Abstract
The development of the HUPO-PSI's (Proteomics Standards Initiative) standard data formats and MIAPE (Minimum Information About a Proteomics Experiment) guidelines should improve proteomics data sharing within the scientific community. Proteomics journals have encouraged the use of these standards and guidelines to improve the quality of experimental reporting and ease the evaluation and publication of manuscripts. However, there is an evident lack of bioinformatics tools specifically designed to create and edit standard file formats and reports, or embed them within proteomics workflows. In this article, we describe a new web-based software suite (The ProteoRed MIAPE web toolkit) that performs several complementary roles related to proteomic data standards. First, it can verify that the reports fulfill the minimum information requirements of the corresponding MIAPE modules, highlighting inconsistencies or missing information. Second, the toolkit can convert several XML-based data standards directly into human readable MIAPE reports stored within the ProteoRed MIAPE repository. Finally, it can also perform the reverse operation, allowing users to export from MIAPE reports into XML files for computational processing, data sharing, or public database submission. The toolkit is thus the first application capable of automatically linking the PSI's MIAPE modules with the corresponding XML data exchange standards, enabling bidirectional conversions. This toolkit is freely available at http://www.proteored.org/MIAPE/.
Despite the current interest in data sharing in the context of collaborative proteomics projects, the large amount of information generated and transferred among specialized laboratories requires agreements on standard exchange data formats. To facilitate data sharing, integration and public dissemination, the Human Proteome Organization Proteomics Standards Initiative (HUPO-PSI) has defined community standards for data representation (1, 2). This group, founded in 2002, has held annual meetings, as well as more frequent workshops, that have contributed to numerous improvements in data sharing (3–15). Briefly, these achievements include several MIAPE (Minimum Information About a Proteomics Experiment) (16–22) reporting guidelines and formal XML schemas that are capable of representing not only the minimal information but also significant additional details in a computationally accessible manner. The MIAPE modules and XML schemas are complementary resources—the MIAPE documents describe what information should be reported about an experimental technique (presented in a human readable format), and the XML schemas describe how such information can be captured in a format open to computational processing.
These contributions have been favorably received by the proteomics community, making data sharing and reporting less time-consuming for both experts and occasional users. Moreover, several proteomics journals (23, 24) encourage submitters to meet these guidelines to ensure both quality control and data reproducibility. However, until now, there has been a lack of bioinformatics tools capable of automatic conversion between MIAPE modules and the XML standards.
Following the initial steps of HUPO-PSI, the EMBL-EBI (Hinxton, UK) developed a centralized, standard-compliant public data repository for protein and peptide identifications (PRIDE)1 (25–27). As an essential complementary tool to the main public repository, an XML schema (PRIDE XML) was defined to describe data in the different phases of a proteomics experiment.
Later, in 2005, The Spanish National Network of Proteomic Facilities (ProteoRed) was created as an initiative for the coordination, integration, and development of proteomics facilities and laboratories distributed throughout Spain (28). One of ProteoRed's main objectives is to support the scientific community, enabling widespread access to emerging proteomics technologies. As a contribution to this goal, the bioinformatics work group was created to improve computational analysis and communication among the different network partners. It accepted HUPO-PSI guidelines regarding data sharing and reporting as the main way to exchange proteomics data among members of the ProteoRed network.
The ProteoRed network configuration made it the ideal setting for testing MIAPE guidelines, and thus contributed to their final agreement and validation (12, 29). As a result, the ProteoRed MIAPE web repository (30) was developed to store and share MIAPE documents generated from data derived from proteomics experiments. Currently, the ProteoRed MIAPE web repository is able to manage MIAPE GE (17) (Gel Electrophoresis), GI (18) (Gel Informatics), MS (19) (Mass Spectrometry), and MSI (20) (Mass Spectrometry Informatics) reports.
Although the repository is widely used, the amount of information required to create a new MIAPE report is a time-consuming task for certain applications. An illustrative example is that MIAPE MSI requests not only the metadata about how peptides and proteins were identified but also the data values (i.e. a complete protein and peptide set). It is unreasonable to expect that a user should enter these manually or that a user produce a specific input format for uploading. In addition, it was previously challenging to automate management of MIAPE reports, making it difficult to embed the reports in third party applications or pipeline software for day to day laboratory management.
To overcome these challenges, we have developed a new web toolkit capable of linking the latest versions of the HUPO-PSI XML schemas to the ProteoRed MIAPE web repository in an automated, accessible, and comprehensive way.
