Abstract
Overexpression represents a principal bottleneck in structural and functional studies of integral membrane proteins (IMPs). Although E. coli remains the leading organism for convenient and economical protein overexpression, many IMPs exhibit toxicity on induction in this host and give low yields of properly folded protein. Different mechanisms related to membrane biogenesis and IMP folding have been proposed to contribute to these problems, but there is limited understanding of the physical and physiological constraints on IMP overexpression and folding in vivo. Therefore, we used a variety of genetic, genomic, and microscopy techniques to characterize the physiological responses of Escherichia coli MG1655 cells to overexpression of a set of soluble proteins and IMPs, including constructs exhibiting different levels of toxicity and producing different levels of properly folded versus misfolded product on induction. Genetic marker studies coupled with transcriptomic results indicate only minor perturbations in many of the physiological systems implicated in previous studies of IMP biogenesis. Overexpression of either IMPs or soluble proteins tends to block execution of the standard stationary-phase transcriptional program, although these effects are consistently stronger for the IMPs included in our study. However, these perturbations are not an impediment to successful protein overexpression. We present evidence that, at least for the target proteins included in our study, there is no inherent obstacle to IMP overexpression in E. coli at moderate levels suitable for structural studies and that the biochemical and conformational properties of the proteins themselves are the major obstacles to success. Toxicity associated with target protein activity produces selective pressure leading to preferential growth of cells harboring expression-reducing and inactivating mutations, which can produce chemical heterogeneity in the target protein population, potentially contributing to the difficulties encountered in IMP crystallization.
Structural studies of integral membrane proteins (IMPs)1 are impeded by many factors, including difficulties associated with their overexpression in simple model organisms like Escherichia coli (1–4). Toxicity during overexpression often reduces cell growth-rate after induction, contributing to low yield of the target IMP. Studies using many different approaches have investigated the physiology of IMP expression (5–8) and overexpression (2, 9, 10) in E. coli. IMP insertion into the cytoplasmic membrane is believed to be coordinated by carefully regulated interactions between translating ribosomes, the bacterial signal recognition particle (SRP) system (i.e. the Ffh and FtsY proteins and 4.5S RNA), and the SecYEG translocon (5–8, 11, 12). However, at least for some IMPs, the SRP system and SecYEG are not essential for proper insertion and folding because they can assemble efficiently into pure lipid membranes after cell-free in vitro translation by E. coli ribosomes (13). Nonetheless, the toxicity frequently observed on IMP overexpression has been attributed to difficulties in accommodating additional IMPs in cellular membranes because of limitations in the capacity of both the enzymes mediating phospholipid biosynthesis and the apparatus mediating IMP insertion (14). Destabilization of membranes because of these limitations has been inferred to cause stress impairing the function of membrane-bound enzymes, especially those involved in aerobic respiration (2).
Methods for obtaining a high yield of a native IMP remain primarily empirical and focus on variations in the sequence of the target protein and changes in the expression host. Whole-genome sequence data have been exploited to identify a wide variety of homologous target proteins for evaluation of their expression and stability properties. Variations in affinity-tag and leader-peptide sequences and fusion to expression-enhancing or solubility-enhancing protein domains have yielded improved results for some specific proteins. Published papers have reviewed these approaches, as well as approaches involving variations in growth medium and the use of different E. coli strains or alternative organisms as expression hosts (1, 4, 9, 10, 15, 16).
The E. coli strains C41λ(DE3) and C43λ(DE3) (17) have been demonstrated to increase the yield of some IMPs as well as some soluble proteins. These strains were selected from the standard BL21λ(DE3) expression host based on their enhanced resistance to the toxicity caused by high-level expression of a specific IMP, the b subunit of the E. coli F1Fo ATPase. Induction of b-subunit expression in these strains causes proliferation of the cytoplasmic membrane (18), which produces spiral membrane invaginations into the cytoplasm that are visible using thin-section electron microscopy (EM). This phenomenon was also reported after overexpression of the fumarate reductase IMP complex (19) or glycerol-3-phosphate acyltransferase (20) in standard E. coli strains, but it has yet to be documented for any IMP other than the b-subunit in C41(DE3) or C43(DE3). Moreover, recent genetic analyses have demonstrated that the enhanced yield of IMPs in these strains is attributable primarily to a promoter mutation reducing transcription of T7 RNA polymerase, which lowers its expression and that of target proteins expressed from T7-polymerase-controlled pET vectors. Such vectors were used in the selection procedure that yielded these strains and the subsequently published physiological analyses. Therefore, their main benefit appears to be attenuated expression of the target protein, which can produce higher net yield when expression of that protein is toxic and inhibits cell growth. Other E. coli strains have been selected to improve expression of specific target proteins (21–23), but their efficacy in improving expression of diverse IMPs has not yet been demonstrated.
Several papers have characterized the influence of IMP overexpression on the expression of specific cellular proteins or vice-versa (24, 25). Other authors have taken a global approach to characterizing the response of E. coli to overexpression of soluble proteins (26, 27) or IMPs (2). Gill et al. (26) reported that cells overexpressing soluble proteins from different phylogenetic sources can activate many stress regulons, but noted that the effects of overexpression on cellular growth rate were protein-specific. A more recent analysis employed a proteomics approach to evaluate the response to overexpression of an IMP with a sizable periplasmic domain (2). These authors propose that the Sec translocon becomes saturated during IMP overexpression, causing accumulation of cytoplasmic aggregates and broad perturbations in the proteome. Some of these perturbations are consistent with inhibition of energy metabolism and cell growth rate due to inefficient oxidative respiration and ATP synthesis in the cytoplasmic membrane. However, these physiological inferences were not evaluated using other methods.
Although these published analyses have provided highly valuable data, many issues remain unresolved concerning the physiology of IMP overexpression in E. coli. Practical experience indicates that a substantial number of membrane proteins can be produced in E. coli at suitable levels for structural studies (1–3 mg per liter of culture) without causing toxicity on induction, whereas others are highly toxic even when expressed at undetectable levels. Therefore, uncertainty remains concerning the generality of the phenomena reported in previous literature on IMP overexpression. Therefore, we undertook a systematic analysis of E. coli cells during attempted overexpression of eight target proteins with different expression, toxicity, and folding properties. Two of these were water-soluble cytoplasmic proteins, whereas six were representative polytopic IMPs lacking periplasmic domains. Like most bacterial IMPs without periplasmic domains, these proteins do not have cleavable signal peptides (supplemental Fig. S1). After target-protein induction, we monitored growth rate, morphology, protein expression level, activity of key transcriptional regulators, and global transcriptional profile. Although we identify a large set of physiological changes that occur during IMP overexpression, many of them are shared by cells overexpressing soluble proteins and reflect a likely blockage of the stationary-phase transcriptional program because of some previously unappreciated form of translational stress. We also present evidence that the toxicity caused by overexpression of several IMPs is associated with the biochemical and biophysical properties of these specific proteins. Although definitive conclusions are not possible based on characterization of a relatively restricted set of IMPs, the totality of our results suggests that there may be no intrinsic barrier to moderate overexpression of IMPs in E. coli and that most problems may be attributable to the target protein itself.
EXPERIMENTAL PROCEDURES
Plasmids
Target proteins were cloned under the control of the IPTG-inducible T5 promoter in pQE-30, pQE-60, or pQE-70 plasmids (Qiagen Inc., Valencia, CA), which were transformed into strains also containing the LacI-expressing pREP4 accessory plasmid. Unless indicated otherwise, constructs retain the native N termini of the target proteins. EcMsbA*, also called EcMsbA-ΔN5, carries an in-frame deletion of residues 2–5 (HNDK). EcYojI** has a nonsense mutation that truncates the protein after residue 492, deleting half of the central β-sheet in its C-terminal nucleotide-binding domain. HP1206* has six missense mutations that arose during cloning: N65S, M342I, S348T, E363K, D383N, and K432E. EcGlpT has a single Ser inserted after the initiator Met in the native sequence. EcEnolase* contains the catalytically inactivating K341A mutation (28). NBD-EcMsbA contains the 245 C-terminal residues comprising the cytoplasmic nucleotide-binding domain of EcMsbA. Wild-type EcMsbA was cloned into the pBAD-Myc-His-A vector, and PCR mutagenesis was used to generate the ΔN5 and Nt-His6 variants. The primers used for cloning, sequencing, and mutagenesis are listed in supplemental Table S1.
E. coli Strains
Except as noted below, strains were obtained from the Yale E. coli Genetic Stock Center. Expression experiments were performed in strain MG1655 (F-, λ−, rph-1) transformed with a pQE-derived protein-expression plasmid and pREP4. For reporter-gene assays, these plasmids were transformed into strain SEA001 (MG1655 ΔlacX74, Φλ[rpoHp3::lacZ]) (29) for σE, strain SEA3122 (MG1655 ΔlacX74, λRS88[cpxP-lacZ]) for CpxR, or strain SEA3084 (MG1655 ΔlacX74, Φλ[htpG P1::lacZ]) for σH. Strain glpR-1 (MG1655 glpR-ΔC150) carries a mutation causing the repressor of the glycerol-degradation regulon to be truncated after residue 44. Strain FB20526 (MG1655 fliA::Tn5KAN-2) was obtained from the University of Wisconsin E. coli Genome Project and shown by DNA sequencing to harbor a cryptic glpR-ΔC150 mutation. Strain W3110A (F-, λ−, IN(rrnD-rrnE)1, rph-1) and its derivative WDS2 carrying a temperature-sensitive msbA mutation (30) were obtained from William T. Doerrler.
Cell Growth and Protein Expression and Fractionation Methods
Cells grown aerobically in Luria broth (LB) at 37 °C were induced at OD600 0.6–0.8 with 1 mM isopropyl β-d-thiogalactoside (IPTG) for 3 h. Cells with pBAD vectors were grown in 0.5% (w/v) glucose before dilution into inducing medium. Small-scale expression experiments employed 8 ml of culture. Membrane solubilization was evaluated using 2% (w/v) βDDM, LDAO, or FC12. See Supplemental Experimental Procedures for details.
β-Galactosidase Assays
Reporter-gene activation was monitored using standard methods (29, 31) to assay the activity of a β-galactosidase fusion carried on a lysogenic λ bacteriophage integrated at a single site on the E. coli chromosome. The σE and CpxR assays employed 0.5 ml samples, while the σH (σ32) assays employed 0.2 ml samples to avoid saturation of OD420 readings.
Microscopy Methods
Cells were fixed with 3.7% formaldehyde, mounted on polylysine-coated coverslips, and visualized using a Photometrix CoolSNAP camera after staining with the fluorescent dyes Mitotracker Green or FM4–64 (Invitrogen, Carlsbad, CA). Cells were thin-sectioned as described (18), stained with uranyl acetate and lead citrate, and imaged using a Philips CM120 transmission electron microscope (FEI, Eindhoven, The Netherlands). Flagella were visualized using a Jeol 100 CX transmission electron microscope (Jeol Ltd., Tokyo, Japan) after staining with 2% (w/v) uranyl acetate. See Supplemental Experimental Procedures for details.
