Abstract
Metal and metal oxide chelating-based phosphopeptide enrichment technologies provide powerful tools for the in-depth profiling of phosphoproteomes. One weakness inherent to current enrichment strategies is poor binding of phosphopeptides containing multiple basic residues. The problem is exacerbated when strong cation exchange (SCX) is used for pre-fractionation, as under low pH SCX conditions phosphorylated peptides with multiple basic residues elute with the bulk of the tryptic digest and therefore require more stringent enrichment. Here, we report a systematic evaluation of the characteristics of a novel phosphopeptide enrichment approach based on a combination of low pH SCX and Ti4+-immobilized metal ion affinity chromatography (IMAC) comparing it one-to-one with the well established low pH SCX-TiO2 enrichment method. We also examined the effect of 1,1,1,3,3,3-hexafluoroisopropanol (HFP), trifluoroacetic acid (TFA), or 2,5-dihydroxybenzoic acid (DHB) in the loading buffer, as it has been hypothesized that high levels of TFA and the perfluorinated solvent HFP improve the enrichment of phosphopeptides containing multiple basic residues. We found that Ti4+-IMAC in combination with TFA in the loading buffer, outperformed all other methods tested, enabling the identification of around 5000 unique phosphopeptides containing multiple basic residues from 400 μg of a HeLa cell lysate digest. In comparison, ∼2000 unique phosphopeptides could be identified by Ti4+-IMAC with HFP and close to 3000 by TiO2. We confirmed, by motif analysis, the basic phosphopeptides enrich the number of putative basophilic kinases substrates. In addition, we performed an experiment using the SCX/Ti4+-IMAC methodology alongside the use of collision-induced dissociation (CID), higher energy collision induced dissociation (HCD) and electron transfer dissociation with supplementary activation (ETD) on considerably more complex sample, consisting of a total of 400 μg of triple dimethyl labeled MCF-7 digest. This analysis led to the identification of over 9,000 unique phosphorylation sites. The use of three peptide activation methods confirmed that ETD is best capable of sequencing multiply charged peptides. Collectively, our data show that the combination of SCX and Ti4+-IMAC is particularly advantageous for phosphopeptides with multiple basic residues.
Reversible protein phosphorylation widely regulates cellular functions through protein kinases and phosphatases (1, 2). Determination and a quantitative analysis of phosphorylation sites are a prerequisite for unraveling regulatory processes and signaling networks (3–6). The analytical methods of choice for characterizing protein phosphorylation have shifted from traditional methods such as radioactive labeling and gel electrophoresis to advanced mass spectrometry, a high-throughput technology (7). It has been estimated that ∼30% of cellular proteins are phosphorylated during the life cycle of the cell (8). There has been a continuing intense focus on developing enrichment and phosphopeptide sequencing strategies to facilitate the large-scale profiling of phosphorylation events. Currently, one of the most commonly adopted strategies is the use of two sequential steps of chromatographic based separations; an initial fractionation step for reducing sample complexity and, subsequently, a more specific enrichment of phosphopeptides.
Typically, low-pH strong cation exchange (SCX)1 chromatography is used as the first step where peptides are fractionated based on their solution net charge (9, 10) and the orientation of peptides to the negatively charged chromatographic material (11, 12). Unlike glutamic and aspartic acid, phosphorylated amino acids are able to retain a negative charge under acidic (pH 2.7) conditions. This property can be exploited in SCX (10) for enrichment of phosphopeptides, which tend to elute earlier and are thus separated from the majority of nonphosphopeptides. Following SCX fractionation, several affinity-based methods have been introduced for improving the level of enrichment including; immobilized metal ion (Fe3+) affinity chromatography (IMAC) (13, 14), and various metal oxides among which TiO2 is the most common (15, 16). Additional enrichment strategies have also been developed applying different metal oxides such as ZrO2 and Nb2O5 (17, 18) or IMAC using alternative metal ions such as Ga3+, Zr4+, and Ti4+ (19–21). Notably, the IMAC technology using Zr4+/Ti4+-metal ions use a phosphate group (as opposed to nitrilotriacetic acid or iminodiacetic acid) as the coordinating ligand that has shown potential to posses superior specificity than traditional metal oxides and Fe3+-IMAC (20, 21) based enrichment strategies. Recently, alternatives to SCX as a first step have also been demonstrated including the use of hydrophilic interaction chromatography (HILIC) (22, 23), electrostatic repulsion liquid chromatography (ERLIC) (24) and strong anion exchange (SAX) (25–27). Although a great number of phosphorylation sites have been identified, it has also been pointed out that each phosphopeptide enrichment technology has inherent biases toward different physiochemical properties of phosphopeptides. For instance, Fe3+-IMAC has been shown to have a more efficient handling of multiply phosphorylated peptides compared with TiO2. This can be rationalized by the weaker binding to phosphopeptides by IMAC than TiO2 (28). The specificity and ability to enrich for each method can vary from nearly 100% to a few percent, depending on sample complexity and peptide composition. One weakness common to most chelation strategies is their poor binding to phosphopeptides that contain multiple basic residues (29–32). We argue that this may lead to an underrepresentation of basophilic kinase substrates in current phosphoproteome studies, as these are characterized by high R/K contents in the neighborhood of the phosphorylated residues. The weak representation of phosphopeptides containing multiple basic residues may be caused by an ionic interaction between the basic and phosphorylated residues that hinders the latter's ability to coordinate with the affinity purification by IMAC or metal oxides. Barnouin et al. demonstrated that issues relating to intramolecular interactions can be alleviated by switching from a water system to a perfluorinated solvent system (31). However, only simple mixtures were used and specificity was not evaluated using a more complex sample. Another possible way of disrupting the ionic interaction is by increasing the strength of the ion pairing reagent. It has been suggested that elevated levels of TFA will preferably interact with basic residues (33).
Under-representation of phosphopeptides containing multiple basic residues in an SCX-chelation strategy can also be explained by additional factors. Phosphorylated peptides containing multiple basic residues co-elute with the bulk of the digested sample and thus their enrichment is hampered because of the higher complexity and dynamic range issue present in these SCX fractions. These fractions also contain highly acidic peptides, which have been demonstrated in the past to bind to chelation material and reduce specificity. Conversion of carboxylate groups to methyl ester derivates has been demonstrated to improve the selectivity of Fe3+-IMAC (34), although the required chemistry can be problematic.
Sequencing of phosphorylated peptides by tandem mass spectrometry is another key issue that may hamper comprehensive analysis (35, 36). Phosphopeptides are traditionally fragmented by collision induced dissociation (CID). However, phospho-serine and phospho-threonine containing peptides often undergo significant neutral loss of phosphate during CID, giving insufficient backbone fragmentation for phosphopeptide identification (35). Methods such as neutral loss-triggered MS3 (10) and multistage activation (37) have been employed to impose additional activation events on pre-selected neutral loss peaks. However, there is still discussion whether it is really advantageous to use neutral loss-driven MS3 and multistage activation scans in phosphoproteomics. Recently, electron capture dissociation (ECD) and electron transfer dissociation (ETD) have shown to be highly complementary to CID because they do not cleave the labile phosphate group, thereby improving the identification of phosphopeptides and localization of phosphorylation sites (38–45).
In this study, we present a systematic evaluation of enrichment of phosphopeptides containing multiple basic residues using SCX-Ti4+-IMAC (21), placing it directly in the context of the SCX-TiO2 enrichment method which we have helped establish (4, 16, 46). We argue, and show by motif analysis of the phosphopeptide datasets, that our approach is in particular beneficial when there is an interest in the analysis of putative basophilic kinases substrates.
EXPERIMENTAL PROCEDURES
Sample Preparation
HeLa and MCF-7 cells were grown on plates until confluence was reached. After harvesting the cells by scraping, lysis was carried out by resuspending them in lysis buffer containing 50 mm ammonium bicarbonate pH 8.0, 8 m urea. Complete EDTA-free protease inhibitors mixture (Roche) and phosSTOP phosphatase inhibitor mixture (Roche). The cell suspension was subjected to ultrasonication for 1 min at 60 W using 30 cycles with 50% duty. Subsequently, cell debris was removed by centrifugation at 1000 × g for 10 min at 4 °C. Six milligrams of total protein from HeLa cell and 1.2 milligrams of MCF-7 cells were re-suspended in a buffer containing 8 m urea, 50 mm ammonium bicarbonate pH 8.0, reduced with 10 mm dithiothreitol for 30 mins at 56 °C and alkylated by addition of to 20 mm iodoacetamide. After 30 min incubation in the dark at room temperature, the first digestion was performed by addition of Lys-C at an enzyme/protein ratio of 1/50 and incubation for 4 h at 37 °C. Subsequently, the digest was diluted to a final urea concentration of 2 m, and a second digestion with trypsin at an enzyme/protein ratio of 1/50 was performed at 37 °C overnight. Finally, the digestion was quenched with 0.1% formic acid. The resulting digest of HeLa cells was desalted using 200cc Sep-Pak C18 cartridges (Waters Corporation), dried in vacuo and stored at −20 °C. The tryptic digest of the MCF-7 cell lysate was split into three aliquots and then desalted using a 50cc Sep-Pack C18 column, and then washing with 0.1% formic acid. On-column stable isotope dimethyl labeling was sequentially performed, as described before (47). The resulting solution was then dried in vacuo and stored at −20 °C. The light, intermediate and heavy dimethyl labeled samples were mixed in a 1:1:1 ratio.
