Abstract
Postsynaptic density (PSD) is a protein supramolecule lying underneath the postsynaptic membrane of excitatory synapses and has been implicated to play important roles in synaptic structure and function in mammalian central nervous system. Here, PSDs were isolated from two distinct regions of porcine brain, cerebral cortex and cerebellum. SDS-PAGE and Western blotting analyses indicated that cerebral and cerebellar PSDs consisted of a similar set of proteins with noticeable differences in the abundance of various proteins between these samples. Subsequently, protein localization in these PSDs was analyzed by using the Nano-Depth-Tagging method. This method involved the use of three synthetic reagents, as agarose beads whose surface was covalently linked with a fluorescent, photoactivable, and cleavable chemical crosslinker by spacers of varied lengths. After its application was verified by using a synthetic complex consisting of four layers of different proteins, the Nano-Depth-Tagging method was used here to yield information concerning the depth distribution of various proteins in the PSD. The results indicated that in both cerebral and cerebellar PSDs, glutamate receptors, actin, and actin binding proteins resided in the peripheral regions within ∼10 nm deep from the surface and that scaffold proteins, tubulin subunits, microtubule-binding proteins, and membrane cytoskeleton proteins found in mammalian erythrocytes resided in the interiors deeper than 10 nm from the surface in the PSD. Finally, by using the immunoabsorption method, binding partner proteins of two proteins residing in the interiors, PSD-95 and α-tubulin, and those of two proteins residing in the peripheral regions, elongation factor-1α and calcium, calmodulin-dependent protein kinase II α subunit, of cerebral and cerebellar PSDs were identified. Overall, the results indicate a striking similarity in protein organization between the PSDs isolated from porcine cerebral cortex and cerebellum. A model of the molecular structure of the PSD has also been proposed here.
Postsynaptic density (PSD)1 is a disk-shaped protein complex lying underneath the postsynaptic membrane and a landmark subcellular structure of most excitatory synapses in mammalian central nervous system (1). Electron microscopic studies have revealed that the PSD consists of globular and filamentous components (1–4). The morphology and protein composition of the PSD in neurons have been reported to undergo development-and activity-dependent changes (5–11). Accumulated evidence has suggested that the PSD plays important roles in organizing the pre-and postsynaptic terminals into an integrated structure, in clustering neurotransmitter receptors to the postsynaptic sites, in signal transduction across synapses, and in the plasticity of synaptic function and structure (12–14).
Biochemical procedures have been developed to isolate samples enriched in PSD from different brain tissues (15–17). The isolated PSD exhibits a morphology closely resembling its counterparts in neurons. However, in contrast to the plastic nature of the PSD structure found in neurons, the biochemically isolated PSD appears to be firmly built and is resistant to various treatments that usually lead to the dissociation of other protein complexes (18). Biochemical analyses have revealed that the PSD-enriched samples consist of hundreds of proteins (19–26). More recently, the abundance of several components in the PSD have been quantified by a variety of techniques (27–30), and the phosphorylation and glycosylation states of PSD proteins have further been studied (31–34).
Compared with the protein constituents of the PSD, our understanding about how this vast number of different proteins is organized in the PSD is still limited. Proteomic analyses of proteins coimmunoprecipitated with various glutamate receptors or PSD-95 have yielded information regarding the potential interactions between various PSD proteins (35–38). CaMKII (calcium, calmodulin-dependent protein kinase II) has been proposed to assemble into tower-like structures in the PSD (39). By means of quantitative immunogold electron microscopy, the distribution of several proteins, including subunits of AMPA and NMDA receptors, proteins associated to NMDA receptors, CaMKII, ProSAP2, Mena, actin, and cortactin, in the PSD has been reported (40, 41). A recent EM tomography study (42) has indicated that the PSD consists of vertically oriented filaments, probably containing PSD-95, which interact with two other types of horizontally oriented filaments also present in the PSD region. The majority of actin molecules in the PSD have been proposed to stay in the polymerized state (43). By using a solid-phase and chemical crosslinking-based methodology (see below), the proteins lining the surface of the PSD have been identified (44). An earlier study has indicated that the PSD isolated from dog cerebral cortex differs from the PSD isolated from dog cerebellum in protein composition, protein phosphorylation, and morphology (17). Recently, a proteomic study has further delineated the molecular heterogeneity between the PSDs isolated from rat forebrain and cerebellum in a quantitative manner (29). It still remains unclear whether the PSDs in different brain regions share a similar protein organization or not.
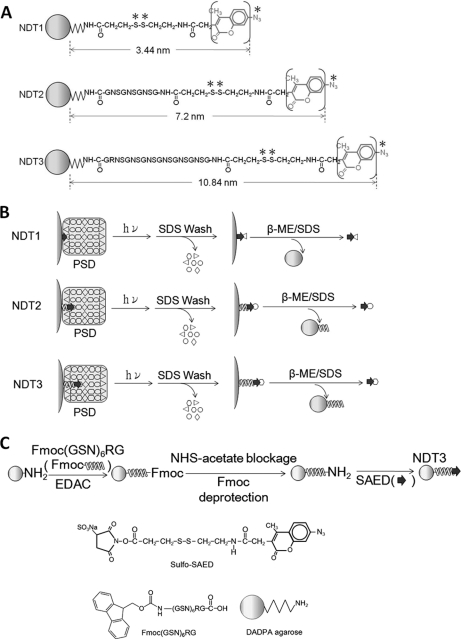
The abovementioned solid-phase and chemical crosslinking-based methodology, called Nano-Depth-Tagging (NDT), has been developed for identifying proteins residing at different depths from the surface of protein supramolecules. This methodology involves the use of several synthetic reagents, called NDT reagents. NDT reagents are agarose beads whose surface is conjugated to SAED (sulfosuccinimidyl-2-(7-azido-4-methylcoumarin-3-acetamido) -ethyl-1,3′-dithiopropionate), a fluorescent, cleavable, and photoactivatable chemical crosslinker (45), or to peptides of different lengths with SAED moiety linked to their N termini (Fig. 1A). On encountering a protein supramolecule like the PSD, the azido groups (indicated by * in Fig. 1A) of SAED and of peptidyl-SAED on bead surface would penetrate into the supramolecule and, when irradiated with UV light, form covalent links with proteins surrounding them (Fig. 1B). After removing noncovalently bound proteins by washing with SDS-containing solution, proteins covalently linked to beads could be subsequently collected from the beads by treatment with solution containing SDS and β-mercaptoethanol (β-ME) that breaks the disulfide bond (indicated by ** in Fig. 1A) in SAED. The released proteins concurrently become fluorescent by acquiring the coumarin fluorophore (in parentheses of Fig. 1A) from the SAED. Because the functional azido groups on the SAED of various NDT reagents are at different distances from the surface of the bead (Fig. 1A), they could thus reach into and interact with proteins residing at different depths in a supramolecule. Quantitative analyses of the abundance of various proteins in the samples isolated from a supramolecule by using different NDT reagents would provide information concerning the depths at which these proteins reside in the supramolecule. An NDT reagent, NDT1 (originally called as NDT3.5), has been used to identify proteins lining the surface of the PSD (44), and a second NDT reagent, NDT2 (originally called as NDT7.0), has been found to be a useful tool to identify proteins residing in regions within 7.2 nm deep from the surface of the PSD (46). Here, we report the synthesis of a third NDT reagent, called NDT3 (Fig. 1C). The distance between the bead surface and the azido group at the end of the peptidyl-SAED on the bead surface has been calculated to be 10.84 nm. The capability of NDT3 of interacting with proteins residing at such a depth in a supramolecule has first been verified by using a synthetic protein complex with known organization and structure. NDT1, NDT2, and NDT3 are used together to study the localization of various proteins in the PSD in a systematic fashion. In addition, interactions between different constituent proteins of the PSD have also been studied by means of immunoabsorption. The PSD samples isolated from the cerebral cortex and cerebellum of porcine brain have been subject to the above analyses. The results indicate that cerebral and cerebellar PSDs share a similar protein organization, despite the differences in their protein composition. A model of the protein organization of the PSD has also been proposed here.
Fig. 1.
Chemical structures, application and syntheses of NDT reagents. A, Chemical structures of NDT1, NDT2, and NDT3. The functional azido group and cleavable disulfide bond are indicated by symbols * and **, respectively. The fluorophore, 4-methylcoumarin, is in parentheses. B, The application of NDT reagents for isolating proteins residing at different depths of the PSD. NDT reagents are incubated with the PSD. The reaction mixtures are subject to UV irradiation to activate the azido groups of these reagents, thereby resulting in the formation of covalent links between the reagents and proteins surrounding the azido groups. The beads are first washed with a SDS-containing solution to remove non-covalently bound proteins and subsequently with a solution containing both SDS and β-ME to release those proteins covalently linked to the beads for further analyses. C, Synthesis of NDT3. The chemical structures of sulfo-SAED, synthetic Fmoc- 20-mer peptide and DADPA agarose bead used in the synthesis are also shown.
MATERIALS AND METHODS
The monoclonal antibodies against JNK3 (05–893), actin (MAB1501), EF1α (05–235), ankyrin (MAB1683), β-tubulin (MAB380), β3-tubulin (MAB1637), AMPA receptor subunit GluR2 (MAB397), kainate receptor subunits GluR6/7 (06–309), and the polyclonal antibodies against nNOS (07–571), kalirin (07–122), spectrin (AB993), calbindin (AB1778), microtubule-associated protein 2 (MAP2) (AB5622), and NMDA receptor subunits NR1 (AB5044P) and NR2C (AB1592P) were purchased from Millipore (Billerica, MA). The monoclonal antibody against CaMKIIα (sc-13141) and the polyclonal antibodies against CaMKIIβ (sc-1540), α-tubulin (sc-12462), cDHC (sc-9115), PSD-95 (sc-8575), and AMPA receptor subunit GluR1 (sc-7609) were purchased from Santa Cruz Biotechnology Inc. (Santa Cruz, CA). The monoclonal antibody against α-actinin (A7811) was obtained from Sigma-Aldrich. The polyclonal antibodies against protein 4.1 (NB100–68234) and homer 1 (NB600–671) were obtained from Novus Biologicals (Littleton, CO). The polyclonal antibody against synaptophysin (L129939) was obtained from Leica Microsystems Inc. (Buffalo Grove, IL). The polyclonal antibodies against HSC70 (GTX111150), cortactin (GTX100253), gelsolin (GTX101185), tropomyosin (GTX113857), Erk2 (GTX113094), and SAP102 (GTX110289) were obtained from GeneTex (Irvine, CA). The Fab fragment of anti-digoxigenin-POD antibody was purchased from Roche (Mannheim, Germany). Mouse anti-idiotypic monoclonal antibody was a generous gift from Dr. Chien-Chung Chang (Institute of Molecular and Cellular Biology, National Tsing Hua University, Hsinchu, Taiwan). Protein G-magnetic beads were obtained from BioLabs (Ipswich, MA). Protein G was purchased from Calbiochem (Darmstadt, Germany). Sulfosuccinimidyl-2-[biotinamido] ethyl-1,3-dithiopropionate (sulfo-NHS-SS-biotin), 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide and disuccinimidy tartarate (DST) were obtained from Pierce (Rockford, IL). Streptavidin (S4762), aldolase (A2714), catalase (C9322), ferritin (F4503), leupeptin (L2884), pepstatin A (P5318), benzamidine (B6506), phenylmethanesulfonyl fluoride (P7626), iodoacetamide (I1149), 3-aminopropyl triethoxysilane (A3648), diethylamine (D0806), dimethylformamide (40255), sodium deoxycholate (D6750), and dimethyl pimelimidate dihydrochloride (DMP) (D8388) were purchased from Sigma-Aldrich. Bovine serum albumin (BSA) was obtained from Affymetrix, Inc. (Santa Clara, CA). Diaminodipropylamine (DADPA)-agarose beads, sulfosuccinimidyl-2-(7-azido-4-methylcoumarin-3-acetamido)-ethyl-1,3′-dithiopropionate (SAED), sulfosuccinimidyl acetate (sulfo-NHS acetate), and sulfosuccinimidyl 6-[α-methyl-α-(2-pyridyldithio)toluamido]hexanoate (sulfo-LC-SMPT) were obtained from Thermo (Rockford, IL). Triethanolamine (A15678) was purchased from Alfa Aesar (Ward Hill, MA). Peptides and peptides with Fmoc-protected N termini (purity> 95%) were ordered from GeneScript (Piscataway, NJ).
Isolation of Cerebral and Cerebellar PSD
PSDs were isolated from porcine cerebral cortex and cerebellum according to the procedures reported previously (44). Briefly, fresh pig brains were obtained from a local slaughterhouse. The cerebrum and cerebellum were isolated from the pig brain, and white matter was removed surgically from the cerebrum. The resultant brain tissue was homogenized in 6 volumes of buffer A (0.32 m sucrose, 0.1 mm EDTA, 0.1 mm EGTA, and 1 mm HEPES at pH 7.4) on ice by a Polytron blender. The homogenate was subject to centrifugation at 4420 × g for 5 min, and the resultant supernatant was taken out and saved on ice. The remaining pellet was resuspended in 4 volumes of buffer A, subject to centrifugation again at 4,420 × g for 5 min, and the resultant supernatant was added to the previous one. The combined supernatants were then subject to centrifugation at 38,900 × g for 45 min. The resultant pellet was resuspended in 2 volumes of 2 mm Tris acetate at pH 8.0 and then placed on the top of a layer of buffer B (1.2 m sucrose, 0.1 mm EDTA, 0.1 mm EGTA, and 1 mm HEPES at pH 7.4). After centrifugation at 245,000 × g for 50 min, the white material in the interface was collected and placed on the top of a layer of buffer C (0.9 m sucrose, 0.1 mm EDTA, 0.1 mm EGTA, and 1 mm HEPES at pH 7.4). After centrifugation at 245,000 × g for 50 min, the white material in the resultant pellet was collected and homogenized in HEPES/Ca2+ buffer (50 mm CaCl2 and 2 mm HEPES at pH 7.4). After centrifugation at 65,000 × g for 20 min, the pellet was homogenized in HEPES/Ca2+ buffer and subject to centrifugation at the same condition again, and the resultant pellet was resuspended in an adequate amount of solution consisting of 1 volume of HEPES/Ca2+ buffer and 2 volumes of a buffer containing 0.4% (v/v) Triton X-100 and 2 mm HEPES at pH 7.4, such that the final protein concentration was 2 mg/ml. The resultant sample was subsequently placed on top of a layer of 1.0 m sucrose buffer (1.0 m sucrose, 0.1 mm EDTA, 0.1 mm EGTA and 2.5 mm HEPES at pH 7.4) and subject to centrifugation at 85,000 × g for 1 h. The resultant pellet was resuspended in Tris acetate buffer solution (50 mm Tris acetate, 0.1 mm EDTA and 0.1 mm EGTA at pH 7.4) and then subject to centrifugation at 50,000 × g for 30 min. After repeating the washing step once more, the resultant pellet, containing mainly the synaptic junction fraction, was diluted to a protein concentration of 4 mg/ml by solution B (50 mm CaCl2 and 6 mm Tris-HCl at pH 8.1), mixed with an equal volume of solution C (0.32 m sucrose, 1% (v/v) Triton X-100 and 12 mm Tris-HCl at pH 8.1), incubated on ice for 15 min and finally subject to centrifugation at 32,800 × g for 20 min. The resultant pellet was resuspended in solution B and placed on the top of a step gradient consisting of 1.0, 1.5, and 2.0 m sucrose in 1 mm NaHCO3 at pH 8.1. After centrifugation at 200,000 × g for 2.5 h, the white material band in the interface between the 1.5 and 2.0 m sucrose layers was collected, mixed with an equal volume of solution D (150 mm KCl and 1% (v/v) Triton X-100) and then subject to centrifugation at 200,000 × g for 20 min. The resultant pellet was resuspended in solution B containing 40% (v/v) glycerol and used as the PSD sample in different experiments. Protease inhibitors, including 1 μg/ml leupeptin, 1 μg/ml pepstatin A, 1 mm benzamidine, and 0.25 mm phenylmethylsulfonyl fluoride, were included in all solutions used in the isolation process.