EXPERIMENTAL PROCEDURES
Handling Laboratory Information Data Sources
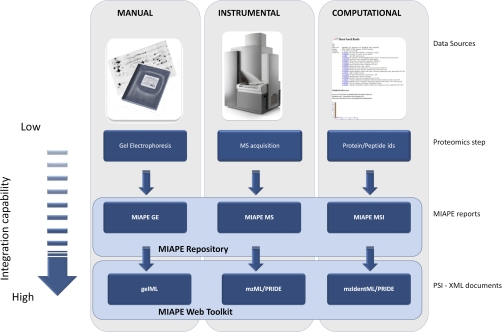
One of the main disadvantages with regard to reporting proteomic experiments is the integration of data from multiple and heterogeneous sources. A basic division of the common laboratory data sources (Fig. 1) would be as follows:
Fig. 1.
Basic division of the common laboratory data sources: Manual (2D-E), Instrumental (MS instruments), and Computational (search engines and software tools). Depending on the previous classification, the MIAPE submission is done by hand, semi-automatically or fully automatically. Although the derived information is conceptually similar, the difficulty for computational tools to understand the semantics and metadata increases as the layers are depicted. Only the PSI-XML layer provides an appropriate structure to automatically capture both the experimental data and metadata with the required precision, whereas the data written in laboratory notebooks must be interpreted and translated before their handling within a computational framework.
1) Manual
This type of information derives from data written in laboratory notebooks. The tasks related to this information source are only minimally supported by either instruments or computers. Gel electrophoresis preparation could be included into this category.
2) Instrumental
The information is generated by analytical equipment such as mass spectrometers, using local computers only as a way to collect, store, and translate the provided data.
3) Computational
The information is both automatically and manually entered, translated, processed, and returned by computational resources (e.g. search engines).
The MIAPE reports offered by the ProteoRed MIAPE web repository contain each of the three types of source data described above. MIAPE GE is filled using manual data such as description of buffer composition and electrophoresis conditions. MIAPE MS mainly gathers the meta-data derived from the mass spectrometer, as instrument components: ion source, analyzer, detector or voltages. Finally, MIAPE MSI requests both search engine submission parameters and protein or peptide results to be reported.
The ProteoRed MIAPE web toolkit
The ProteoRed MIAPE web toolkit (version 1.0) has been implemented using Java and ASP languages, and made accessible through Microsoft Internet Explorer 6.0 or higher, Mozilla Firefox 2.0 or higher, or Safari 4.0. It is freely available at the ProteoRed website (http://www.proteored.org/MIAPE/).
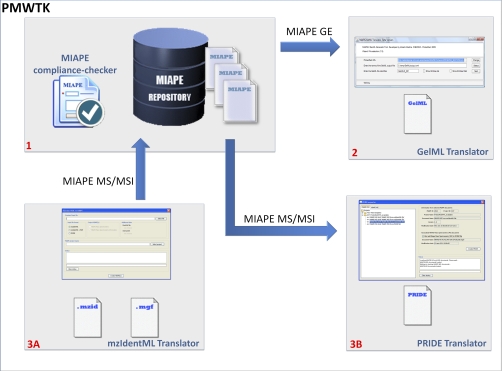
The following three modules (Fig. 2) are included in the ProteoRed MIAPE web toolkit: (1) a MIAPE Compliance Checker, (2) a GelML Translator, and (3) an mzIdentML and PRIDE Translator. To create the rules underlying these tools, specific mappings were created manually between items within each MIAPE report type and elements in the corresponding XML document. The MIAPE web toolkit thus works in a bidirectional manner: assisting in creating a MIAPE report directly from XML data, for example to fulfill a journal's requirement, and also incorporating metadata and data into PSI XML exchange documents from manually entered information in MIAPE reports, as illustrated below.
Fig. 2.
