Abstract
The purpose of this study was to use metabonomic profiling to identify a potential specific biomarker pattern in urine as a noninvasive bladder cancer (BC) detection strategy. A liquid chromatography-mass spectrometry based method, which utilized both reversed phase liquid chromatography and hydrophilic interaction chromatography separations, was performed, followed by multivariate data analysis to discriminate the global urine profiles of 27 BC patients and 32 healthy controls. Data from both columns were combined, and this combination proved to be effective and reliable for partial least squares-discriminant analysis. Following a critical selection criterion, several metabolites showing significant differences in expression levels were detected. Receiver operating characteristic analysis was used for the evaluation of potential biomarkers. Carnitine C9:1 and component I, were combined as a biomarker pattern, with a sensitivity and specificity up to 92.6% and 96.9%, respectively, for all patients and 90.5% and 96.9%, respectively for low-grade BC patients. Metabolic pathways of component I and carnitine C9:1 are discussed. These results indicate that metabonomics is a practicable tool for BC diagnosis given its high efficacy and economization. The combined biomarker pattern showed better performance than single metabolite in discriminating bladder cancer patients, especially low-grade BC patients, from healthy controls.
Bladder cancer (BC)1 is one of the most commonly occurring tumors in the urinary system. The incidence of BC continues to rise, and mortality rates have not changed significantly in the past three decades. The success of BC treatment depends mainly on early detection (1). Currently, cystoscopy and urinary cytology are the standard diagnostic tools for BC (2, 3). However, cystoscopy is invasive and labor-intensive, and carcinoma in situ (CIS) of the bladder may easily be missed, given the similarity in appearance of red mucosal spots in inflammatory lesions and CIS of the bladder (4). Urinary cytology is an adjunctive noninvasive detection technique that can detect high-grade lesions with high accuracy but cannot efficiently detect low-grade BC patients (2, 5). Statistically, urinary cytology can only reach a median sensitivity of 35% (6). Screening BC patients through biomarker technology is a promising strategy to improve detection and diagnosis.
Most of the current strategies for cancer biomarker detection use a proteomic approach. A series of important proteomic biomarkers have been reported in recent years, including telomerase, hyaluronidase, CEACAM1 (carcinoembryonic antigen-related cell adhesion molecule 1), nuclear matrix protein 22, cystatin B, cytokeratins, growth factors, and surviving (1, 6, 7). All these urinary markers present challenges for obtaining both high specificity and high sensitivity at the same time (6). Urinary peptides were also evaluated as diagnostic biomarkers, but the reported panel of peptides showed low specificity (8). Although proteins or peptides seem to be promising biomarkers, proteomics or peptidomics approaches are time-consuming, labor-intensive, and costly. Thus, economic, convenient, and noninvasive methods for BC detection should be explored.
Cellular tumor genes linked to molecular pathway alterations produce or secrete specific metabolites into biofluids (9). These metabolites can potentially serve as biomarkers for cancer diagnosis (10). Metabonomics is defined by Nicholson et al. as “quantitative measurement of the dynamic multi-parametric metabolic response of living systems to pathophysiological stimuli or genetic modification” (11). It provides information that cannot be obtained directly from the genotype, gene expression profiles, or even the proteome of an individual (1, 12, 13). Of all human malignancies, BC seems to be ideally suited for carrying out a urinary diagnostic assay because the bladder is a temporary biofluid container (13–16). However, metabolic research about BC is rare and incomplete. Pasikanti et al. used gas chromatography mass spectrometry (GC-MS) for urinary metabolic profiling of BC patients and non-BC controls and demonstrated that urinary metabonomics is amenable to the noninvasive diagnosis of human BC (2). Issaq et al. also reported that metabonomics using RPLC-MS had the potential to be a noninvasive early detection strategy for BC (17). Unfortunately, however, they did not further investigate the biomarkers related to bladder tumorigenesis.
It is well known that several metabolites cannot be analyzed by GC-MS because they are prone to thermal decomposition or are unable to be volatilized. In contrast, a LC-based platform can detect a wider range of chemical species, and reversed phase liquid chromatography (RPLC)-MS is the most widely used platform in metabonomic studies (14). Urine is predominantly aqueous and may contain a large proportion of polar compounds, which would typically be unretained on RP systems (18). To make up for this technical deficiency, extensive hydrophilic interaction chromatography (HILIC) separations should be performed (19, 20). HILIC has a separation principle similar to normal phase chromatography, as it utilizes a polar stationary phase (21). Furthermore, HILIC allows the use of aqueous solvents, which is fully compatible with an electrospray ionization (ESI) source (22, 23).
In this study, a LC-MS based method that utilized both RPLC and HILIC separations was carried out, followed by multivariate data analysis to discriminate the global urine profiles of BC patients and healthy controls. Data from both columns were combined and evaluated by internal permutation tests and external validation tests. The clinical utility of potential biomarkers was evaluated using ROC analysis. To our knowledge, no studies have previously used two complementary chromatographic techniques to construct a screening model for the noninvasive detection of BC. The purpose of this study was to identify a potential biomarker pattern in urine using metabonomics to aid noninvasive BC detection.
EXPERIMENTAL PROCEDURES
Chemicals
HPLC-grade acetonitrile (ACN) was obtained from Tedia (USA). HPLC-grade formic acid (FA) was purchased from Sigma/Fluka (Germany). Distilled water (18.2 mΩ) for chromatographic separation was prepared using a Milli-Q water purification system (Millipore, Billerica, MA). Ammonium acetate was purchased from Tedia (Fairfield, OH, USA). The standard hippuric acid and phenylacetylglutamine (PAGN) was purchased from MaJin (China). Leucylproline, acetylcarinitine, were obtained from Acros (Fairlawn, NJ).
Clinical Samples
Twenty-seven BC patients and 32 healthy volunteers from the First Hospital of Xiamen were enrolled in this study. All the patients were diagnosed by histopathology examination, and none had received chemotherapy or radiation before sample collection. Informed consent was obtained from each participant before sample collection, and the procedures were approved by the institutional reviewer board. Detailed information of the clinicopathogical characteristics of the BC patients and healthy controls is provided in Table I. Tumors were graded according to the criteria recommended by the World Health Organization (24). All urine samples were collected in the morning before breakfast and stored at −80 °C until analysis.