Strong Cation Exchange Chromatography for Peptide Fractionation
The peptides or dimethyl labeled peptides were reconstituted in 10% formic acid and then loaded onto C18 cartridge (Aqua, Phenomenex) using an Agilent 1100 HPLC system. The flow rate applied was 100 μL/min using 0.05% formic acid (pH 2.7) as solvent. Peptides were eluted from the C18 cartridge with 80% acetonitrile (ACN)/0.05% formic acid (pH 2.7); onto a Polysulfoethyl A column (200 × 2.1 mm) (PolyLC) for 10 min at the same flow rate. Separation of peptides was performed at using a nonlinear 65 min gradient: from 0 to 10 min, 100% solvent A (5 mm KH2PO4, 0.05% formic acid, 30% acetonitrile, pH 2.7); from 10 min to 15 min, to 26% solvent B (5 mm KH2PO4, 0.05% formic acid, 30% acetonitrile, 350 mm KCl, pH 2.7); from 15 min to 40 min, to 35% solvent B; and from 40 to 45 min, to 60% solvent B. At 49 min, the concentration of solvent B was 100%. The column was subsequently washed for 6 min with 100% solvent B and finally equilibrated with 100% solvent A for 9 min. The flow rate applied during the SCX gradient was 300 μL/min. Fractions were collected every minute for 40 min. Each fraction was lyophilized and then desalted by resuspension in 2% acetic acid using 50cc Sep-Pak C18 cartridges (Water Corporation). The eluted peptides from each fraction were split into five aliquots, lyophilized and stored at −20 °C. The dimethyled labled peptides from each fraction were also desalted, dried in vacuo and stored at −20 °C.
Phosphopeptide Enrichment
Ti4+-IMAC material was prepared and used essentially as previously described by us (21, 48). Titansphere (GL Science) microcolumns were prepared as previously described (16, 49). Affinity material (TiO2 and Ti4+-IMAC resin) was loaded onto Gel-loader tip microcolumns using a C8 plug and ∼1–2 cm length of material. The enrichment procedures for “positive” SCX fractions of tryptic digest of HeLa cell were as follows: first, Pre-equilibrated by 2 × 30 μL of TiO2 loading buffer (Approach TiO2a: 80% ACN, 0.1% TFA with 50 mg/ml DHB or approach TiO2b: 80% ACN, 6% TFA with 50 mg/ml DHB) for TiO2 columns, 2 × 30 μL of Ti4+-IMAC loading buffer (Approach Ti4+-IMACa: 80% ACN, 6% TFA) to Ti4+-IMAC column, 2 × 30 μL of Ti4+-IMAC loading buffer (Approach Ti4+-IMAC b: 60% HFP, 0.1% TFA) for Ti4+-IMAC column,. Next, four aliquots of each SCX fraction were, re-suspended in 60 μL of loading buffer each and loaded onto the equilibrated gel-loader tip microcolumns. Columns were sequentially washed with 60 μL of loading buffer, followed by washing with 60 μL of 50% ACN/0.1% TFA for TiO2 columns, 60 μL of 50% ACN/0.5% TFA containing 200 mm NaCl for Ti4+-IMACa column and additional washing by 60 μL of 50% ACN/0.1% TFA for Ti4+-IMACa and Ti4+-IMACb columns, respectively. The bound peptides were eluted by 20 μL of 5% ammonia into 20 μL of 10% formic acid and then stored at −20 °C for LC-MS/MS analysis. The same procedures were performed for phosphopeptide enrichment from neutral phosphopeptide-enriched SCX fractions and whole unfractionated digests of HeLa lysate. For the SCX fractions of the dimethyl labeled of MCF-7 cell lysate enrichment was performed using the Ti4+-IMACa strategy.
Mass Spectrometry
The analysis of the peptides was performed on a reversed phase (RP) nano-LC-coupled LTQ Orbitrap XL ETD (Thermo Fisher Scientific) as described previously with minor modifications (50). An Agilent 1200 series HPLC system was equipped with a 20 mm Aqua C18 (Phenomenex, Torrance, CA) trapping column (packed in-house, 100 μm i.d., 5 μm particle size) and a 400 mm ReproSil-Pur 120 C18-AQ (Dr. Maisch-GmbH) analytical column (packed in-house, 50 μm i.d., 3 μm particle size). Trapping was performed at 5 μl/min solvent C (0.1 m acetic acid in water) for 10 min, and elution was achieved with a gradient of 10–25% (v/v) solvent D (0.1 m acetic acid in 80% ACN) in 90 min with a total analysis time of 120 min. The flow rate was passively split from 0.60 ml/min to 100 nL/min when performing the elution. Nanospray was achieved using a distally coated fused silica emitter (360 μm o.d., 20 μm i.d., 10 μm tip i.d.; constructed in-house) biased to 1.7 kV. The LTQ Orbitrap ETD was operated in the data dependent mode to automatically switch between MS and MS/MS. Survey full scan MS spectra were acquired from m/z 350 to m/z 1500 in the Orbitrap with a resolution of 60,000 at m/z 400 after accumulation to a target value of 500,000 in the linear ion trap. Supplementary activation was enabled for ETD (51). The five most intense peaks at a threshold of above 500 were alternatively fragmented in the linear ion trap using CID/ETD at a target value of 30,000. The ETD reagent target value was set to 100,000 and the reaction time to 50 ms.
The enriched phosphopeptides from dimethyl labeled samples were analyzed on an ETD equipped Orbitrap Velos instrument (Thermo Fisher Scientific, Bremen) connected to an LC system as described above. All instrument methods for the Orbitrap Velos were set up in the data dependent acquisition mode. Each sample was analyzed with either a Top10 HCD or Top10 CID/ETD method. For the low-resolution CID/ETD-MS/MS method, full scan MS spectra (from m/z 350–1500) were acquired in the Orbitrap analyzer after accumulation to a target value of 5e5 in the linear ion trap. Resolution in the Orbitrap system was set to 60,000. The ten most intense peptide ions were sequentially isolated to a target value of 5000 and fragmented in the high-pressure linear ion trap by low-energy CID with normalized collision energy of 35%. The ETD reagent target value was set to 2e5 and the reaction time to 100 ms. For the HCD method, survey full scan MS spectra (from m/z 350–1500) were acquired in the Orbitrap system with resolution 30,000. The ten most intense peptide ions were sequentially isolated to a target value of 3e4 and fragmented in the HCD collision cell with normalized energy of 35%. The resulting fragments were detected in the Orbitrap with resolution 7500.
Data Analysis
From each raw data file recorded by the mass spectrometer, representing the enriched phosphopeptide from each SCX fraction, peak lists containing CID, ETD and HCD fragmentation data were generated using Proteome Discoverer (Version 1.3, Thermo Fisher Scientific) with a signal-to-noise threshold of 3 and the following settings for the ETD nonfragment filter: precursor peak removal within a 4 Da, charge-reduced precursor removal within a 2 Da, and removal of known neutral losses from charge-reduced precursor within a 2 Da (the maximum neutral loss mass was set to 120 Da). Single-fraction peak lists generated from each method were then merged into one larger peak list for database searching using in-house scripts, in which CID peak lists was further filtered as minimum fragment ions was set to 100, maximum number of fragment ion count was set to 100, maximum number of fragment ions was set to 100. Peak list files from HCD were deisotoped and charge deconvoluted as described (52, 53). Peak lists were searched against a IPI human database (version 3.52, 69,164 sequences; 29,064,824 residues) and its decoy database created by Mascot using Mascot software version 2.3.02 (Matrix Science, UK). The database search was performed with the following parameters: a mass tolerance of ±50 ppm for precursor masses and ±0.6 Da for CID/ETD fragment ions and ±0.05 Da for HCD fragments, allowing two missed cleavages, cysteine carbamidomethylation as fixed modification and methionine oxidation, phosphorylation of serine, threonine, and tyrosine as variable modifications. When the data was triple dimethyl labeled peptides, additional variable modifications relating to the N terminus and Lysine residues were also added. The enzyme was specified as trypsin and the fragment ion type was specified as electrospray ionization (ESI)-TRAP, ETD-TRAP, and ESI-quadrupole-time-of-flight (Q-TOF) for the corresponding mass spectra. The resulting .dat files were exported and filtered for a <1% false discovery rate (FDR) at the peptide level using in-house developed software “Rockerbox” (Version 1.2.6) (54) utilizing the percolator-based algorithm (55). Note, only PSMs with Mascot score ≥20 were accepted and then exported using “Rockerbox” (54). Site localization confidence was calculated using mascot delta score (43), which has been reported to be a viable method by Savitski and coworkers (56). The frequency of amino acids within identified phosphopeptides was performed using the IceLogo program (57), which builds on probability theory to visualize significant conserved sequence patterns in multiple peptide sequence alignments against background sequence sets and has a more dynamic nature and is correct and completer in the analysis of conserved sequence patterns, was used to analyze the frequency of amino acids. Note: considering the possibility of missed cleaved peptides, the peptide sequences were extended to 21 amino residues window surrounding the center of phosphorylation S/T/Y sites.