ProteoRed MIAPE web toolkit (PMWTK) modules: (1) MIAPE Compliance Checker: The MIAPE Compliance Checker has been added to ensure an appropriate conversion of MIAPE reports into a set of formal HUPO-PSI XML documents. The MIAPE validation is performed in three stages: contents, semantic, and internal references. (2) Exporting GE data: GelML translator (MIAPE GE → GelML): From a previously created MIAPE GE report, this module retrieves the data derived from a gel-based experiment to create a valid and well-formed GelML document. (3) Importing/Exporting MS and MSI data: mzIdentML and PRIDE translator. A) Import flow (mzIdentML → MIAPE): The MIAPE web repository converts mzIdentML (and optionally .mgf peaklist files) into MIAPE MSI (and MS compliant reports). B) Export flow (MIAPE MS/MSI → PRIDE): From a validated MIAPE MSI (and optionally MS) the export utility will generate a valid PRIDE report containing all the information required by the PRIDE schema definition. Both workflows ensure an appropriate connection between the data contained in the XML documents and required by the MIAPE guidelines.
1) MIAPE Validation: MIAPE Compliance Checker
One of the most critical issues regarding data sharing and reporting is that data sets are accompanied with accurate metadata to describe in sufficient detail how they were generated, and potentially to allow results to be reproduced. The MIAPE Compliance Checker (Fig. 2–1) has been developed as an aid for metadata validation to ensure an appropriate conversion of MIAPE reports (written using natural language) into a set of formal PSI XML documents, including syntax validation. The Compliance Checker runs during file import (checking external documents to ensure that values required by the MIAPE specification have been provided) and during file export (checking the metadata required for the PSI XML output) from or to the MIAPE database.
Although the Compliance Checker included in the release (version.1.0) described in this article only contains the validation between the Gel Electrophoresis MIAPE (MIAPE GE) reports and PSI standard GelML (31), further versions of the toolkit will include the remaining PSI exchange formats: mzML (32) and mzIdentML (33).
To provide comprehensive validation, the MIAPE Compliance Checker performs this complex task in the following three steps:
First, validation of the contents is done to check for contextual information related to the experiment type. As an example, in a two-dimensional gel separation, details should be provided for both electrophoresis runs. The rules underlying the MIAPE (minimum information) module are also checked in the XML data file, because an XML file can be valid against the schema without being MIAPE-compliant. Second, semantic validation is performed using a set of specialized controlled vocabularies (CVs). In this stage each CV term is evaluated using the validation rules defined in the PSI validator framework (34), to ensure that only semantically valid CV terms have been included in each location. Third and finally, the Compliance Checker will also check that links have been correctly created among related elements in different parts of the document, such as linking the protocol of one image acquisition step back to the description of the gel from which the image was produced.
All validation steps are automatically and sequentially performed by a single routine. If the assessment does not result in errors, the document will be accepted as a valid MIAPE report. Otherwise the MIAPE Compliance Checker will indicate how to correct it and, in most cases, a link to the precise location in the report to edit and replace the invalid data.
2) Exporting GE Data: GelML Translator
To improve the capabilities of the MIAPE web repository for data sharing in the context of gel electrophoresis experiments, one of the modules of the new MIAPE web toolkit exports the GelML format automatically from a MIAPE GE report.
This module can be run directly from the MIAPE web site. Starting from a MIAPE GE document that has been validated by the MIAPE Compliance Checker, the GelML instance will be created (Fig. 2–2) according to the three following steps that compose the main algorithm:
First, the application loads all the information contained in the MIAPE document and extracts the MIAPE identifiers such as the gel matrix, buffer components, and gel images. Second, all the necessary information required by GelML and not included in the MIAPE report, will be automatically named and incorporated for the user to edit manually if necessary. Finally, the last step will formalize the relationships among the different elements within GelML, for instance completing rules such as: “link the sample element to the gel matrix, including a reference to the element that captures the loading buffer.” This transforms the newly created document into a linked report, using unique identifiers and foreign keys to ensure unambiguous references among elements.
3) Importing/Exporting MS and MSI Data: mzIdentML and PRIDE Translator
In contrast to data resulting from gel electrophoresis experiments, both mass spectrometry (instrument) and protein/peptide identification (computational) derived data can be more easily exported automatically. The MIAPE web repository minimizes the effort required to convert MS and MSI standard output file formats (in this case PRIDE XML and mzIdentML) into MIAPE compliant reports. These are stored in the MIAPE document repository to allow users to edit these reports further (if necessary to complete missing metadata) via a web interface (30).