Table I. Clinicopathogical characteristics of BC patients and healthy controls.
| Characteristics | BC patients | Healthy controls |
|---|---|---|
| No. of subjects | 27 | 32 |
| Age (mean, range) | 56, 42–71 | 53, 46–67 |
| Male | 19 (70.4%) | 18 (56.2%) |
| Female | 8 (29.6%) | 14 (43.8%) |
| Cancer grade | ||
| Low grade (LG) | 21 (77.8%) | \ |
| High grade (HG) | 6 (22.2%) | \ |
| BMI (median, range) | 23.3, 16.4–26.8 | 24.4, 18.1–27.4 |
| Hematuria | 3 (11.1%) | 0 |
| Medications | 0 | 0 |
| Smoking habit | ||
| Nonsmokers | 13 (48.1%) | 18 (56.3%) |
| Ex-smokers | 3 (11.1%) | 0 |
| smokers | 10 (37.0%) | 12 (37.5%) |
| information not available | 1 (3.7%) | 2 (6.2%) |
| Race | Chinese | Chinese |
Urine Preparation
Urine samples were thawed at room temperature before analysis. For RPLC separation, a volume of 800 μl distilled water was added to 200 μl urine and then centrifuged at 13,000 rpm for 10 min at 4 °C. For HILIC separation, a volume of 800 μl of ACN was added to 200 μl urine. After vortexing, the mixture was set aside at 4 °C for 10 min, and then centrifuged at 13,000 rpm for 10 min at 4 °C. The addition of ACN is necessary to avoid the creation of a water “plug,” which would greatly affect the chromatograph. The supernatants from the two separation modes were filtered through 0.22 μm regenerated cellulose filters before HPLC-MS analysis. An in-house quality control (QC) was prepared by pooling and mixing the same volume of each sample. A subset of six samples (three BC patients and three healthy controls) was randomly selected to form an independent test set.
HPLC-MS Analysis
All chromatographic separations were performed using an Ultimate-3000 HPLC system (Dionex, USA). A 2.1 × 150 mm Acclaim C18 3 μm column (Dionex, Sunnyvale, CA) was used for reversed phase separation. The column was maintained at 30 °C. The mobile phase was a mixture of (A) H2O with 0.1% FA and (B) ACN with 0.1% FA, with a programmed gradient as follows: initial 15% B maintained for 3 min, then increased to 60% in 9 min, increased to 95% in 4 min, held at 95% for 10 min, decreased to 15% in 0.1 min, and finally maintained at 15% for 4 min. HILIC separation was performed using a 2.1 × 150 mm Atlantis HILIC Silica 3 μm column (Waters, Milford, MA). The mobile phases were (A) H2O containing 10 mm of ammonium acetate and 0.1% FA and (B) ACN. The gradient started with an isocratic run at 95% B for 5 min, decreased to 80% in 7 min, proceeded to 50% in 6 min, maintained with 50% in 7 min, and returned to 95% for column equilibration for 5 min. The injection volume was 15 μl for both separation modes.
The chromatograph was coupled directly to the mass spectrometer at a flow rate of 200 μl min−1 without splitting. The data were acquired on a mass spectrometer (MicrOTOF-QII, Bruker Daltonics, Billerica, MA) using positive ESI over the scan range of 50 to 1000 m/z. The acquisition rate was set at 1 spectrum per second. In the source, the capillary voltage was set at −4500 V, the end plate offset potential was set at −500 V, the nebulizer gas pressure was set at 0.7 bar, and the dry gas flow rate was set at 6 L min−1 at a temperature of 180 °C. A blank and a QC sample were injected every 10 samples to monitor the stability of the system. MS/MS experiments were carried out to identify potential biomarkers. Argon was used as collision gas, and the collision energy was adjustable from 10 eV to 30 eV.
Raw Data Pretreatment and Chemometric Analyses
All chromatograms were evaluated by ProfileAnalysis 1.1 software (Bruker), which performs peak alignment, background noise subtraction, and data reduction in an automated and unbiased way. Only peaks with signal-to-noise ratio (S/N) greater than 5 were utilized for further analysis. The main parameters were set as follows: retention time range 2–20 min for RPLC and 2–25 min for HILIC, mass window 0.5 Dalton, and retention window of 1 min. To correct enrichment factors for different samples and the MS shift during long analysis duration, the data of each sample were normalized to the total area. Another advantage of normalizing to the total area is that inaccuracies caused by injection can be eliminated (25). After correction, the data were exported to SIMCA-P v11.5 software (Umetrics AB, Sweden) for multivariate data analysis. Partial least squares-discriminant analysis (PLS-DA) was used for modeling the difference between the BC patients and healthy controls. Potential biomarkers were selected based on the S-plot and Variable Importance in the Project (VIP) column plot. Furthermore, unpaired Student's t-tests, with a Bonferonni correction for multiple comparisons, were employed to ensure the selected metabolites significantly differentially expressed between the BC patients and the controls. p value threshold of 0.001 was used to define the significance. For the identification of potential biomarkers, the following databases were used: HMDB (http://www.hmdb.ca/), METLIN (http://metlin.scripps.edu/), MassBank (http://www.massbank.jp), PubChem (http://ncbi.nim.nih.gov/), and KEGG (http://www.kegg.com/). A summary of the analysis strategy utilized in this study is shown in supplemental Fig. S1.