Chemical Syntheses of NDT1, NDT2, and NDT3 Reagents
The NDT1 and NDT2 reagents were synthesized according to the procedures reported earlier (44, 46). For the synthesis of NDT3 (Fig. 1C), Fmoc-20-mer-peptide (Fmoc-(GSN)6RG, 97.8% purity) was first conjugated to DADPA beads by using CarboxyLinkTM Kit (Pierce). Thirty-two mg of Fmoc-20-mer-peptide was mixed with 32 mg of 1-ethyl-2-(2-dimethylaminopropyl)carbodiimide in 0.8 ml conjugation buffer (25 mm MES at pH 6.0). Five minutes later, 0.8 ml of 50% (v/v) DADPA beads that had been washed extensively with the conjugation buffer was added to the reaction mixture. After incubation at room temperature for 3 h, the beads were washed with conjugation buffer containing 1 m NaCl. By fluorescamine test (47), it was calculated that 61.6 ± 7.3% (n = 5) of the amino groups on the bead surface were conjugated with the peptide. To block the residual un-reacted amino groups on the bead surface, these beads were incubated with 10 mm sulfo-NHS acetate in conjugation buffer at room temperature for 30 min, followed by extensive washing with conjugation buffer to remove unreacted sulfo-NHS acetate. By fluorescamine test, it was determined that 6.0 ± 1.6% (n = 3) of the amino groups originally present on the DADPA beads still remained free at this stage. The peptides on the beads were then deprotected (i.e. to remove the Fmoc) by incubating with piperidine (final concentration of 20% (v/v)) at room temperature for 20 min (48). The resulting beads were washed with conjugation buffer five times and then with borate buffer (20 mm sodium borate at pH 8.2) five times. Peptide-conjugated beads in borate buffer (0.8 ml) were mixed with 20 μl SAED stock solution (50 mm in dimethylformamide) at room temperature in the dark, and aliquots of 20 μl SAED stock solution were added to reaction mixture 5, 10, and 15 min later. After the final addition of SAED, the reaction mixture was cooled to 4 °C, incubated in the dark for 1 h, and Tris stock solution (1 m at pH 8.4) was added to the reaction mixture to a final Tris concentration of 50 mm to block any unreacted SAED. Unreacted SAED was removed from the beads by filtering the reaction mixture through a sintered glass filter. By quantifying the SAED released from the beads after the treatment with β-ME, it was determined that 29.8% of the amino groups originally present on the DADPA beads were conjugated with 20-mer peptidyl SAED. The length of the 20-mer peptide in NDT3 was estimated to be 7.27 nm, according to the crystal structure of amino acids (49). The distances between the terminal azido groups and the surface of agarose beads of NDT3 was calculated to be 10.84 nm by using the RasMol Molecular Visualization Software (Version 2.5, enhanced by the MultiCHEM Facility, University of California, Berkeley, CA) (Fig. 1A).
Chemical Synthesis and Application of a Four-layered Protein Complex (FPC) on Glass Coverslip
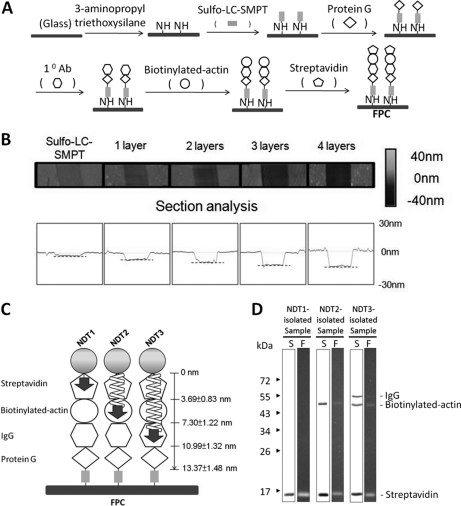
For the synthesis of FPC (Fig. 2A), glass coverslips (18 mm × 18 mm, #1 thickness; Matsunami Glass Ind., Ltd., Kishiwada City, Osaka, Japan) were cleaned by immersion in piranha solution (70:30 (v/v) a mixture of concentrated H2SO4 to H2O2) overnight at room temperature. The coverslips were washed thoroughly with distilled water for 12 h and then dried in air. Each air-dried coverslip was treated with 1 ml of 2% 3-aminopropyl triethoxysilane in 95% ethanol for 45 min at room temperature according to the procedure of Matsuzawa (50). Subsequently, the coverslips were washed twice with 100% ethanol for 1 min each and then baked in an oven at 115 °C for 1 h. After cooling to room temperature, the coverslips were washed several times with 95% ethanol and then with water. The resulting amine-modified coverslips were incubated with 0.2 mm sulfo-LC-SMPT (sulfosuccinimidyl 6-[α-methyl-α-(2-pyridyldithio)toluamido]hexanoate) in phosphate-buffered saline (PBS) at room temperature for 1.5 h. The reaction was stopped by adding 50 mm Tris-HCl (pH 8.0). The coverslips were washed several times with PBS (0.1 m sodium phosphate and 0.15 m NaCl at pH 7.2) to remove any uncoupled sulfo-LC-SMPT and then incubated with protein G (1 mg/ml in PBS containing 10 mm EDTA) for 12 h at 4 °C. After being washed twice with the PBS-EDTA solution, the coverslips were further washed with Tris-buffered saline (TBS) (20 mm Tris-HCl and 50 mm NaCl at pH 7.4) and then incubated with the mouse monoclonal anti-actin antibody (1:200 in TBS) for 1 h at room temperature. After being washed 3 times with TBS for 5 min each, the coverslips were incubated with biotinylated actin (1 mg/ml in TBS) for 1 h at room temperature. Biotinylated actin was generated by incubating sulfo-NHS-SS-biotin and actin, purified from porcine skeletal muscles according to the procedure of Pardee (51), at the ratio of 10:1 (mol/mol) for 1 h at 4 °C. After being washed three times with TBS for 5 min each, the coverslips were then incubated with streptavidin (1 mg/ml in TBS) for 1 h at room temperature, followed by washing three times with TBS for 5 min each. To analyze the composition of the proteins on the coverslips, each coverslip was covered with 100 μl of a solution containing 3% (w/v) SDS, 10% (v/v) glycerol, 125 mm Tris-HCl at pH 6.8, and 5% (v/v) β-mercaptoethanol (ME). Ten minutes later, the solution was collected and then subject to SDS-PAGE analysis. To determine the thickness of the protein layers on the coverslips, coverslips containing the bottom single protein layer, the bottom two protein layers, the bottom three protein layers, and all four protein layers were treated with 0.5 mm DST at room temperature for 30 min. After washing with water and air-dried, linear cuts were made by a steel surgical blade (#10, Feather Safety Razor Co., Ltd., Kita-ku, Osaka, Japan) on the surface of these glass coverslips. Cuts were also made on the surface of coverslips containing the sulfo-LC-SMPT layer. The depths of the cuts on glass coverslips were then measured by an Atomic Forced Microscope (AFM) (NS3a controller with D3100 stage, Digital Instruments, Santa Barbara, CA, USA) and analyzed by NanoScope (version 5.12r5 software, Digital Instruments).
Fig. 2.
Synthesis of four-layer protein complex (FPC) and the use of FPC in validating the applicability of NDT reagents. A, Synthesis of FPC on the surface of glass coverslip. B, Upper panels: Atomic Forced Microscopic (AFM) images of scratches made on coverslips coated with sulfo-LC-SMPT, with protein G (1 layer), with protein G/anti-actin antibody (2 layers), with protein G/anti-actin antibody/biotinylated actin (3 layers), and with protein G/anti-actin antibody/biotinylated actin/streptavidin (4 layers). The height signals are indicated by a gray-colored gradient on the right. Lower panels: Quantification of the depths of the scratches made on the above samples. The broken lines indicate the positions inside the scratches. C, Depth ranges of the four protein layers on the surface of glass coverslip in FPC as determined by AFM. Also indicated are the depths at which the azido groups on the SAED moieties of NDT reagents, as indicated by arrows, can reach in the FPC. D, SDS-PAGE analysis of the proteins in the samples isolated from FPC by using NDT reagents. The resultant polyacrylamide gels were examined on top of a UV illuminator (lanes F) and visualized by silver staining (lanes S). Molecular weight markers are shown to the left.
To validate the applicability of NDT reagents, each of the coverslips containing FPC was covered with 40 μl NDT beads and 360 μl PBS. After being irradiated with UV light (365 nm wavelength, TLW-20 transilluminator, UVP Inc., Upland, CA) at a distance of 1 cm for 10 min, SDS (10% (w/v)), glycerol and Tris stock solution (2 m at pH 6.8) at final concentrations of 3% (w/v), 10% (v/v), and 125 mm, respectively, were added. Beads were then collected by filtration and washed extensively by the same solution. Finally, the beads were incubated with the same solution containing β-ME (5%) in a boiling water bath for 5 min. The samples collected from the beads were subject to SDS-PAGE analysis.
Application of NDT1, NDT2, and NDT3 to Study the Localization of Various Proteins in the PSD
NDT1, NDT2, or NDT3 beads (0.4 ml, 50% (v/v)) in PBS containing 1 mm iodoacetamide were incubated with PSD (2 mg of protein in 6 mm Tris-HCl at pH 8.1, 50 μm CaCl2, 40% (v/v) glycerol, and 2 mm iodoacetamide) at 4 °C in the dark with gentle mixing. Two minutes later, the samples were placed on ice and irradiated with u.v. light for 10 min. After the photolytic reaction, Tris-HCl (2 m at pH 6.8) and SDS (10%) were added to the samples at the final concentration of 125 mm and 3%, respectively. The samples were heated in a boiling water bath for 5 min. The beads were collected by filtration and washed repeatedly with a solution containing 3% (w/v) SDS, 10% (v/v) glycerol, and 125 mm Tris-HCl at pH 6.8. Finally, the beads were resuspended in a solution containing 3% (w/v) SDS, 10% (v/v) glycerol, 125 mm Tris-HCl at pH 6.8 and 5% (v/v) β-ME and heated in a boiling water bath for 5 min. After removing the beads by filtration, the filtrates were subjected to SDS-PAGE and quantitative Western blotting analyses.
Chemical Syntheses and Application of 10-mer Peptidyl-SAED and 20-mer Peptidyl-SAED
Samples containing 1 mg of 10-mer peptide ((GSN)3G, 95.3% purity) or 2 mg of 20-mer peptide ((GSN)6RG, 95.4% purity) in 0.1 ml of a borate buffer (50 mm sodium borate at pH 8.4) were mixed with 25 μl SAED stock solution (25 mm in dimethylformamide) at room temperature. Aliquots of 25 μl SAED stock solution were added to the reaction mixture 5, 10, and 15 min later. After the final addition of SAED, the reaction mixture was incubated at 4 °C for 1 h and then clarified by centrifugation at 16,000 × g for 30 min. All of the procedures described above were performed in the dark. The resultant supernatants were subject to high-performance liquid chromatography separation with a gradient between 0–50% acetonitrile (Waters 2796 Bioseparations Module equipped with a Waters Atlantis dC18 column) while monitoring the fluorescence (λex = 355 nm and λem = 460 nm) by VICTOR3 Multilabel Readers (PerkinElmer Inc. Waltham, MA). The collected fractions were further subject to analysis with an electrospray ionization quadrupole-time-of-flight (ESI Q-TOF) mass spectrometer (Waters Corporation, Milford, Massachusetts; Mass Spectrometry Center of NTHU). The fractions containing 10-mer peptidyl-SAED and 20-mer peptidyl-SAED, as species of m/z 1254 and m/z 2183 respectively, were collected and lyophilized. The resultant powder was dissolved in PBS and incubated with 200 μg PSD. After gentle mixing for 5 min, the reaction mixture was subject to UV irradiation for 10 min on ice, treated with 5% (v/v) β-ME in a boiling water bath for 5 min and finally subject to SDS-PAGE analysis with 8.5% polyacrylamide gels. The fluorescence photographs of the gels containing the peptidyl-SAED-treated proteins were taken on top of a transilluminator (365 nm) through a 435 nm cutoff filter (CVI Laser Corp., Albuquerque, NM) after the gels were washed four times with 10% (v/v) acetic acid each for 15 min. Gels were subsequently stained by Coomassie blue.