RNA Extraction and Microarray Analyses
RNA extracted with the RNeasy Mini Kit (Qiagen, Valencia, CA) 3 h after induction of protein expression was used to synthesize biotinylated cDNA, which was hybridized on Affymetrix E. coli 2.0 arrays by the Gene Expression Center at the University of Wisconsin Biotechnology Center. Raw data (.cel) files were analyzed using the RMA (Robust Multi-chip Average) algorithm in the Affymetrix Expression Console. The transcription-factor-finder software at www.prfect.org used a Fisher's Exact Test with a linear threefold threshold. See Supplemental Experimental Procedures for details.
RESULTS
Overview of Experimental Design
To investigate the effects of IMP overexpression on E. coli physiology, we induced the expression of six inner membrane proteins and two soluble proteins in strain MG1655 (Table I). We selected polytopic IMPs without large periplasmic domains to focus on factors involved in membrane biogenesis and insertion of transmembrane (TM) α-helices. This experimental design isolated factors involved in membrane biogenesis from those involved in secretion of periplasmic protein domains. Typical of bacterial IMPs lacking periplasmic domains, our target proteins do not possess cleavable N-terminal signal peptides based on either predictive algorithms (supplemental Fig. S1) or experimental observations (32, 33). We chose two structurally unrelated E. coli IMPs with known crystal structures, GlpT (EcGlpT) and MsbA (EcMsbA*), the Salmonella typhimurium ortholog of MsbA (StMsbA), plus two functionally uncharacterized homologs of MsbA, E. coli YojI (EcYojI), and Helicobacter pylori HP1206*.
Table I. Summary of protein constructs and expression results.
| Proteina | Species | Length | Locationb | His-tagc | mRNA Inductiond | Protein Expression | Solubility/Ni-NTA yielde |
Toxicity | ||
|---|---|---|---|---|---|---|---|---|---|---|
| βDDM | LDAO | FC12 | ||||||||
| EcMsbA* | E. coli | 582 | IM | C | 12×, 12× | ++ | M/M | S/M | A/M | Medium |
| StMsbA | S. typhimurium | 582 | IM | C | n.d. | ++ | Medium | |||
| EcYojI | E. coli | 547 | IM | C | 32× | +++ | S/S | S/S | M/S | Non-toxic |
| EcYojI** | E. coli | 493 | IM | - | 31×, 37× | +++ | S/n.d. | S/n.d. | M/n.d. | Non-toxic |
| HP1206* | H. pylori | 578 | IM | C | n.d. | - | Toxic | |||
| EcGlpT | E. coli | 452 | IM | C | 31× | + | Af/M | Mf/M | Mf/M | Toxic |
| NBD-EcMsbA | E. coli | 245 | Cytosol | C | n.d. | ++ | H/n.d. | Non-toxic | ||
| EcEnolase* | E. coli | 432 | Cytosol | N | 4×, 4× | +++ | M/n.d. | Non-toxic | ||
a The asterisks indicate proteins harboring mutations (a 5-residue N-terminal truncation for EcMsbA*, a 55-residue C-terminal truncation for EcYojI**, seven missense mutations for HP1206*, and a single missense mutation blocking catalytic activity for EcEnolase*.
b IM stands for inner membrane.
c All proteins except EcYojI** were engineered to have hexa-histidine tags at either their amino (N) or carboxy (C) termini; tag sequences are not included in the length indicated in the third column.
d Fold-change results from microarray analyses are reported for all replicates for each target protein. Transcripts for NBD-EcMsbA and for proteins from organisms other than E. coli were not detected (n.d.), as explained in the Results section.
e Yields were estimated as all (A), most (M), roughly half (H), or slight (S) based on visual inspection of Coomasie-stained SDS-PAGE gels (supplemental Fig. S1). Extraction was performed without detergent for the soluble proteins or in the presence of the indicated detergents for the membrane proteins. Detergent extracts underwent to microscale batch Ni-NTA purification, except for YojI** because it does not have a his-tag.
f These estimate are less accurate than the others because of low expression and overlap with other proteins on the SDS-PAGE gel.
EcGlpT transports glycerol 3-phosphate across the inner membrane; it contains 12 TM α-helices (supplemental Fig. S1) and has short extramembranous loops except for a 54-residue cytoplasmic domain that is disordered in its crystal structure (32). EcMsbA*, StMsbA, EcYojI, and HP1206* belong to the ATP-binding cassette (ABC) transporter superfamily, which uses stereotyped cytoplasmic ATPase domains to drive a wide variety of cellular processes, including TM transport of diverse inorganic, organic, and polymeric substrates. The homodimeric ABC transporters included in our study have six TM α-helices in each protomer (supplemental Fig. S1), and they contain exclusively short extrahelical loops except for their C-terminal cytoplasmic ABC domains (∼250 residues in length). EcMsbA, essential for the viability of E. coli, is believed to translocate Lipid A molecules across the inner membrane (33). We found that full-length EcMsbA with a C-terminal hexahistidine (his) tag could not be maintained stably in IPTG-inducible expression plasmids (i.e. either pET vectors or pQE vectors), although it could be maintained stably in the presence of the catabolite-repressor glucose in the more tightly controlled arabinose-inducible pBAD plasmid. The most common variant recovered after cloning this construct into pET or pQE vectors harbors a deletion of residues 2–5. We call this construct EcMsbA* or alternatively EcMsbA-ΔN5. The functional consequences of this deletion are explored in detail below (see Fig. 7B and supplemental Fig. S10). In contrast, StMsbA (96% identical to EcMsbA) could be maintained stably in IPTG-inducible plasmids with a fully wild-type sequence, which was an important reason for including this protein in our overexpression studies. Full-length EcYojI seems to accumulate in a partially misfolded form on overexpression even though it is an endogenous E. coli protein (see below). We also analyzed overexpression of a mutant variant of this protein (EcYojI**) that is incapable of forming the native cytoplasmic ABC domain structure because of truncation of its 55 C-terminal amino acids. This protein segment contains approximately half of the mostly parallel β-sheet that forms the core of the ABC domain. This mutant protein was included as an example of an improperly folding IMP that accumulates in cells after induction. The final ABC transporter, HP1206*, was included as an example of an IMP that is rapidly degraded on induction. This protein is presumably destabilized by six missense mutations that arose during cloning of the gene.
Fig. 7.
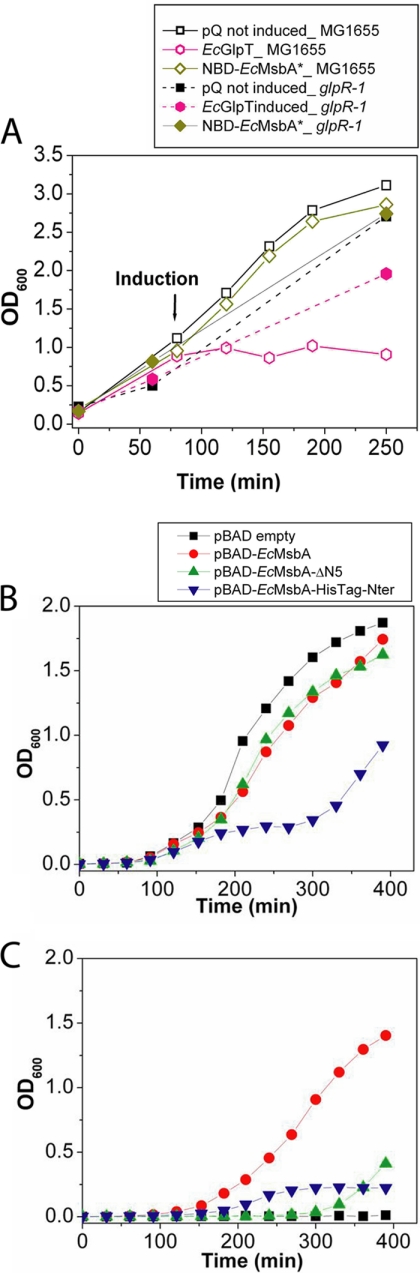
Toxicity is controlled by the biochemical and biophysical properties of the target IMP. A, OD600 was used to monitor cell growth during induction of expression of EcGlpT or NBD-EcMsbA in E. coli MG1655 cells without (empty symbols) or with (filled symbols) the glpR-1 mutation that produces constitutive expression of the enzymes mediating glycerol degradation. These experiments employed the same pQE-derived expression vectors used in Fig. 1 and elsewhere in this paper. B, Cell growth was monitored during induction of several different EcMsbA constructs from arabinose-controlled pBAD plasmids in E. coli W3110A cells: pBAD-EcMsbA expresses the full-length protein (red); pBAD-EcMsbA-ΔN5 expresses the ΔN5 construct which is missing residues 2–5 and equivalent to the EcMsbA* construct in the pQE60 vector (green); and EcMsbA-HisTag-Nter expresses the full-length protein with an N-terminal hexahistidine tag (blue). Induction was carried out at 42 °C in presence of 0.02% arabinose. C, Growth of E. coli strain WD2S containing the indicated pBAD expression plasmid at 42 °C, the nonpermissive temperature for the temperature-sensitive mutation in the chromosomally encoded msbA gene in this derivative of strain W3110A. For the experiments in panels B and C, the growth medium contained 0.02% arabinose to induce MsbA construct expression starting at zero time.
To distinguish specific cellular responses to membrane protein overexpression from general consequences of protein overexpression, we also included two soluble proteins. One of these was the cytoplasmic ABC domain or nucleotide-binding domain of EcMsbA (NBD-EcMsbA), which is expressed in approximately equal amounts in a soluble form and in cytoplasmic inclusion bodies (see below). The other was a catalytically inactive variant of the E. coli enolase enzyme (EcEnolase*), which is one of the most abundant proteins in the cytoplasm under normal growth conditions. A previously characterized active-site mutation (K341A) was introduced into this protein to prevent perturbations in intermediary metabolism.
All protein constructs were cloned under IPTG-inducible promoter control into small ampicillin-resistant plasmids (∼3.4 kB without insert) bearing the ColE1 origin of replication (pQE series vectors from Qiagen). These vectors were chosen because they employ a bacteriophage T5 promoter to drive strong mRNA expression using the endogenous E. coli RNA polymerase and thereby avoid potential physiological complications arising from the use of an exogeneous RNA polymerase (e.g. as employed in pET-series expression vectors). All expressed proteins included a C-terminal his-tag, except for EcEnolase* which included an N-terminal his-tag and YojI** which did not include any tag (because of its C-terminal truncation).
E. coli strain MG1655 was chosen as the overexpresion host because it is widely used for genetic studies and has a completely sequenced genome. All overexpression strains also contained the pREP4 plasmid that expresses the LacI repressor protein to tighten control of the IPTG-inducible promoter controlling target-gene transcription. The resulting overexpression strains were stable in glycerol stocks and gave toxicity and protein-expression results comparable to those observed using the same plasmids in the commonly used BL21λ(DE3)-pLysS and C43 λ(DE3) host strains (data not shown). Overexpression experiments employed a 3-hour IPTG-induction period in LB at 37 °C (Fig. 1), a standard condition used for IMP overexpression. Overexpressing cells were compared with both noninduced and IPTG-induced empty-vector control cells (i.e. containing pQE-60 without an insert).