RESULTS
Metabonomic Profiling of RPLC-MS and HILIC-MS
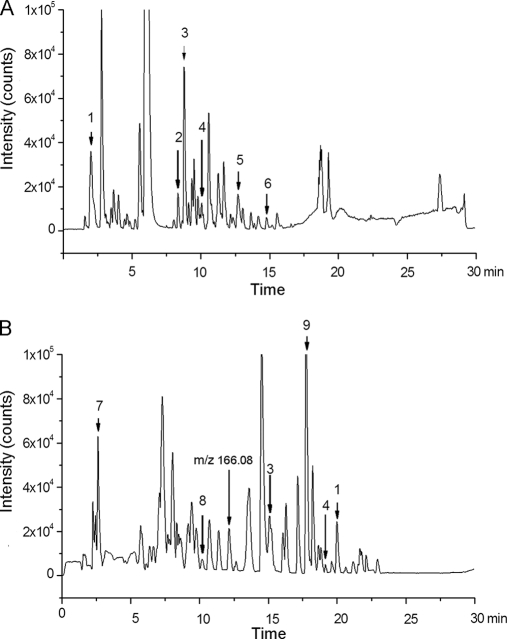
Both the RPLC and HILIC showed satisfactory chromatography, and their typical chromatograms are shown in Fig. 1. For metabonomic analysis, the stability of the analytical method is important to obtain valid data. Three compounds, retained on both columns, and six other compounds, retained only on one column, were selected to verify the stability of the method. The selection of nine representative peaks follows fair principles of covering a range of intensities and retention times across the QC samples. The peaks are shown with arrows in the chromatograms in Fig. 1. The variations of m/z values for all nine compounds were less than 10 mDa. Standard deviations (S.D.) of retention times (tR) were less than 0.7 min for both chromatographic modes. RSDs of peak areas ranged from 4.8% to 16.7% for RPLC and 8.4% to 19.8% for HILIC (Table II). These results demonstrated the acceptable stability and reproducibility of the chromatographic separation and mass measurement. HILIC offers superb retention of the polar analytes that elute quickly from RP column (22). For example, ions of m/z 144.10 (peak No. 1 in Fig. 1) were eluted at 2.1 min for RPLC, but have an tR of 20.3 min for HILIC; ions of m/z 166.08, which cannot be observed in RPLC, show a clear peak from the HILIC column (Fig. 1B). A high proportion of organic solvent present in the mobile phase of HILIC greatly improved the MS response (18, 23). For example, for ions of m/z 180.07, the response in HILIC is nearly twofold larger in area than that of RPLC (supplemental Fig. S2). In total, 703 compounds were detected by HILIC, compared with 417 compounds by RPLC.
Fig. 1.
Typical base peak chromatograms of urine from (A) RPLC and (B) HILIC. The chromatograms from two separation modes cover the detection of both polar and apolar metabolites. Peak numbers refer to the corresponding peaks in Table II.
Table II. Reproducibility evaluation based on selected peaks.
| Peak No.a | RPLC |
HILIC |
||||||
|---|---|---|---|---|---|---|---|---|
| tR | m/z | S.D. tRb | RSDc | tR | m/z | S.D. tRb | RSDc | |
| 1 | 2.1 | 144.10 | 0.19 | 4.8% | 20.3 | 144.10 | 0.21 | 11.4% |
| 2 | 8.4 | 376.14 | 0.08 | 9.8% | \ | \ | \ | |
| 3 | 9.0 | 288.28 | 0.13 | 16.7% | 15.1 | 288.28 | 0.35 | 11.4% |
| 4 | 10.8 | 286.19 | 0.23 | 5.2% | 19.2 | 286.19 | 0.46 | 10.8% |
| 5 | 12.8 | 314.23 | 0.41 | 10.6% | \ | \ | \ | \ |
| 6 | 14.9 | 274.26 | 0.38 | 14.2% | \ | \ | \ | \ |
| 7 | \ | \ | \ | \ | 2.5 | 284.28 | 0.31 | 9.8% |
| 8 | \ | \ | \ | \ | 10.2 | 312.11 | 0.69 | 19.8% |
| 9 | \ | \ | \ | \ | 17.9 | 407.77 | 0.33 | 8.4% |
a Peak No. refers to the corresponding peaks in Fig. 1.
b S.D. tR: standard deviation of retention times in six QC samples.
c RSD: relative stand deviation of peak areas in six QC samples.
Multivariate Statistical Analysis
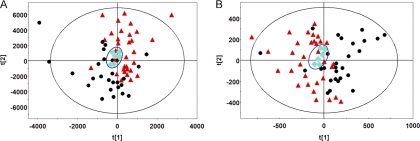
To further determine the overall performance of the method, principal component analysis (PCA) was performed on all samples, including the 6 QC samples. As shown in Figs. 2A and 2B, the QC samples are clustered together in both separation modes, indicating satisfactory stability and reproducibility of the chromatographic separation and mass measurement during the whole sequence. However, the BC group and the healthy controls are not well distinguished in the PCA score plot. This is likely because human urine samples are extremely complex, and the unsupervised PCA data analysis technique separates samples based on random, BC-irrelevant variation of metabolites. PLS-DA, in contrast, is an extension of PCA that makes use of class information to attempt to maximize the separation among classes of observations (10, 26). Thus, PLS-DA was used to identify potential biomarkers related to BC.
Fig. 2.
PCA score plots based on the data from (A) RP separation and (B) HILIC separation (● BC patients, ▴ controls, and ♦ QC). The QC cluster is highlighted with blue diamonds. QC samples are clustered together in both separation modes, indicating the satisfactory stability. The BC group and the healthy controls cannot be well distinguished in PCA score plots.
The PLS-DA results are shown in Figs. 3A and 3B. Both RPLC and HILIC data were mean-centered and Pareto scaled. Distinct clustering between the BC patients and controls was achieved with both columns. In PLS-DA, the R2Y (cum) and Q2 (cum) parameters were used for the evaluation of the models, indicating the fitness and prediction ability, respectively (27). For RPLC, two principal components were calculated with R2Y (cum) and Q2 (cum) of 0.878 and 0.572, respectively, whereas three principal components were extracted from HILIC with R2Y (cum) and Q2 (cum) of 0.979 and 0.676, respectively. These parameters demonstrate that HILIC can generate a more robust model than RPLC, thus indicating a strong contribution of polar molecules in the model set-up and in disease onset. To construct a model containing as much information on metabolites as possible, the datasets from the two separation techniques were combined and analyzed by PLS-DA. As shown in Fig. 3C, the combined dataset depicts a more clear separation between the BC and the controls than those built on the dataset from a single column. The values of R2Y (cum) and Q2 (cum) of the combined model are 0.995 and 0.751, respectively, indicating that a combination of datasets from the two columns provides better classification and prediction. To guard against model overfitting, permutation tests with 100 iterations were performed (Fig. 3D) to compare the goodness of fit of the original model with that of randomly permuted models (28). The criteria for validity included the following: all the permuted R2 and Q2 values to the left were lower than the original points to the right and the regression line of the Q2 points intersects the vertical axis (on the left) at or below zero (29). To evaluate the predictive ability of the combined model, an external test using urine samples from three BC patients (two LG and one HG) and three healthy controls was performed, and satisfactory results were obtained (Fig. 3E). None of those samples had been previously included in the supervised analysis, which therefore allowed for the estimation of true predictive accuracy. The combined PLS-DA model correctly predicted all BC patients and healthy controls with 100% sensitivity and specificity. This result shows the great potential for the combined dataset using PLS-DA analysis as a viable technique for noninvasive BC screening. Therefore, the combined dataset was used in the subsequent study.