Quantitative Analysis of the Interactions Between NDT Reagents and FPC
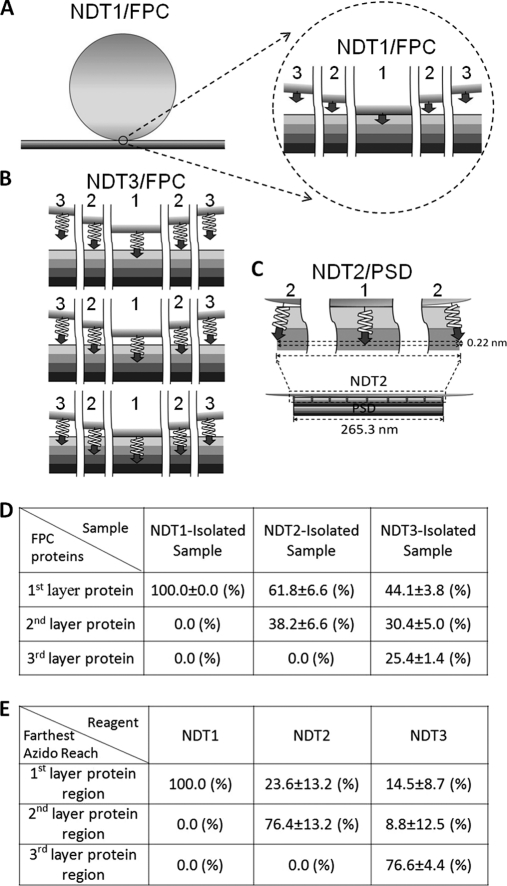
When NDT beads with different spacers' lengths encountered the FPC, the interactions between the former's azido groups and the latter's proteins could occur in different fashions. These interactions were assumed to occur in three modes. The first mode is that the farthest depth to which the azido groups could reach was in the first layer region of FPC (Fig. 3A and top row panels of Fig. 3B), and the abundance of the first layer protein in the sample isolated by the NDT in such a reaction mode would be 100%. In the second mode where the farthest depth was in the second layer region (middle row panels of Fig. 3B), because of the curved nature of the bead surface both the proteins in the top and second layers could interact with azido groups occupying different regions of bead surface and thus be isolated. The amounts of the top and second layer proteins interacting with the crosslinkers were assumed to be proportional to the amounts of azido groups residing in the regions occupied respectively by these two layers of proteins. These latter values could be calculated as the areas of the spherical bead surface crossing these regions and were found to be equal. Thus, the abundances of the top and second layer proteins in the sample isolated by NDT in such an interaction mode would be 50% each. In the third mode where the farthest depth of NDT's azido was in the third layer region (bottom row panel of Fig. 3B), the relative areas of bead surface crossing the top, second, and third layer regions were calculated to be 33.0/33.8/33.2. Therefore, the abundances of the top, second, and third layer proteins in the sample isolated by NDT in such an interaction mode would be 33.0%, 33.8%, and 33.2%, respectively. When the abundances of different layer proteins in a NDT-isolated sample were known, the proportions of this NDT reagent in different modes during photolysis whereas encountering FPC could be calculated by the following simplified equations: P1 = 1-P2-P3; P2 = 2×A2-2×A3; P3 = 3×A3, where P1, P2, and P3 were the proportions of the NDT beads interacting with FPC in the first, second and third modes, respectively, and A1, A2, and A3 were the abundances of the first, second and third layer proteins respectively in the sample isolated by the NDT reagents.
Fig. 3.
Modes of interaction of NDT reagents with FPC and PSD. A, The interaction between NDT1 and FPC. The NDT1 is depicted as a sphere sitting on top of FPC, as four consecutive planar layers in different shades. The portion enclosed by the circle is shown at higher magnification to the right. Position 1 indicates the region of NDT1 surface where the azido group of SAED ([GRAPHIC]) reaches farthest, 3.44 nm, in FPC. Positions 2 and 3 indicate respectively the regions 537 nm and 770 nm horizontally away from the position 1 on the bead surface. B, Three modes of interaction between NDT3 and FPC. Top, middle, and bottom rows: The interaction mode that the farthest depth where azido group of the 20-mer peptidyl-SAED ([GRAPHIC]) penetrates is in the first, second, and third protein layer region, respectively. In each of these interaction modes, position 1 indicates the region of the bead where the azido groups reach farthest into the FPC, and positions 2 and 3 respectively indicate the azido groups in areas 537 nm and 770 nm horizontally away from position 1 on the bead surface. C, The interaction between NDT2 and PSD. Bottom panel: The azido groups in the center region of NDT2 reach into the PSD structure, depicted as a multilayered structure of 265.3 nm wide at the bottom, to a depth similar to its spacer's length, 7.2 nm. Top panels: Enlarged views of the azido groups in the center region (position 1) and azido groups in areas on the bead surface 133 nm horizontally away from position 1 (position 2). The difference in the depths where the azido groups in regions depicted in positions 1 and 2 reside has been calculated to be 0.22 nm. D, Quantification of the abundances of the proteins in different layers of the FPC in the NDT1-, NDT2-, and NDT3-isolated samples from the densitometric scans of the gels (Fig. 2D). Data are the means ±S.D. from three independent experiments. E, The proportions of NDT beads in different interaction modes with FPC during the UV-activated crosslinking reaction as calculated by the equations described in Materials and Methods.
Immunoabsorption of PSD Proteins
Antibodies against various PSD proteins were covalently immobilized on protein G-magnetic beads according to the methods published earlier with some modifications (52, 53). One hundred μl of protein G-magnetic beads (50% v/v) was washed three times with Dulbecco's phosphate buffered saline (d-PBS) containing 8 mm Na2HPO4, 2 mm K2HPO4, 140 mm NaCl, and 10 mm KCl at pH 7.4. Antibody (15 μg) in 100 μl d-PBS was then added to the washed beads. After incubation at room temperature for 1 h, the beads were washed with 375 μl cross-linking buffer (0.2 m triethanolamine at pH 8.2) once and incubated with 375 μl cross-linking buffer containing 25 mm DMP at room temperature for 30 min. The resultant antibody-conjugated beads were washed with 375 μl low pH elution buffer (0.1 m glycine-HCl at pH 2.5) to remove antibodies not covalently linked to the beads.
Immunoabsorption of the PSD proteins was performed according to the procedures reported earlier (54) with some modifications. The PSD was first incubated with a solution containing 7 m urea and 2 m thiourea at room temperature for 20 min. SDS and Nonidet P-40 were subsequently added to the solution to the final concentrations of 0.1% (w/v) and 0.5% (v/v), respectively. After incubation at room temperature for 3 min, the sample was dialyzed against the RIA solution (0.5% (v/v) Nonidet P-40, 0.1% (w/v) SDS, 200 mm NaCl, 10 mm EDTA, 1 μg/ml leupeptin, 1 μg/ml pepstatin, 1 mm benazmidine, 0.25 mm phenylmethylsulfonyl fluoride, 2 mm iodoacetamide, and 10 mm sodium phosphate at pH 6.3) at 4 °C overnight with three changes of the dialyzing solution. The resulting PSD sample (henceforth called uuPSD) was first adjusted to a protein concentration of 1 mg/ml, precleared twice for 30 min each by incubating with 1/8 volume of protein G-magnetic beads that had been washed with the high-pH elution solution (50 mm diethylamine and 0.5% (w/v) sodium deoxycholate at pH 11.5) and then washed with the RIA solution three times for 1 min each at room temperature. The precleared uuPSD sample was incubated with the abovementioned antibody-conjugated protein G-magnetic beads at 4 °C overnight. The beads were then collected by the aid of a magnet and washed six times with 200 μl RIA solution for 1 min each. Finally, proteins immunoabsorbed to the magnetic beads were released twice by incubating with 100 μl high-pH elution solution for 2 min each. The collected samples were neutralized immediately by adding 1/10 volume of 0.5 m NaH2PO4.
SDS-PAGE, Western Blotting and Sucrose-Density-Gradient Centrifugation
SDS-PAGE analysis was carried out according to the procedure of Laemmli (55) by using a minigel apparatus (Mini-Protean 3; Bio-Rad Laboratories Inc., Hercules, CA). Western blotting was performed as described previously (56). Proteins in the gel were visualized by silver (57) or Coomassie-blue staining method. The relative intensities of silver-stained or immunostained bands were quantified by the UN-SCAN-IT Gel Automated Digitizing System (Silk Scientific Inc., Orem, Utah). Sucrose-density-gradient centrifugation analyses were performed by the procedures described earlier (54). Briefly, samples (0.5 ml) were loaded onto 0–70% or 0–30% (m/v) sucrose gradients, and the gradients were centrifuged at 200,000 × g at 4 °C for 1 h in a SW-41 rotor (Beckman Instruments, Palo Alto, CA). Fractions of 1 ml were collected from the bottom of the gradients. Aliquots of 200 μl were removed from the collected fractions, concentrated by using the choloform/methanol method and subject to SDS-PAGE analysis using 9% polyacrylamide gel. Proteins on the resultant gels were either visualized by silver-staining or electrotransferred to polyvinylidene difluoride membrane for Western blotting analysis.
Scanning Electron Microscopy
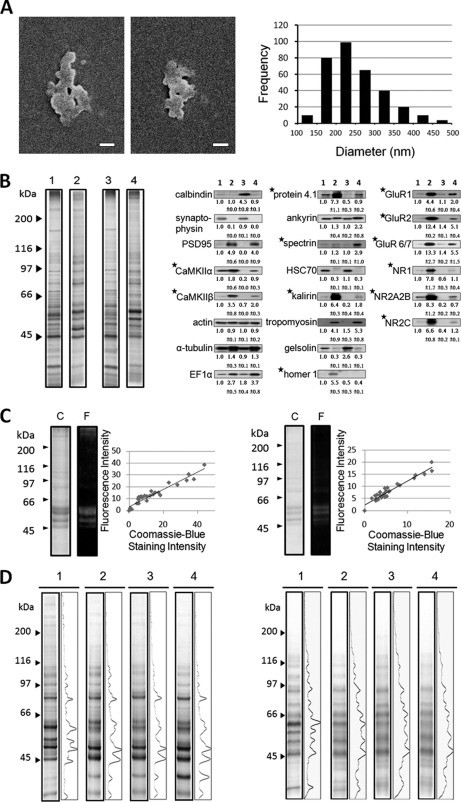
Cerebellar PSD sample (0.06 mg of protein/ml in 5 mm Tris-HCl at pH 7.2) was dropped onto the surface of 18 mm × 18 mm glass coverslips. After removing the excessive liquid by blotting with filter paper, the coverslip was air-dried, coated with platinum by an ion coater (E-1030; Hitachi, Tokyo, Japan), and examined under a scanning electron microscope (Hitachi, Model S-4700). All data were collected at a magnification of 30,000. The areas of the PSDs in EM micrographs were determined by the image quantification software ImageJ 1.44P (National Institutes of Health, Washington, DC)). The diameter of a PSD was calculated from its area by assuming that the PSD was circular in shape.
RESULTS
Synthesis of NDT3 and Four-layered Protein Complex
The synthesis and application of NDT3 were described in the Materials and Methods section (Figs. 1B and 1C). To verify that the azido group of the SAED of this reagent could indeed reach and form covalent links with proteins residing at a depth similar to its peptidyl spacer's length, 10.84 nm, in a supramolecule, a protein complex with known organization and structure was synthesized and used as a model. The synthetic complex, which was termed as the four-layered protein complex (FPC), was made on the surface of glass coverslip. It consisted of four layers of proteins: protein G, anti-actin antibody, biotinylated actin, and streptavidin, from bottom to top (Fig. 2A). The protein composition of this model complex was confirmed by SDS-PAGE analyses of the samples collected from the FPC and of the samples collected as the intermediates obtained during the synthetic process, i.e. the complex containing the protein of the bottom layer, the complex containing the proteins of the bottom two layers, and the complex containing the proteins of the bottom three layers (data not shown). The thicknesses of these intermediates and complete FPC were determined by AFM (Figs. 2B). The depths for the sulfo-LC-SMPT layer, the bottom layer, the bottom two layers, the bottom three layers and the complete FPC were 1.34 ± 0.60 (n = 9), 3.73 ± 0.28 (n = 9), 7.41 ± 0.42 (n = 4), 11.02 ± 0.79 (n = 4), and 14.71 ± 0.26 nm (n = 6), respectively. Based on these data, the thickness of each protein layer, from top to bottom, were calculated to be 3.69 ± 0.83, 3.61 ± 0.89, 3.68 ± 0.50, and 2.39 ± 0.66 nm. Therefore, the thicknesses of the top layer, top two layers, top three layers, and all four layers were calculated to be 3.69 ± 0.83, 7.30 ± 1.22, 10.99 ± 1.32, and 13.37 ± 1.48 nm, respectively (Fig. 2C). The results indicated that each of these layers consisted of a sheet of proteins with a diameter similar to that of globular proteins with sizes between 30 and 60 kDa.
Interactions Between NDT Reagents and the FPC
NDT3, along with NDT1 and NDT2, were then used to interact with the FPC by the procedure depicted in Fig. 1B. SDS-PAGE analysis indicated that the samples isolated from the FPC by using NDT1, NDT2, and NDT3 contained the proteins of the top-most layer (streptavidin), the proteins of the top two layers (streptavidin and biotinylated actin) and the proteins of the top three layers (streptavidin, biotinylated actin, and anti-actin IgG) of the FPC, respectively (Fig. 2D, lanes indicated by S). When the same SDS-gel was examined under UV light, these protein bands were fluorescent (Fig. 2D, lanes indicated by F), indicating the transferring of the coumarin fluorophore from the SAED on NDT reagents to these proteins when the disulfide bond of SAED was reduced (Fig. 1B). These results were consistent with our earlier findings that NDT1 and NDT2 could interact with proteins lining the surface and with proteins residing within 8.0 nm deep from the surface in a protein complex, respectively (44, 46). The results also indicated that the azido group of NDT3 could indeed penetrate into a region between 7.30 and 10.99 nm deep in a protein complex and form covalent links with proteins residing in this depth range.
The abundances of proteins residing at different layers of the FPC in different NDT-isolated samples were further used to gain insights regarding the proportions of NDT reagents with their azido groups positioning in different layer regions of the FPC when the photoactivated crosslinking reaction took place. Because of the hindrance of the agarose bead, the azido groups on the surface of NDT1 could only reach in the top layer region of FPC (Fig. 3A), and the abundance of the top layer protein in the NDT1-isolated sample from FPC was expected to be 100%. On the other hand, there were at least three modes that could occur when a NDT3 bead encountered FPC where the farthest depth region the azido groups could reach was in the first, second and third layers (Fig. 3B). By analogy, when a NDT2 bead encountered the FPC, there were two modes that could occur where the farthest depth region the azido groups could reach was in the first and second layers (not shown). When the abundances of the proteins of the different FPC layers in NDT-isolated samples were obtained experimentally (Fig. 2D), the proportions of NDT reagents that interacted with FPC in different modes during the photolytic reaction could be calculated (Fig. 3E, by equations in the Material and Method section). The results indicated that during the photolytic reaction, the azido groups of NDT1 reagent could only penetrate into the top layer protein region. For NDT2, 23.6 and 76.4% of these beads would have their azido groups penetrate as far as to the first and second layer protein regions, respectively. For NDT3, 14.5, 8.8, and 76.6% of these beads would have their azido groups penetrate as far as to the first, second, and third layer protein regions, respectively. It was thus concluded that the majority (>76%) of the NDT reagents could have their azido groups penetrate to depths similar to their spacers' lengths when interacting with FPC.