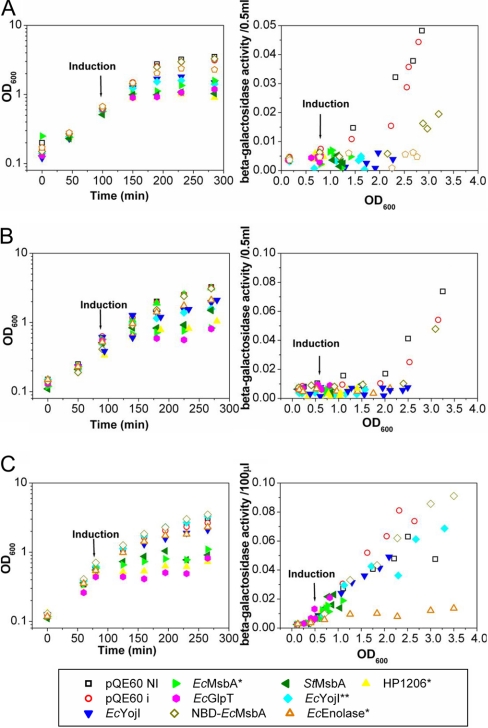
Fig. 1.
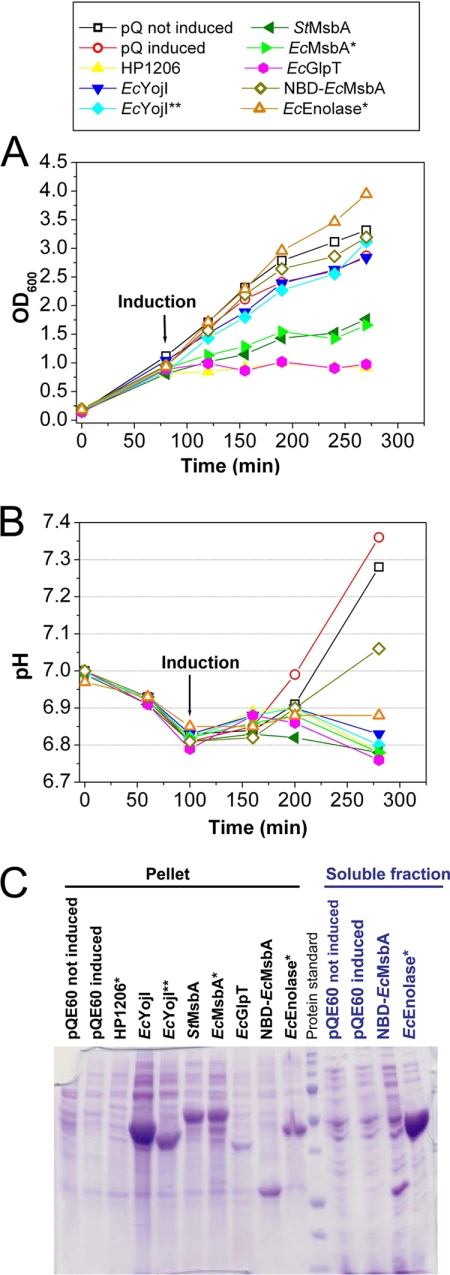
Influence of protein overexpression on cell growth and culture pH. A, Growth curves in LB medium at 37 °C. E. coli MG1655 cells harboring the indicated pQE-derived expression vector and the pREP4 LacI-expressing plasmid were induced with 1 mm IPTG at an OD600 of 0.4–0.6, and growth was continued for 3 h. Measurements were conducted on culture aliquots diluted with LB to OD600 < 1.0. Two control cultures harboring the empty pQE-60 vector were evaluated, one induced with IPTG and the other not induced. B, Culture pH was measured using a standard electrode at the indicated times. C, Coomassie-Blue-stained 15% SDS-PAGE gel of cellular fractions at the end of the induction period. After lysis by sonication with a microtip probe, the extract was centrifuged for 1 h at 14,000 rpm to separate pellet and soluble fractions.
Characterization of Cell Growth
OD600 was used to monitor growth before and after protein induction (Fig 1A). Growth was significantly inhibited by induction of four of the six IMPs (all but the two EcYojI variants) but neither soluble protein. Induction of the MsbA orthologs inhibited growth only slightly, whereas induction of EcGlpT or HP1206* inhibited growth strongly. Induction of some of the toxic proteins, especially EcGlpT, gave variable growth profiles with a tendency to switch to a wild-type growth rate after a lag period (supplemental Fig. S2), presumably because of genetic selection of toxicity-reducing mutations. Growth profiles were monitored explicitly during all physiology experiments reported in this paper, and cells were discarded without analysis if a transition to a higher growth-rate was observed. The very strong toxicity of HP1206* likely accounts for the low transformation efficiency of the corresponding expression plasmid (data not shown).
We also monitored the pH of the cultures, which changed in a reproducible manner during the course of E. coli growth in LB (Fig. 1B). Although variations in pH reflect in part the level of acetate secretion and oxidative respiration in the culture, the cause of these changes and the factors controlling them are more complex and not fully characterized in rich growth media like LB. Slight acidification occurred in all cultures during the early logarithmic-phase growth, and pH was then stable for about 100 min. Although the pH of the control cultures rose to ∼7.3 during the transition into stationary phase (i.e. toward the end of the growth period in Fig. 1A, 1B), this alkalinization was suppressed in the cells overexpressing IMPs. However, whereas this effect was stronger for the IMPs, the cells overexpressing soluble proteins gave intermediate values of pH (6.9 for EcEnolase* and 7.1 for NBD-EcMsbA). Therefore, the lack of alkalinization of the culture medium is at least in part a general consequence of protein overexpression and not a unique feature of IMP overexpression. Based on this observation combined with several analyses presented below, we infer that the maintenance of a more acidic pH in the protein-overexpressing cells is likely attributable to blockage of the stationary-phase transcriptional program and not to inhibition of oxidative respiration.
Comparison of mRNA and Protein Expression Levels
For most of the E. coli target proteins, the levels of their mRNAs at the end of the induction period were characterized in the microarray profiling experiments reported below (as summarized in Table I). These measurements indicate that the EcYojI, EcYojI**, and EcGlpT transcripts were expressed at ∼30-fold higher levels compared with control cells, whereas EcMsbA* was expressed at a 12-fold higher level, and EcEnolase* at a fourfold higher level. The relatively low increase in expression of the mRNA for EcEnolase* was verified not to be influenced by the missense mutation in the expression construct because the homologous probes on the microarray chip do not include the mutated region. The modest fold-change is likely to be attributable either to its high endogenous expression level or perhaps to partial saturation of the chip because the EcEnolase signal was among the 13 strongest measured for all strains. The microarray did not yield data for NBD-EcMsbA because five out of nine probes for msbA matched gene sequences missing in this construct. Real-time PCR measurements were employed to characterize the expression level of the mRNA for HP1206*, which shows minimal protein expression. These measurements indicated that the level of its transcript was higher than that for EcYojI in cells overexpressing that protein (data not shown).
Protein expression levels were evaluated via Coomasie-stained SDS-PAGE gels (Fig. 1C and Table I) and Western blots (supplemental Fig. S3). After a low-speed spin to clear unlysed cells, the supernatant and pellet from a high-speed spin were analyzed by SDS-PAGE. Very high expression was observed for EcEnolase*, the two EcYojI variants, and NBD-MsbA, whereas moderate expression was observed for the two MsbA orthologs, low expression was observed for EcGlpT, and minimal expression was observed for HP1206*. This last protein was not detectable via Coomasie staining but could be detected as a series of faint bands in the pellet fraction in a Western blot with an antibody against the C-terminal his-tag (supplemental Fig. S3). Overexpressed soluble proteins were detected in both supernatant (soluble) and pellet (membrane) fractions, whereas overexpressed IMPs were detected exclusively in the pellet fraction. EcEnolase* was recovered primarily in the supernatant, indicating that most of this protein is soluble, whereas NBD-EcMsbA was distributed approximately equally between the supernatant and pellet. Morphological studies (see below) show that this protein forms inclusion bodies (IBs), which likely accounts for its high level in the pellet. Extraction and batch Ni-NTA purification assays in three nonionic detergents (supplemental Fig. S4 and Table I) demonstrate that most of the expressed EcGlpT and MsbA proteins can be extracted in mild detergents and recovered during Ni-NTA chromatography. The expressed EcYojI variants can be extracted only in the harsh detergent fos-choline 12 (FC12) and are mostly lost during Ni-NTA chromatography even in this detergent. These observations suggest that both the full-length and truncated EcYojI variants are mostly misfolded, despite their high expression, whereas EcGlpT and the MsbA orthologs are mostly properly folded and membrane-integrated.
Coomassie-Blue-stained SDS-PAGE gels show that the overexpressed target proteins other than EcGlpT and HP1206* are among the most abundant visualized in the respective ITPG-induced cells (Fig. 1C). EcEnolase* and full-length EcYojI levels are so high that they appear to represent roughly half or perhaps even more of all proteins visualized in SDS-PAGE analyses of whole-cell extracts (data not shown). The total amount of Coomassie Blue staining observed in SDS-PAGE analyses of such extracts is substantially greater than that in extracts prepared from equivalent volumes of control cultures, suggesting that cells overexpressing EcEnolase* or full-length EcYojI may have a higher total protein content. After observing qualitatively different staining of the cytoplasm in negatively stained thin-section EM images of these cells (see below), which would be consistent with a significant difference in their chemical content, we attempted to quantify total cell protein content using the Coomassie-Blue-based Bradford assay. These assays gave results correlated with the OD600 of the cultures but not with the relative amount of Coomassie-Blue staining observed when a given volume of culture was analyzed by SDS-PAGE. Therefore, it remains unclear whether the cells overexpressing EcEnolase* or full-length EcYojI have a higher total protein content. Further research will be needed to explain the apparent discrepancy among the different Coomassie-Blue-based assays for cellular protein content. Possible explanations include interference with the Bradford assay by other cellular constituents or domination of whole-cell protein content by low molecular-weight polypeptides too small to be visualized in SDS-PAGE.
Visible and Electron Microscopy of Induced Cells
E. coli cell morphology during protein overexpression was evaluated using several methods. Visible microscopy was performed using differential interference contrast (DIC) optics (Figs. 2A and supplemental Fig. S5) as well as fluorescent staining with FM4–64 (Fig. 2A), a dye that labels the outer leaflet of the outer membrane (34), or Mitotracker Green (supplemental Fig. S6), a membrane-permeant lipophilic dye that labels both external and internal membranes (35). Whole cells (supplemental Fig. S7) or thin-sections of cells (Fig. 3) were also imaged using negative-stain EM (Fig. 3).
Fig. 2.