The translator contains a partial mapping between PRIDE XML elements (initially based on the previous HUPO-PSI mass spectrometry standard, mzData) and the MIAPE MS specification. It is difficult to create a fully automatic mapping between PRIDE XML and MIAPE MS for certain types of metadata, because the PRIDE XML schema offers a high number of combinations for capturing metadata regarding the mass spectrometry acquisition using a large number of possible CV terms. Because of this fact, the translator associates as many of the data acquisition parameters as possible, and the user must verify or edit the resulting MIAPE MS report to complete any missing fields. The mapping from mass spectrometry data, i.e. spectra, and identified proteins and peptides within PRIDE XML to MIAPE MS/MSI is straightforward and can be achieved in a fully automated way by the translator. In addition, mzIdentML, the latest HUPO-PSI standard for mass spectrometry informatics, contains a similar underlying model to the corresponding MIAPE module (MIAPE MSI) and hence a bidirectional mapping between mzIdentML and MIAPE MSI reports can be achieved relatively simply. The PSI group responsible for mzIdentML and MIAPE MSI has produced a document to illustrate the correspondence: http://psidev.info/index.php?q=node/386.
The translator starts mapping the elements and attributes from the uploaded files (PRIDE XML or mzIdentML) to the MIAPE data model (MIAPE MS and/or MSI) according to the underlying mappings previously described. Once all the information is processed, the document is stored in the MIAPE web repository, for further processing and visualization (Fig. 2–3).
RESULTS
The following example uses as input a standard peak list generated in a proteomics experiment. It has been included to show the performance of the MIAPE web toolkit. Although the MIAPE web toolkit can handle complete proteomics workflows, including separation related metadata (Gel and LC based), only the generation of MIAPE reports and XML standards for mass spectrometry and protein/peptide identification are illustrated here. Implementation of standard reports for Gel-based experiments has been described in more detail elsewhere (31).
LC-ESI Analysis: ABRF sPRG 2010 Sample
To validate the MIAPE web toolkit, data obtained in our contribution to the ABRF sPRG2010 study were used. The goal of the study (described at ABRF's Proteomics Standards Research Group website—http://www.abrf.org/index.cfm/group.show/proteomicsstandardsresearchgroup.47.htm) was the identification and characterization of phosphorylation sites present in the corresponding synthetic peptides (n = 23). The study sample was a mixture containing equimolar quantities of a tryptic digest of six proteins (5 pmol), with singly and multiply phosphorylated residues. The sample was analyzed using an Ultimate 3000 nano high performance liquid chromatography (HPLC) (Dionex) coupled to a HCT Ultra Ion Trap mass spectrometer (Bruker Daltonics, Bremen, Germany). The analysis was based on a data-dependent experiment of the two most abundant ions in the survey scan in MS mode, alternating collision-induced dissociation (CID) and electron-transfer dissociation (ETD) fragmentation techniques in MS/MS mode. Both CID and electron-transfer dissociation (ETD) fragmentation modes generated two peak lists containing 753 and 604 MS/MS spectra respectively, which we refer to as peaklistA and peaklistB.
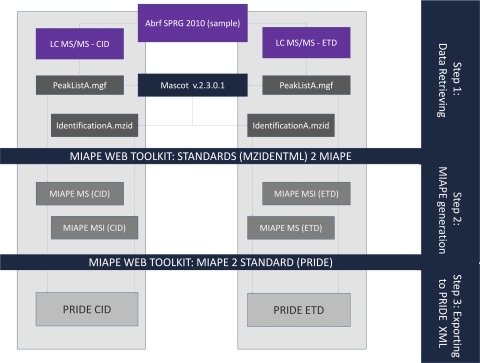
The strategy followed in this example (Fig. 3) was divided in three steps
Fig. 3.
ABRF sPRG2010 experiment: MIAPE and PRIDE XML creation pipeline. Step 1) Both peaklists (peaklistA.mgf and peaklistB.mgf) derived from the two experimental approaches (CID and ETD fragmentation modes) are submitted to Mascot. The final results provided by the search engine are exported using mzIdentML format (identificationA.mzid and identificationB.mzid). Step 2) Automatic generation of MIAPE documents: the source peaklist and resulting mzIdentML identification file (i.e. peaklist A.mgf in addition to identificationA.mzid) are submitted to the ProteoRed MIAPE web toolkit (PMWTK) to create the corresponding MIAPE reports (MIAPE MS from the MS peak lists and MIAPE MSI from the mzIdentML). Step 3) Export to PRIDE XML from MIAPE MS and MSI. Finally two PRIDE XML documents (CID and ETD modes) are generated and were submitted to the central repository (http://www.ebi.ac.uk/pride) for public access.
(1) Data retrieval: MS peak lists and MSI protein/peptide identification results (Mascot submission).