Characterization of Cerebral and Cerebellar PSDs
The diameter and thickness of cerebral PSDs were have been calculated to be 265.3 nm and 41 nm, respectively, in a previously study (44). The thickness of cerebellar PSD has been reported to be 33 nm (17). By means of scanning electron microscopy, the diameter of cerebellar PSD was determined here to be 248.2 ± 72.7 nm (mean ± S.D., n = 328) (Fig. 4A). SDS-PAGE analyses indicated that cerebral and cerebellar PSD samples consisted of a similar set of proteins (lanes 2 and 4 of the left panel of Fig. 4B). However, the abundance of various proteins in these two samples differed. Consistent with the findings of an earlier comparative study of PSD samples isolated from dog cerebral cortex and cerebellum, the content of a 51 kDa protein band, previously called the “major PSD protein” (15) that was identified later as the α-subunit of calcium, calmodulin-dependent protein kinase II (CaMKIIα) (58), in the cerebral PSD sample was much higher than its counterpart in cerebellar PSD. Western blotting analyses indicated that CaMKIIα and CaMKIIβ were enriched in cerebral and cerebellar PSD compared with the homogenates of these brain regions, respectively. However, the abundances of CaMKIIα and CaMKIIβ in cerebral PSD were much higher than those in cerebellar PSD, respectively (right panel of Fig. 4B). The cerebellar homogenate was enriched with calbindin, a protein marker of cerebellar Purkinje cells (59). Both cerebral and cerebellar PSD samples were enriched with proteins known to reside at postsynaptic terminals, including various subtypes of glutamate receptors and PSD95. In addition, α-tubulin, EF1α, protein 4.1, kalirin, and tropomyosin were also enriched in cerebral and cerebellar PSD samples. On the other hand, cerebral and cerebellar PSD samples contained very low levels of synaptophysin, a presynaptic marker. Both cerebral and cerebellar PSD samples contained actin at levels similar to those of cerebral and cerebellar homogenates. Cerebral PSD was found to contain ankyrin and spectrin at levels similar to those found in cerebral homogenates, whereas these proteins were enriched in the cerebellar PSD sample when compared with cerebellar homogenates. Homer 1 was enriched in cerebral PSD and, however, undetected in cerebellar homogenates or cerebellar PSD. The results thus indicated that the cerebral and cerebellar PSD samples consisted of a similar set of proteins with abundance differences of many proteins between these two samples.
Fig. 4.
Characterization of PSD and of the interactions between PSD and NDT reagents. A, Scanning electron microscopic examination of cerebellar PSD. Left and middle panels: images of cerebellar PSD. Scale bars: 100 nm. Right panel: histogram of the diameters of cerebellar PSDs. Diameters are calculated from circles with areas equivalent to the sizes of the PSD as determined from scanning electron micrographs. The average diameter of cerebellar PSD is 248.2 ± 72.7 nm (mean ± S.D., n = 328). B, Biochemical analyses of cerebral and cerebellar PSDs. Left panel: The protein compositions of cerebral homogenate (lane 1), cerebral PSD (lane 2), cerebellar homogenate (lane 3), and cerebellar PSD (lane 4) by SDS-PAGE and silver-staining. Molecular weights markers are shown to the left. Right panel: Western blotting analyses of cerebral homogenate (lane 1), cerebral PSD (lane 2), cerebellar homogenate (lane 3), and cerebellar PSD (lane 4) with the antibodies to various proteins. The first and second rows of numbers under the immunostained bands are respectively the means and standard deviations of the relative intensities of these bands from 3 to 6 independent experiments with the intensities of the bands of cerebral homogenate being set as 1. Proteins with the contents in cerebral and cerebellar PSDs being statistically different (p < 0.05, Student t test) are indicated by asterisks. Samples containing 8 μg protein are subject to these analyses. C, Interactions between free peptidyl-SAED with cerebral PSD. Synthetic 10-mer peptidyl-SAED (left panel) and 20-mer peptidyl-SAED (right panel) were incubated with cerebral PSD. After UV irradiation, the samples were subject to SDS-PAGE analysis using 8.5% polyacrylamide gels. The resultant gels were first examined over a UV illuminator (lanes F) and then stained by Coomassie blue (lanes C). The fluorescence intensities of the various bands found in the fluorographs exhibit a linear relationship with their intensities in the Coomassie-blue-stained gels (right panels) with a correlation coefficient of 0.96 and 0.95, respectively. D, SDS-PAGE analysis of the original PSD sample (lanes 1) and the samples isolated from cerebral PSD (left panel) and from cerebellar PSD (right panel) by using NDT1, NDT2, and NDT3 (lanes 2, lanes 3, and lanes 4, respectively) with 8.5% polyacrylamide gels and visualized by silver staining. Figures to the right of the lanes of different samples in silver-stained gels are the densitometric scans of the silver-stained lanes. Molecular weight markers are shown to the left.
Free Crosslinkers of NDT Reagents Could Interact with Various PSD Proteins Nondiscriminatively
A prerequisite for the applicability of NDT reagents to probe the depths of various protein components in the PSD was that the cross-linker parts of NDT (i.e. the SAED of NDT1 and the peptide-SAED conjugates of NDT2 and NDT3), when not bound to agarose beads, could penetrate freely into different regions of the PSD structure and interact equally well with various PSD proteins. SAED, the crosslinker part of NDT1, has already been demonstrated to meet this prerequisite. To test if 10-mer and 20-mer peptidyl-SAEDs, the crosslinker parts of NDT2 and NDT3, respectively, could also do so, these crosslinkers were synthesized and incubated with cerebral PSD. After UV irradiation, the resultant samples were subject to SDS-PAGE analyses. It was found that the intensities of the various bands in Coomassie-blue-stained gel (lanes labeled by C in Fig. 4C) exhibited a linear correlation (right panels in Fig. 4C) with their fluorescence intensities in the fluorography (lanes labeled by F in Fig. 4C). The results indicated that both 10-mer and 20-mer peptidyl-SAEDs could interact with various proteins in the PSD in a nondiscriminative fashion.
Studying the Protein Distribution in Cerebral and Cerebellar PSDs
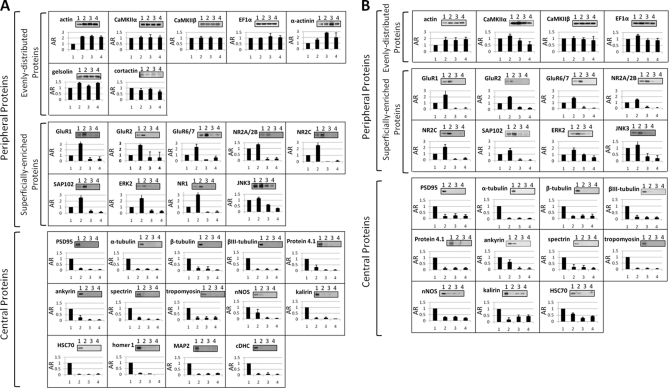
NDT1, NDT2, and NDT3 reagents were used to interact with the PSD samples isolated from cerebral cortex and cerebellum according to the procedure as depicted in Fig. 1B. The protein amounts of the samples isolated from cerebral PSD by using NDT1, NDT2, and NDT3 were 2.8 ± 0.67%, 1.02 ± 0.2%, and 0.81 ± 0.04% (mean ± S.D., n = 3), respectively, of the protein amount of the PSD used for isolation. The protein amounts of the NDT1-, NDT2-, and NDT3-isolated samples from cerebellar PSD were 1.54 ± 0.09%, 0.79 ± 0.19%, and 0.47 ± 0.12% (mean ± S.D., n = 3), respectively, of the protein amount of the PSD used for isolation. The PSD and NDT-isolated samples were subject to SDS-PAGE analyses. After silver-staining, densitometric scans of the resultant gels indicated that the concentrations of many proteins in the NDT-isolated samples were different from those of their counterparts in the original PSD samples (Fig. 4D). Equal amounts of NDT-isolated and original PSD proteins were subsequently subject to Western blotting analysis with commercially available antibodies to various PSD proteins. Out of the 42 antibodies originally used, 30 were chosen (top panels of Figs. 5A and 5B) for this study because each of these antibodies resulted in a prominent immunostained band with a size matching that of the antibody's antigen in Western blotting analyses of these samples. The intensities of the immunostained bands in the resultant blots were quantified. The abundance ratio (AR) was the ratio of the intensity of an immunostained band of the sample isolated by using a NDT reagent to the intensity of its counterpart in the original PSD sample (bottom panels of Figs. 5A and 5B). The AR's of 30 cerebral PSD proteins in the NDT1-, NDT2-, and NDT3-isolated samples, designated as AR1, AR2, and AR3, are shown in Table I. Because of the low abundance of α-actinin, gelsolin, cortactin, cDHC, MAP2, homer 1, and NR1 in cerebellar PSD, the AR values of 23 cerebellar PSD proteins in NDT-isolated samples were calculated (Table II). These AR values were then used to calculate parameters relating to the localizations of various proteins in cerebral and cerebellar PSDs by the equations described below.
Fig. 5.
Western blotting analyses of the samples isolated from cerebral PSD (A) and cerebellar PSD (B) by using NDT reagents. Equal amounts (0.8 μg of protein in cerebral PSD experiments and 0.5 μg of protein in cerebellar PSD experiments) of the original PSD and NDT1-, NDT2-, and NDT3-isolated samples, labeled by 1, 2, 3, and 4, respectively, were subject to Western blotting analysis using antibodies against various indicated PSD proteins (top of each panel). The intensities of the immunostained bands were quantified from the densitometric scans of the resultant blots. The abundance ratios (ARs) are the ratios of the intensities of the immunostained bands in the NDT1-, NDT2-, and NDT3-isolated samples to those of their counterparts in the PSD sample with the latter's intensities being set to 1 (bottom of each panel). Data are presented as mean ± S.D. from at least three independent experiments.
Table I. Localization parameters of various proteins in cerebral PSD. AR, abundance ratio; n1/N, the proportion of a protein in the region between the surface and 3.69 nm deep; n2/N, the proportion of a protein in the region between 3.69 and 7.30 nm deep; n3/N, the proportion of a protein in the region between 7.30 and 10.99 nm deep; I/N, the proportion of a protein in the region deeper than 10.99 nm from the surface.
| Protein | AR1 | AR2 | AR3 | n1/N (%) | n2/N (%) | n3/N (%) | I/N (%) | ||
|---|---|---|---|---|---|---|---|---|---|
| Peripheral Proteins | Evenly-distributed Proteins | Actin | 2.25 | 2.33 | 2.18 | 50.54 | 46.56 | 40.24 | 0.00a |
| CaMKIIα | 1.07 | 1.05 | 1.05 | 24.01 | 21.05 | 19.42 | 35.52 | ||
| CaMKIIβ | 1.03 | 1.03 | 1.00 | 23.15 | 20.58 | 18.46 | 37.81 | ||
| EF1α | 1.00 | 1.14 | 1.06 | 22.48 | 22.87 | 19.55 | 35.09 | ||
| α-actinin | 1.60 | 2.81 | 1.97 | 35.91 | 56.27 | 36.33 | 0.00a | ||
| Gelsolin | 1.42 | 1.18 | 1.43 | 31.95 | 23.61 | 26.38 | 18.05 | ||
| Cortactin | 0.81 | 0.92 | 0.70 | 18.31 | 18.31 | 12.87 | 50.51 | ||
| Superficially-enriched Proteins | GluR1 | 3.14 | 0.35 | 0.38 | 70.57 | 7.07 | 7.02 | 15.34 | |
| GluR2 | 2.73 | 0.62 | 0.58 | 61.32 | 12.33 | 10.63 | 15.73 | ||
| GluR6/7 | 2.34 | 0.24 | 0.56 | 52.64 | 4.74 | 10.30 | 32.33 | ||
| NR2A/2B | 1.36 | 0.09 | 0.14 | 30.54 | 1.89 | 2.58 | 64.99 | ||
| NR2C | 2.56 | 0.04 | 0.12 | 57.60 | 0.83 | 2.15 | 39.42 | ||
| SAP102 | 2.57 | 0.35 | 0.17 | 57.81 | 6.96 | 3.22 | 32.01 | ||
| Erk2 | 2.45 | 0.43 | 0.32 | 55.18 | 8.61 | 5.86 | 30.35 | ||
| NR1 | 3.08 | 0.12 | 0.18 | 69.26 | 2.36 | 3.29 | 25.10 | ||
| JNK3 | 1.15 | 0.60 | 0.30 | 25.88 | 12.02 | 5.59 | 56.51 | ||
| Central Proteins | PSD-95 | 0.15 | 0.08 | 0.05 | 3.28 | 1.67 | 1.00 | 94.05 | |
| α-tubulin | 0.10 | 0.11 | 0.06 | 2.36 | 2.27 | 1.16 | 94.20 | ||
| β-tubulin | 0.19 | 0.15 | 0.06 | 4.37 | 2.94 | 1.08 | 91.61 | ||
| βIII-tubulin | 0.07 | 0.06 | 0.05 | 1.64 | 1.13 | 0.88 | 96.35 | ||
| Protein 4.1 | 0.30 | 0.07 | 0.05 | 6.81 | 1.41 | 1.00 | 90.78 | ||
| Ankyrin | 0.25 | 0.08 | 0.06 | 5.59 | 1.51 | 1.07 | 91.84 | ||
| Spectrin | 0.20 | 0.04 | 0.04 | 4.49 | 0.86 | 0.79 | 93.86 | ||
| Tropomyosin | 0.18 | 0.17 | 0.21 | 4.09 | 3.46 | 3.80 | 88.65 | ||
| NNOS | 0.54 | 0.09 | 0.04 | 12.07 | 1.78 | 0.75 | 85.40 | ||
| Kalirin | 0.08 | 0.11 | 0.05 | 1.91 | 2.19 | 0.92 | 94.98 | ||
| HSC70 | 0.04 | 0.03 | 0.05 | 0.90 | 0.56 | 0.90 | 97.64 | ||
| Homer 1 | 0.12 | 0.06 | 0.01 | 2.60 | 1.25 | 0.18 | 95.98 | ||
| MAP2 | 0.08 | 0.11 | 0.15 | 1.91 | 2.25 | 2.71 | 93.13 | ||
| cDHC | 0.09 | 0.09 | 0.05 | 2.05 | 1.71 | 0.88 | 95.36 |
a Negative I/N values.