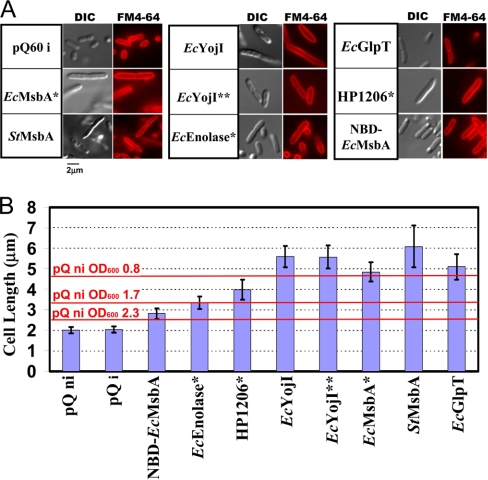
Cellular morphology analyzed by visible microscopy. A, At the end of the 3-hour induction period, cells were fixed with formaldehyde and treated with the fluorescent dye FM4–64, which stains the outer membrane. Differential interference contrast (DIC) and FM4–64 fluorescence images are shown for the same field-of-view containing both dividing and nondividing cells. B, Mean cell length in each culture, with error bars representing the standard deviation. The double asterisks (**) indicate a probability of less than 0.0001 that the distribution is the same as that in the control cells, according to the Student t test (n > 50). Red lines show the mean length of uninduced empty-vector control cells at the indicated OD600 (supplemental Fig. S5); induced cells gave equivalent results (data not shown).
Fig. 3.
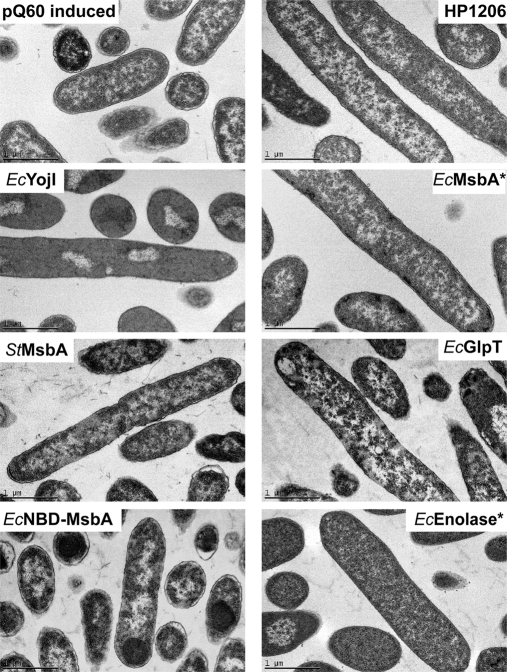
Cellular morphology analyzed by thin-section electron microscopy. At the end of the 3-h induction period, cells were fixed, embedded, sectioned, stained with uranyl acetate, and imaged at 31,000 × magnification using a Philips CM12 microscope. The scale bar represents 1 μm. The cells exhibiting the highest cytosolic electron density contain the greatest amount of the overexpressed target proteins (EcEnolase* and EcYojI) based on Coomassie-Blue-stained SDS-PAGE analysis (Fig. 1C). Inner-membrane invaginations were not observed in any sample.
All methods yielded consistent results, showing that, three hours after IPTG induction, the protein-overexpressing cells are substantially elongated compared with controls (Fig. 2B). Control cells showed a mean length of 2 μm, typical of E. coli in early stationary phase. An increase of about 50% in mean length was observed in cells overexpressing EcEnolase* or NBD-EcMsbA. Cytoplasmic inclusion bodies were visible in DIC (Figs. 2A and supplemental Fig. S6), Mitotracker Green (supplemental Fig. S6), and thin-section EM (Fig. 3) images of most cells overexpressing NBD-EcMsbA and some overexpressing EcEnolase*. Cells overexpressing IMPs were generally even longer, with mean lengths from 4–6 μm (Fig. 2B). However, their length distribution is similar to that of control cells growing in logarithmic phase, which are systematically longer than in stationary phase (Figs. 2B and supplemental Fig. S5). The mean length of the control cells decreased from 4.6 μm to 2.5 μm as culture OD600 rose from 0.8 to 2.5 (supplemental Fig. S5). Considered together with the gene-expression studies presented below, the elongated size of the protein-overexpressing cells is likely to reflect a blockage of their entry into stationary phase, which is somewhat more severe for cells overexpressing IMPs compared with cells expressing soluble proteins. Notably, none of the protein-overexpressing populations showed evidence of impaired cell division or filamentation (i.e. multiple cells attached to each other or multiple septa), which is readily detected via the staining of septa by FM4–64 (Fig. 2A) (34).
Negatively stained thin-section EM images show equivalent ultrastructure in the control cells and most of the overexpressing cells (Fig. 3). No evidence of proliferation or invagination of the cytoplasmic membrane (18, 19) was observed in any cells even though the images have sufficient resolution. These structures should be visible if present. Electron-dense spheroidal IBs were visible near the poles of the cells expressing NBD-EcMsbA, which showed objects with similar dimensions and subcellular localization in DIC and Mitotracker Green fluorescence images. The cells expressing EcEnolase* and EcYojI, which produced the highest levels of the target proteins as visualized via Coomassie-Blue-stained SDS-PAGE gels, displayed uniformly higher cytoplasmic electron density in negative-stain EM. This pattern reflects a higher content of some species (e.g. total protein or ribosome particles) that binds one or both of the electron-dense uranyl acetate and lead citrate compounds used for staining. However, as discussed above, Bradford assays for total protein content failed to show a difference relative to control cells, whereas microarray analyses (see below) of ribosomal RNAs and mRNAs encoding ribosomal proteins failed to show any difference in the levels of these species. However microarray data on rRNA could have been compromised by saturation because of the high concentration of rRNA in the samples. Therefore, the explanation remains unclear for the uniformly high cytoplasmic electron density in EM images of the cells expressing EcEnolase* or EcYojI.
Overexpression Activates neither Envelope-stress nor Folding-stress Transcription Factors
We used reporter-gene assays to evaluate the activity in the protein-overexpressing cells of three stress-related transcription factors (Fig. 4). In these assays, the lacZ gene encoding the enzyme β-galactosidase is fused to a promoter that is strongly modulated by the activity of a specific transcription factor so that the observed level of β-galactosidase activity reflects the net activity of that transcription factor. We assayed reporters for the CpxR transcriptional dual regulator and for σE (σ24) and σH (σ32), so-called sigma-factors that bind to E. coli RNA polymerase to modulate its activity on large sets of promoters comprising transcriptional “regulons.” The σE system helps maintain the integrity of the cell envelope (36). It is activated in a guanosine-tetraphosphate ((p)ppGpp) dependent manner on entry into stationary phase or by several kinds of envelope stress, including overexpression of outer membrane proteins (37). Genes regulated by σE include a variety of cell-envelope proteases and chaperones as well as enzymes required for the biosynthesis of fatty acids and lipopolysaccharide (38). CpxR is a second global regulator of envelope stress that also influences transcription of σE and σH as well as genes involved in motility and biofilm formation (39). The σH system mediates the bacterial heat-shock response and can be activated by unfolded proteins in the cytoplasm (40). It controls the expression of a wide variety of chaperones and proteases as well as other genes.
Fig. 4.
Reporter-gene assays of the activity of selected stress-response regulators. Cell growth was monitored via OD600 (left) in parallel with β-galactosidase reporter-gene activity (right) in MG1655-derived E. coli strains harboring a pQE-derived expression plasmid together with the pREP4 accessory plasmid. Expression of the target protein was induced with 1 mm IPTG at an OD600 of 0.4–0.6. The β-galactosidase reporter gene is fused to the σE-dependent promoter for the rpoH gene in strain SEA001 (A), to the CpxR-dependent promoter for the cpxP gene in strain SEA3122 (B), or to the σH-dependent promoter for the htpG gene in strain SEA3084 (C). Reporter-gene activity is displayed as a differential rate plot (29) showing β-galactosidase activity as a function of culture density (i.e. OD600).
The control cells in our study recapitulated earlier results for all three of these stress-response transcription factors. Specifically, σE (Fig. 4A) (29) and CpxR (Fig. 4B) (41) activity is low during logarithmic-phase growth but rises as cells transition into early stationary phase (i.e. at OD600 ≥ 1.5 for σE and OD600 ≥ 2.0 for CpxR). The activity of σH (Fig. 4C) is significant and steady throughout most of the characterized growth period.
Strikingly, none of the protein-overexpressing cells shows enhanced activity from any of these stress-related transcription factors, despite the accumulation of significant amounts of misfolded protein in the cells expressing NBD-EcMsbA, EcYojI, and EcYojI**. Instead, σE and CpxR activity is suppressed in all of the protein-overexpressing cells (Figs. 4A, 4B), slightly in the cells expressing NBD-EcMsbA and strongly in the cells expressing the other proteins. These results provide further evidence of blocked entry into stationary phase in protein-overexpressing cells. The activity of σH is similar in the control and protein-overexpressing cells except for those expressing EcEnolase*, which surprisingly suppresses σH activity.
Transcriptional Microarrays Confirm Inferences from Reporter-gene Studies
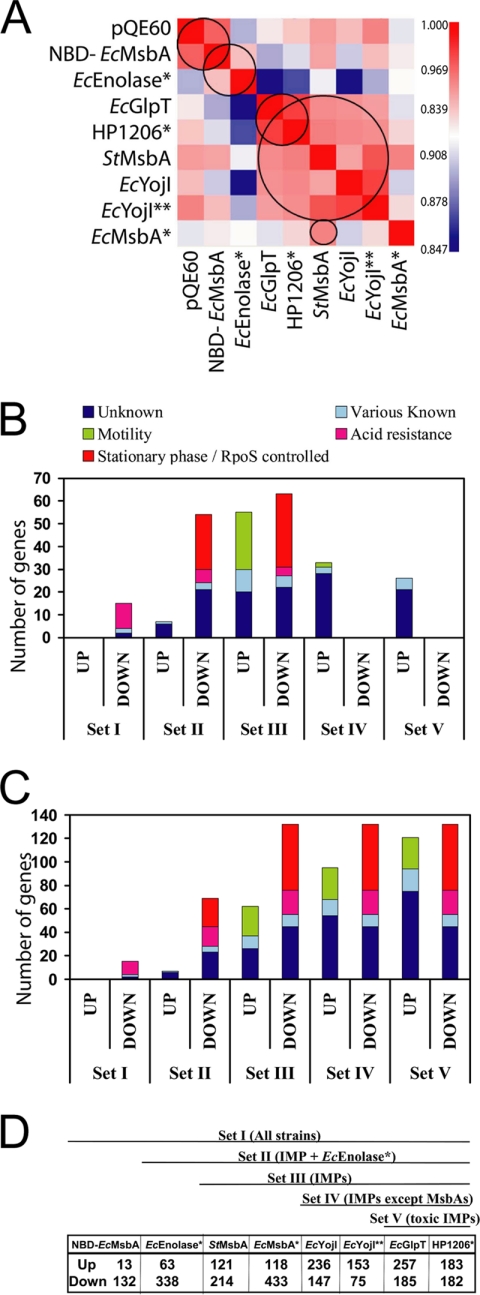
Microarray profiling was used to characterize global RNA content in the protein-overexpressing cells 3 h after IPTG induction (Figs. 5, 6 and supplemental Fig. S8, Tables I and supplemental Table S2, and supplemental file GubelliniMicroarrayDataMCP.xls). Complete biological replicates (three for controls and two for all overexpressing strains except EcGlpT and StMsbA) showed excellent reproducibility with a minimal number of genes exhibiting a significant change in one microarray experiment while not changing in its replicate (supplemental Fig. S8 and additional data not shown). Five groups of strains with strongly correlated expression profiles were identified based on analysis of the Spearman rank correlation coefficients for all MG1655 transcripts (Fig. 5A): (1) empty-vector controls and cells overexpressing NBD-EcMsbA; (2) cells overexpressing soluble proteins; (3) cells overexpressing IMPs except EcMsbA*; (4) cells overexpressing strongly toxic IMPs; and (5) cells overexpressing EcMsbA* or StMsbA. These correlations indicate global similarities in the transcriptional changes in cells overexpressing the corresponding proteins. The observation that cells overexpressing NBD-EcMsbA have a very similar transcriptional profile to empty-vector controls indicates that inclusion-body formation is physiologically protective. Given the high expression level of NBD-EcMsbA (Fig. 1C), this observation suggests that enhanced metabolic flux into the overexpressed protein population is not a significant contributor to the observed changes in gene expression. The higher correlation between cells expressing the strongly toxic IMPs EcGlpT and HP1206* compared with cells overexpressing the other IMPs suggests that some transcriptional changes are related to their toxicity. Finally, the correlation between EcMsbA* and StMsbA suggests that some transcriptional effects are attributable to their biochemical activity; cells overexpressing these orthologous proteins share nine genes showing at least threefold reductions that are not significantly reduced in the other overexpressing strains (arnB and arnC encoding lipid-A-modification enzymes, plus asiA, yoaD, cadC, yjhS, ybiO, yjcC, and ycbC).