All of the data sets can be accessed as scaffold data files (www.proteomesoftware.com) at the publicly available repository Tranche (https://proteomecommons.org/) using the following hash code: p9mbfyL44qFNOl4ejNDmC+/wwGbPvnSQVsj6yueyNfOElaq84opm09URtLWGtaaqulI+c6pQjMK+5F2zghoogxtOJ/IAAAAAAAADKA = = .
RESULTS
Effect of Organic Additives on Phosphopeptides Enrichment by TiO2 and Ti4+-IMAC
Our main objective was to develop a phosphoproteomics method specifically improving enrichment of phosphopeptides containing multiple basic residues, enabling the expansion of global phosphoproteome profiles. Applying a low pH (2.7) SCX separation on a lysate allowed the fractionation of peptides based on charge state, resulting from protonation and deprotonation of basic and acidic groups. It is estimated that ∼68% of an in silico tryptic digest of the human IPI protein database generates peptides with a net charge of 1+ under pH 2.7 assuming protonation of both the N terminus and the basic residue and deprotonation of the C terminus (9). Such tryptic peptides are often referred to as “2+” in the literature (ourselves guilty) which ignores the deprotonation of the C terminus that has an important impact on the separation (12). Phosphopeptides containing only one basic residue will have a net charge of zero (protonation of the N terminus and the basic residue combined with deprotonation of the C terminus and phosphorylated residue) and will elute earlier in an SCX based separation. For the sake of simplicity we will refer to these as “neutral phosphopeptides”. Phosphopeptides containing more than one basic residue will have a net charge corresponding to the number of basic residues minus one and for simplicity these positively charged phosphopeptides will be referred to as “positive phosphopeptides”. Note that the “positive phosphopeptides” will co-elute with regular tryptic peptides that also possess a net positive charge. The reference sample we tested was a HeLa cell lysate digest (6 mg), which was initially fractionated by SCX. Each of these SCX fractions was divided into five aliquots for further experiments. Initially, single aliquots of the fractions containing positively charged peptides were pooled together and used as an initial test bed for optimizing enrichment protocols. It is noteworthy that all SCX fractions were desalted before enrichment to eliminate any deleterious effects caused by the phosphate present in the SCX solvents. Previously, Cramer and coworkers suggested that fluorinated solvents such as HFP can significantly improve “positive phosphopeptides” enrichment by reducing the level of intra-molecular ionic interaction (31). Using this work as a reference, where it was suggested 40% HFP would be beneficial, we tested a range of organic solvents differing in their HFP (40, 60, and 80%) and TFA content (0 and 0.1%). Both TiO2 and Ti4+-IMAC enrichment protocols were tested. It was observed that the optimal loading buffer is 60% HFP with 0.1% TFA for Ti4+-IMAC whereas the results for TiO2 were much poorer than the classical solvent system (See supplemental Table S1). These initial findings were then used for a much larger and refined experiment.
Comparative Evaluation of TiO2 and Ti4+-IMAC Enrichment of Phosphopeptides
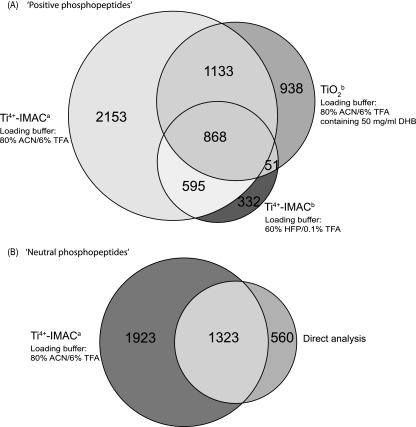
Aliquots of each of the SCX fractions corresponding to positively charged peptides were subjected to enrichment by TiO2 and Ti-IMAC using several solvent systems that will help evaluate the use of TFA, DHB and HFP. In order to be consistent we created two enrichment methods for TiO2, one containing DHB (the classic strategy and termed TiO2a) and one containing 6% TFA (to mimic the classical conditions used for Ti-IMAC, termed TiO2b). Two Ti-IMAC methods were also used; Ti4+-IMAC with 6% TFA (termed Ti4+-IMACa) and Ti4+-IMAC with 60% HFP and 0.1% TFA (termed Ti4+-IMACb). The SCX fractions were subjected to enrichment by each method and then one-third of the enriched eluent (corresponding to a total lysate level of 400 μg) was analyzed by LC-MS/MS where each precursor was subjected to both CID and ETD fragmentation events. An overview of the experimental strategy is shown in Fig. 1. The four methods TiO2a, TiO2b, Ti4+-IMACa, and Ti4+-IMACb yielded 588, 2990, 4749, and 1846 unique phosphopeptide identifications originating from 430, 1534, 1939, and 929 phosphoproteins, respectively (Table I and supplemental Tables S2, S3, S4 and S5). In addition, the specificity of phosphopeptide enrichment was in the order of Ti4+-IMACa > TiO2b > Ti4+-IMACb > TiO2a (Table I). Considering that the TiO2b method identified more phosphopeptides than the TiO2a method, only the phosphopeptides identified using the TiO2b method were further analyzed and compared with the Ti4+-IMAC approaches. Comparing overlap of the 3 methods clearly indicated that 6% TFA in conjunction with Ti4+-IMAC, Ti4+-IMACa, was superior and most comprehensive (Fig. 2A). 1321 unique phosphopeptides representing 21.7% of all phosphopeptides were solely identified by the TiO2b and Ti4+-IMACa approaches. It should be pointed out that the use of SCX and TiO2/Fe3+-IMAC for the enrichment of “neutral phosphopeptides” is superb, leading to the generation of nearly pure pools of phosphopeptides (13–15, 58). In contrast, TiO2 performs here relatively poorly for “positive phosphopeptides,” in line with what has been previously pointed out (29, 32). In order to generate a reference to allow a more in-depth evaluation of the positive phosphopeptide pools and to confirm that Ti4+-IMACa can also generate similar results to TiO2, a portion of the SCX fractions corresponding to where “neutral phosphopeptides” are found were also subjected to enrichment. Four of these SCX fractions were further purified by TI4+-IMACa approach, generating a data set of 3246 unique phosphopeptides from 1743 phosphoproteins (supplemental Table S6).
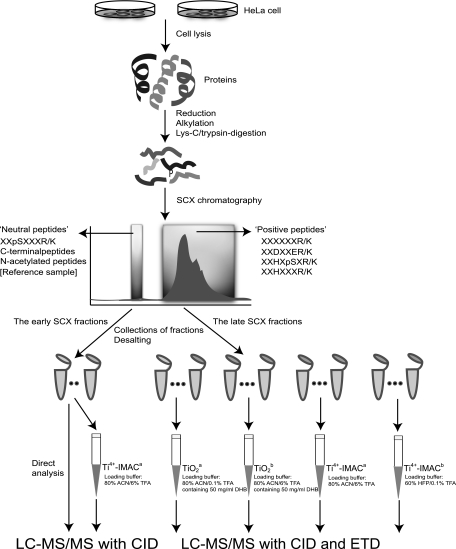
Fig. 1.