Fig. 3.
PLS-DA score plots obtained from (A) RPLC, (B) HILIC, and (C) a combined dataset. D, Validation plot obtained from 100 permutation tests. E, T-predicted scatter plot obtained from the combined PLS-DA model (● BC, ▴ healthy controls, [daif2] BC patient prediction set, and ■ control prediction set). In total, 24 BC patients and 29 controls participated in the PLS-DA modeling process. Urine samples from three BC patients (two LG and one HG) and three healthy controls were used to test the predictive accuracy of the combined model.
Metabonomic Biomarker Determination and Evaluation
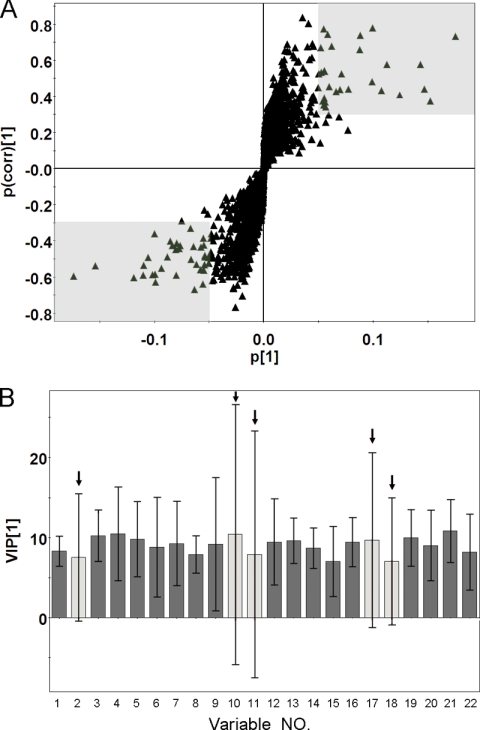
Metabolites were carefully screened before being approved as potential biomarkers. First, significant original variables were extracted from the S-plot, which is a covariance-correlation-based procedure, and thus the risk of false positives in metabolite selection was reduced (30). The S-plot (Fig. 4A), derived from the first component of the combined model, explains most of the variables in data set. The markers with higher p and p(corr) values were the most relevant metabolites for the separation between the two groups. Sixty-six variables (in two shaded areas of Fig. 4A) with absolute p > 0.5 and p(corr) > 0.3 were selected in the S-plot (29). These are potentially interesting biomarker candidates, reflecting discrepant metabolic traits. Second, the VIP values of the biomarker candidates were checked. Variables with VIP scores >1 have above average influence on the explanation of the classification (29). Fifty-nine variables with VIP scores >1 were picked in the second step. Third, variables without the support of the necessary confidence interval were rejected (31). For example, on the VIP column plot of some variables shown in Fig. 4B, variables highlighted with an arrow could not be considered as biomarkers because of their negative confidence intervals (29). Fifteen unqualified variables were eliminated in this step. Unpaired Student's t-tests were performed as the final testing procedure, and variables without significant differences (p value > 0.001) between BC patients and the controls were eliminated. Only 20 variables of the original 66 passed the above criteria. Of these 20 variables, 12 of them corresponded to 6 metabolites retained on both columns, and the other 8 variables were retained on only one column. Therefore, 14 variables were considered meaningful metabolites in discriminating the BC patients from controls.
Fig. 4.
A, S-plot of the combined PLS-DA model. B, VIP column plot with jack-knifed confidence intervals. The variables in two shaded areas are considered significant because of their high p and p(corr) values. The variables highlighted with arrows were eliminated from the potential biomarker list because of their negative confidence intervals.
In the metabonomic analysis workflow, low molecular weight biomarker confirmation and further structure elucidation are considered to be the most challenging steps. A molecular formula was determined according to the exact mass and the isotope pattern. Additional MS/MS experiments were carried out to identify potential biomarkers. Hippuric acid, PAGN, leucylproline, and acetylcarinitine were confirmed by standard compounds, whereas other metabolites were identified based on database information, from the literature, or through structure elucidation. The identifications of carnitine C9:1 and component I are illustrated in supplemental Fig. S3. What we need to emphasize is that the identity of component I as BAHE is tentative because BAHE standard is not available for the confirmation. In fact, component I has not been observed in nature previously. Additional effort is required to confirm the identification. These potential biomarkers are listed in Table III.
Table III. Potential urine biomarkers for BC.
| NO. | m/z | Metabolite | Element composition | Column | Peak area ratio (B/N) | AUCa | VIP valuea | p valuea | Trend | %RSDb (RP & HILIC) |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 318.30 | Component I | C18H39NO3 | RP&HILIC | 2.81 | 0.900 | 3.73 | 2.88E-07 | ↑ | 6.3 and 13.8 |
| 2 | 300.20 | Carnitine C9:1 | C16H29NO4 | RPLC | 0.39 | 0.881 | 10.52 | 3.00E-04 | ↓ | 10.2 |
| 3 | 180.07 | Hippuric acid | C9H9NO3 | RP & HILIC | 0.18 | 0.867 | 11.46 | 6.70E-05 | ↓ | 4.9 and 22.1 |
| 4 | 105.03 | Fragment of hippuric acid | C9H9NO3 | RP & HILIC | 0.27 | 0.852 | 8.24 | 1.48E-06 | ↓ | 4.6 and 28.2 |
| 5 | 326.19 | Unidentified | unidentified | RPLC | 1.17 | 0.834 | 6.11 | 2.67E-06 | ↑ | 5.5 |
| 6 | 138.07 | Trigonelline | C7H7NO2 | HILIC | 0.34 | 0.808 | 3.21 | 4.44E-04 | ↓ | 12.4 |
| 7 | 302.23 | 2, 6-Dimethylheptanoyl carnitine | C16H31NO4 | HILIC | 0.48 | 0.796 | 9.98 | 7.56E-05 | ↓ | 8.5 |
| 8 | 314.23 | Carnitine C10:1 | C17H31NO4 | RPLC | 0.48 | 0.785 | 4.07 | 8.62E-04 | ↓ | 10.7 |
| 9 | 265.12 | PAGN | C13H16N2O4 | RP & HILIC | 0.45 | 0.774 | 16.18 | 9.13E-04 | ↓ | 8.4 and 21.3 |
| 10 | 286.19 | Carnitine C8:1 | C15H27NO4 | RP & HILIC | 0.49 | 0.723 | 4.41 | 9.51E-04 | ↓ | 5.2 and 10.8 |
| 11 | 229.16 | Leucylproline | C11H20N2O3 | RP & HILIC | 0.80 | 0.711 | 8.72 | 5.31E-05 | ↓ | 10.9 and 6.0 |
| 12 | 185.08 | Phosphorylcholine | C5H15NO4P | HILIC | 0.41 | 0.647 | 7.34 | 9.40E-04 | ↓ | 14.1 |
| 13 | 126.01 | Fragment of phosphorylcholine | C5H15NO4P | HILIC | 0.34 | 0.608 | 6.18 | 1.79E-04 | ↓ | 10.1 |
| 14 | 204.14 | Acetylcarinitine | C9H17NO4 | HILIC | 2.16 | 0.598 | 6.27 | 1.06E-05 | ↑ | 14.7 |
a Data given from the column with smaller RSD.