Fig. 5.
Transcriptional microarray analyses. At the end of the 3-h induction period, RNA was isolated, reverse-transcribed, and the resulting fluorescent cDNA pools were hybridized to E. coli 2.0 microarray chips (Affymetrix Inc., Santa Clara, CA). Data were scaled using the RMA algorithm in the Affymetrix Expression Console (producing the values given in the supplemental data file entitled GubelliniMicroarrayDataMCP.xls). A, Heat map of Spearman rank correlation coefficients of the scaled expression levels observed in all pairs of arrays. Red and blue indicate the highest and lowest correlations, respectively. Five strongly correlated expression profiles were identified, as indicated by the black circles: IPTG-induced empty pQE-60 vector and overexpressed NBD-EcMsbA; overexpressed soluble proteins; strongly toxic IMPs; all IMPs except EcMsbA*; and EcMsbA* and StMsbA. (B–D) Incremental (B) and cumulative (C) counts of transcripts with significantly changed expression levels in progressively more restricted sets of protein-overexpressing cells (proceeding from left to right), colored according to membership in the functional categories indicated on the plot. The cumulative number of changes for each set (C) is equal to the sum of the incremental changes (B) in that set and all sets further to the left. A threefold change in expression level (i.e. a 1.58 increment in log2) relative to the IPTG-induced empty-vector control was used as the threshold for significance, although transcripts with greater than twofold changes were included in the counts if the majority of the samples in the corresponding of IPMs exceeded the threefold threshold. The schematic diagram at the bottom (D) defines the samples included in each of the five progressively restricted sets; the most inclusive (Set I) contains all overexpressing cells, whereas the most restricted (Set V) contains only the cells expressing the two strongly toxic IMPs (EcGlpT and HP1206*). A list of the genes included in each set is given in supplemental Table S2.
Fig. 6.
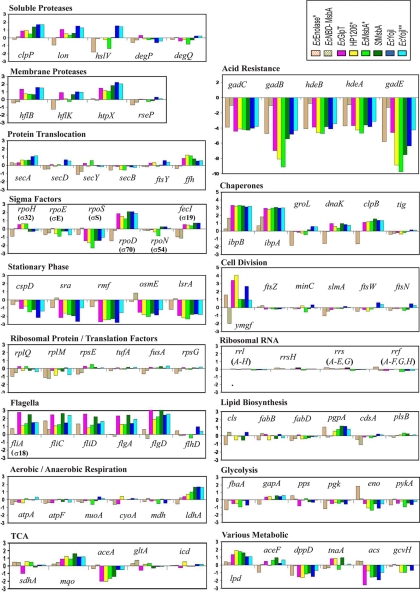
Expression changes in representative genes in various physiological systems. Changes are specified as log2 of the ratio of the transcript level in cells overexpressing the indicated protein to that in the IPTG-induced empty-vector control. Note that the scale on the ordinate is identical in all of the plots (i.e., the increment in log2 per unit of length on the page).
Transcripts showing significant changes in their expression levels could be grouped into five functional categories (Figs. 5B, 5C, and supplemental Table S2): (1) genes expressed under control of the stationary-phase sigma-factor σS (σ38, product of the rpoS gene) (red); (2) genes involved in the acid stress-response, which also systematically increase in expression in stationary-phase (purple); (3) genes involved in motility, which systematically decrease in stationary-phase (green); (4) miscellaneous genes of known biochemical function (cyan); and (5) miscellaneous genes of unknown biochemical function that are not readily assignable to any coherent biochemical pathway (blue). Most significant transcriptional changes were shared by multiple overexpressing strains. Fig. 5B-D organizes the strains into progressively more restricted sets sharing a larger number of transcripts with significant changes in expression level. Set I includes all overexpressing strains. These share 15 down-regulated transcripts, 11 of which typically increase in expression level in stationary phase, but they do not share any up-regulated transcripts. Set V is the most restricted and includes just the strains overexpressing the strongly toxic IMPs EcGlpT and HP1206*. These share 132 down-regulated transcripts, 77 of which typically increase in expression level in stationary phase, and 121 up-regulated targets, 27 of which typically decrease in expression level in stationary phase (Fig. 5C).
Based on this analysis, the vast majority of the shared transcriptional changes (Figs. 5B, 5C, Fig. 6, and supplemental Table S2) are related to the stationary-phase developmental program of E. coli. Although the IMP-overexpressing cells showed a larger number of significant expression changes than the EcEnolase*-overexpressing cells, the affected transcripts have an equivalent distribution among the functional categories shown in Fig. 5B-C (i.e. comparing sets III and IV with set II). The single largest category of down-regulated transcripts contains genes controlled by σS that normally increase in stationary phase. Combining these genes with those related to acid-resistance (Fig. 6), which also increase in stationary phase, accounts for 58% (77/132) of the shared down-regulated transcripts (Fig. 5C). Similarly, 22% (27/121) of the shared up-regulated transcripts are involved in flagellar motility and normally decrease in stationary phase; these constitute 66% (27/41) of the up-regulated genes with known biochemical functions. Consistent with this trend, EM images of negatively stained IPTG-induced cells (supplemental Fig. S7) show long flagella on the IMP-overexpressing cells, and these are not observed on empty-vector control cells. An intermediate phenotype is observed for cells overexpressing EcEnolase* and NBD-EcMsbA. Moreover, in the IMP-overexpressing cells (Fig. 6), the transcript encoding σS is consistently down whereas the transcript encoding the logarithmic phase “housekeeping” sigma-factor σD (σ70) is consistently up. We conclude that all protein-overexpressing cells experience a blockage of the stationary-phase transcriptional program, but that this blockage is stronger in the cells overexpressing IMPs compared with those overexpressing soluble proteins.
A smaller but still significant number of transcriptional changes appear to be related to overexpression of toxic IMPs. The changes shared exclusively by the strains overexpressing the strongly toxic IMPs (i.e. sets V versus IV), comprise 26 up-regulated transcripts and no down-regulated transcripts (Fig. 5B). Among the up-regulated transcripts, 81% (21/26) encode proteins of unknown function (supplemental Table S2). These observations suggest that overexpression of at least some toxic IMPs may elicit a physiological response involving uncharacterized cellular systems.
Fig. 6 presents an analysis of the expression of transcripts of various metabolic and cell biological systems related to protein folding and membrane biogenesis, as well as several systems showing systematic differences in our microarray studies. Even in the most strongly perturbed strains, only minimal differences are observed in the expression of genes involved in the metabolic pathways that mediate phospholipid biosynthesis, glycolysis, and aerobic and anaerobic respiration. No consistent changes of significant magnitude are observed in the components of the SecYEG translocon (6, 8) or the SRP system (i.e. ffh, 4.5S RNA, and ftsY) (11, 12, 25), both of which are believed to play a role in integrating transmembrane α-helices into the membrane. Similarly, no consistent changes of significant magnitude are observed in the widely studied cytoplasmic chaperones GroEL (Hsp60), DnaK (Hsp70), HtpG (Hsp90), or trigger factor (tig). The failure to see changes in the transcripts encoding these heat-shock proteins, except for a reduction in cells overexpresssing EcEnolase*, is consistent with the results from the σH-dependent reporter gene assays reported above (Fig. 4C). Notably, the Hsp20-family chaperones IbpA and IbpB both showed a ∼threefold increase in the cells overexpressing NBD-EcMsbA and a stronger ∼eightfold increase in all IMP-overexpressing cells. These observations suggest that IbpA/B may play a role in assisting IMP biogenesis or in responding to related translational stress (2). The IMP-overexpressing cells also showed consistent ∼twofold increases in transcripts for several soluble (ClpP and Lon) and membrane-bound (HflB/FtsH and HtpX) proteases. Although expression of these up-regulated chaperones and proteases can be stimulated by σH, their transcription in this case seems likely to be driven by other sigma factors based on the overall behavior of the σH regulon. Transcripts for proteins involved in cell division were unaffected except for that encoding YmgF, whose exact molecular function is unclear.
Inferences Concerning Transcription Factor Activity
RegulonDb was used to explore the potential contribution of previously characterized transcription factors (TFs) to the observed physiological responses. Table II shows TF activities with significant correlations to the responses in multiple protein-overexpressing strains as assessed using a Fisher's Exact Test with a 3-fold linear threshold. Only activation by FlhDC, a dual regulator that enhances expression of genes related to flagellar motility (42), is correlated with the up-regulated genes in multiple strains. In contrast, 13 different TF activities are significantly correlated with the down-regulated genes in multiple strains. The TF activities correlated with down-regulation in all overexpressing strains are related to acid response (activation by GadE and GadX and repression by GadW and MarA). Six additional TF activities are significantly correlated with observed changes in cells overexpressing IMPs or EcEnolase* (activation by AppY, ArgR, CysB, and IscR and repression by Fis and LsrR), and three further TF activities are significantly correlated with observed changes in just the IMP-overexpressing cells (activation by NtrC and OmpR and repression by CsiR). Many of these effects are consistent with blocked entry into stationary phase (i.e. the FlhDC, GadE, GadX, GadW, AppY, Fis, and NtrC effects). Several others reflect suppressed expression of nitrogen-scavenging systems (ArgR, NtrC, and CsiR), suggesting that the overexpressing strains are not nitrogen-limited despite their high protein-production levels.
