancer: Body Mass Index and Prognosis. Am. J. Gastroenterol. 104, 2047–2056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Liu R., Li Z., Bai S. J., Zhang H., Tang M., Lei Y., Chen L., Liang S., Zhao Y. L., Wei Y., Huang C. (2009) Mechanism of Cancer Cell Adaptation to Metabolic Stress: Proteomics identification of a novel thyroid hormone-mediated gastric carcinogenic singaling pathway. Mol. Cell Proteomics 8, 70–85 [DOI] [PubMed] [Google Scholar]
- 31. Tong A., Wu L., Lin Q., Lau Q. C., Zhao X., Li J., Chen P., Chen L., Tang H., Huang C., Wei Y. Q. (2008) Proteomic analysis of cellular protein alterations using a hepatitis B virus-producing cellular model. Proteomics 8, 2012–2023 [DOI] [PubMed] [Google Scholar]
- 32. Liu R., Wang K., Yuan K., Wei Y., Huang C. (2010) Integrative oncoproteomics strategies for anticancer drug discovery. Expert Rev. Proteomics 7, 411–429 [DOI] [PubMed] [Google Scholar]
- 33. Leibovitz A., Stinson J. C., McCombs W. B., 3rd, McCoy C. E., Mazur K. C., Mabry N. D. (1976) Classification of human colorectal adenocarcinoma cell lines. Cancer Res. 36, 4562–4569 [PubMed] [Google Scholar]
- 34. Hewitt R. E., McMarlin A., Kleiner D., Wersto R., Martin P., Tsokos M., Stamp G. W., Stetler-Stevenson W. G., Tsoskas M. (2000) Validation of a model of colon cancer progression. J. Pathol. 192, 446–454 [DOI] [PubMed] [Google Scholar]
- 35. Subauste M. C., Kupriyanova T. A., Conn E. M., Ardi V. C., Quigley J. P., Deryugina E. I. (2009) Evaluation of metastatic and angiogenic potentials of human colon carcinoma cells in chick embryo model systems. Clin. Exp. Metastas 26, 1033–1047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Stein U., Walther W., Arlt F., Schwabe H., Smith J., Fichtner I., Birchmeier W., Schlag P. M. (2009) MACC1, a newly identified, key regulator of HGF-Met signaling predicts colon cancer metastasis. Nat. Med. 15, 59–67 [DOI] [PubMed] [Google Scholar]
- 37. Katayama M., Nakano H., Ishiuchi A., Wu W., Oshima R., Sakurai J., Nishikawa H., Yamaguchi S., Otsubo T. (2006) Protein pattern difference in the colon cancer cell lines examined by two-dimensional differential in-gel electrophoresis and mass spectrometry. Surg. Today 36, 1085–1093 [DOI] [PubMed] [Google Scholar]
- 38. Zhao L., Liu L., Wang S., Zhang Y. F., Yu L., Ding Y. Q. (2007) Differential proteomic analysis of human colorectal carcinoma cell lines metastasis-associated proteins. J. Cancer Res. Clin. Oncol. 133, 771–782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zhao L., Wang H., Liu C., Liu Y., Wang X., Wang S., Sun X., Li J., Deng Y., Jiang Y., Ding Y. (2010) Promotion of colorectal cancer growth and metastasis by the LIM and SH3 domain protein 1. Gut. 59, 1226–1235 [DOI] [PubMed] [Google Scholar]
- 40. Ma C., Rong Y., Radiloff D. R., Datto M. B., Centeno B., Bao S., Cheng A. W. M., Lin F., Jiang S., Yeatman T. J., Wang X. F. (2008) Extracellular matrix protein beta ig-h3/TGFBI promotes metastasis of colon cancer by enhancing cell extravasation. Genes & Development 22, 308–321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kubens B. S., Zänker K. S. (1998) Differences in the migration capacity of primary human colon carcinoma cells (SW480) and their lymph node metastatic derivatives (SW620). Cancer Letters 131, 55–64 [DOI] [PubMed] [Google Scholar]
- 42. Gerold G., Ajaj K. A., Bienert M., Laws H. J., Zychlinsky A., de Diego J. L. (2008) A Toll-like receptor 2-integrin beta(3) complex senses bacterial lipopeptides via vitronectin. Nat. Immunol. 9, 761–768 [DOI] [PubMed] [Google Scholar]
- 43. Singer S., Ehemann V., Brauckhoff A., Keith M., Vreden S., Schirmacher P., Breuhahn K. (2007) Protumorigenic overexpression of Stathmin/Op18 by gain-of-function mutation in p53 in human hepatocarcinogenesis. Hepatology 46, 759–768 [DOI] [PubMed] [Google Scholar]