b RSD of peak areas derived from the QCs.
Potential biomarkers were selected after applying PLS-DA algorithms. The next step toward clinical utility is validation of the markers. One popular variable ranking criterion is AUC (area under the curve), which is also called area under the “receiver operating characteristic ” (ROC) curve (32) AUC combines the sensitivity and specificity of a given marker for disease diagnosis (33). Similar utilizations have been reported previously. Adam et al. showed that the power of a single variable in discriminating prostate cancer from normal patients could be determined by AUC, which ranges from 0.5 (no discriminating power) to 1.0 (complete separation) (34). Another study demonstrated that metabolite significance in differentiating weight gain group during early breast cancer chemotherapy could also be revealed by AUC (35).
It is worth noting that VIP values cannot be used to evaluate clinical utility because they mainly reflect the importance of the variables in the PLS model (29). A variable may have a large VIP value, indicating its high weight on the corresponding PLS model, because of large absolute variation between the two groups. Small peaks will be easily ranked behind by the PLS method. Component I, for example, has a VIP value of 3.73, but its AUC reaches 0.900. Another example is PAGN, a common component of urine, which has the largest VIP value (16.2) but a low AUC value in terms of clinical utility.
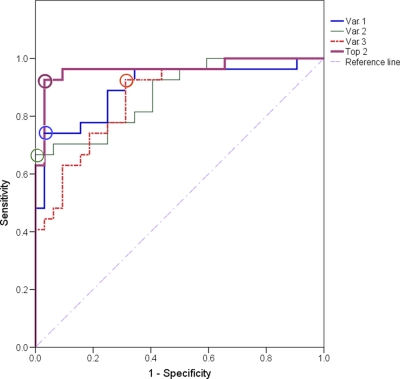
Univariate ROC analysis of potential biomarkers was carried out to identify the important metabolites and examine their utility for the prediction of BC (36). Table III shows the result of ROC analysis, and the variables are ranked according to their AUC. All samples in the training set and predictive set were used in the test. Component I had the highest AUC of 0.900. At the best cut-off point (Fig. 5), 74.1% sensitivity and 96.6% specificity were obtained. The specificity for component I was high but the sensitivity was unsatisfactory. Considering that cancer is a complex disease involving systemic deregulation of cell proliferation, survival, apoptosis, and cell cycle,(37) a “biomarker pattern” containing a group of biomarkers could be more effective for discrimination and more informative for elucidating the pathophysiology of cancer (38, 39). Thus, metabolites (excluding fragments) with AUC > 0.85 were combined for the purpose of building a clinically valuable panel. Permutations and combinations of the top three variables in Table III were fully processed, and the result of ROC analysis for each combination is shown in Table IV. All sensitivities and specificities were calculated at their best cut-off points. An extremely promising result was achieved with the combination of the top two variables (component I and carnitine C9:1). The AUC reached 0.963, and the sensitivity and specificity were improved to 92.6% and 96.9%, respectively. Other combinations did not increase overall sensitivities and specificities as well as this combination. We further used ROC analysis to test the ability of the combination of the top two variables for LG-BC detection by excluding data from six HG-BC patients and obtained a sensitivity and specificity of 90.5% and 96.9%, respectively. These results are shown in Fig. 6 and Table V. The high sensitivity and specificity achieved by the combined use of multiple urinary biomarkers suggests that component I and carnitine C9:1 give an optimal biomarker pattern that could be used in clinical applications for diagnosing BC, including LG-BC detection. Future studies will include much larger urine sample sets to verify these conclusions.
Fig. 5.
Diagnostic efficacy evaluation using ROC curves for potential biomarkers. Var. 1, 2, and 3 correspond to variable No. 1, 2, and 3 in Table III, respectively. Top 2 represents the combination of top two variables in Table III. The open circles denote best cut-off points. The single biomarkers (Var. 1, 2, and 3) cannot achieve both good sensitivity and specificity. The AUC of the biomarker pattern containing component I and carnitine C9:1 (Top 2) reached 0.963, and the sensitivity and specificity were improved to 92.6% and 96.9%, respectively.
Table IV. Results of ROC analysis of variables in Table III.
| Var No.a | AUC | Sensitivity | Specificity |
|---|---|---|---|
| 1 | 0.900 | 74.1% | 96.9% |
| 2 | 0.881 | 66.7% | 100% |
| 3 | 0.867 | 92.6% | 68.8% |
| 1 + 2 | 0.963 | 92.6% | 96.9% |
| 1 + 3 | 0.926 | 88.9% | 87.5% |
| 2 + 3 | 0.951 | 96.3% | 81.3% |
| 1 + 2 + 3 | 0.962 | 96.3% | 90.6% |
a Var No. corresponds to var No. in Table III.
Fig. 6.
Diagnostic efficacy evaluation using ROC curves for potential biomarkers after excluding data from six HG-BC patients. Var. 1, 2, and 3 correspond to variable No. 1, 2, and 3 in Table III, respectively. Top 2 represents the combination of top two variables in Table III. The open circles denote best cut-off points. The biomarker pattern was used for LG-BC detection, which gave good sensitivity (90.5%) and specificity (96.9%).