Table II. Fisher's exact text on transcription factors correlations.
| Transcription factora | Direction of regulationb | # Targetsc | HP1206* |
EcMsbA* |
EcGlpT |
EcYojI** |
EcYojI |
StMsbA |
EcEnolase* |
NBD-EcMsbA |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Log(p)d | #e | Log(p) | # | Log(p) | # | Log(p) | # | Log(p) | # | Log(p) | # | Log(p) | # | Log(p) | # | |||
| Above Positive Thresholdf | ||||||||||||||||||
| F1hDC | A | 76 | −9 | 26 | −23 | 34 | −29 | 49 | −26 | 40 | −16 | 36 | −35 | 46 | −5 | 8 | − | − |
| Below Negative Thresholdg | ||||||||||||||||||
| GadE | A | 31 | −6 | 12 | −3 | 12 | −6 | 12 | −8 | 12 | −6 | 12 | −6 | 12 | −4 | 13 | −6 | 7 |
| GadW | R | 5 | −6 | 5 | −4 | 5 | −6 | 5 | −7 | 5 | −6 | 5 | −6 | 5 | −4 | 5 | −3 | 2 |
| GadX | A | 21 | −11 | 14 | −7 | 14 | −10 | 14 | −12 | 13 | −11 | 14 | −11 | 14 | −8 | 14 | −7 | 14 |
| MarA | R | 7 | −5 | 5 | −3 | 5 | −4 | 5 | −5 | 5 | −5 | 5 | −5 | 5 | −3 | 5 | −4 | 3 |
| AppY | A | 10 | −9 | 9 | −6 | 9 | −9 | 9 | −11 | 9 | −9 | 9 | −9 | 9 | −7 | 9 | − | − |
| CysB | A | 17 | −6 | 9 | −5 | 11 | −5 | 8 | −8 | 9 | −7 | 10 | −6 | 9 | −3 | 8 | − | − |
| Fis | R | 74 | −9 | 24 | −13 | 40 | −6 | 20 | −5 | 15 | −10 | 25 | −5 | 18 | −5 | 26 | − | − |
| LsrR | R | 9 | −10 | 9 | −7 | 9 | −10 | 9 | −12 | 9 | −10 | 9 | −10 | 9 | −6 | 8 | − | − |
| IscR | A | 7 | −6 | 6 | −3 | 5 | −6 | 6 | − | − | − | − | −6 | 6 | −4 | 6 | − | − |
| CsiR | R | 5 | −6 | 5 | −4 | 5 | −6 | 5 | −7 | 5 | −6 | 5 | −6 | 5 | − | − | − | − |
| NtrC | A | 44 | −5 | 13 | −4 | 17 | −8 | 18 | −6 | 12 | −6 | 14 | −6 | 14 | − | − | − | − |
| OmpR | A | 10 | −4 | 5 | −3 | 6 | − | − | −4 | 5 | −4 | 5 | −5 | 6 | − | − | − | − |
| ArgR | A | 5 | −6 | 5 | −4 | 5 | −6 | 5 | −7 | 5 | −6 | 5 | −6 | 5 | − | − | − | − |
a Systematic correlations exist between the activities of some transcription factors due to overlapping target-gene specificity. One such cross-correlated set includes GadE, GadW, GadX, and MarA. A second cross-correlated set includes Fis, AppY, GadW, and GadE, while a third includes ArgR and NtrC.
b Influence of the transcription factor on the controlled operon, with “A” standing for activator and “R” standing for repressor.
c Total number of target genes controlled by the transcription factor as according to RegulonDb.
d Log(p) indicates the base-10 logarithm of the probability for the observed number of changes to occur at random among targets of the indicated transcription factor, based on the number of changes observed in the entire E. coli transcriptome.
e # indicates the number of genes regulated by the transcription factor showing changes above or below the threefold lineaer threshold in each array data set.
f Transcription factors regulating transcripts showing at least threefold linear increases compared to the control cells.
g Transcription factors regulating transcripts showing is at least threefold linear decreases compared to the control cells.
Target Protein Properties Control the Toxicity of GlpT and MsbA
The data presented above demonstrate that IMP overexpression does not induce established stress-response systems. Combined with the observation that several IMPs can be overexpressed at moderate levels without inhibiting cell growth, these data suggested that the toxicity caused by the induction of some IMPs may derive primarily from their own specific biochemical and biophysical properties. Consistent with this inference, HP1206*, the single most toxic protein in our study, carries six missense mutations and appears to be rapidly degraded on overexpression. The mutations in HP1206* seem likely to cause aberrant folding in the membrane, which could be responsible for its toxicity. The data presented above suggest that EcGlpT and EcMsbA, the other toxic proteins in our study, are both properly folded after overexpression, an inference supported by the fact that their crystal structures were solved from protein samples overexpressed in E. coli. Therefore, we investigated whether the toxicity caused by overexpression of these proteins is related to their biochemical activities.
For EcGlpT, we examined whether its toxicity is modulated by constitutive induction of the enzymes involved in glycerol degradation (Fig. 7A). Some sugar phosphates are known to exert toxic effects in the E. coli cytoplasm, suggesting that the activity of EcGlpT in transporting glycerol-3-phosphate could contribute to its toxicity. Therefore, we induced its overexpression in MG1655 cells carrying the glpR-1 mutation, which truncates the GlpR repressor and leads to constitutive expression of the glycerol degradation regulon (data not shown). In the glpR-1 strain, the toxicity caused by overexpression of EcGlpT is strongly attenuated in a consistent way (Fig. 7A), supporting the inference that its toxicity is attributable in part to its transport activity. Equivalent results were obtained in an MG1655 strain obtained from the University of Wisconsin Genome Center (strain FB205226 from UW Genome Project), which carries a disruption of the fliA gene and also the glpR-1 mutation (supplemental Fig. S9).
The spontaneous deletion we repeatedly observed in IPTG-inducible plasmids of the five N-terminal residues in EcMsbA suggested that the structure of its N terminus might modulate the toxicity caused by its overexpression. Fig. 7B shows growth curves during induction of EcMsbA constructs with the native N terminus, the ΔN5 mutation at the N terminus, or an N-terminal his-tag. These experiments were conducted with the constructs cloned under control of the very tightly regulated arabinose promoter in the pBAD plasmid, in which they all can be stably maintained in the presence of the catabolite-repressor glucose. Comparable toxicity was observed on induction of the constructs with either the native N terminus or the ΔN5 mutation (which have C-terminal his-tags), whereas much stronger toxicity was observed on induction of the construct with the N-terminal his-tag (Fig. 7B). Therefore, the toxicity caused by EcMsbA overexpression is clearly modulated by its sequence and very likely by its own biochemical properties. To dissect the mechanism of the observed effects, we examined the expression levels of the three MsbA constructs (supplemental Fig. S10), and we also examined their abilities to complement a temperature-sensitive msbA mutation at the nonpermissive temperature (Fig. 7C). The N-terminally his-tagged construct showed a greatly reduced capacity to rescue growth, whereas the ΔN5 construct was able to rescue growth only at higher inducer concentration compared with the construct with the native N terminus (Fig. 7 and additional data not shown). The expression level of the ΔN5 construct is substantially lower than that of the other two constructs (supplemental Fig. S10). Because reduced expression is likely to relieve toxicity associated with unregulated expression, this property seems likely to explain spontaneous selection of the ΔN5 mutation in leaky IPTG-inducible plasmids.
These data demonstrate that the N-terminally his-tagged MsbA construct is functionally deficient. Because MsbA is essential for cell growth (30) and active as a homodimer, the strong toxicity of the N-terminally his-tagged construct is likely to be attributable to subunit mixing causing trans-dominant inhibition of chromosomally encoded MsbA molecules. Further research will be required to dissect the mechanistic basis of this effect because the existing crystal structure of MsbA shows its N terminus to be disordered and remote from the homodimer interface. However, this structure was produced using the N-terminally his-tagged protein construct. Most importantly relative to the topic of the current paper, these studies on MsbA provide another example where the toxicity caused by IMP overexpression derives at least in part from the biochemical properties of the target protein itself.
The transcriptional changes occurring on overexpression of EcMsbA* are most highly correlated with those occurring on overexpression of the orthologous StMsbA protein (Fig. 5A). The protein-specific transcriptional changes observed on overexpression of the other IMPs (data not shown) might similarly be associated with their biochemical activities. Although definitive conclusions are not possible pending analysis of a more diverse set of wild-type target IMPs, the consistency of the trends observed for all of the IMPs included in the current study suggests there may be no intrinsic barrier to moderate overexpression of IMPs in E. coli. Most problems encountered during IMP overexpression could be attributable to the biochemical and biophysical properties of the target protein itself.
DISCUSSION
Overview of Conclusions
The results presented above have significant implications for E. coli biology in addition to providing insight into the obstacles to successful IMP overexpression. They reveal several unexpected physiological effects: blockage of the stationary-phase transcriptional program during overexpression of soluble proteins or IMPs; a failure to induce the heat-shock response on overexpression of misfolded proteins; and a failure to induce established envelope-stress systems on overexpression of toxic IMPs. Moreover, they suggest that there may not be any intrinsic barrier to moderate overexpression of IMPs in E. coli and that the toxicity sometimes observed during this process generally may be attributable to the biochemical and biophysical properties of the target IMPs themselves, rather than to inherent limitations in the capacity for membrane-biogenesis or IMP insertion. Toxicity caused by protein expression leads to selective pressure for mutations improving cell growth. The variability in growth rate frequently observed during induction of toxic IMPs suggests that the selection of such mutations is common. Although toxicity-suppressing mutations might occur in genes encoding other proteins, they can potentially occur in the target IMP itself, which would produce chemical and potentially conformational heterogeneity in the purified IMP population. Mutational variations in target IMPs during overexpression may be an unrecognized obstacle to their successful crystallization.
Implications for the Systems Biology of Protein Translation
Our results suggest that there are important gaps in current understanding of the systems biology of protein translation in E. coli. Based on the themes emphasized in previous literature, pushing cells to enhance protein translation might have been expected to induce the well-characterized “stringent response,” in which accumulation of uncharged tRNAs in the cytosol induces transcriptional changes associated with entry into stationary phase. Such a response could have occurred because of a kinetic lag in ramping-up related biochemical resources, or it could have occurred because of metabolic depletion of the chemical precursors and energy needed to support high-level protein biosynthesis. Instead, the exact opposite of the stringent response is observed in all of the protein-overexpressing cells that we characterized, i.e. no evidence is observed of metabolic depletion, and the transcriptional program associated with stationary-phase entry is impeded even though the period of protein overexpression overlaps the time of this physiological transition.