Table II. Localization parameters of various proteins in cerebellar PSD. AR, abundance ratio; n1/N, the proportion of a protein in the region between the surface and 3.69 nm deep; n2/N, the proportion of a protein in the region between 3.69 and 7.30 nm deep; n3/N, the proportion of a protein in the region between 7.30 and 10.99 nm deep; I/N, the proportion of a protein in the region deeper than 10.99 nm from the surface.
| Protein | AR1 | AR2 | AR3 | n1/N (%) | n2/N (%) | n3/N (%) | I/N (%) | ||
|---|---|---|---|---|---|---|---|---|---|
| Peripheral proteins | Evenly distributed Proteins | Actin | 1.78 | 1.76 | 1.88 | 47.89 | 41.70 | 40.60 | 0.00a |
| CaMKIIα | 1.21 | 0.83 | 0.55 | 32.55 | 19.67 | 11.88 | 35.90 | ||
| CaMKIIβ | 1.03 | 0.95 | 0.86 | 27.71 | 22.51 | 18.57 | 31.21 | ||
| EF1α | 1.26 | 0.90 | 0.90 | 33.90 | 21.33 | 19.44 | 25.34 | ||
| Superficially enriched Proteins | GluR1 | 2.34 | 0.08 | 0.08 | 62.96 | 1.90 | 1.73 | 33.42 | |
| GluR2 | 2.02 | 0.36 | 0.25 | 54.35 | 8.53 | 5.40 | 31.72 | ||
| GluR6/7 | 1.81 | 0.19 | 0.05 | 48.70 | 4.50 | 1.08 | 45.72 | ||
| NR2A/2B | 1.51 | 0.18 | 0.05 | 40.63 | 4.27 | 1.08 | 54.03 | ||
| NR2C | 2.10 | 0.09 | 0.26 | 56.50 | 2.13 | 5.61 | 35.75 | ||
| SAP102 | 1.58 | 0.18 | 0.07 | 42.51 | 4.27 | 1.51 | 51.71 | ||
| Erk2 | 1.73 | 0.99 | 0.58 | 46.55 | 23.46 | 12.53 | 17.47 | ||
| JNK3 | 1.24 | 0.36 | 0.20 | 33.36 | 8.53 | 4.32 | 53.79 | ||
| Central proteins | PSD-95 | 0.20 | 0.25 | 0.22 | 5.38 | 5.92 | 4.75 | 83.94 | |
| α-tubulin | 0.07 | 0.08 | 0.07 | 1.88 | 1.90 | 1.51 | 94.71 | ||
| β-tubulin | 0.19 | 0.24 | 0.21 | 5.11 | 5.69 | 4.54 | 84.67 | ||
| βIII-tubulin | 0.18 | 0.15 | 0.12 | 4.84 | 3.55 | 2.59 | 89.01 | ||
| Protein 4.1 | 0.11 | 0.13 | 0.13 | 2.96 | 3.08 | 2.81 | 91.15 | ||
| Ankyrin | 0.62 | 0.11 | 0.06 | 16.68 | 2.61 | 1.30 | 79.42 | ||
| Spectrin | 0.22 | 0.21 | 0.12 | 5.92 | 4.98 | 2.59 | 86.51 | ||
| Tropomyosin | 0.08 | 0.04 | 0.07 | 2.15 | 0.95 | 1.51 | 95.39 | ||
| nNOS | 0.30 | 0.32 | 0.22 | 8.07 | 7.58 | 4.75 | 79.60 | ||
| Kalirin | 0.18 | 0.41 | 0.43 | 4.84 | 9.71 | 9.29 | 76.16 | ||
| HSC70 | 0.60 | 0.27 | 0.41 | 16.14 | 6.40 | 8.85 | 68.61 |
a Negative I/N values.
Calculation of Localization Parameters of PSD Proteins
The PSD was assumed to be a disk-shaped object consisting of spherical proteins of ∼50 kDa in size, the average size of PSD proteins (60). The diameters of cerebral and cerebellar PSDs (265.3 and 248.2 nm, respectively) are much smaller than that of the NDT bead (∼80 μm in diameter). In solution, the PSD would collide with a NDT bead in all orientations, thereby forming contacts between the bead surface and various positions of the PSD's surface, including the cytoplasm- and exoplasm-facing surface and exposed lateral side. The differences between the depths to which the azido groups on different parts of a bead could penetrate in the PSD, because of the curvature of bead surface, was calculated from the interaction between the NDT and largest surface, the exoplasm- or cytoplasm-facing surface of cerebral PSD, of a PSD was calculated and found to be negligible (Fig. 3C). On the basis of the above analyses over the interaction modes between NDT and FPC (Fig. 3E), it was further assumed that when a NDT reagent encountered a PSD, most of the NDT beads would have the azido groups on their surface reach to a region of the PSD at a depth similar to this reagent's spacer length.
The AR values of a protein a could be described by the following equation
where (AR)a,i was the AR of protein a in the sample isolated by using NDTi reagent, na,i was the number of protein a in the NDTi-isolated sample, vi was the volume of the region reachable by the azido groups of the NDTi reagent, Na was the total number of protein a in the original PSD sample and vT was the total PSD volume. Equation 1 could be re-arranged into:
where the v1/vT, v2/vT, and v3/vT values were the proportions of the volumes between the surface and 3.69 nm deep, between 3.69 nm and 7.30 nm and between 7.30 and 10.99 nm deep, from all sides of PSD structure, of the total volume of the PSD, respectively. Assuming that the structure of cerebral PSD was a circular disk of 265.3 nm in diameter and 41 nm in thickness, the v1/vT, v2/vT, and v3/vT values of cerebral PSD proteins were calculated to be 0.22, 0.20, and 0.18, respectively. Assuming that the structure of cerebellar PSD was also a circular disk of 248.2 nm in diameter and 33 nm in thickness, the v1/vT, v2/vT, and v3/vT values of cerebellar PSD proteins were calculated to be 0.27, 0.24, and 0.22, respectively. Finally, the Ia/Na value, the proportion of protein a residing in the region of the PSD that was unreachable by any of the three NDT reagents could be calculated as:
where Ia is the number of protein a residing in the interiors of the PSD unreachable by any of the three NDT reagents. By using Equations 2 and 3 and the AR values obtained from Western blotting analyses (Figs. 5A and 5B), the na,i/Na and (Ia/Na)values of a total of 30 proteins in cerebral PSDs and those of 23 proteins in cerebellar PSD were calculated (Tables I and II).
According to the ARi, ni/N and I/N values, the proteins in cerebral and cerebellar PSDs subject to study here could be grouped into two categories: central and peripheral proteins (Figs. 5A and 5B; Tables I and II). Central proteins are those residing primarily in regions deeper than 10.99 nm from the surface of the PSD (with an arbitrarily chosen I/N threshold value of 75%, except for HSC70 with an I/N value of 68.61 in cerebellar PSD), and this latter region is called henceforth as the central region. Peripheral proteins are those residing primarily in regions within 10.99 nm from the surface of the PSD, and these regions are called henceforth as the peripheral regions. According to the distribution pattern, peripheral proteins are further divided into two groups: superficially enriched and evenly distributed peripheral proteins. Superficially enriched peripheral proteins are those proteins with their protein masses being concentrated in a superficial layer of ∼3.69 nm thick of the PSD. NR2A/2B and JNK3 were also classified to this category because of their protein masses residing predominantly in the outmost layer of ∼3.69 nm in thickness and in regions deeper than 10.99 nm from the surface and rarely residing in between. Evenly distributed peripheral proteins are those with at least 50% of their masses residing in peripheral regions (except for cortactin with only 49.49% of it residing in this region of cerebral PSD), and a significant amount of each peripheral protein could be found in the three regions between the surface and 3.69 nm deep, between 3.69 and 7.30 nm deep, and between 7.30 and 10.99 nm deep.
Studying Protein-Protein Interactions in the PSD by Immunoabsorption
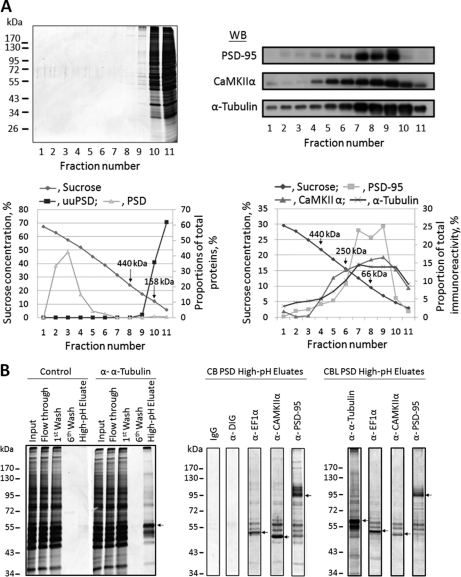
The interactions between different constituent proteins in cerebral and cerebellar PSDs were further studied by immunoabsorption. The large PSD structure was first reduced to smaller protein aggregates, yet leaving some local protein-protein interactions intact, by a procedure similar to that reported earlier (54). The resultant sample, designated as uuPSD, was subject to sucrose-density-gradient centrifugation and SDS-PAGE analyses (Fig. 6A). The absence of clear protein bands in regions where the original PSD would appear in the gradient indicated that the organized structure of the original PSD was disrupted nearly completely in the uuPSD sample. The results also indicated that nearly all proteins in the uuPSD migrated in sucrose gradients as individual proteins or aggregates with sizes smaller than 440 kDa and were found in fractions near the top of the gradients (left panels of Fig. 6A). The uuPSD sample was then subject to immunoabsorption by using protein G-magnetic beads containing covalently linked antibodies to CaMKIIα, EF1α, PSD-95, and α-tubulin. It has been reported that CaMKIIα and α-tubulin are abundant in the PSD (61) and that PSD-95 is a major scaffold protein of the PSD (62). EF1α, a moonlighting protein proposed to regulate the actin and microtubule cytoskeleton (63, 64), is also a prominent component in the PSD (19, 22, 65). Immunoabsorption of the uuPSD was also performed by using free protein G-magnetic beads or the same beads covalently linked with anti-DIG Fab fragments or mouse anti-idiotypic antibody as control experiments. After washing extensively, proteins bound to the magnetic beads were eluted by a high-pH buffer solution. SDS-PAGE analysis (Fig. 6B) indicated that each of these high-pH-eluates contained a major protein band with the size corresponding to that of the antigen of the particular antibody used (indicated by arrows). Many minor proteins were co-immunoabsorbed with these antigens. On the other hand, the high-pH-eluates of the control experiments only exhibited few very faint protein bands (Fig. 6B).
Fig. 6.
Immunoabsorption of PSD proteins. A, Sucrose-density-gradient centrifugation analysis of the uuPSD and original PSD samples isolated from cerebral cortex. Top left panel: SDS-PAGE analysis of the fractions collected from the 0–70% gradient of uuPSD (0.2 mg protein) as visualized by silver staining. The molecular weight markers are shown to the left. Bottom left panel: Protein contents of the fractions collected from the gradients of the uuPSD (■) and PSD (▴) samples. The protein content of each fraction is expressed as the percentage proportion of the fraction's integrated protein intensity against the sum of the integrated intensities of all collected fractions as calculated from the densitometric scan of the gel after silver staining. The positions of two standards, ferritin (440 kDa) and aldolase (158 kDa) are indicated by arrows. Top right panel: The high-pH-eluates collected from immunoabsorption experiments using the antibodies against PSD-95, CaMKIIα and α-tubulin were subject to sucrose-density-gradient centrifugation analysis, and the collected fractions were subject to Western blotting analysis by using antibodies against PSD-95, CaMKIIα and α-tubulin. Bottom right panel: Quantification of the intensities of the inmmunostained bands found in the top panel. The intensity of an immunostained band in a fraction is expressed as the proportion of its intensity against the sum of the intensities of all fractions. The positions of ferritin (440 kDa), catalase (250 kDa), and bovine serum albumin (66 kDa) are indicated by arrows. B, SDS-PAGE analyses of the fractions collected in immunoabsorption experiments. Left panel: Aliquots of 38 μl were removed from the indicated fractions of the immunoabsorption experiments conducted with cerebral PSD wherein no antibody (control) or the anti-α-tubulin antibody was used for SDS-PAGE analyses. Molecular weight markers are shown to the left. Middle and right panels: Aliquots of 38 μl were removed from the high-pH-eluates obtained from the immunoabsorption experiments of cerebral (CB) and cerebellar (CBL) PSDs, respectively, using mouse idiotypic monoclonal antibody (IgG) and antibodies against DIG, EF1α, CaMKIIα, and PSD-95 and subject to SDS-PAGE analysis. Arrows indicate the major protein bands found in the high-pH-eluates, which are of the same sizes as the antigens of the antibodies used in the immunoabsorption experiments. The results are from a representative experiment out of a total of 3 to 6 experiments.
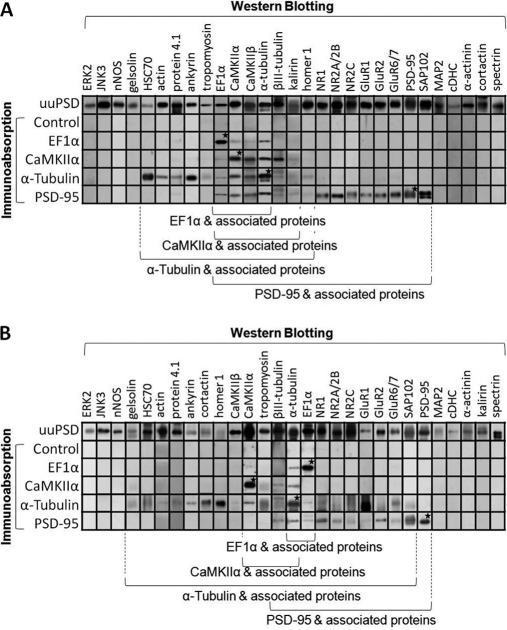
To identify proteins co-immunoabsorbed with the abovementioned four antigens, the high-pH-eluates were subject to Western blotting with the antibodies to 29 PSD proteins (Fig. 7). The results indicated that the major proteins found in the high-pH-eluates (as indicated by arrows in Fig. 6B) were recognized by the antibodies used in the respective immunoabsorption experiments (as indicated by stars in Fig. 7). On the other hand, the blots of the high-pH-eluates of the control experiments did not show any immunostained bands when antibodies to these PSD proteins were used. The results further indicated that 14, 11, 5, and 3 of cerebral PSD proteins were co-immunoabsorbed with PSD-95, α-tubulin, CaMKIIα, and EF-1α, respectively (Fig. 7A) and that 10, 19, 3, and 1 of cerebellar PSD proteins were co-immunoabsorbed with PSD-95, α-tubulin, CaMKIIα, and EF-1α, respectively (Fig. 7B). Sucrose-density-gradient (0–30%) centrifugation analysis (right panels of Fig. 6A) indicated that the antigens in the high-pH-eluates of the three immunoabsorption experiments, where antibodies to PSD-95, α-tubulin, and CaMKIIα were used, migrated in the gradients as individual proteins or as components of protein aggregates with sizes smaller than 440 kDa. This result indicated that the proteins co-immunoabsorbed with these antigens might bind directly or be linked to these antigens indirectly via other proteins.