Table V. Results of ROC analysis of variables in Table III with six samples from HG-BC patients excluded.
| Var No.a | AUC | Sensitivity | Specificity |
|---|---|---|---|
| 1 | 0.875 | 66.7% | 96.9% |
| 2 | 0.871 | 66.7% | 100% |
| 3 | 0.871 | 81.0% | 81.3% |
| 1 + 2 | 0.954 | 90.5% | 96.9% |
| 1 + 3 | 0.911 | 85.7% | 87.5% |
| 2 + 3 | 0.946 | 95.2% | 81.3% |
| 1 + 2 + 3 | 0.953 | 95.2% | 90.6% |
a Var No. corresponds to Var No. in Table III.
DISCUSSION
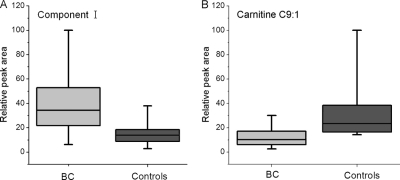
To find a specific potential biomarker pattern in urine that can aid noninvasive detection of BC, we performed LC-MS-based metabolic profiling using both RPLC and HILIC to identify abnormal levels of metabolites in BC patients. As shown in Fig. 7A, the component I levels in the BC group were significantly higher than those of healthy controls. Regrettably, no report was found about component I. The function of component I and the reason why it is elevated in BC urine are unclear. Further structure identification and the biological function of this compound are well worth studying in future work.
Fig. 7.
Variations of urine (A) Component I and (B) Carnitine C9:1. Boxes are drawn from the 25th to 75th percentiles in the intensity distribution. The median, or 50th percentile, is drawn as a black horizontal line inside the box.
The carnitine C9:1 level in the BC group was lower than those of healthy controls, as shown in Fig. 7B. Among the 14 potential biomarkers identified, five of them were carnitine species, including carnitine C10:1, carnitine C8:1, carnitine C9:1, acetylcarnitine, and 2, 6-dimethylheptanoyl carnitine. As a group, carnitines function as important intermediates in fatty acid transport across mitochondrial membranes. We found that urinary acylcarnitine deficiency was prevalent in BC patients (supplemental Fig. S4). Acylcarnitines are generated when corresponding fatty acids are activated into acyl-coenzyme A (acyl-CoA) and then participate in a carnitine palmitoyl transferase I (CPT I)-catalyzed reaction. Afterward, acylcarnitines are catalyzed to carnitine and acyl-CoA by carnitine palmitoyl transferase II (CPT II). In this way, fatty acids penetrate the membrane of mitochondria, become substrates of β-oxidation, and produce several ATP molecules (40). Fatty acids can be oxidized in several ways, but mainly undergo β-oxidation, producing two carbon unit acetyl-coenzyme A (acetyl-CoA). Acetyl-CoA successively participates in energy metabolism and produces acetylcarnitine. In healthy subjects, lipid metabolism is constantly in dynamic equilibrium, which means that some fatty acids are being oxidized in mitochondria to satisfy the energy needs, whereas others are being synthesized and stored in the cytoplasm. Notably, we observed an increased amount of acetylcarnitine concentration in BC patients. The finding of varying levels of carnitine species not only provides a better understanding of the pathophysiological changes of BC but also indicates possible nutritional treatment and chemotherapy for patients. McClinton et al. demonstrated significant differences in the plasma phospholipid profiles of BC patients when compared with the controls (41). They raised one possibility that fatty acid abnormalities might be involved in the pathogenesis of the tumor. We are inclined to take the same point of view, that is, dysregulation of lipid metabolism might provide an environment which is beneficial to development of BC tumor. Disturbed fatty acid transportation, fatty acid β-oxidation, or energy metabolism might suggest one possible reason that patients with BC are prone to feel fatigue.
In summary, with the combined use of two highly sensitive and complementary separation techniques, we were able to construct a combined PLS-DA model with 100% predictive accuracy and further identify metabolic differences between BC patients and controls. Two urinary metabolites, component I and carnitine C9:1, give an effective BC diagnostic indicator biomarker pattern. ROC analysis shows that the biomarker pattern achieves a sensitivity and specificity up to 92.6% and 96.9%, respectively, for all patients and up to 90.5% and 96.9%, respectively, for LG patients. This potential specific biomarker pattern may thus be an effective choice for the diagnosis of BC in addition to conventional screening tools.
Further study toward clinical applications is under consideration as possible extensions of our work. The biomarker pattern containing component I and carnitine C9:1 implementation should undergo a vigorous process of initial disease-specificity confirmation, quantitation assay establishment, and multi-institutional cross-validation. Ideal disease biomarkers or biomarker panels should exhibit high specificity and high sensitivity. Therefore, larger and more diverse patient population, including various cancer patients and other noncancer patients, will be enrolled in the study to further verify the specificity of this newly discovered biomarker pattern.
Footnotes
* This work was supported by the Xiamen University, Department of Science and Technology of Fujian Province (2009D023), and Medical Center Construction Foundation of Xiamen.
 This article contains supplemental Figs. S1 to S4.
This article contains supplemental Figs. S1 to S4.
1 The abbreviations used are:
- BC
- bladder cancer
- CIS
- carcinoma in situ
- RPLC
- reversed phase liquid chromatography
- HILIC
- hydrophilic interaction chromatography
- ESI
- electrospray ionization
- PCA
- principal component analysis
- PLS-DA
- partial least squares-discriminant analysis
- LG
- low grade
- HG
- high grade
- ROC
- receiver operating characteristic
- QC
- quality control
- VIP
- variable importance in the project
- AUC
- area under the curve
- BAHE
- 2-(1-(7-(butyl(ethyl)amino)heptyloxy)propan-2-yloxy)ethanol
- PAGN
- phenylacetylglutamine
- CoA
- coenzyme
- CPT
- carnitine palmitoyl transferase
- and CEACAM1
- carcinoembryonic antigen-related cell adhesion molecule 1.