These observations suggest that the protein translation activity of E. coli cells plays an important role in determining its physiological state and developmental program. The stringent response provides a well-established paradigm by which ribosomes participate in controlling cellular physiology. In this pathway, the ribosome acts as an allosteric regulator of the enzymes RelA and SpoT that control the level of the “alarmones” guanosine tetra and penta-phosphate ((p)ppGpp). These alarmones bind to RNA polymerase to modulate the transcriptional level of many different genes involved in amino acid metabolism and control of entry into stationary phase (43, 44). It is believed that this regulatory activity is controlled by the docking of uncharged tRNAs to ribosomes. Thus, the ribosome is believed to act as a sensor of related metabolic resources. The results presented here suggest that ribosomes are not acting merely as a sensor of metabolic state and that instead their translation activity may itself play an important role in controlling the developmental program of E. coli cells, including the manner in which they use available metabolic resources. Empty-vector control cells induced with IPTG have an equivalent behavior to noninduced cells in our experiments. Therefore, the altered developmental program is driven either by the genetically engineered mRNA transcription, by the resulting protein translation, or by some related but yet-uncharacterized stress-response system. Although suppression of (p)ppGpp synthesis because of ribosomal modulation of the RelA/SpoT system could contribute to the altered developmental program observed in our experiments, such an effect would require either a significant increase in the overall level of translation activity or a previously uncharacterized linkage of RelA/SpoT to some sensor of translational stress. Further research will be required to dissect the physiological circuitry accounting for suppression of the stationary-phase transcriptional program in protein-overexpressing E. coli cells.
One issue to be addressed in such studies is whether protein-overexpressing cells have an enhanced overall level of mRNA or protein synthesis. The magnitude of the blockage in the stationary-phase transcriptional program is clearly not correlated with the net accumulation of the overexpressed target proteins in our studies. However, because of degradation of nascent proteins, net accumulation of the target protein may not track the instantaneous rate of mRNA or protein synthesis, and there could be a hidden correlation between one of these parameters and the magnitude of the blockage. We observe the mildest transcriptional perturbations in the cells expressing NBD-EcMsbA, which produce the third highest net yield of target protein (after EcEnolase* and EcYojI), whereas we observe the strongest perturbations is in the cells producing EcGlpT and HP1206*, which give the lowest net yield of target protein. The production of HP1206* can be detected using Western blotting but not via Coomassie staining of SDS-PAGE gels, presumably because of rapid degradation of this mutationally destabilized protein. Thus, HP1206* provides a pointed example of why instantaneous protein synthesis rate cannot be assessed via net accumulation level. The other target proteins that do accumulate to moderate-to-high levels still could be partially degraded contemporaneously with translation. Moreover, as discussed in detail in the Results section, there are technical ambiguities in quantifying the total protein concentration in E. coli cells, and it is unclear whether the strongly overexpressing cells have an elevated content of either protein or ribosomes. More accurate quantification of both instantaneous protein biosynthesis rate and total cellular protein content would provide important baseline data to guide efforts to understand the physiological circuitry controlling the blockage of the stationary-phase transcriptional program in protein-overexpressing E. coli cells.
Cellular Metabolism and pH Homeostasis During Protein Overexpression
Related issues are discussed in more detail in the online Supplemental Information for this paper. The control cells and protein-overexpressing cells in our study generally display similar levels of transcripts for metabolic enzymes, suggesting that the latter are executing the standard program for utilization of metabolic resources (45–47) even though there is obvious blockage of many of the transcriptional and physiological changes associated with entry into stationary phase. Notably, there are no systematic perturbations in the expression of operons encoding enzymes involved in phospholipid biosynthesis, protein biosynthesis, nucleic acid biosynthesis, carbohydrate or amino acid catabolism, or aerobic or anaerobic respiration (Fig. 6). These observations suggest that all of these core metabolic processes are proceeding similarly in protein-overexpressing cells versus control cells, which implies that there are no metabolic barriers to high-level expression of either soluble or membrane proteins for cells growing with standard aeration in LB. Similarly, it is clear that the altered pH homeostasis in the protein-overexpressing cells is not an impediment to protein overexpression, although additional research will be required to understand this effect (as discussed in the Supplemental Information). The totality of our results raises questions concerning manner in which metabolic resources control the developmental program of E. coli. Control cells and those overexpressing the highest levels of the induced target proteins display indistinguishable growth properties (Fig. 1A), despite large differences in gene-expression profile, striking alterations in cytoplasmic morphology in negatively stained thin-section EM images, and significant differences in at least the pH of the growth medium. These observations suggest the possibility of a complex interplay in which developmental processes significantly influence the utilization of metabolic resources rather than being directly controlled by their availability.
Implications for Soluble and Membrane Protein Folding in E. coli
Surprisingly, reporter-gene studies (Fig. 4) and microarray profiling consistently show no activation of the σH-controlled heat-shock regulon in the protein-overexpressing cells, even in those expressing partially (NBD-EcMsbA) or mostly (EcYojI) misfolded proteins. Remarkably, overexpression of EcEnolase* strongly suppresses expression of σH-dependent genes, presumably reflecting a uncharacterized feature of the biology of this protein that has been reported previously to have functions beyond its enzymatic activity in glycolysis (48, 49). However, expression of σH-dependent genes including dnaK (Hsp70) and groELS (Hsp60) continues unchanged in cells overexpressing the other target proteins, demonstrating that the heat-shock regulon remains active but is not enhanced in its activity by misfolding of some of our target proteins at 37 °C in vivo. In contrast, all overexpressing cells show enhanced expression of the ibpA and ibpB genes, which encode Hsp20-family chaperones. These genes may play a larger role in controlling protein folding in vivo in E. coli than previously appreciated and might participate directly in the biogenesis polytopic IMPs. For example, they could bind nascent IMPs during synthesis in the cytosol and deliver them to the membrane for insertion. Although Gross and coworkers found that overexpression of several inner membrane proteins induced σH-dependent transcription (50), they noted that not all IMPs do so. This observation, along with our results, suggests that activation of σH may not be a general consequence of protein overproduction. Instead, we propose that specific features of individual target proteins determine whether or not overexpression induces σH (26, 27).
It is noteworthy but nonetheless difficult to interpret the observation that IMP overexpression does not consistently influence expression of the SecYEG translocon (6, 8) or the SRP system (i.e. the ffh and ftsY proteins and 4.5S RNA) (11, 12, 25). There does not appear to be a significant increase in the amount of cytosolic membrane in our overexpressing cells, and it is unclear whether any have an elevated net content of properly folded IMPs. Therefore, normal levels of the translocon and SRP system components may be sufficient to mediate biogenesis of the overexpressed IMPs that are properly inserted into the membrane. In any event, limitations in these components do not appear to be an obstacle to moderate overexpression of IMPs.
E. coli Response to Attempted Overexpression of Toxic IMPs
It is noteworthy that the expression levels of a set of specific genes are consistently enhanced exclusively in cells expressing the two strongly toxic IMPs included in our study, EcGlpT and HP1206*. In contrast, there is no comparable set of genes whose expression is consistently suppressed exclusively in these strains (Fig. 5). This pattern suggests that cells may be able to sense and respond to the stresses caused by overexpression of IMPs, although it remains unclear what specific conditions they are sensing, what transcription factors they are activating, or what physiological consequences are caused by the transcriptional response. Although the induced transcripts encode a mixture of proteins of characterized and uncharacterized functions (supplemental Table S2), few of them are known to function in common pathways, and none has an obvious relationship to IMP biogenesis based on prior literature. Much of the toxicity caused by EcGlpT expression is related to its biochemical activity transporting glycerol-3-phosphate into the cytosol (Fig. 7A), which is not likely to be related to the toxicity caused by HP1206*. Nonetheless, it seems likely that EcGlpT overexpression causes some additional stress that parallels that caused by attempted overexpression of HP1206*. This stress might be related to problems with IMP translation or folding. However, additional research will be required to understand its nature and the physiological purpose of the common response observed on induction of the two toxic IMPs.
Many factors make E. coli the most commonly used host for protein overexpression, including inexpensive and rapid cell growth, extensive genetic and physiological characterization, and the availability of a wide range of expression vectors using different inducers and producing different levels of protein expression. However, other host organisms may give superior results, especially for specific target proteins. To the extent that the toxicity caused by IMP overexpression in E. coli is caused by the biochemical activity of the target protein itself, organisms insensitive to that activity may give improved results. For example, Gram-positive bacteria might be better hosts for overespression of MsbA orthologs because these organisms do not make Lipid A, the likely transport substrate for MsbA. Future research should address whether alternative expression hosts can give improved overexpression of some or all IMPs. The results reported in this paper provide a foundation for such studies and suggest that they need to take into account the general effects of protein overexpression on host-cell physiology and also the specific biochemical and biophysical properties of the target IMPs.
Acknowledgments
We thank S. Kachan and M. Repka for assistance with preliminary microarray experiments, N. Minc for advice and assistance with microscopy experiments, and C. R. Sanders for a critical review of the manuscript. We thank W. T. Doerrler for the gift of E. coli strains W3110A and WD2S. We are grateful to S. Gottesman and N. Majdalani, T. G. Bernhardt, K. Ochi, and R. L. Gourse and T. Gaal for advice on the execution and interpretation of E. coli physiology experiments.
Footnotes
* This work was supported by National Institutes of Health grants 1R21GM07595933 and 1R01GM072867 to J. F. Hunt, National Institutes of Health grants 1U54GM75026 and 5U54GM075026 to W. A. Hendrickson, and National Science Foundation grant MCB-0347302 to S. E. Ades.
 This article contains supplemental Information, Figs. S1 to S10, and Tables S1 and S2.
This article contains supplemental Information, Figs. S1 to S10, and Tables S1 and S2.
1 The abbreviations used are:
- IMP
- integral membrane protein
- SRP
- signal recognition particle
- EM
- electron microscopy
- LB
- Luria broth
- IPTG
- isopropyl β-d-thiogalactoside
- TM
- transmembrane
- ABC
- ATP-binding cassette
- NBD
- nucleotide-binding domain
- IBs
- inclusion bodies
- DIC
- differential interference contrast
- TF
- transcription factor.