Fig. 7.
Western blotting analyses of proteins co-immunoabsorbed with EF1α, CaMKIIα, α-tubulin and PSD-95 in cerebral PSD (A) and cerebellar PSD (B). The uuPSD (10 μg protein) and the high-pH-eluates obtained in the immunoabsorption experiments using no antibody (control) or the antibodies against EF1α, CaMKIIα, α-tubulin and PSD-95 (1/8 of the collected samples) were subject to Western blotting analysis by using antibodies against 29 PSD proteins. The gels shown are from a representative experiment out of at least three independent experiments. Asterisks indicate the immunostained bands that colocalize with the major proteins found in the high-pH-eluates of immunoabsorption experiments (as indicated by arrows in Fig. 6B) by using the same antibodies. Proteins associated to the above antigens are indicated by the brackets at the bottom.
DISCUSSION
Here, the samples isolated from cerebral and cerebellar PSDs by using different NDT reagents were subject to quantitative Western blotting analyses. From these results, parameters pertaining the depth distribution of a protein in these PSDs, namely (na,i/Na) and (Ia/Na) values, were evaluated. On the basis of the localization information derived from the resultant parameters, these proteins are divided into two categories: central and peripheral proteins (Fig. 5; Tables I and II).
Central proteins reside primarily in the region deeper than 10.99 nm from the surface of the PSD. In 30 cerebral and 23 cerebellar PSD proteins subject to this analysis, 14 and 11 proteins, respectively, belong to this category. Central proteins found in both cerebral and cerebellar PSDs include (a) the building blocks of microtubules, α-and β-tubulin, (b) protein constituents of the membrane cytoskeleton underlying the plasma membrane of mammalian erythrocytes, including spectrin, tropomyosin, ankyrin, and protein 4.1, (c) PSD95, a scaffold protein, and (d) other proteins including a nonconstitutive heat shock protein, HSC70, and two proteins that are known to interact with PSD-95, kalirin and nNOS. Kalirin has been reported to involve in the maintenance of the structure of dendritic spines (66), and nNOS is an intracellular signaling molecule (67). In the cerebral PSD, two microtubule-associated proteins, cDHC and MAP2, and another scaffold protein, homer 1, were also found to reside mainly in the central region.
Peripheral proteins reside primarily in the regions within 10.99 nm deep from the surface of the PSD. According to the distribution pattern in the peripheral region, these proteins are further divided into evenly distributed and superficially enriched peripheral proteins. The evenly distributed peripheral proteins in cerebral PSD include cortactin, gelsolin, EF1α, CaMKIIα, CaMKIIβ, actin, and α-actinin, and the evenly distributed peripheral proteins in cerebellar PSD include EF1α, CaMKIIα, CaMKIIβ, and actin. Actin is the building block of microfilament, and the remainder of these proteins has been reported to bind directly or indirectly to actin microfilament. In cerebral PSD, cortactin, EF1α, CaMKIIα, and CaMKIIβ also reside partly in the central region because their relatively high I/N values, 35.09–50.51%. Similarly, EF1α, CaMKIIα, and CaMKIIβ reside partly in the central region of cerebellar PSD. That the sums of ni/N values of actin and α-actinin in cerebral PSD and that the sum of ni/N values of actin in cerebellar PSD are larger than 1 indicate the enrichment of these proteins in the peripheral region. That the sums of the ni/N values of these proteins are larger than 1 is most likely because of the overestimation of these ni/N values by assuming that all azido groups of NDT2 and NDT3 penetrate into the PSD to depths similar to their spacers' lengths. In fact, only about 76% of these NDT beads could do that according to our study by using FPC as a model (Fig. 3).
Superficially enriched peripheral proteins are found primarily in a narrow strip of 3.69 nm thick on the surface of the PSD. In both cerebral and cerebellar PSDs, this group includes the NR2C and NR2A/2B subunits of NMDA receptor, the GluR1 and GluR2 subunits of AMPA receptor, the GluR6/7 subunits of kainate receptor, and three cytosolic proteins, Erk2, JNK3, and SAP102. In cerebral PSD, this group also includes the NR1 subunit of NMDA receptor.
The ni/N and I/N value of a protein are basically the probabilities of finding it in different depth regions of the PSD. For large proteins like glutamate receptors, which are tetrameric protein assemblies with extracellular domains measuring ∼14 nm in length, ∼15 nm in width, and ∼9 nm in thickness (68–71), their ni/N and I/N values are also likely to be related to the distribution of their protein masses at different depth regions in the PSD. The n1/N and I/N values of various glutamate receptor subunits are much larger than the n2/N and n3/N values, suggesting that the protein mass of glutamate receptors resides predominantly in the outmost layer of ∼3.69 nm in thickness and in regions deeper than 10.99 nm from the surface and rarely in between. In both cerebral and cerebellar PSDs, the I/N values of NR2A/2B and NR2C are larger than those of NR1, GluR1, GluR2, and GluR6/7. The differences in I/N values seem to reflect the differences in the size of these receptors' cytosolic domains. The cytosolic domains of NR2A/2B and NR2C account for 45–47 and 36% of their protein masses, respectively (72), whereas the cytosolic domains of NR1, GluR1, GluR2, and GluR6/7 account for only 16%, 13%, 10%, and 12–13% of these subunits' masses, respectively (73, 74). PSD-95, a protein interacting with the C termini of NR2 subunits from the cytosolic side and anchoring NMDA receptors to the postsynaptic site (62, 75), is found to reside exclusively in the central region. The results are consistent with that the proteins of glutamate receptor subunits span the peripheral region with their cytosolic domains residing in the central region.
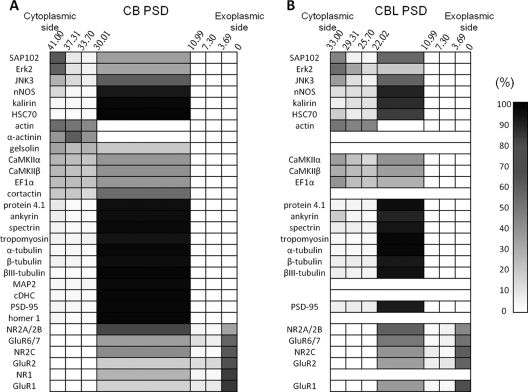
The conventional definition of the PSD is “the accumulation of dense material on the postsynaptic membrane's cytosolic face” (1). Quantitative immunogold electron microscopic studies have indicated that glutamate receptor subunits NR2 and GluR1 reside in regions where PSD joins the postsynaptic membrane, that PSD-95, nNOS, shank, CRIPT, and GKAP reside in the middle of the PSD and that actin, Ena/VASP, CaMKIIα, and cortactin reside in the PSD half close to the cytoplasm (39, 41). Earlier EM studies have indicated that actin microfilaments may be part of the PSD structure (3, 16) and that actin microfilaments are associated with the cytosol-facing surface of the PSD (76, 77). These results indicate that glutamate receptors and actin along with various microfilament-associated proteins occupy respectively the peripheral regions on the exoplasm- and cytoplasm-facing sides of the PSD. On the basis of the above EM studies and the localization information of various proteins in the PSD obtained in this study, the localization of various proteins in cerebral and cerebellar PSDs is shown as cell plots in Fig. 8. In these plots, all glutamate receptor subunits are placed in the exoplasm-facing peripheral region, and the remaining peripheral proteins are placed in the cytoplasm-facing peripheral region of the PSD. All of the central proteins listed in Tables I and II are placed in the central region.
Fig. 8.
Distribution of various proteins in cerebral PSD (A) and cerebellar PSD (B). The distribution of various proteins in cerebral and cerebellar PSDs is shown in cell plots with the ni/N and I/N values, as indicated by a gray-colored gradient shown to the right, of a particular protein (y axis) residing in different depth ranges in the PSD (x axis, with the edges on the exoplasmic and cytoplasmic sides being at 0 and 41 nm respectively in cerebral PSD and with the edges on the exoplasmic and cytoplasmic sides being at 0 and 33 nm respectively in cerebellar PSD).
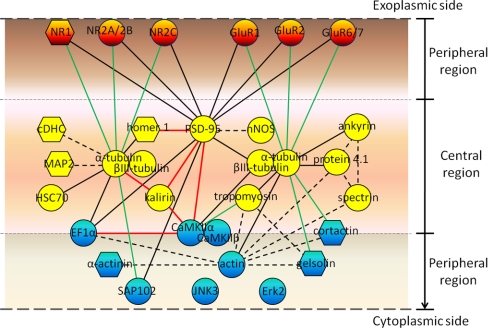
The results of immunoabsorption experiments have revealed interactions between various proteins in cerebral and cerebellar PSDs. A model (Fig. 9) is proposed here to show these interactions with the participating proteins being placed at their respective positions as indicated in Fig. 8, with all glutamate receptors (in color orange) being placed in the exoplasm-facing peripheral regions and the remainder of peripheral proteins in the cytoplasm-facing peripheral regions (in color blue). All central proteins (in color yellow) are placed randomly in the central region. In this model, α-tubulin and PSD-95, both residing exclusively in the central region, interact not only with other proteins in the same region, but also interact with proteins residing in the peripheral regions. Interestingly, α-tubulin was co-immunoabsorbed with PSD-95, but PSD-95 was not co-immunoabsorbed with α-tubulin. This is likely because of the fact that the amount of α-tubulin molecules, accounting for 8% of the PSD protein mass (18), greatly exceeds the amount of PSD-95 molecules, which account for only ∼0.8% of the protein mass of the PSD isolated from brain tissues by a similar procedure (27). Conceivably, only a small fraction of α-tubulin would bind to PSD-95, whereas the majority of PSD-95 may be associated to α-tubulin. CaMKIIα, CaMKIIβ, EF1α, and cortactin are placed on the border between the central and cytoplasm-facing peripheral regions to indicate that substantial proportions of these proteins are found in both of these regions. CaMKIIα and EF1α interact mainly with other proteins also residing in these regions. Numerous studies have already indicated that PSD-95 plays important roles as a major scaffold protein in building the PSD structure (62). CaMKIIα may assemble into a tower-like structure in the PSD (39) and interact with other PSD proteins, including NR2B, densin-180, and α-actinin, thereby contributing to the organization of the PSD structure (78). Some of the proteins found to interact with α-tubulin, PSD-95, and CaMKIIα in this study have been implicated previously to play important roles in organizing the PSD structure. For example, ankyrin, protein 4.1, and tropomyosin interact with α-tubulin (Fig. 7), and these three proteins are three major components of the spectrin-based membrane cytoskeleton underlying the cytoplasmic membrane of mammalian erythrocytes (79, 80). Ankyrin and protein 4.1 have been proposed to take part in the PSD structure organization by their interactions with spectrin, actin, neurofilaments, and AMPA receptors (81–83) (Fig. 9, indicated by broken lines). Homer 1, which is found to interact with α-tubulin and PSD-95 in cerebral PSD here, has been reported to interact with metabotropic glutamate receptors and IP3 receptors (84) as well as shank, and the interaction between homer and shank has been proposed to form a polymeric network structure for the recruitment of other PSD proteins (85). The protein-protein interactions as described in Fig. 9 may work in concert with others, such as those found in the proteomic studies of proteins associated to various glutamate receptors, PSD-95 and ProSAP/shank proteins (35–38, 86), during the process of PSD structure organization. Recent quantitative analyses have consistently indicated that the proteins that have been implicated earlier to participate in the organization of PSD structures account for small fractions of the mass of the PSD (27, 29, 87). For example, the two most abundant ones of these proteins, CaMKIIα and PSD-95, account for 3.6% and 0.8% of the protein mass of the PSD isolated by conventional biochemical procedures, respectively (27). It is thus proposed here that actin, tubulins, and their associated proteins, which together account for more than 25% of the PSD protein mass (18), may build several interconnected frameworks within the PSD. Proteins like CaMKII and scaffold proteins are recruited to these frameworks, and in turn, these latter proteins bring in their various binding partners and/or link the abovementioned frameworks together, thereby forming the PSD structure as observed in EM studies.
Fig. 9.
A model of the protein organization of the PSD. The proteins subject to study here are placed in the positions as in Fig. 8, and interactions between different protein pairs are connected by solid or broken lines of different colors. Symbols: solid lines, protein-protein interactions identified in this study; broken lines, protein-protein interactions reported earlier in the literature; black lines, protein-protein interactions identified in both cerebral and cerebellar PSDs; red lines, protein-protein interactions identified in cerebral PSD only; green lines, protein-protein interactions identified in cerebellar PSD only; circles, proteins with localization identified in both cerebral and cerebellar PSDs; hexagons, proteins with localization identified in cerebral PSD only. The interactions between proteins could be direct or indirect. Peripheral proteins facing the exoplasmic side and cytoplasmic side of the PSD are colored in orange and blue, respectively. Central proteins are colored in yellow. EF1α, CaMKIIα, CaMKIIβ, and cortactin are placed on the border between the central and cytoplasm-facing peripheral regions to indicate that these proteins reside in both regions.
A comparison between the protein localizations and protein-protein interactions in cerebral and cerebellar PSDs indicates a high similarity in their basic protein organization (Fig. 9). Differences, however, are also found between these PSDs. First, there are noticeable differences in the abundances of various proteins between cerebral and cerebellar PSDs. These differences are evident in the silver-stained SDS-PAGE gels and Western blotting analyses of these PSD samples conducted here (Fig. 4). Consistently, differences in the abundances of various proteins in cerebral (or forebrain) and cerebellar PSDs have also been reported in earlier studies (17, 19). Second, some protein-protein interactions, such as those linking PSD-95 to kalirin, CaMKIIα, and homer 1, those linking kalirin to CaMKIIα and α-tubulin and that between CaMKIIα and EF1α (as indicated by red lines in Fig. 9) are only detected in cerebral PSD, but not in cerebellar PSD. On the other hand, the interactions between α-tubulin and various glutamate receptors, cortactin, gelsolin, and SAP102 and between tropomyosin and CaMKIIα are found in cerebellar PSD, but not in cerebral PSD (as indicated by green lines in Fig. 9). These differences may, at least partly, account for the differences in morphology between cerebral and cerebellar PSDs (17).