REFERENCES
- 1. Mitra A. P., Cote R. J. (2010) Molecular screening for bladder cancer: progress and potential. Nat. Rev. Urol. 7, 11–20 [DOI] [PubMed] [Google Scholar]
- 2. Pasikanti K. K., Esuvaranathan K., Ho P. C., Mahendran R., Kamaraj R., Wu Q. H., Chiong E., Chan E. C. Y. (2010) Noninvasive Urinary Metabonomic Diagnosis of Human Bladder Cancer. J. Proteome Res. 9, 2988–2995 [DOI] [PubMed] [Google Scholar]
- 3. Chen Y. T., Chen C. L., Chen H. W., Chung T., Wu C. C., Chen C. D., Hsu C. W., Chen M. C., Tsui K. H., Chang P. L., Chang Y. S., Yu J. S. (2010) Discovery of Novel Bladder Cancer Biomarkers by Comparative Urine Proteomics Using iTRAQ Technology. J. Proteome Res. 9, 5803–5815 [DOI] [PubMed] [Google Scholar]
- 4. Cauberg E. C., de Bruin D. M., Faber D. J., van Leeuwen T. G., de la Rosette J. J., de Reijke T. M. (2009) A New Generation of Optical Diagnostics for Bladder Cancer: Technology, Diagnostic Accuracy, and Future Applications. Eur. Urol. 56, 287–296 [DOI] [PubMed] [Google Scholar]
- 5. Schwamborn K., Krieg R. C., Grosse J., Reulen N., Weiskirchen R., Knuechel R., Jakse G., Henkel C. (2009) Serum Proteomic Profiling in Patients with Bladder Cancer. Eur. Urol. 56, 989–996 [DOI] [PubMed] [Google Scholar]
- 6. Vrooman O. P., Witjes J. A. (2008) Urinary Markers in Bladder Cancer. Eur. Urol. 53, 909–916 [DOI] [PubMed] [Google Scholar]
- 7. Feldman A. S., Banyard J., Wu C. L., McDougal W. S., Zetter B. R. (2009) Cystatin B As a Tissue and Urinary Biomarker of Bladder Cancer Recurrence and Disease Progression. Clin. Cancer Res. 15, 1024–1031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Schiffer E., Vlahou A., Petrolekas A., Stravodimos K., Tauber R., Geschwend J. E., Neuhaus J., Stolzenburg J. U., Conaway M. R., Mischak H., Theodorescu D. (2009) Prediction of Muscle-invasive Bladder Cancer Using Urinary Proteomics. Clin. Cancer Res. 15, 4935–4943 [DOI] [PubMed] [Google Scholar]
- 9. Mitra A. P., Bartsch C. C., Cote R. J. (2009) Strategies for molecular expression profiling in bladder cancer. Cancer Metastasis Rev. 28, 317–326 [DOI] [PubMed] [Google Scholar]
- 10. Nicholson J. K., Lindon J. C. (2008) Systems biology: Metabonomics. Nature 455, 1054–1056 [DOI] [PubMed] [Google Scholar]
- 11. Nicholson J. K., Lindon J. C., Holmes E. (1999) ‘Metabonomics’: understanding the metabolic responses of living systems to pathophysiological stimuli via multivariate statistical analysis of biological NMR spectroscopic data. Xenobiotica 29, 1181–1189 [DOI] [PubMed] [Google Scholar]
- 12. Holmes E., Wilson I. D., Nicholson J. K. (2008) Metabolic Phenotyping in Health and Disease. Cell 134, 714–717 [DOI] [PubMed] [Google Scholar]
- 13. Spratlin J. L., Serkova N. J., Eckhardt S. G. (2009) Clinical Applications of Metabolomics in Oncology: A Review. Clin. Cancer Res. 15, 431–440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kind T., Tolstikov V., Fiehn O., Weiss R. H. (2007) A comprehensive urinary metabolomic approach for identifying kidney cancer. Anal. Biochem. 363, 185–195 [DOI] [PubMed] [Google Scholar]
- 15. Slupsky C. M., Rankin K. N., Fu H., Chang D., Rowe B. H., Charles P. G. P., McGeer A., Low D., Long R., Kunimoto D., Sawyer M. B., Fedorak R. N., Adamko D. J., Saude E. J., Shah S. L., Marrie T. J. (2009) Pneumococcal Pneumonia: Potential for Diagnosis through a Urinary Metabolic Profile. J. Proteome Res. 8, 5550–5558 [DOI] [PubMed] [Google Scholar]
- 16. Claudino W. M., Quattrone A., Biganzoli L., Pestrin M., Bertini I., Di Leo A. (2007) Metabolomics: Available Results, Current Research Projects in Breast Cancer, and Future Applications. J. Clin. Oncol. 25, 2840–2846 [DOI] [PubMed] [Google Scholar]
- 17. Issaq H. J., Nativ O., Waybright T., Luke B., Veenstra T. D., Issaq E. J., Kravstov A., Mullerad M. (2008) Detection of Bladder Cancer in Human Urine by Metabolomic Profiling Using High Performance Liquid Chromatography/Mass Spectrometry. J. Urol. 179, 2422–2426 [DOI] [PubMed] [Google Scholar]
- 18. Cubbon S., Bradbury T., Wilson J., Thomas-Oates J. (2007) Hydrophilic Interaction Chromatography for Mass Spectrometric Metabonomic Studies of Urine. Anal. Chem. 79, 8911–8918 [DOI] [PubMed] [Google Scholar]
- 19. Wang Y., Lehmann R., Lu X., Zhao X., Xu G. (2008) Novel, fully automatic hydrophilic interaction/reversed-phase column-switching high-performance liquid chromatographic system for the complementary analysis of polar and apolar compounds in complex samples. J. Chromatogr. A 1204, 28–34 [DOI] [PubMed] [Google Scholar]
- 20. Qin F., Zhao Y. Y., Sawyer M. B., Li X. F. (2008) Hydrophilic Interaction Liquid Chromatography-Tandem Mass Spectrometry Determination of Estrogen Conjugates in Human Urine. Anal. Chem. 80, 3404–3411 [DOI] [PubMed] [Google Scholar]
- 21. Cubbon S., Antonio C., Wilson J., Thomas-Oates J. (2010) Metabolomic Applications of HILIC-LC-MS. Mass Spectrom. Rev. 29, 671–684 [DOI] [PubMed] [Google Scholar]