REFERENCES
- 1. Gordon E., Horsefield R., Swarts H. G., de Pont J. J., Neutze R., Snijder A. (2008) Effective high-throughput overproduction of membrane proteins in Escherichia coli. Protein Expr. Purif. 62, 1–8 [DOI] [PubMed] [Google Scholar]
- 2. Wagner S., Baars L., Ytterberg A. J., Klussmeier A., Wagner C. S., Nord O., Nygren P. A., van Wijk K. J., de Gier J. W. (2007) Consequences of membrane protein overexpression in Escherichia coli. Mol. Cell Proteomics 6, 1527–1550 [DOI] [PubMed] [Google Scholar]
- 3. Wagner S., Klepsch M. M., Schlegel S., Appel A., Draheim R., Tarry M., Högbom M., van Wijk K. J., Slotboom D. J., Persson J. O., de Gier J. W. (2008) Tuning Escherichia coli for membrane protein overexpression. Proc. Natl. Acad. Sci. U.S.A. 105, 14371–14376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wang D. N., Safferling M., Lemieux M. J., Griffith H., Chen Y., Li X. D. (2003) Practical aspects of overexpressing bacterial secondary membrane transporters for structural studies. Biochim. Biophys. Acta 1610, 23–36 [DOI] [PubMed] [Google Scholar]
- 5. White S. H. (2009) Biophysical dissection of membrane proteins. Nature 459, 344–346 [DOI] [PubMed] [Google Scholar]
- 6. White S. H., von Heijne G. (2008) How translocons select transmembrane helices. Annu. Rev. Biophys. 37, 23–42 [DOI] [PubMed] [Google Scholar]
- 7. von Heijne G. (2011) Introduction to theme “membrane protein folding and insertion”. Annu. Rev. Biochem. 80, 157–160 [DOI] [PubMed] [Google Scholar]
- 8. Dalbey R. E., Wang P., Kuhn A. (2011) Assembly of bacterial inner membrane proteins. Annu. Rev. Biochem. 80, 161–187 [DOI] [PubMed] [Google Scholar]
- 9. Junge F., Schneider B., Reckel S., Schwarz D., Dötsch V., Bernhard F. (2008) Large-scale production of functional membrane proteins. Cell Mol. Life Sci. 65, 1729–1755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Schlegel S., Klepsch M., Gialama D., Wickström D., Slotboom D. J., de Gier J. W. (2010) Revolutionizing membrane protein overexpression in bacteria. Microb. Biotechnol. 3, 403–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Yosef I., Bochkareva E. S., Adler J., Bibi E. (2010) Membrane protein biogenesis in Ffh- or FtsY-depleted Escherichia coli. PLoS One 5, e9130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Yosef I., Bochkareva E. S., Bibi E. (2010) Escherichia coli SRP, Its Protein Subunit Ffh, and the Ffh M Domain Are Able To Selectively Limit Membrane Protein Expression When Overexpressed. MBio 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Katzen F., Peterson T. C., Kudlicki W. (2009) Membrane protein expression: no cells required. Trends Biotechnol. 27, 455–460 [DOI] [PubMed] [Google Scholar]
- 14. Drew D., Fröderberg L., Baars L., de Gier J. W. (2003) Assembly and overexpression of membrane proteins in Escherichia coli. Biochim. Biophys. Acta 1610, 3–10 [DOI] [PubMed] [Google Scholar]
- 15. Geertsma E. R., Poolman B. (2010) Production of membrane proteins in Escherichia coli and Lactococcus lactis. Methods Mol. Biol. 601, 17–38 [DOI] [PubMed] [Google Scholar]
- 16. Laage R., Langosch D. (2001) Strategies for prokaryotic expression of eukaryotic membrane proteins. Traffic 2, 99–104 [DOI] [PubMed] [Google Scholar]
- 17. Miroux B., Walker J. E. (1996) Over-production of proteins in Escherichia coli: mutant hosts that allow synthesis of some membrane proteins and globular proteins at high levels. J. Mol. Biol. 260, 289–298 [DOI] [PubMed] [Google Scholar]
- 18. Arechaga I., Miroux B., Karrasch S., Huijbregts R., de Kruijff B., Runswick M. J., Walker J. E. (2000) Characterisation of new intracellular membranes in Escherichia coli accompanying large scale over-production of the b subunit of F(1)F(o) ATP synthase. FEBS Lett. 482, 215–219 [DOI] [PubMed] [Google Scholar]
- 19. Weiner J. H., Lemire B. D., Elmes M. L., Bradley R. D., Scraba D. G. (1984) Overproduction of fumarate reductase in Escherichia coli induces a novel intracellular lipid-protein organelle. J. Bacteriol. 158, 590–596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wilkison W. O., Bell R. M., Taylor K. A., Costello M. J. (1992) Structural characterization of ordered arrays of sn-glycerol-3-phosphate acyltransferase from Escherichia coli. J. Bacteriol. 174, 6608–6616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Chen Y., Song J., Sui S. F., Wang D. N. (2003) DnaK and DnaJ facilitated the folding process and reduced inclusion body formation of magnesium transporter CorA overexpressed in Escherichia coli. Protein Expr. Purif. 32, 221–231 [DOI] [PubMed] [Google Scholar]
- 22. Link A. J., Skretas G., Strauch E. M., Chari N. S., Georgiou G. (2008) Efficient production of membrane-integrated and detergent-soluble G protein-coupled receptors in Escherichia coli. Protein Sci. 17, 1857–1863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Massey-Gendel E., Zhao A., Boulting G., Kim H. Y., Balamotis M. A., Seligman L. M., Nakamoto R. K., Bowie J. U. (2009) Genetic selection system for improving recombinant membrane protein expression in E. coli. Protein Sci. 18, 372–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Zweers J. C., Wiegert T., van Dijl J. M. (2009) Stress-responsive systems set specific limits to the overproduction of membrane proteins in Bacillus subtilis. Appl. Environ. Microbiol. 75, 7356–7364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ulbrandt N. D., Newitt J. A., Bernstein H. D. (1997) The E. coli signal recognition particle is required for the insertion of a subset of inner membrane proteins. Cell 88, 187–196 [DOI] [PubMed] [Google Scholar]
- 26. Gill R. T., Valdes J. J., Bentley W. E. (2000) A comparative study of global stress gene regulation in response to overexpression of recombinant proteins in Escherichia coli. Metab. Eng. 2, 178–189 [DOI] [PubMed] [Google Scholar]
- 27. Smith H. E. (2007) The transcriptional response of Escherichia coli to recombinant protein insolubility. J. Struct. Funct. Genomics 8, 27–35 [DOI] [PubMed] [Google Scholar]
- 28. Boël G., Pichereau V., Mijakovic I., Mazé A., Poncet S., Gillet S., Giard J. C., Hartke A., Auffray Y., Deutscher J. (2004) Is 2-phosphoglycerate-dependent automodification of bacterial enolases implicated in their export? J. Mol. Biol. 337, 485–496 [DOI] [PubMed] [Google Scholar]
- 29. Costanzo A., Ades S. E. (2006) Growth phase-dependent regulation of the extracytoplasmic stress factor, sigmaE, by guanosine 3′,5′-bispyrophosphate (ppGpp). J. Bacteriol. 188, 4627–4634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Doerrler W. T., Reedy M. C., Raetz C. R. (2001) An Escherichia coli mutant defective in lipid export. J. Biol. Chem. 276, 11461–11464 [DOI] [PubMed] [Google Scholar]
- 31. Miller J. (1972) Experiments in Molecular Genetics. Cold Spring Harbor Laboratory, NY [Google Scholar]
- 32. Huang Y., Lemieux M. J., Song J., Auer M., Wang D. N. (2003) Structure and mechanism of the glycerol-3-phosphate transporter from Escherichia coli. Science 301, 616–620 [DOI] [PubMed] [Google Scholar]
- 33. Ward A., Reyes C. L., Yu J., Roth C. B., Chang G. (2007) Flexibility in the ABC transporter MsbA: Alternating access with a twist. Proc. Natl. Acad. Sci. U.S.A. 104, 19005–19010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sharp M. D., Pogliano K. (1999) An in vivo membrane fusion assay implicates SpoIIIE in the final stages of engulfment during Bacillus subtilis sporulation. Proc. Natl. Acad. Sci. U.S.A. 96, 14553–14558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Aung S., Shum J., Abanes-De Mello A., Broder D. H., Fredlund-Gutierrez J., Chiba S., Pogliano K. (2007) Dual localization pathways for the engulfment proteins during Bacillus subtilis sporulation. Mol. Microbiol. 65, 1534–1546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ades S. E. (2008) Regulation by destruction: design of the sigmaE envelope stress response. Curr. Opin. Microbiol. 11, 535–540 [DOI] [PubMed] [Google Scholar]
- 37. Mecsas J., Rouviere P. E., Erickson J. W., Donohue T. J., Gross C. A. (1993) The activity of sigma E, an Escherichia coli heat-inducible sigma-factor, is modulated by expression of outer membrane proteins. Genes Dev. 7, 2618–2628 [DOI] [PubMed] [Google Scholar]
- 38. Hayden J. D., Ades S. E. (2008) The extracytoplasmic stress factor, sigmaE, is required to maintain cell envelope integrity in Escherichia coli. PLoS One 3, e1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Raivio T. L. (2005) Envelope stress responses and Gram-negative bacterial pathogenesis. Mol. Microbiol. 56, 1119–1128 [DOI] [PubMed] [Google Scholar]
- 40. Parsell D. A., Sauer R. T. (1989) Induction of a heat shock-like response by unfolded protein in Escherichia coli: dependence on protein level not protein degradation. Genes Dev. 3, 1226–1232 [DOI] [PubMed] [Google Scholar]
- 41. De Wulf P., Kwon O., Lin E. C. (1999) The CpxRA signal transduction system of Escherichia coli: growth-related autoactivation and control of unanticipated target operons. J. Bacteriol. 181, 6772–6778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Anderson J. K., Smith T. G., Hoover T. R. (2010) Sense and sensibility: flagellum-mediated gene regulation. Trends Microbiol. 18, 30–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sharma U. K., Chatterji D. (2010) Transcriptional switching in Escherichia coli during stress and starvation by modulation of sigma activity. FEMS Microbiol. Rev. 34, 646–657 [DOI] [PubMed] [Google Scholar]
- 44. Srivatsan A., Wang J. D. (2008) Control of bacterial transcription, translation and replication by (p)ppGpp. Curr. Opin. Microbiol. 11, 100–105 [DOI] [PubMed] [Google Scholar]
- 45. Baev M. V., Baev D., Radek A. J., Campbell J. W. (2006) Growth of Escherichia coli MG1655 on LB medium: monitoring utilization of sugars, alcohols, and organic acids with transcriptional microarrays. Appl. Microbiol. Biotechnol. 71, 310–316 [DOI] [PubMed] [Google Scholar]
- 46. Baev M. V., Baev D., Radek A. J., Campbell J. W. (2006) Growth of Escherichia coli MG1655 on LB medium: determining metabolic strategy with transcriptional microarrays. Appl. Microbiol. Biotechnol. 71, 323–328 [DOI] [PubMed] [Google Scholar]
- 47. Baev M. V., Baev D., Radek A. J., Campbell J. W. (2006) Growth of Escherichia coli MG1655 on LB medium: monitoring utilization of amino acids, peptides, and nucleotides with transcriptional microarrays. Appl.Microbiol. Biotechnol. 71, 317–322 [DOI] [PubMed] [Google Scholar]
- 48. Vanderpool C. K. (2007) Physiological consequences of small RNA-mediated regulation of glucose-phosphate stress. Curr. Opin. Microbiol. 10, 146–151 [DOI] [PubMed] [Google Scholar]
- 49. Kaberdin V. R., Lin-Chao S. (2009) Unraveling new roles for minor components of the E. coli RNA degradosome. RNA Biol. 6, 402–405 [DOI] [PubMed] [Google Scholar]
- 50. Nonaka G., Blankschien M., Herman C., Gross C. A., Rhodius V. A. (2006) Regulon and promoter analysis of the E. coli heat-shock factor, sigma32, reveals a multifaceted cellular response to heat stress. Genes Dev. 20, 1776–1789 [DOI] [PMC free article] [PubMed] [Google Scholar]