The presence of tubulins in the PSD in neurons has remained controversial thus far. Reports have indicated that the content of tubulin subunits in the PSD increases if the brain tissues used for isolating PSD have been kept on ice for longer periods (88), but decreases if the isolated PSD sample is washed repeatedly with detergent-containing solutions (89). EM studies have seldom revealed microtubule-like structures in dendritic spines of neurons (90). These observations lead to the supposition that the presence of tubulins in the isolated PSD may be resulting from nonspecific association of tubulins to the authentic PSD during the isolation process. However, more recently, microtubules have been implicated to play important roles in dendritic spine development (91). The present results further indicate that tubulin subunits reside in the central region of the PSD and interact with many other PSD proteins. These latter findings are consistent with the notion that tubulins may play important roles in the organization of PSD structure.
It has been recently reported that certain PSD proteins, including tubulins, CaMKIIα, CaMKIIβ, MAP2, and cDHC, would exit from dendritic spines of cultured rat hippocampal neurons when these neurons are kept at low temperature for a few minutes or treated with drugs either disrupting actin cytoskeleton (87) or with drugs inducing calcium release from internal stores (Cheng and Chang unpublished results). On the other hand, the same treatments do not affect the localization of PSD-95 or glutamate receptors in dendritic spines. Local depolymerization of F-actin has been proposed to be necessary for the translocation of CaMKIIα into spines (92). It is thus likely that the recruitment of various proteins to the PSD in neurons is regulated not only by the protein-protein interactions among PSD proteins, but that the mechanisms controlling the movement of various PSD proteins in and out of dendritic spines also play important roles in regulating the assembly of the PSD. The notion that PSD assembly is regulated by multiple mechanisms may account for the dynamic nature of PSD structure and composition under in vivo conditions. In the absence of such regulatory machinery, e.g. under in vitro conditions after isolation, the PSD structure would become very firmly built and difficult to be disrupted. The mechanisms underlying in vivo PSD structure assembly will require further investigation in the future.
The present study demonstrates the usefulness of a combination of the NDT and immunoabsorption methods in investigating the protein organization of the PSD. By using the three NDT reagents available presently, we have obtained information concerning protein distribution in regions between the surface and ∼10 nm deep in the PSD. In the future, by synthesizing new NDT reagents that contain longer linkers, such as oligopeptides containing more than 30 amino acid residues and other linear and hydrophilic chemicals, the interiors of the PSD could be explored. By utilizing the various quantitative mass-spectrometry-based strategies developed recently (93–96), the relative contents of more PSD constituent proteins in the samples isolated from different NDT reagents could be calculated. These latter results would provide us with new information about the localization of many more proteins in the PSD. The methodologies described here also offer a tool for studying the protein organization of other supramolecules.
Footnotes
1 The abbreviations used are:
- PSD
- postsynaptic density
- NDT
- Nano-Depth-Tagging
- CaMKII
- calcium, calmodulin-dependent protein kinase II
- cDHC
- cytoplasmic dynein heavy chain
- NMDA
- N-methyl-D-aspartate
- AMPA
- α-amino-3-hydroxy-5-methyl-4-isoxazole propionate
- SAED
- sulfosuccinimidyl-2-(7-azido-4-methylcoumarin-3-acetamido) -ethyl-1,3′-dithiopropionate
- SDS
- sodium dodecyl sulfate
- PAGE
- polyacrylamide gel electrophoresis
- sulfo-NHS-SS-biotin
- sulfosuccinimidyl-2-[biotinamido] ethyl-1,3-dithiopropionate
- DST
- disuccinimidyl tartarate
- DMP
- dimethyl pimelimidate dihydrochloride
- DADPA
- diaminodipropylamine
- sulfo-NHS acetate
- sulfosuccinimidyl acetate
- sulfo-LC-SMPT
- sulfosuccinimidyl 6-[α-methyl-α-(2-pyridyldithio)toluamido]hexanoate
- EDAC
- 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide
- AFM
- atomic forced microscopy
- SEM
- scanning electron microscopy.
REFERENCES
- 1. Peters A., Palay S. L., Webster H. D. (1991) The Fine Structure of the Nervous System, 3rd. ed Oxford University Press, NY., pp. 160–166 [Google Scholar]
- 2. Cohen R. S., Siekevitz P. (1978) Form of the postsynaptic density. A serial section study. J. Cell Biol. 78, 36–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Matus A. I., Taff-Jones D. H. (1978) Morphology and molecular composition of isolated postsynaptic junctional structures. Proc. R. Soc. Lond. B Biol. Sci. 203, 135–151 [DOI] [PubMed] [Google Scholar]
- 4. Landis D. M., Weinstein L. A., Reese T. S. (1987) Substructure in the postsynaptic density of Purkinje cell dendritic spines revealed by rapid freezing and etching. Synapse 1, 552–558 [DOI] [PubMed] [Google Scholar]
- 5. Toni N., Buchs P. A., Nikonenko I., Bron C. R., Muller D. (1999) LTP promotes formation of multiple spine synapses between a single axon terminal and a dendrite. Nature 402, 421–425 [DOI] [PubMed] [Google Scholar]
- 6. Lüscher C., Nicoll R. A., Malenka R. C., Muller D. (2000) Synaptic plasticity and dynamic modulation of the postsynaptic membrane. Nat. Neurosci. 3, 545–550 [DOI] [PubMed] [Google Scholar]
- 7. Toni N., Buchs P. A., Nikonenko I., Povilaitite P., Parisi L., Muller D. (2001) Remodeling of Synaptic Membranes after Induction of Long-Term Potentiation. J. Neurosci. 21, 6245–6251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Dosemeci A., Tao-Cheng J. H., Vinade L., Winters C. A., Pozzo-Miller L., Reese T. S. (2001) Glutamate-induced transient modification of the postsynaptic density. Proc. Natl. Acad. Sci. U.S.A. 98, 10428–10432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Marrs G. S., Green S. H., Dailey M. E. (2001) Rapid formation and remodeling of postsynaptic densities in developing dendrites. Nat. Neurosci. 4, 1006–1013 [DOI] [PubMed] [Google Scholar]
- 10. Murthy V. N., Schikorski T., Stevens C. F., Zhu Y. (2001) Inactivity produces increases in neurotransmitter release and synapse size. Neuron 32, 673–682 [DOI] [PubMed] [Google Scholar]
- 11. Ehlers M. D. (2003) Activity level controls postsynaptic composition and signaling via the ubiquitin-proteasome system. Nat. Neurosci. 6, 231–242 [DOI] [PubMed] [Google Scholar]
- 12. Siekevitz P. (1985) The postsynaptic density: a possible role in long-lasting effects in the central nervous system. Proc. Natl. Acad. Sci. U.S.A. 82, 3494–3498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ziff E. B. (1997) Enlightening the postsynaptic density. Neuron 19, 1163–1174 [DOI] [PubMed] [Google Scholar]
- 14. Kennedy M. B. (2000) Signal-processing machines at the postsynaptic density. Science 290, 750–754 [DOI] [PubMed] [Google Scholar]
- 15. Cohen R. S., Blomberg F., Berzins K., Siekevitz P. (1977) The structure of postsynaptic densities isolated from dog cerebral cortex. I. Overall morphology and protein composition. J. Cell Biol. 74, 181–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Blomberg F., Cohen R. S., Siekevitz P. (1977) The structure of postsynaptic densities isolated from dog cerebral cortex. II. Characterization and arrangement of some of the major proteins within the structure. J. Cell Biol. 74, 204–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Carlin R. K., Grab D. J., Cohen R. S., Siekevitz P. (1980) Isolation and characterization of postsynaptic densities from various brain regions: enrichment of different types of postsynaptic densities. J. Cell Biol. 86, 831–845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lai S. L., Chiang S. F., Chen I. T., Chow W. Y., Chang Y. C. (1999) Interprotein disulfide bonds formed during isolation process tighten the structure of the postsynaptic density. J. Neurochem. 73, 2130–2138 [PubMed] [Google Scholar]
- 19. Walsh M. J., Kuruc N. (1992) The postsynaptic density: constituent and associated proteins characterized by electrophoresis, immunoblotting, and peptide sequencing. J. Neurochem. 59, 667–678 [DOI] [PubMed] [Google Scholar]
- 20. Walikonis R. S., Jensen O. N., Mann M., Provance D. W., Jr., Mercer J. A., Kennedy M. B. (2000) Identification of Proteins in the Postsynaptic Density Fraction by Mass Spectrometry. J. Neurosci. 20, 4069–4080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Satoh K., Takeuchi M., Oda Y., Deguchi-Tawarada M., Sakamoto Y., Matsubara K., Nagasu T., Takai Y. (2002) Identification of activity-regulated proteins in the postsynaptic density fraction. Genes Cells 7, 187–197 [DOI] [PubMed] [Google Scholar]
- 22. Li K. W., Hornshaw M. P., Van Der Schors R. C., Watson R., Tate S., Casetta B., Jimenez C. R., Gouwenberg Y., Gundelfinger E. D., Smalla K. H., Smit A. B. (2004) Proteomics analysis of rat brain postsynaptic density. Implications of the diverse protein functional groups for the integration of synaptic physiology. J. Biol. Chem. 279, 987–1002 [DOI] [PubMed] [Google Scholar]
- 23. Yoshimura Y., Yamauchi Y., Shinkawa T., Taoka M., Donai H., Takahashi N., Isobe T., Yamauchi T. (2004) Molecular constituents of the postsynaptic density fraction revealed by proteomic analysis using multidimensional liquid chromatography-tandem mass spectrometry. J. Neurochem. 88, 759–768 [DOI] [PubMed] [Google Scholar]
- 24. Peng J., Kim M. J., Cheng D., Duong D. M., Gygi S. P., Sheng M. (2004) Semiquantitative proteomic analysis of rat forebrain postsynaptic density fractions by mass spectrometry. J. Biol. Chem. 279, 21003–21011 [DOI] [PubMed] [Google Scholar]
- 25. Jordan B. A., Fernholz B. D., Boussac M., Xu C., Grigorean G., Ziff E. B., Neubert T. A. (2004) Identification and verification of novel rodent postsynaptic density proteins. Mol. Cell Proteomics 3, 857–871 [DOI] [PubMed] [Google Scholar]
- 26. Hahn C. G., Banerjee A., Macdonald M. L., Cho D. S., Kamins J., Nie Z., Borgmann-Winter K. E., Grosser T., Pizarro A., Ciccimaro E., Arnold S. E., Wang H. Y., Blair I. A. (2009) The post-synaptic density of human postmortem brain tissues: an experimental study paradigm for neuropsychiatric illnesses. PLoS One 4, e5251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Chen X., Vinade L., Leapman R. D., Petersen J. D., Nakagawa T., Phillips T. M., Sheng M., Reese T. S. (2005) Mass of the postsynaptic density and enumeration of three key molecules. Proc. Natl. Acad. Sci. U.S.A. 102, 11551–11556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sugiyama Y., Kawabata I., Sobue K., Okabe S. (2005) Determination of absolute protein numbers in single synapses by a GFP-based calibration technique. Nat. Methods 2, 677–684 [DOI] [PubMed] [Google Scholar]
- 29. Cheng D., Hoogenraad C. C., Rush J., Ramm E., Schlager M. A., Duong D. M., Xu P., Wijayawardana S. R., Hanfelt J., Nakagawa T., Sheng M., Peng J. (2006) Relative and Absolute Quantification of Postsynaptic Density Proteome Isolated from Rat Forebrain and Cerebellum. Mol. Cell. Proteomics 5, 1158–1170 [DOI] [PubMed] [Google Scholar]
- 30. Sheng M., Hoogenraad C. C. (2007) The postsynaptic architecture of excitatory synapses: a more quantitative view. Annu. Rev. Biochem. 76, 823–847 [DOI] [PubMed] [Google Scholar]
- 31. Trinidad J. C., Specht C. G., Thalhammer A., Schoepfer R., Burlingame A. L. (2006) Comprehensive identification of phosphorylation sites in postsynaptic density preparations. Mol. Cell. Proteomics 5, 914–922 [DOI] [PubMed] [Google Scholar]
- 32. Vosseller K., Trinidad J. C., Chalkley R. J., Specht C. G., Thalhammer A., Lynn A. J., Snedecor J. O., Guan S., Medzihradszky K. F., Maltby D. A., Schoepfer R., Burlingame A. L. (2006) O-linked N-acetylglucosamine proteomics of postsynaptic density preparations using lectin weak affinity chromatography and mass spectrometry. Mol. Cell. Proteomics 5, 923–934 [DOI] [PubMed] [Google Scholar]
- 33. Trinidad J. C., Thalhammer A., Specht C. G., Schoepfer R., Burlingame A. L. (2005) Phosphorylation state of postsynaptic density proteins. J. Neurochem. 92, 1306–1316 [DOI] [PubMed] [Google Scholar]
- 34. Thalhammer A., Trinidad J. C., Burlingame A. L., Schoepfer R. (2009) Densin-180: revised membrane topology, domain structure and phosphorylation status. J. Neurochem. 109, 297–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Husi H., Ward M. A., Choudhary J. S., Blackstock W. P., Grant S. G. (2000) Proteomic analysis of NMDA receptor-adhesion protein signaling complexes. Nat. Neurosci. 3, 661–669 [DOI] [PubMed] [Google Scholar]
- 36. Farr C. D., Gafken P. R., Norbeck A. D., Doneanu C. E., Stapels M. D., Barofsky D. F., Minami M., Saugstad J. A. (2004) Proteomic analysis of native metabotropic glutamate receptor 5 protein complexes reveals novel molecular constituents. J. Neurochem. 91, 438–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Collins M. O., Husi H., Yu L., Brandon J. M., Anderson C. N., Blackstock W. P., Choudhary J. S., Grant S. G. (2006) Molecular characterization and comparison of the components and multiprotein complexes in the postsynaptic proteome. J. Neurochem. 97, 16–23 [DOI] [PubMed] [Google Scholar]