- 44. Danen E. H. J., vanKraats A. A., Cornelissen I., Ruiter D. J., vanMuijen G. N. P. (1996) Integrin beta 3 cDNA transfection into a highly metastatic alpha v beta 3-negative human melanoma cell line inhibits invasion and experimental metastasis, Biochem. Biophys. Res. Commun. 226, 75–81 [DOI] [PubMed] [Google Scholar]
- 45. International Union Against Cancer (UICC) (2009) TNM Classification of malignant tumours. 7th ed., Sobin LH, Wittekind Ch. eds. Wiley, New York [Google Scholar]
- 46. Li Z., Huang C., Bai S., Pan X., Zhou R., Wei Y., Zhao X. (2008) Prognostic evaluation of epidermal fatty acid-binding protein and calcyphosine, two proteins implicated in endometrial cancer using a proteomic approach. Int. J. Cancer 123, 2377–2383 [DOI] [PubMed] [Google Scholar]
- 47. Kaur S., Kenny H. A., Jagadeeswaran S., Zillhardt M. R., Montag A. G., Kistner E., Yamada S. D., Mitra A. K., Lengyel E. (2009) beta(3)-integrin expression on tumor cells inhibits tumor progression, reduces metastasis, and is associated with a favorable prognosis in patients with ovarian cancer. Am. J. Pathol. 175, 2184–2196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Agrez M. V., Bates R. C., Mitchell D., Wilson N., Ferguson N., Anseline P., Sheppard D. (1996) Multiplicity of fibronectin-binding alpha v integrin receptors in colorectal cancer. Br. J. Cancer 73, 887–892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Rolli M., Fransvea E., Pilch J., Saven A., Felding-Habermann B. (2003) Activated integrin alpha v beta 3 cooperates with metalloproteinase MMP-9 in regulating migration of metastatic breast cancer cells. Proc. Natl. Acad. Sci. U. S. A. 100, 9482–9487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Leavesley D. I., Ferguson G. D., Wayner E. A., Cheresh D. A. (1992) Requirement of the integrin beta 3 subunit for carcinoma cell spreading or migration on vitronectin and fibrinogen. J. Cell Biol. 117, 1101–1107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Trikha M., Zhou Z., Timar J., Raso E., Kennel M., Emmell E., Nakada M. T. (2002) Multiple roles for platelet GPIIb/IIIa and alpha v beta 3 integrins in tumor growth, angiogenesis, and metastasis. Cancer Res. 62, 2824–2833 [PubMed] [Google Scholar]
- 52. Cheresh D. A., Spiro R. C. (1987) Biosynthetic and functional properties of an Arg-Gly-Asp-directed receptor involved in human melanoma cell attachment to vitronectin, fibrinogen, and von Willebrand factor. J. Biol. Chem. 262, 17703–17711 [PubMed] [Google Scholar]
- 53. Steeg P. S. (2006) Tumor metastasis: mechanistic insights and clinical challenges, Nat. Med. 12, 895–904 [DOI] [PubMed] [Google Scholar]
- 54. Hegde P., Qi R., Gaspard R., Abernathy K., Dharap S., Earle-Hughes J., Gay C., Nwokekeh N. U., Chen T., Saeed A. I., Sharov V., Lee N. H., Yeatman T. J., Quackenbush J. (2001) Identification of tumor markers in models of human colorectal cancer using a 19,200-element complementary DNA microarray, Cancer Res. 61, 7792–7797 [PubMed] [Google Scholar]
- 55. Reinmuth N., Liu W. B., Ahmad S. A., Fan F., Stoeltzing O., Parikh A. A., Bucana C. D., Gallick G. E., Nickols M. A., Westlin W. F., Ellis L. M. (2003) alpha(v) beta(3) Integrin antagonist S247 decreases colon cancer metastasis and angiogenesis and improves survival in mice. Cancer Res. 63, 2079–2087 [PubMed] [Google Scholar]
- 56. Taira T., Saito Y., Niki T., Iguchi-Ariga S. M., Takahashi K., Ariga H. (2004) DJ-1 has a role in antioxidative stress to prevent cell death. EMBO Rep. 5, 213–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Miao L., St Clair D. K. (2009) Regulation of superoxide dismutase genes: Implications in disease. Free Radic. Biol. Med. 47, 344–356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Nagakubo D., Taira T., Kitaura H., Ikeda M., Tamai K., Iguchi-Ariga S. M., Ariga H. (1997) DJ-1, a novel oncogene which transforms mouse NIH3T3 cells in cooperation with ras. Biochem. Biophys. Res. Commun. 231, 509–513 [DOI] [PubMed] [Google Scholar]