Experimental workflow used for the selective enrichment and identification of phosphopeptides. A HeLa lysate tryptic digest was initially fractionated by SCX. After desalting and aliquoting the SCX fractions, the late SCX fractions (containing the bulk of the tryptic peptides, and the phosphopeptides with multiple basic residues), were subjected to TiO2 (Approach TiO2a: 80% ACN/0.1% TFA containing 50 mg/ml DHB used as loading buffer; Approach TiO2b: 80% ACN/6% TFA containing 50 mg/ml DHB used as loading buffer), Ti4+-IMAC (Approach Ti4+-IMACa: 80% ACN/6% TFA used as loading buffer; Approach Ti4+-IMACb: 60% HFP/0.1% TFA used as loading buffer) enrichment. The eluant was, subsequently, analyzed by LC-MS/MS utilizing both CID and ETD. The early SCX fractions were also analyzed, either directly but also after Ti4+-IMACa enrichment, albeit by using CID only.
Table I. Overview of phosphorylated peptides identified with each of the used methods, for the analysis of the late (positive peptides) and early (neutral peptides) SCX fractions. The late SCX fractions contain the bulk of “normal” tryptic peptides.
| Sample used | Approach | Phosphopeptides |
Summary |
||||
|---|---|---|---|---|---|---|---|
| CID | ETD | Unique phosphopeptides | Unique phosphorylation sites | Phosphoproteins | Specificity | ||
| The late SCX Fraction | TiO2a | 435 | 366 | 588 | 633 | 430 | 3% |
| TiO2b | 2177 | 1287 | 2990 | 3075 | 1534 | 46% | |
| Ti4+-IMACa | 2814 | 3890 | 4749 | 4814 | 1939 | 63% | |
| Ti4+-IMACb | 832 | 1558 | 1846 | 1980 | 929 | 38% | |
| The early SCX Fraction | Ti4+-IMACa | 3246 | 3246 | 3354 | 1743 | 98% | |
| Direct analysis | 1883 | 1883 | 2054 | 1145 | 88% | ||
* The threshold for accepted phosphopeptide identifications was taken at mascot score at least 20 corresponding to an FDR of <1%.
Fig. 2.
Multipronged phosphoproteomics analysis of a HeLa lysate using the workflow described in Fig. 1. Venn diagrams displaying the overlap in detected (unique) phosphopeptides for (A) phosphopeptides detected in the late SCX fractions corresponding to peptides with a positive charge using the enrichment methods TiO2a, Ti4+-IMACa and Ti4+-IMACb, respectively (B) phosphopeptides detected in the early SCX fractions containing peptides with a net neutral charge either through direct analysis or after the additional enrichment using Ti4+-IMACa.
We further compared the number of unique phosphopeptides recovered from the early SCX fractions and those originating from later SCX fractions after enrichment with TI4+-IMACa approach. We found, as anticipated, little overlap in phosphopeptide identifications in between these early and late SCX fractions because of the separation power of SCX. In addition, direct analysis of these four fractions without enrichment resulted in the identification of 1883 unique phosphopeptides originating from 1145 phosphoproteins (supplemental Table S7). When comparing the data set obtained by direct analysis after SCX enrichment and that obtained after subsequent enrichment by Ti4+-IMACa, it became apparent that the additional enrichment is also beneficial for the early SCX fractions, nearly doubling the amount of detected phosphopeptides (Fig. 2B). The level of results for the direct analysis are largely made possible by the separation of the N-acetylated peptides away from the phosphorylated peptides by SCX (12). Interestingly, the enrichment factor without Ti4+-IMAC was close to 90% and with enrichment was close to 100%, according to the results generated by Mascot.
ETD Fragmentation Improves Phosphopeptide Identification, in Particular, of Positive Phosphopeptides
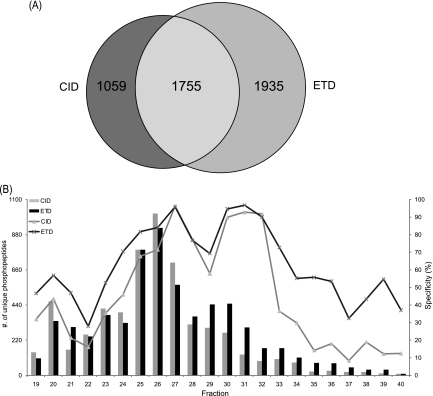
All enriched fractions were analyzed by LC-MS/MS with alternating CID and ETD fragmentation. For the Ti4+-IMACa experiment, CID and ETD contributed to the identification of 2814 and 3690 unique phosphopeptides, respectively (Fig. 3A). A total of 1755 unique phosphopeptides were commonly identified by both fragmentation techniques, corresponding to an overlap of only 37%. ETD generated more (41%) identifications than CID. When combined, the two dissociation approaches resulted in the identification of 4749 unique phosphopeptides, corresponding to 4814 unique phosphorylation sites. We further investigated the performance of CID and ETD by breaking down results obtained per individual SCX fraction (Fig. 3B). Our analysis indicates that, for individual fractions, CID and ETD identified differing numbers of phosphopeptides as well as generating different ‘apparent’ phosphopeptide specificities, ranging from 10% percent to 100%. Not only did ETD contributed to more identifications, it also resulted in a higher specificity being calculated for positive phosphopeptide enrichment by the Ti4+-IMACa method. An observation that can be rationalized by the known higher efficiency of ETD in fragmenting phosphopeptides with a higher charge where the higher charge is a product of the presence of multiple basic residues (38, 39, 45).
Fig. 3.
Comparison of the identified “positive phosphopeptides” using CID and ETD as observed in the HeLa lysate Ti4+-IMACa data set. A, Venn diagram indicating the overlap between the phosphorylated peptides identified by CID and ETD (B) Bar diagram representing the unique phosphopeptides detected by CID and ETD for each SCX fraction analyzed, coupled to the left y axis. Gray and black bars indicate the number of phosphopeptides identified by CID and ETD, respectively. In addition, the line diagrams represent the specificity of identified phosphopeptides per SCX fraction, by CID (gray) and ETD (black), coupled to the right y axis.
Comparison of Amino Acid Frequency of the Identified Positive Phosphopeptides and Neutral Phosphopeptides
To investigate the amino acid composition of phosphopeptides obtained for each method, we plotted the frequency of amino acids surrounding the phosphorylation site against a pooled data set consisting of all five data sets as background (Table. 1) using the freeware program Ice Logo (57). This background pooled data set contained a total of 8924 unique phosphorylated peptides.
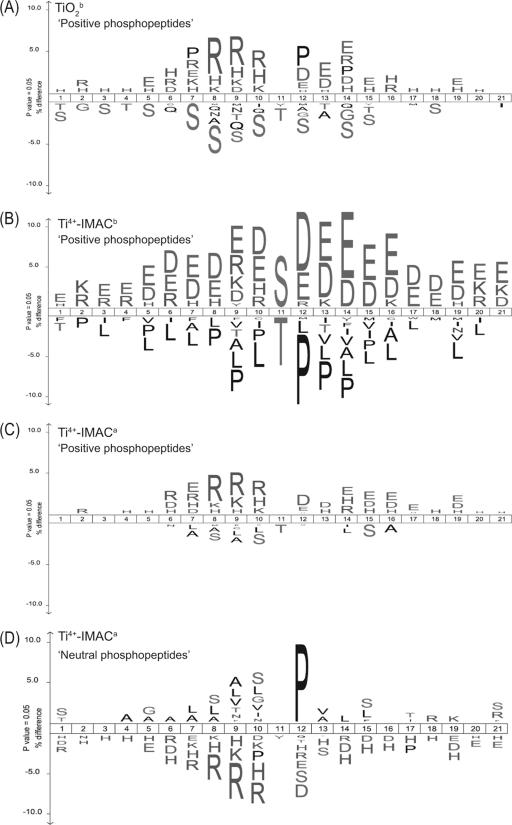
Tryptic phosphopeptides have been reported to have an average length of 16 amino acids and also tend to harbor more missed cleavages. It has been reported that on average 1.1 missed cleavages per phosphopeptide are observed whereas for regular peptides the average is 0.3 (38). To accommodate for these trends, sequences of 21 amino acids surrounding all identified phosphorylated S/T/Y residues were analyzed. In Fig. 4 each panel represents a data set from a single experimental approach, against a background data set containing all datasets. Ice Logo plots represent enrichment of amino acids as positive values, whereas underrepresented amino acids occur as negative values, keeping in mind that for these data sets there will be two basic amino acids present in each sequence. Each data set generated a unique frequency for amino acids surrounding the phosphorylation site. We found overrepresentation of the basic amino acids R, K and H in proximity to phosphorylated residues for the TiO2b strategy (Fig. 4A). The frequency of arginine occurrence at positions P-2, P-3, and P-4 was increased more than 5% (p < 0.01) and also the basic amino acids K and H were overrepresented. A possible reason for these observations is that the proximity of phosphorylated residues to K/R can hamper the efficiency of trypsin digestion as suggested by others (38, 59, 60). For the Ti4+-IMACb strategy (Fig. 4B), surprisingly, acidic amino acids of aspartic acid and glutamic acid are more frequently overrepresented (p < 0.05) along the peptide backbone. The basic amino acids are marginally overrepresented. Interestingly, proline which is a common member for several kinase substrate motifs, is markedly underrepresented in the position of p + 1. If a proline is adjacent to a phosphosite which is not close to a basic residue then trypsin is unhindered whereas if a proline is adjacent to a lysine or arginine then trypsin maybe hindered. Our data suggests that prolines are often close to phosphosites and sufficiently far away from basic residues leading to the generation of a neutral phosphopeptide. The logo generated by Ti4+-IMACa approach has a similar pattern to the TiO2b strategy, in which the basic amino acids of H, R and K and acidic amino acids of D and E are overrepresented (Fig. 4C). On closer inspection, the occurrence of basic amino acids is slightly higher than that for acidic amino acids (5% to 3%). It was also observed that Rs are more frequently enriched at P-2, P-3, and P-4. However, D and E have higher occurrence in the proximity on the right side (p + 1…p + 5) of phosphorylation sites. Strikingly, there is also a clear enrichment of phosphopeptides containing histidines, a pool of peptides that are not a by-product of an incomplete digestion and thus potentially represent a new set of phosphopeptides. However, the neutral phosphopeptide pool generated with Ti4+-IMACb enrichment produced a different pattern with the residue proline overrepresented (Fig. 4D). Proline direction is common in phosphorylation motifs and, as expected, it is enriched at p + 1. Finally, both basic residues and acidic residues showed depletion.