- 38. Dosemeci A., Makusky A. J., Jankowska-Stephens E., Yang X., Slotta D. J., Markey S. P. (2007) Composition of the synaptic PSD-95 complex. Mol. Cell. Proteomics 6, 1749–1760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Petersen J. D., Chen X., Vinade L., Dosemeci A., Lisman J. E., Reese T. S. (2003) Distribution of postsynaptic density (PSD)-95 and Ca2+/calmodulin-dependent protein kinase II at the PSD. J. Neurosci. 23, 11270–11278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Valtschanoff J. G., Weinberg R. J. (2001) Laminar Organization of the NMDA Receptor Complex within the Postsynaptic Density. J. Neurosci. 21, 1211–1217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Rostaing P., Real E., Siksou L., Lechaire J. P., Boudier T., Boeckers T. M., Gertler F., Gundelfinger E. D., Triller A., Marty S. (2006) Analysis of synaptic ultrastructure without fixative using high-pressure freezing and tomography. Eur. J. Neurosci. 24, 3463–3474 [DOI] [PubMed] [Google Scholar]
- 42. Chen X., Winters C., Azzam R., Li X., Galbraith J. A., Leapman R. D., Reese T. S. (2008) Organization of the core structure of the postsynaptic density. Proc. Natl. Acad. Sci. U.S.A. 105, 4453–4458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ratner N., Mahler H. R. (1983) Structural organization of filamentous proteins in postsynaptic density. Biochemistry 22, 2446–2453 [DOI] [PubMed] [Google Scholar]
- 44. Liu S. H., Cheng H. H., Huang S. Y., Yiu P. C., Chang Y. C. (2006) Studying the Protein Organization of the Postsynaptic Density by a Novel Solid Phase- and Chemical Cross-linking-based Technology. Mol. Cell. Proteomics 5, 1019–1032 [DOI] [PubMed] [Google Scholar]
- 45. Thevenin B. J., Shahrokh Z., Williard R. L., Fujimoto E. K., Kang J. J., Ikemoto N., Shohet S. B. (1992) A novel photoactivatable cross-linker for the functionally-directed region-specific fluorescent labeling of proteins. Eur. J. Biochem. 206, 471–477 [DOI] [PubMed] [Google Scholar]
- 46. Horng W. C., Yen Y. H., Chang Y. C. (2008) A novel solid phase- and chemical crosslinking-based technology for determining protein localization in biological supramolecules. Proteomics 8, 4642–4646 [DOI] [PubMed] [Google Scholar]
- 47. Udenfriend S., Stein S., Böhlen P., Dairman W., Leimgruber W., Weigele M. (1972) Fluorescamine: A reagent for assay of amino acids, peptides, proteins, and primary amines in the picomole range. Science 178, 871–872 [DOI] [PubMed] [Google Scholar]
- 48. Sheppeck J. E., Kar H., Hong H. (2000) A convenient and scaleable procedure for removing the Fmoc group in solution. Tetrahedron Letters 41, 5329–5333 [Google Scholar]
- 49. Pauling L., Corey R. B., Branson H. R. (1951) The structure of proteins; two hydrogen-bonded helical configurations of the polypeptide chain. Proc. Natl. Acad. Sci. U.S.A. 37, 205–211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Matsuzawa M., Liesi P., Knoll W. (1996) Chemically modifying glass surfaces to study substratum-guided neurite outgrowth in culture. J. Neurosci. Methods 69, 189–196 [DOI] [PubMed] [Google Scholar]
- 51. Pardee J. D., Aspudich J. (1982) [18] Purification of muscle actin. In: Dixie W., Frederiksen L. W. C. ed. Methods in Enzymology, pp. 164–181, Academic Press; [DOI] [PubMed] [Google Scholar]
- 52. Schneider C., Newman R. A., Sutherland D. R., Asser U., Greaves M. F. (1982) A one-step purification of membrane proteins using a high efficiency immunomatrix. J. Biol. Chem. 257, 10766–10769 [PubMed] [Google Scholar]
- 53. Qoronfleh M. W., Ren L., Emery D., Perr M., Kaboord B. (2003) Use of immunomatrix methods to improve protein-protein interaction detection. J. Biomed. Biotechnol. 2003. (5), 291–298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Chang C. W., Peng S. C., Cheng W. Y., Liu S. H., Cheng H. H., Huang S. Y., Chang Y. C. (2007) Studying the protein–protein interactions in the postsynaptic density by means of immunoabsorption and chemical crosslinking. Proteomics 1, 1499–1512 [DOI] [PubMed] [Google Scholar]
- 55. Laemmli U. K. (1970) Cleavage of Structural Proteins during the Assembly of the Head of Bacteriophage T4. Nature 227, 680–685 [DOI] [PubMed] [Google Scholar]
- 56. Wu T. Y., Liu C. I., Chang Y. C. (1996) A study of the oligomeric state of the alpha-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid-preferring glutamate receptors in the synaptic junctions of porcine brain. Biochem. J. 319, 731–739 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Wray W., Boulikas T., Wray V. P., Hancock R. (1981) Silver staining of proteins in polyacrylamide gels. Anal. Biochem. 118, 197–203 [DOI] [PubMed] [Google Scholar]
- 58. Kennedy M. B., Bennett M. K., Erondu N. E. (1983) Biochemical and immunochemical evidence that the “major postsynaptic density protein” is a subunit of a calmodulin-dependent protein kinase. Proc. Natl. Acad. Sci. U.S.A. 80, 7357–7361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Barski J. J., Hartmann J., Rose C. R., Hoebeek F., Mörl K., Noll-Hussong M., De Zeeuw C. I., Konnerth A., Meyer M. (2003) Calbindin in Cerebellar Purkinje Cells Is a Critical Determinant of the Precision of Motor Coordination. J. Neurosci. 23, 3469–3477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Ku M. Y. (2003) Analysis of postsynaptic density and α-CaMKII in rat brain by two-dimensional gel electrophoresis. M. Sc. thesis, National Tsing Hua University, Hsinchu, Taiwan, Republic of China [Google Scholar]
- 61. Cheng H. H., Liu S. H., Lee H. C., Lin Y. S., Huang Z. H., Hsu C. I., Chen Y. C., Chang Y.-C. (2006) Heavy chain of cytoplasmic dynein is a major component of the postsynaptic density fraction. J. Neurosci. Res. 84, 244–254 [DOI] [PubMed] [Google Scholar]
- 62. Sheng M., Sala C. (2001) PDZ domains and the organization of supramolecular complexes. Annu. Rev. Neurosci. 24, 1–29 [DOI] [PubMed] [Google Scholar]
- 63. Moore R. C., Durso N. A., Cyr R. J. (1998) Elongation factor-1α stabilizes microtubules in a calcium/calmodulin-dependent manner. Cell Motility Cytoskeleton 41, 168–180 [DOI] [PubMed] [Google Scholar]
- 64. Gross S. R., Kinzy T. G. (2005) Translation elongation factor 1A is essential for regulation of the actin cytoskeleton and cell morphology. Nat. Struct. Mol. Biol. 12, 772–778 [DOI] [PubMed] [Google Scholar]
- 65. Cho S. J., Jung J. S., Ko B. H., Jin I., Moon I. S. (2004) Presence of translation elongation factor-1A (eEF1A) in the excitatory postsynaptic density of rat cerebral cortex. Neurosci. Lett. 366, 29–33 [DOI] [PubMed] [Google Scholar]
- 66. Penzes P., Johnson R. C., Sattler R., Zhang X., Huganir R. L., Kambampati V., Mains R. E., Eipper B. A. (2001) The neuronal Rho-GEF Kalirin-7 interacts with PDZ domain-containing proteins and regulates dendritic morphogenesis. Neuron 29, 229–242 [DOI] [PubMed] [Google Scholar]
- 67. Ko G. Y., Kelly P. T. (1999) Nitric Oxide Acts as a Postsynaptic Signaling Molecule in Calcium/Calmodulin-Induced Synaptic Potentiation in Hippocampal CA1 Pyramidal Neurons. J. Neurosci. 19, 6784–6794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Wu T. Y., Chang Y. C. (1994) Hydrodynamic and pharmacological characterization of putative alpha-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid/kainate-sensitive L-glutamate receptors solubilized from pig brain. Biochem. J. 300, 365–371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Laube B., Kuhse J., Betz H. (1998) Evidence for a tetrameric structure of recombinant NMDA receptors. J. Neurosci. 18, 2954–2961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Rosenmund C., Stern-Bach Y., Stevens C. F. (1998) The tetrameric structure of a glutamate receptor channel. Science 280, 1596–1599 [DOI] [PubMed] [Google Scholar]
- 71. Sobolevsky A. I., Rosconi M. P., Gouaux E. (2009) X-ray structure, symmetry and mechanism of an AMPA-subtype glutamate receptor. Nature 462, 745–756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Kutsuwada T., Kashiwabuchi N., Mori H., Sakimura K., Kushiya E., Araki K., Meguro H., Masaki H., Kumanishi T., Arakawa M., et al. (1992) Molecular diversity of the NMDA receptor channel. Nature 358, 36–41 [DOI] [PubMed] [Google Scholar]
- 73. Bettler B., Egebjerg J., Sharma G., Pecht G., Hermans-Borgmeyer I., Moll C., Stevens C. F., Heinemann S. (1992) Cloning of a putative glutamate receptor: A low affinity kainate-binding subunit. Neuron 8, 257–265 [DOI] [PubMed] [Google Scholar]
- 74. Keinänen K., Wisden W., Sommer B., Werner P., Herb A., Verdoorn T. A., Sakmann B., Seeburg P. H. (1990) A family of AMPA-selective glutamate receptors. Science 249, 556–560 [DOI] [PubMed] [Google Scholar]
- 75. Kornau H. C., Schenker L. T., Kennedy M. B., Seeburg P. H. (1995) Domain interaction between NMDA receptor subunits and the postsynaptic density protein PSD-95. Science 269, 1737–1740 [DOI] [PubMed] [Google Scholar]
- 76. Fifková E., Delay R. J. (1982) Cytoplasmic actin in neuronal processes as a possible mediator of synaptic plasticity. J. Cell Biol. 95, 345–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Landis D. M., Reese T. S. (1983) Cytoplasmic organization in cerebellar dendritic spines. J. Cell Biol. 97, 1169–1178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Robison A. J., Bass M. A., Jiao Y., MacMillan L. B., Carmody L. C., Bartlett R. K., Colbran R. J. (2005) Multivalent interactions of calcium/calmodulin-dependent protein kinase II with the postsynaptic density proteins NR2B, densin-180, and alpha-actinin-2. J. Biol. Chem. 280, 35329–35336 [DOI] [PubMed] [Google Scholar]
- 79. Bretscher A. (1991) Microfilament structure and function in the cortical cytoskeleton. Annu. Rev. Cell Biol. 7, 337–374 [DOI] [PubMed] [Google Scholar]
- 80. Bennett V., Baines A. J. (2001) Spectrin and ankyrin-based pathways: metazoan inventions for integrating cells into tissues. Physiol. Rev. 81, 1353–1392 [DOI] [PubMed] [Google Scholar]
- 81. Shen L., Liang F., Walensky L. D., Huganir R. L. (2000) Regulation of AMPA receptor GluR1 subunit surface expression by a 4. 1N-linked actin cytoskeletal association. J. Neurosci. 20, 7932–7940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Baines A. J., Keating L., Phillips G. W., Scott C. (2001) The postsynaptic spectrin/4.1 membrane protein “accumulation machine”. Cell. Mol. Biol. Lett. 6, 691–702 [PubMed] [Google Scholar]
- 83. Scott C., Keating L., Bellamy M., Baines A. J. (2001) Protein 4.1 in forebrain postsynaptic density preparations: enrichment of 4.1 gene products and detection of 4.1R binding proteins. Eur. J. Biochem. 268, 1084–1094 [DOI] [PubMed] [Google Scholar]
- 84. Shiraishi-Yamaguchi Y., Furuichi T. (2007) The Homer family proteins. Genome Biol. 8, 206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Hayashi M. K., Tang C., Verpelli C., Narayanan R., Stearns M. H., Xu R. M., Li H., Sala C., Hayashi Y. (2009) The postsynaptic density proteins Homer and Shank form a polymeric network structure. Cell 137, 159–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Baron M. K., Boeckers T. M., Vaida B., Faham S., Gingery M., Sawaya M. R., Salyer D., Gundelfinger E. D., Bowie J. U. (2006) An architectural framework that may lie at the core of the postsynaptic density. Science 311, 531–535 [DOI] [PubMed] [Google Scholar]
- 87. Cheng H. H., Liu S. H., Lee H. C., Lin Y. S., Huang Z. H., Hsu C. I., Chen Y. C., Chang Y. C. (2006) Heavy chain of cytoplasmic dynein is a major component of the postsynaptic density fraction. J. Neurosci. Res. 84, 244–254 [DOI] [PubMed] [Google Scholar]
- 88. Carlin R. K., Grab D. J., Siekevitz P. (1982) Postmortem accumulation of tubulin in postsynaptic density preparations. J. Neurochem. 38, 94–100 [DOI] [PubMed] [Google Scholar]
- 89. Cho K. O., Hunt C. A., Kennedy M. B. (1992) The rat brain postsynaptic density fraction contains a homolog of the Drosophila discs-large tumor suppressor protein. Neuron 9, 929–942 [DOI] [PubMed] [Google Scholar]
- 90. Sorra K. E., Harris K. M. (2000) Overview on the structure, composition, function, development, and plasticity of hippocampal dendritic spines. Hippocampus 10, 501–511 [DOI] [PubMed] [Google Scholar]
- 91. Gu J., Firestein B. L., Zheng J. Q. (2008) Microtubules in dendritic spine development. J. Neurosci. 28, 12120–12124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Ouyang Y., Wong M., Capani F., Rensing N., Lee C. S., Liu Q., Neusch C., Martone M. E., Wu J. Y., Yamada K., Ellisman M. H., Choi D. W. (2005) Transient decrease in F-actin may be necessary for translocation of proteins into dendritic spines. Eur. J. Neurosci. 22, 2995–3005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Li J., Steen H., Gygi S. P. (2003) Protein Profiling with Cleavable Isotope-coded Affinity Tag (cICAT) Reagents. Mol. Cell. Proteomics 2, 1198–1204 [DOI] [PubMed] [Google Scholar]
- 94. Han D. K., Eng J., Zhou H., Aebersold R. (2001) Quantitative profiling of differentiation-induced microsomal proteins using isotope-coded affinity tags and mass spectrometry. Nat. Biotech. 19, 946–951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Hansen K. C., Schmitt-Ulms G., Chalkley R. J., Hirsch J., Baldwin M. A., Burlingame A. L. (2003) Mass Spectrometric Analysis of Protein Mixtures at Low Levels Using Cleavable 13C-Isotope-coded Affinity Tag and Multidimensional Chromatography. Mol. Cell. Proteomics 2, 299–314 [DOI] [PubMed] [Google Scholar]
- 96. Gerber S. A., Rush J., Stemman O., Kirschner M. W., Gygi S. P. (2003) Absolute quantification of proteins and phosphoproteins from cell lysates by tandem MS. Proc. Natl. Acad. Sci. U. S. A. 100, 6940–6945 [DOI] [PMC free article] [PubMed] [Google Scholar]