- 59. Kim R. H., Peters M., Jang Y., Shi W., Pintilie M., Fletcher G. C., DeLuca C., Liepa J., Zhou L., Snow B., Binari R. C., Manoukian A. S., Bray M. R., Liu F. F., Tsao M. S., Mak T. W. (2005) DJ-1, a novel regulator of the tumor suppressor PTEN. Cancer Cell 7, 263–273 [DOI] [PubMed] [Google Scholar]
- 60. Pardo M., Garcia A., Thomas B., Piñeiro A., Akoulitchev A., Dwek R. A., Zitzmann N. (2006) The characterization of the invasion phenotype of uveal melanoma tumour cells shows the presence of MUC18 and HMG-1 metastasis markers and leads to the identification of DJ-1 as a potential serum biomarker. Int. J. Cancer 119, 1014–1022 [DOI] [PubMed] [Google Scholar]
- 61. Weydert C. J., Waugh T. A., Ritchie J. M., Iyer K. S., Smith J. L., Li L., Spitz D. R., Oberley L. W. (2006) Overexpression of manganese or copper-zinc superoxide dismutase inhibits breast cancer growth. Free Radic. Biol. Med. 41, 226–237 [DOI] [PubMed] [Google Scholar]
- 62. Doñate F., Juarez J. C., Burnett M. E., Manuia M. M., Guan X., Shaw D. E., Smith E. L., Timucin C., Braunstein M. J., Batuman O. A., Mazar A. P. (2008) Identification of biomarkers for the antiangiogenic and antitumour activity of the superoxide dismutase 1 (SOD1) inhibitor tetrathiomolybdate (ATN-224). Br. J. Cancer 98, 776–783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Saldanha R. G., Xu N., Molloy M. P., Veal D. A., Baker M. S. (2008) Differential proteome expression associated with urokinase plasminogen activator receptor (uPAR) suppression in malignant epithelial cancer. J. Proteome Res. 7, 4792–4806 [DOI] [PubMed] [Google Scholar]
- 64. Yu G. Z., Chen Y., Long Y. Q., Dong D., Mu X. L., Wang J. J. (2008) New insight into the key proteins and pathways involved in the metastasis of colorectal carcinoma. Oncol. Rep. 19, 1191–1204 [PubMed] [Google Scholar]
- 65. Liu L., Wang S., Zhang Q., Ding Y. (2008) Identification of potential genes/proteins regulated by Tiam1 in colorectal cancer by microarray analysis and proteome analysis. Cell Biol. Int. 32, 1215–1222 [DOI] [PubMed] [Google Scholar]
- 66. Kondoh H., Lleonart M. E., Bernard D., Gil J. (2007) Protection from oxidative stress by enhanced glycolysis; a possible mechanism of cellular immortalization. Histology and Histopathology 22, 85–90 [DOI] [PubMed] [Google Scholar]
- 67. Liu Z., Lu H., Shi H., Du Y., Yu J., Gu S., Chen X., Liu K. J., Hu C. A. (2005) PUMA overexpression induces reactive oxygen species generation and proteasome-mediated stathmin degradation in colorectal cancer cells. Cancer Res. 65, 1647–1654 [DOI] [PubMed] [Google Scholar]
- 68. Bates R. C., Bellovin D. I., Brown C., Maynard E., Wu B., Kawakatsu H., Sheppard D., Oettgen P., Mercurio A. M. (2005) Transcriptional activation of integrin beta 6 during the epithelial-mesenchymal transition defines a novel prognostic indicator of aggressive colon carcinoma. Journal of Clinical Investigation 115, 339–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Hsu M. Y., Shih D. T., Meier F. E., Van Belle P., Hsu J. Y., Elder D. E., Buck C. A., Herlyn M. (1998) Adenoviral gene transfer of beta 3 integrin subunit induces conversion from radial to vertical growth phase in primary human melanoma. Am. J. Pathol. 153, 1435–1442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Vonlaufen A., Wiedle G., Borisch B., Birrer S., Luder P., Imhof B. A. (2001) Integrin alpha(v)beta(3) expression in colon carcinoma correlates with survival. Modern Pathology 14, 1126–1132 [DOI] [PubMed] [Google Scholar]
- 71. Morozevich G. E., Kozova N. I., Chubukina A. N., Berman A. E. (2003) Role of integrin alphavbeta3 in substrate-dependent apoptosis of human intestinal carcinoma cells. Biochemistry 68, 416–423 [DOI] [PubMed] [Google Scholar]
- 72. Cully M., You H., Levine A. J., Mak T. W. (2006) Beyond PTEN mutations: the PI3K pathway as an integrator of multiple inputs during tumorigenesis. Nat. Rev. Cancer 6, 184–192 [DOI] [PubMed] [Google Scholar]