Fig. 4.
Frequency distribution of specifically enriched (+) or depleted (-) amino acids, surrounding the site of phosphorylation (position 11), in the HeLa lysate phosphopeptide datasets. To display theses distributions Ice Logos were generated for each data set using all our phosphoproteomics data as background. The data sets originating from the late SCX fractions are represented in (A) TiO2a, (B) Ti4+-IMACb, and (C) Ti4+-IMACa. For comparison, in (D) the IceLogo for the Ti4+-IMACa data set obtained from the early SCX fractions is represented.
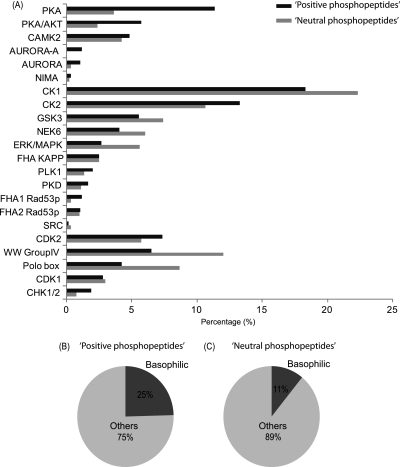
Specific Enrichment of Phosphopeptides Originating From Putative Basophilic Kinase Substrates
Protein kinase target substrates typically depends on the primary amino acid sequence proximal to the site that will be modified (3). The identification of kinase specific substrates can be evaluated using such sequence motifs. Here we performed such a motif-dependent categorization of the detected phosphopeptides for both, albeit separately, the “positive” and “neutral” phosphopeptides enriched by Ti4+-IMACa using the motif classification tool implemented in Maxquant (61). In the positive phosphopeptide and neutral phosphopeptide data sets, the algorithm available within Maxquant defined a preferred kinase to 1832 and 985 phosphopeptides, respectively. In Fig. 5A, a summary is displayed of the relative contribution of each kinase to the total data set, ignoring kinases that contributed less than 1% to the full data set. Significantly, substrates of different kinases are overrepresented when comparing these two data sets. Most obviously, there is an ∼2-fold higher enrichment of peptides originating from putative basophilic kinase substrates such as PKA, PKA/AKT and AURORA in the positive phosphopeptide data set (Fig. 5B and 5C). Basophilic kinase substrates only contributed to a mere 11% in the neutral phosphopeptide data set (Fig. 5B and 5C).
Fig. 5.
Putative kinase specific substrates present in the phosphopeptide data sets generated by Ti4+-IMACa enrichment. A, The bar chart shows the classification and distribution of substrates of different kinases found in the “positive phosphopeptide” pool (displayed with black bar) whereas the gray bars indicate those found in the “neutral” phosphopeptide pool. B and C, The pie chart shows the distributions of basophilic (black) and other kinases (gray) in the “positive” and “neutral” phosphopeptide data set, respectively.
An Evaluation of the SCX/Ti4+-IMACa Strategy Using a Triple Dimethyl Labeled Lysate
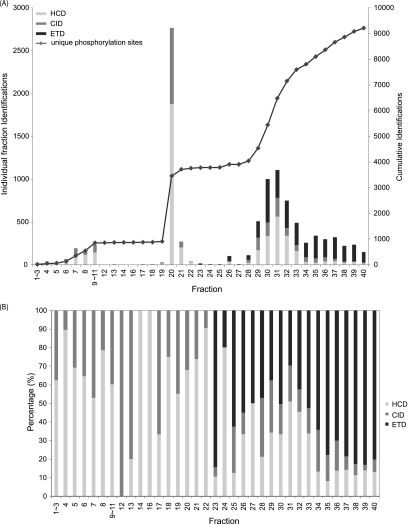
To further evaluate the use of SCX/Ti4+-IMACa approach for quantitative phosphoproteomics where complexity is increased because of the introduction of isotope labels, we performed a phosphorylation analysis on a total of 400 μg of dimethyl labeled tryptic digests of MCF-7 cell lysate. A total of 35 fractions were generated using SCX, each of which was subjected to phosphopeptide enrichment via the Ti4+-IMACa approach. After enrichment, the eluate of each fraction was analyzed on an LTQ-Orbitrap Velos mass spectrometer by using either HCD or alternating CID and ETD. The data was filtered to <1% FDR using “Rockerbox” (54), which contains percolator (55). The data was further filtered to solely possess PSMs with a minimum mascot score of 20. Next, if a phosphosite was identified more than once, we performed two stages of filtering. Initially, the PSMs corresponding to the earliest fraction were kept. Subsequently, the PSM corresponding to the highest score was kept regardless of the activation technique. supplemental Table S8 contains the final filtered results, summarized in Fig. 6. A total of 9117 unique phosphorylation sites on 9678 unique phosphopeptides was identified (supplemental Table S8). Strikingly, 5300 unique phosphorylation sites (58% of the total) were identified from the later SCX fractions (28–40) corresponding to the positive phosphopeptides. The filtering applied means any site observed in an earlier fraction would be the only site reported. Thus, if we observed the same site in two distinct peptides the peptide observed in the later fraction would be removed regardless of its score. A side effect of this filtering strategy is that it discriminates against phosphopeptides containing miss cleavages. The contribution to the unique phosphopeptide identifications per fraction broken down by activation technique are shown in Fig. 6B. As we have recently reported for regular peptides (53), HCD and CID were most effective for the earlier SCX fraction which contain peptides of lower net charge state, and the peptides carrying on 2+ charge. For the late SCX fractions, particularly fractions 34–40, ETD outperformed HCD and CID, contributing 76% of the phosphopeptide identifications.
Fig. 6.
Evaluation of the SCX/Ti4+-IMAC approach applied to a triple dimethyl labeled MCF-7 cell lysate digest. A, Bar diagram representing the individual phosphopeptides identified per fraction and the fragmentation method by which it was identified. The accumulation of unique phosphorylation sites resolved by fraction are plotted as a dotted line using the right hand y axis. B, The contribution of phosphopeptides identified by HCD, CID, or ETD in each fraction. HCD identifications are shown in light gray. CID identifications are shown in gray. ETD identifications are shown in black.