- 22. Gika H. G., Theodoridis G. A., Wilson I. D. (2008) Hydrophilic interaction and reversed-phase ultra-performance liquid chromatography TOF-MS for metabonomic analysis of Zucker rat urine. J. Sep. Sci. 31, 1598–1608 [DOI] [PubMed] [Google Scholar]
- 23. Mohamed R., Varesio E., Ivosev G., Burton L., Bonner R., Hopfgartner G. (2009) Comprehensive Analytical Strategy for Biomarker Identification based on Liquid Chromatography Coupled to Mass Spectrometry and New Candidate Confirmation Tools. Anal. Chem. 81, 7677–7694 [DOI] [PubMed] [Google Scholar]
- 24. Epstein J. I., Amin M. B., Reuter V. R., Mostofi F. K., Committee T. B. C. C. (1998) The World Health Organization/International Society of Urological Pathology Consensus Classification of Urothelial (Transitional Cell) Neoplasms of the Urinary Bladder. Am. J. Surg. Pathol. 22, 1435–1448 [DOI] [PubMed] [Google Scholar]
- 25. Chen J., Wang W., Lv S., Yin P., Zhao X., Lu X., Zhang F., Xu G. (2009) Metabonomics study of liver cancer based on ultra performance liquid chromatography coupled to mass spectrometry with HILIC and RPLC separations. Anal. Chim. Acta 650, 3–9 [DOI] [PubMed] [Google Scholar]
- 26. Fukui Y., Kato M., Inoue Y., Matsubara A., Itoh K. (2009) A metabonomic approach identifies human urinary phenylacetylglutamine as a novel marker of interstitial cystitis. J. Chromatogr. 877, 3806–3812 [DOI] [PubMed] [Google Scholar]
- 27. Qiu Y., Cai G., Su M., Chen T., Zheng X., Xu Y., Ni Y., Zhao A., Xu L. X., Cai S., Jia W. (2009) Serum Metabolite Profiling of Human Colorectal Cancer Using GC-OFMS and UPLC-TOFMS. J. Proteome Res. 8, 4844–4850 [DOI] [PubMed] [Google Scholar]
- 28. Wiklund S., Nilsson D., Eriksson L., Sjöström M., Wold S., Faber K. (2007) A randomization test for PLS component selection. J. Chemom. 21, 427–439 [Google Scholar]
- 29. (2005) User's Guide to SIMCA-P, SIMCA-P+ version 11.5 [Google Scholar]
- 30. Wiklund S., Johansson E., Sjöstrom L., Mellerowicz E. J., Edlund U., Shockcor J. P., Gottfries J., Moritz T., Trygg J. (2008) Visualization of GC/TOF-MS-Based Metabolomics Data for Identification of Biochemically Interesting Compounds Using OPLS Class Models. Anal. Chem. 80, 115–122 [DOI] [PubMed] [Google Scholar]
- 31. Yin P., Wan D., Zhao C., Chen J., Zhao X., Wang W., Lu X., Yang S., Gu J., Xu G. (2009) A metabonomic study of hepatitis B-induced liver cirrhosis and hepatocellular carcinoma by using RP-LC and HILIC coupled with mass spectrometry. Mol. Biosyst. 5, 868–876 [DOI] [PubMed] [Google Scholar]
- 32. Hilario M., Kalousis A., Prados J., Binz P. A. (2004) Data mining for mass-spectra based diagnosis and biomarker discovery. Drug Discov. Today 2, 214–222 [Google Scholar]
- 33. Poynard T., Halfon P., Castera L., Munteanu M., Imbert-Bismut F., Ratziu V., Benhamou Y., Bourliere M., de Ledinghen V. (2007) Standardization of ROC Curve Areas for Diagnostic Evaluation of Liver Fibrosis Markers Based on Prevalences of Fibrosis Stages. Clin. Chem. 53, 1615–1622 [DOI] [PubMed] [Google Scholar]
- 34. Adam B. L., Qu Y., Davis J. W., Ward M. D., Clements M. A., Cazares L. H., Semmes O. J., Schellhammer P. F., Yasui Y., Feng Z., Wright G. L., Jr. (2002) Serum Protein Fingerprinting Coupled with a Pattern-matching Algorithm Distinguishes Prostate Cancer from Benign Prostate Hyperplasia and Healthy Men. Cancer Res. 62, 3609–3614 [PubMed] [Google Scholar]
- 35. Keun H. C., Sidhu J., Pchejetski D., Lewis J. S., Marconell H., Patterson M., Bloom S. R., Amber V., Coombes R. C., Stebbing J. (2009) Serum Molecular Signatures of Weight Change during Early Breast Cancer Chemotherapy. Clin. Cancer Res. 15, 6716–6723 [DOI] [PubMed] [Google Scholar]
- 36. Odunsi K., Wollman R. M., Ambrosone C. B., Hutson A., McCann S. E., Tammela J., Geisler J. P., Miller G., Sellers T., Cliby W., Qian F., Keitz B., Intengan M., Lele S., Alderfer J. L. (2005) Detection of epithelial ovarian cancer using H-1-NMR-based metabonomics. Int. J. Cancer 113, 782–788 [DOI] [PubMed] [Google Scholar]
- 37. Evan G. I., Vousden K. H. (2001) Proliferation, cell cycle and apoptosis in cancer. Nature 411, 342–348 [DOI] [PubMed] [Google Scholar]
- 38. Ludwig J. A., Weinstein J. N. (2005) Biomarkers in Cancer Staging, Prognosis and Treatment Selection. Nat. Rev. Cancer 5, 845–856 [DOI] [PubMed] [Google Scholar]
- 39. Theodorescu D., Wittke S., Ross M. M., Walden M., Conaway M., Just I., Mischak H., Frierson H. F. (2006) Discovery and validation of new protein biomarkers for urothelial cancer: a prospective analysis. Lancet Oncol. 7, 230–240 [DOI] [PubMed] [Google Scholar]
- 40. Cruciani R. A., Dvorkin E., Homel P., Culliney B., Malamud S., Lapin J., Portenoy R. K., Esteban-Cruciani N. (2009) L-Carnitine Supplementation in Patients with Advanced Cancer and Carnitine Deficiency: A Double-Blind, Placebo-Controlled Study. J. Pain Symptom Manage. 37, 622–631 [DOI] [PubMed] [Google Scholar]
- 41. McClinton S., Moffat L. E., Horrobin D. F., Manku M. S. (1991) Abnormalities of essential fatty acid distribution in the plasma phospholipids of patients with bladder cancer. Br. J. Cancer 63, 314–316 [DOI] [PMC free article] [PubMed] [Google Scholar]