DISCUSSION
In recent years the combination of SCX and metal chelating enrichment strategies have enabled the large scale analysis of phosphoproteomes, identifying several thousands of phosphorylation sites (3–5, 38, 39). Notwithstanding this success, several issues hampering comprehensive analysis have remained relatively unaddressed. First of all, different affinity-based enrichment technologies have inherent biases often dependent on certain physicochemical properties of the diverse pool of phosphorylated peptides. Another issue is the relatively poor enrichment of phosphopeptides containing multiple basic residues. Poor binding of positive phosphopeptides was suggested to be caused by the intra-peptide interaction between the basic and phosphorylated residues (31), which hinders the ability to coordinate with the affinity material. For instance, Klemm and coworkers found drastically reduced recovery for synthetic phosphopeptides with multiple basic amino acids using TiO2 enrichment (29). Comparative evaluation of the recovery of a basic tyrosine-phosphorylated angiotensin II peptide by Iliuk and coworkers yielded only 25% recovery from a Fe3+-IMAC resin and 50% after TiO2 enrichment (32). The problem in enriching these phosphopeptides containing multiple basic residues is further exacerbated because these phosphopeptides require typically the most stringent enrichment as they, in SCX, co-elute with the bulk of the regular tryptic peptide population. Importantly, many basophilic kinases such as those in the AGC kinase family (e.g. PKA, PKG, Aurora) are to a great extent directed by flanking basic amino acids. The current lack of a solution to enrich these basic phosphopeptides can restrict the profiling of phosphorylation sites in particular related to these cellular pathways. Earlier, Barnouin et al. has proposed the use of the perflourinated solvent HFP to improve the enrichment of basic phosphopeptides by Fe3+-IMAC (31). However, a systematic evaluation of HFP on enrichment for phosphopeptides containing multiple basic residues at the proteome level was not performed. A recently introduced enrichment technology known as Ti4+-IMAC was shown to have a higher inherent specificity for phosphopeptide enrichment using solely a high level of TFA (21) and without the need of additives, in contrast to the case of TiO2 (21). In this study, we sought to evaluate Ti4+-IMAC and TiO2 in conjunction with the perfluorinated solvent (HFP) and/or TFA for improving the enrichment for positive phosphopeptides eluting in the late SCX fractions. Our results showed that Ti4+-IMACa (containing 6% TFA) is capable of achieving both the largest number of identification of positive phosphopeptides and the highest specificity (63%) of enrichment for the late SCX fractions compared with Ti4+-IMACb (38%) and TiO2b (46%). Interestingly, TiO2b (containing 6% TFA) outperformed TiO2a (the classical DHB centric approach). To investigate the characteristic of identified phosphopeptides by different approaches and from different phosphopeptide populations, we initially performed a pI analysis for all datasets, however; each data set revealed a similar pI profile (supplemental Fig. S1), reflecting pattern similar to those of others who also applied SCX/TiO2 for phosphoproteomics (5, 27). The lack of insight generated by the pI plots led us to investigate amino acid frequency graphs to distinguish the characteristics for each data set. In the TiO2b approach, basic amino acids R, K, and H were found to be overrepresented with varying degrees (Fig. 4A). In the Ti4+-IMACb data set, acidic amino acids D and E were more frequently enriched than the basic amino acids R, K, and H (Fig. 4B). One possible explanation is that HFP stretches the peptide backbone by breaking the intra-peptide ion interaction between negatively charged phosphate group and positively charged basic residues (31) such that the phosphate group can be more easily accessed by Ti4+-IMAC. As a result, the chance of affinity interaction of acidic amino acids with Ti4+-IMAC is also similarly increased. Apart from this, acidic residues such as aspartic acid and glutamic acid adjacent to tryptic cleavage sites can reduce the efficiency of trypsin digestion (38, 59, 60), resulting in miss-cleavages that increase the net charges of peptides. This also explains our observation that Ti4+-IMACb data set has the highest proportion of phosphopeptides with at least 1 mis-cleavage (62.1%) compared with Ti4+-IMACa (55.1%) and TiO2b (52.4%). The basic amino acids R, K and H as well as acidic amino acids D and E are overrepresented in the pool after Ti4+-IMACa enrichment (Fig. 4C). Among the three methods, Ti4+-IMACa appears to contain to a large degree all populations enriched by the use HFP and TiO2 displaying thus the least bias. As mentioned earlier, the high content of TFA (6%) itself can reduce the level of intrapeptide bonding between positively charged basic residues and negative phosphate group (33) and aid coordination of the phosphate group to Ti4+. The high level of TFA also aided the enrichment levels achieved by TiO2 but not to the same extent. The superior results for positive phosphopeptides by Ti4+-IMAC may be related to the use of a phosphate scaffold to immobilize the Ti4+ metal ion which is more resilient to the high levels of TFA and may also restrict the type of ligand that can coordinate and allow phosphate groups to be favored.
Identifying specific substrates of kinases is essential for constructing phosphorylation networks and understanding signal transduction in complex biological systems (62). Most interestingly, our data set (Fig. 5) shows that positive phosphopeptides recovered from the late SCX fractions contain relative higher occurrences of phosphopeptides originating from putative basophilic kinase substrates (63). Overall, the relative contribution of basophilic kinase substrate phosphopeptides was ∼25% in the positive phosphopeptide data set compared with only 11% in the neutral phosphopeptide data set. In the positive phosphopeptides we detected the majority of putative PKA, AKT and AURORA substrates. These findings suggest that in SCX based large scale phosphoproteomics analysis, the late SCX fractions, which require high level of phosphopeptide enrichment, cannot be ignored especially not when one is interested for instance in cAMP/PKA (64), cGMP/PKG, AKT, or Aurora involved signaling. A bias toward analyzing only the early SCX fractions, which is quite common in current phosphoproteomic analysis, will negatively influence the outcome of such analysis.
For the systematic evaluation, all enriched positive phosphopeptides were analyzed by alternating CID and ETD and we also performed HCD, ETD, and CID for the triple dimethyl labeled sample. Using these datasets we were able to assess whether HCD, CID, or ETD is better-suited for phosphopeptide containing multiple basic residues. Our data revealed that, in particular, for the late SCX fractions ETD boosted the number of phosphopeptide identifications significantly (Fig. 3A, Fig. 6), in line with previous reports (38, 39, 45, 65, 66).
CONCLUSIONS
The data presented here indicate that a combination of SCX fractionation and Ti4+-IMAC based enrichment, provides one of the most comprehensive phosphopeptide enrichment and analysis technologies up to date, enabling to further boost the number of phosphopeptide identifications in large scale phosphoproteomics analyses. The Ti4+-IMAC method enhances in particular the recovery of positive phosphopeptides, containing multiple basic residues, but performed also well in neutral phoshopeptide enrichment. We exemplify our strategy by being able to generate over 9000 phosphorylation sites from a single experiment consisting of a triple dimethyl labeled lysate. Therefore, we conclude that the Ti4+-IMAC based enrichment technology may be an extremely valuable tool in the ever expanding field of phosphoproteomics.
Acknowledgments
We thank Dr. Dave Siak-Wei Ow and Dr. Nikolai Mischerikow for their critical and insightful comments on the manuscript. We like to acknowledge Pieter Neerincx for bioinformatics support.
Footnotes
* This work was supported in part by the PRIME-XS project, grant agreement number 262067, funded by the European Union 7th Framework Programme; The Netherlands Proteomics Centre, embedded in the Netherlands Genomics Initiative, and the Centre for Biomedical Genetics; the Netherlands Organization for Scientific Research (NWO) with the VIDI grant (700.10.429).
 This article contains supplemental Fig. S1 and Tables S1 to S8.
This article contains supplemental Fig. S1 and Tables S1 to S8.
1 The abbreviations used are:
- SCX
- strong cation exchange chromatography
- Ti4+-IMAC
- immobilized titanium (IV) ion affinity chromatography
- HFP
- 1,1,1,3,3,3-hexafluoroisopropanol
- DHB
- 2,5-dihydroxybenzoic acid
- FDR
- false discovery rate
- ETD
- electron transfer dissociation with supplementary activation
- HCD
- higher energy collision induced dissociation
- CID
- collision induced dissociation
- PSM
- peptide to spectrum match
- MS/MS
- tandem mass spectrometry.
REFERENCES
- 1. Hunter T. (2000) Signaling–2000 and beyond. Cell. 100, 113–127 [DOI] [PubMed] [Google Scholar]
- 2. Pawson T., Scott J. D. (2005) Protein phosphorylation in signaling–50 years and counting. Trends. Biochem. Sci. 30, 286–290 [DOI] [PubMed] [Google Scholar]
- 3. Dephoure N., Zhou C., Villén J., Beausoleil S. A., Bakalarski C. E., Elledge S. J., Gygi S. P. (2008) A quantitative atlas of mitotic phosphorylation. Proc. Natl. Acad. Sci. U.S.A. 105, 10762–10767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Van Hoof D., Muñoz J., Braam S. R., Pinkse M. W., Linding R., Heck A. J., Mummery C. L., Krijgsveld J. (2009) Phosphorylation dynamics during early differentiation of human embryonic stem cells. Cell. Stem. Cell 5, 214–226 [DOI] [PubMed] [Google Scholar]
- 5. Olsen J. V., Blagoev B., Gnad F., Macek B., Kumar C., Mortensen P., Mann M. (2006) Global, in vivo, and site-specific phosphorylation dynamics in signaling networks. Cell. 127, 635–648 [DOI] [PubMed] [Google Scholar]
- 6. White F. M. (2008) Quantitative phosphoproteomic analysis of signaling network dynamics. Curr. Opin. Biotechnol. 19, 404–409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Johnson S. A., Hunter T. (2005) Kinomics: methods for deciphering the kinome. Nat. Methods. 2, 17–25 [DOI] [PubMed] [Google Scholar]
- 8. Cohen P. (2000) The regulation of protein function by multisite phosphorylation–a 25 year update. Trends. Biochem. Sci. 25, 596–601 [DOI] [PubMed] [Google Scholar]
- 9. Ballif B. A., Villén J., Beausoleil S. A., Schwartz D., Gygi S. P. (2004) Phosphoproteomic analysis of the developing mouse brain. Mol. Cell. Proteomics. 3, 1093–1101 [DOI] [PubMed] [Google Scholar]
- 10. Beausoleil S. A., Jedrychowski M., Schwartz D., Elias J. E., Villén J., Li J., Cohn M. A., Cantley L. C., Gygi S. P. (2004) Large-scale characterization of HeLa cell nuclear phosphoproteins. Proc. Natl. Acad. Sci. U.S.A. 101, 12130–12135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gauci S., Helbig A. O., Slijper M., Krijgsveld J., Heck A. J., Mohammed S. (2009) Lys-N and trypsin cover complementary parts of the phosphoproteome in a refined SCX-based approach. Anal. Chem. 81, 4493–4501 [DOI] [PubMed] [Google Scholar]
- 12. Alpert A. J., Petritis K., Kangas L., Smith R. D., Mechtler K., Mitulović G., Mohammed S., Heck A. J. (2010) Peptide orientation affects selectivity in ion-exchange chromatography. Anal. Chem. 82, 5253–5259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gruhler A., Olsen J. V., Mohammed S., Mortensen P., Faergeman N. J., Mann M., Jensen O. N. (2005) Quantitative phosphoproteomics applied to the yeast pheromone signaling pathway. Mol. Cell. Proteomics. 4, 310–327 [DOI] [PubMed] [Google Scholar]
- 14. Villén J., Gygi S. P. (2008) The SCX/IMAC enrichment approach for global phosphorylation analysis by mass spectrometry. Nat. Protoc. 3, 1630–1638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Pinkse M. W., Mohammed S., Gouw J. W., van Breukelen B., Vos H. R., Heck A. J. (2008) Highly robust, automated, and sensitive online TiO2-based phosphoproteomics applied to study endogenous phosphorylation in Drosophila melanogaster. J. Proteome Res. 7, 687–697 [DOI] [PubMed] [Google Scholar]
- 16. Benschop J. J., Mohammed S., O'Flaherty M., Heck A. J., Slijper M., Menke F. L. (2007) Quantitative phosphoproteomics of early elicitor signaling in Arabidopsis. Mol. Cell. Proteomics. 6, 1198–1214 [DOI] [PubMed] [Google Scholar]
- 17. Kweon H. K., Håkansson K. (2006) Selective zirconium dioxide-based enrichment of phosphorylated peptides for mass spectrometric analysis. Anal. Chem. 78, 1743–1749 [DOI] [PubMed] [Google Scholar]
- 18. Ficarro S. B., Parikh J. R., Blank N. C., Marto J. A. (2008) Niobium(V) oxide (Nb2O5): application to phosphoproteomics. Anal. Chem. 80, 4606–4613 [DOI] [PubMed] [Google Scholar]
- 19. Posewitz M. C., Tempst P. (1999) Immobilized gallium(III) affinity chromatography of phosphopeptides. Anal. Chem. 71, 2883–2892 [DOI] [PubMed] [Google Scholar]
- 20. Zhou H., Xu S., Ye M., Feng S., Pan C., Jiang X., Li X., Han G., Fu Y., Zou H. (2006) Zirconium phosphonate-modified porous silicon for highly specific capture of phosphopeptides and MALDI-TOF MS analysis. J. Proteome Res. 5, 2431–2437 [DOI] [PubMed] [Google Scholar]
- 21. Zhou H., Ye M., Dong J., Han G., Jiang X., Wu R., Zou H. (2008) Specific phosphopeptide enrichment with immobilized titanium ion affinity chromatography adsorbent for phosphoproteome analysis. J. Proteome Res. 7, 3957–3967 [DOI] [PubMed] [Google Scholar]
- 22. McNulty D. E., Annan R. S. (2008) Hydrophilic interaction chromatography reduces the complexity of the phosphoproteome and improves global phosphopeptide isolation and detection. Mol. Cell. Proteomics. 7, 971–980 [DOI] [PubMed] [Google Scholar]
- 23. Albuquerque C. P., Smolka M. B., Payne S. H., Bafna V., Eng J., Zhou H. (2008) A multidimensional chromatography technology for in-depth phosphoproteome analysis. Mol. Cell. Proteomics. 7, 1389–1396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Alpert A. J. (2008) Electrostatic repulsion hydrophilic interaction chromatography for isocratic separation of charged solutes and selective isolation of phosphopeptides. Anal. Chem. 80, 62–76 [DOI] [PubMed] [Google Scholar]
- 25. Nühse T. S., Stensballe A., Jensen O. N., Peck S. C. (2003) Large-scale analysis of in vivo phosphorylated membrane proteins by immobilized metal ion affinity chromatography and mass spectrometry. Mol. Cell. Proteomics. 2, 1234–1243 [DOI] [PubMed] [Google Scholar]
- 26. Wiśniewski J. R., Nagaraj N., Zougman A., Gnad F., Mann M. (2010) Brain phosphoproteome obtained by a FASP-based method reveals plasma membrane protein topology. J. Proteome Res. 9, 3280–3289 [DOI] [PubMed] [Google Scholar]
- 27. Dai J., Wang L. S., Wu Y. B., Sheng Q. H., Wu J. R., Shieh C. H., Zeng R. (2009) Fully automatic separation and identification of phosphopeptides by continuous pH-gradient anion exchange online coupled with reversed-phase liquid chromatography mass spectrometry. J. Proteome Res. 8, 133–141 [DOI] [PubMed] [Google Scholar]
- 28. Thingholm T. E., Jensen O. N., Robinson P. J., Larsen M. R. (2008) SIMAC (sequential elution from IMAC), a phosphoproteomics strategy for the rapid separation of monophosphorylated from multiply phosphorylated peptides. Mol. Cell. Proteomics. 7, 661–671 [DOI] [PubMed] [Google Scholar]
- 29. Klemm C., Otto S., Wolf C., Haseloff R. F., Beyermann M., Krause E. (2006) Evaluation of the titanium dioxide approach for MS analysis of phosphopeptides. J. Mass. Spectrom. 41, 1623–1632 [DOI] [PubMed] [Google Scholar]
- 30. Wilson-Grady J. T., Villén J., Gygi S. P. (2008) Phosphoproteome analysis of fission yeast. J. Proteome Res. 7, 1088–1097 [DOI] [PubMed] [Google Scholar]
- 31. Barnouin K. N., Hart S. R., Thompson A. J., Okuyama M., Waterfield M., Cramer R. (2005) Enhanced phosphopeptide isolation by Fe(III)-IMAC using 1,1,1,3,3,3-hexafluoroisopropanol. Proteomics. 5, 4376–4388 [DOI] [PubMed] [Google Scholar]
- 32. Iliuk A. B., Martin V. A., Alicie B. M., Geahlen R. L., Tao W. A. (2010) In-depth analyses of kinase-dependent tyrosine phosphoproteomes based on metal ion functionalized soluble nanopolymers. Mol. Cell. Proteomics 2162–2172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Mysling S., Palmisano G., Højrup P., Thaysen-Andersen M. (2010) Utilizing ion-pairing hydrophilic interaction chromatography solid phase extraction for efficient glycopeptide enrichment in glycoproteomics. Anal. Chem. 82, 5598–5609 [DOI] [PubMed] [Google Scholar]
- 34. Ficarro S. B., McCleland M. L., Stukenberg P. T., Burke D. J., Ross M. M., Shabanowitz J., Hunt D. F., White F. M. (2002) Phosphoproteome analysis by mass spectrometry and its application to Saccharomyces cerevisiae. Nat. Biotechnol. 20, 301–305 [DOI] [PubMed] [Google Scholar]
- 35. Boersema P. J., Mohammed S., Heck A. J. (2009) Phosphopeptide fragmentation and analysis by mass spectrometry. J. Mass. Spectrom. 44, 861–878 [DOI] [PubMed] [Google Scholar]
- 36. Grimsrud P. A., Swaney D. L., Wenger C. D., Beauchene N. A., Coon J. J. (2010) Phosphoproteomics for the masses. ACS. Chem. Biol. 5, 105–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Schroeder M. J., Shabanowitz J., Schwartz J. C., Hunt D. F., Coon J. J. (2004) A neutral loss activation method for improved phosphopeptide sequence analysis by quadrupole ion trap mass spectrometry. Anal. Chem. 76, 3590–3598 [DOI] [PubMed] [Google Scholar]
- 38. Molina H., Horn D. M., Tang N., Mathivanan S., Pandey A. (2007) Global proteomic profiling of phosphopeptides using electron transfer dissociation tandem mass spectrometry. Proc. Natl. Acad. Sci. U.S.A. 104, 2199–2204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Swaney D. L., Wenger C. D., Thomson J. A., Coon J. J. (2009) Human embryonic stem cell phosphoproteome revealed by electron transfer dissociation tandem mass spectrometry. Proc. Natl. Acad. Sci. U.S.A. 106, 995–1000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Chi A., Huttenhower C., Geer L. Y., Coon J. J., Syka J. E., Bai D. L., Shabanowitz J., Burke D. J., Troyanskaya O. G., Hunt D. F. (2007) Analysis of phosphorylation sites on proteins from Saccharomyces cerevisiae by electron transfer dissociation (ETD) mass spectrometry. Proc. Natl. Acad. Sci. U.S.A. 104, 2193–2198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Swaney D. L., McAlister G. C., Coon J. J. (2008) Decision tree-driven tandem mass spectrometry for shotgun proteomics. Nat. Methods. 5, 959–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Taouatas N., Altelaar A. F., Drugan M. M., Helbig A. O., Mohammed S., Heck A. J. (2009) Strong cation exchange-based fractionation of Lys-N-generated peptides facilitates the targeted analysis of post-translational modifications. Mol. Cell. Proteomics. 8, 190–200 [DOI] [PubMed] [Google Scholar]
- 43. Mischerikow N., Altelaar A. F., Navarro J. D., Mohammed S., Heck A. J. (2010) Comparative assessment of site assignments in CID and electron transfer dissociation spectra of phosphopeptides discloses limited relocation of phosphate groups. Mol. Cell. Proteomics. 9, 2140–2148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Mohammed S., Heck A., Jr. (2011) Strong cation exchange (SCX) based analytical methods for the targeted analysis of protein post-translational modifications. Curr. Opin. Biotechnol. 22, 9–16 [DOI] [PubMed] [Google Scholar]
- 45. Domon B., Bodenmiller B., Carapito C., Hao Z., Huehmer A., Aebersold R. (2009) Electron transfer dissociation in conjunction with collision activation to investigate the Drosophila melanogaster phosphoproteome. J. Proteome Res. 8, 2633–2639 [DOI] [PubMed] [Google Scholar]
- 46. Pinkse M. W., Uitto P. M., Hilhorst M. J., Ooms B., Heck A. J. (2004) Selective isolation at the femtomole level of phosphopeptides from proteolytic digests using 2D-NanoLC-ESI-MS/MS and titanium oxide precolumns. Anal. Chem. 76, 3935–3943 [DOI] [PubMed] [Google Scholar]
- 47. Boersema P. J., Raijmakers R., Lemeer S., Mohammed S., Heck A. J. (2009) Multiplex peptide stable isotope dimethyl labeling for quantitative proteomics. Nat. Protoc. 4, 484–494 [DOI] [PubMed] [Google Scholar]
- 48. Yu Z., Han G., Sun S., Jiang X., Chen R., Wang F., Wu R., Ye M., Zou H. (2009) Preparation of monodisperse immobilized Ti(4+) affinity chromatography microspheres for specific enrichment of phosphopeptides. Anal. Chim. Acta 636, 34–41 [DOI] [PubMed] [Google Scholar]
- 49. Thingholm T. E., Jørgensen T. J., Jensen O. N., Larsen M. R. (2006) Highly selective enrichment of phosphorylated peptides using titanium dioxide. Nat. Protoc. 1, 1929–1935 [DOI] [PubMed] [Google Scholar]
- 50. Raijmakers R., Berkers C. R., de Jong A., Ovaa H., Heck A. J., Mohammed S. (2008) Automated online sequential isotope labeling for protein quantitation applied to proteasome tissue-specific diversity. Mol. Cell. Proteomics 7, 1755–1762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Swaney D. L., McAlister G. C., Wirtala M., Schwartz J. C., Syka J. E., Coon J. J. (2007) Supplemental activation method for high-efficiency electron-transfer dissociation of doubly protonated peptide precursors. Anal. Chem. 79, 477–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Savitski M. M., Mathieson T., Becher I., Bantscheff M. H-score, a mass accuracy driven rescoring approach for improved peptide identification in modification rich samples. J. Proteome Res. 9, 5511–5516 [DOI] [PubMed] [Google Scholar]
- 53. Frese C. K., Altelaar A. F., Hennrich M. L., Nolting D., Zeller M., Griep-Raming J., Heck A. J., Mohammed S. (2011) Improved peptide identification by targeted fragmentation using CID, HCD and ETD on an LTQ-Orbitrap Velos. J. Proteome Res. 10, 2377–2388 [DOI] [PubMed] [Google Scholar]
- 54. van den Toorn H. W., Muñoz J., Mohammed S., Raijmakers R., Heck A. J., van Breukelen B. (2011) RockerBox: analysis and filtering of massive proteomics search results. J. Proteome Res. 10, 1420–1424 [DOI] [PubMed] [Google Scholar]
- 55. Käll L., Canterbury J. D., Weston J., Noble W. S., MacCoss M. J. (2007) Semi-supervised learning for peptide identification from shotgun proteomics datasets. Nat. Methods. 4, 923–925 [DOI] [PubMed] [Google Scholar]
- 56. Savitski M. M., Lemeer S., Boesche M., Lang M., Mathieson T., Bantscheff M., Kuster B. (2011) Confident phosphorylation site localization using the mascot delta score. Mol. Cell. Proteomics 10, 1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Colaert N., Helsens K., Martens L., Vandekerckhove J., Gevaert K. (2009) Improved visualization of protein consensus sequences by iceLogo. Nat. Methods. 6, 786–787 [DOI] [PubMed] [Google Scholar]
- 58. Villén J., Beausoleil S. A., Gerber S. A., Gygi S. P. (2007) Large-scale phosphorylation analysis of mouse liver. Proc. Natl. Acad. Sci. U.S.A. 104, 1488–1493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Schlosser A., Pipkorn R., Bossemeyer D., Lehmann W. D. (2001) Analysis of protein phosphorylation by a combination of elastase digestion and neutral loss tandem mass spectrometry. Anal. Chem. 73, 170–176 [DOI] [PubMed] [Google Scholar]
- 60. Knight Z. A., Schilling B., Row R. H., Kenski D. M., Gibson B. W., Shokat K. M. (2003) Phosphospecific proteolysis for mapping sites of protein phosphorylation. Nat. Biotechnol. 21, 1047–1054 [DOI] [PubMed] [Google Scholar]
- 61. Cox J., Mann M. (2008) MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372 [DOI] [PubMed] [Google Scholar]
- 62. Linding R., Jensen L. J., Ostheimer G. J., van Vugt M. A., Jørgensen C., Miron I. M., Diella F., Colwill K., Taylor L., Elder K., Metalnikov P., Nguyen V., Pasculescu A., Jin J., Park J. G., Samson L. D., Woodgett J. R., Russell R. B., Bork P., Yaffe M. B., Pawson T. (2007) Systematic discovery of in vivo phosphorylation networks. Cell. 129, 1415–1426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Hanks S. K., Hunter T. (1995) Protein kinases 6. The eukaryotic protein kinase superfamily: kinase (catalytic) domain structure and classification. FASEB J. 9, 576–596 [PubMed] [Google Scholar]
- 64. Oberprieler N. G., Lemeer S., Kalland M. E., Torgersen K. M., Heck A. J., Taskén K. (2010) High-resolution mapping of prostaglandin E2-dependent signaling networks identifies a constitutively active PKA signaling node in CD8+CD45RO+ T cells. Blood. 116, 2253–2265 [DOI] [PubMed] [Google Scholar]
- 65. Frese C. K., Altelaar A. F., Hennrich M. L., Nolting D., Zeller M., Griep-Raming J., Heck A. J., Mohammed S. (2011) Improved Peptide Identification by Targeted Fragmentation Using CID, HCD and ETD on an LTQ-Orbitrap Velos. J. Proteome Res. 10, 2377–2388 [DOI] [PubMed] [Google Scholar]
- 66. Aguiar M., Haas W., Beausoleil S. A., Rush J., Gygi S. P. (2010) Gas-phase rearrangements do not affect site localization reliability in phosphoproteomics data sets. J. Proteome Res. 9, 3103–3107 [DOI] [PMC free article] [PubMed] [Google Scholar]