(2) Automatic generation of MIAPE documents: MIAPE MS from the MS peak lists, and MIAPE MSI from the mzIdentML files produced by Mascot.
(3) Exporting to PRIDE XML from documents created in step 2. PRIDE XML visualization and submission to the central repository (http://www.ebi.ac.uk/pride).
1) Data Retrieval
To use the MIAPE web toolkit, protein identification data should be formatted as an mzIdentML standard file. To date, only the Mascot search engine is able to export mzIdentML files directly, but tools are under development or released in beta versions for some of the most widely used search engines like Phenyx (http://www.genebio.com/products/phenyx/), X!Tandem (http://www.thegpm.org/tandem/), or OMSSA (http://pubchem.ncbi.nlm.nih.gov/omssa/) for export to mzIdentML. An extensive list of the current mzIdentML implementations is described in more detail at the HUPO PSI web site (http://www.psidev.info/index.php?q=node/408).
Database searches were done using a licensed version of Mascot version 2.3.0.1. for peak lists obtained from both CID and ETD fragmentations, corresponding to the liquid chromatography-electrospray ionization tandem MS (LC-ESI MS/MS) analysis (files: peaklistA.mgf and peaklistB.mgf). Search parameters were set as follows: database: UniProt/SwissProt Release: 2010_12 (30-Nov-2010) (523,151 sequences; 184,678,199 residues), Taxonomy: Mammalia (65,293 sequences), Variable modifications: methionine oxidation and phosphorylation (serine, threonine or tyrosine), 2 trypsin missed cleavages, Mass tolerance for precursor mass: ± 0.5 Da and Mass tolerance for fragment masses: ± 0.6 Da. In most cases, an accuracy of ± 0.1–0.2 Da was found for both MS and MS/MS spectra. Finally, results were exported as mzIdentML format (files: identificationA.mzid and identificationB.mzid) directly from Mascot.
2) MIAPE MS and MSI Document Generation
Even though the mzIdentML file is the only mandatory format for the MIAPE web toolkit, the additional submission of the peak list file is strongly encouraged, because mzIdentML only contains the results and not the spectra that were searched. Thus, the inclusion of the mascot generic file (.mgf) format obtained from the source peak list together with the resulting mzIdentML identification file (i.e. peaklistA.mgf in addition to identificationA.mzid), will lead to a comprehensive report of the whole experiment containing data from both mass spectrometry and protein identification steps.
To create the MIAPE MSI report, and optionally the MS, the option “Standard to MIAPE” should be selected in the tool. The tool then requests the mzIdentML (and .mgf) files to be uploaded. After the initial uploading step, the MIAPE web toolkit validates the precision and suitability according to the MIAPE requirements. Subsequently, the corresponding MIAPE MS and MIAPE MSI documents are created. Because neither mzIdentML nor mgf files capture MS instrument settings, the user must complete some metadata for the MS report, describing the instrument and optionally data acquisition parameters (for further information see supplementary MIAPE files).
3) Exporting to PRIDE XML: Visualization and Submission
The PRIDE XML file is created in a single step by the user selecting the “MIAPE to Standard” option in the toolkit menu. Then, the user will choose which MIAPE MSI report will be exported. If a MIAPE MS instance was attached to a MIAPE MSI report during the previous step, both documents will be enclosed to create a complete and valid PRIDE XML file. If not, only the information related to the protein identification will be exported. This stage does not require additional input from the user.
Finally, submission and verification of the new PRIDE XML files (CID and ETD runs) can be carried out through two different and complementary approaches. First, a review can be performed using the PRIDEViewer tool (35), which provides a user-friendly graphical interface allowing to browse and visualize both metadata and data enclosed in the PRIDE XML files. Second, the experiments will be validated using the EBI PRIDE website, and submitted for public access (accession numbers 16437 and 16438; reviewer login: user: review75163, password YM#T7sTQ).
DISCUSSION
The ProtoRed MIAPE web toolkit is a free bioinformatics tool designed to cater to small, medium, or large proteomics projects. Its main characteristic is its ability to automatically translate and link data derived from proteomics experiments, using the current HUPO-PSI standards and MIAPE guidelines, saving significant effort and time for users. Moreover, the ProteoRed MIAPE web toolkit offers the capability to be included in third-party applications embedding it in the daily workflow for reporting experiments.
The main advantage of the toolkit is that it is designed for day to day use by laboratory scientists, i.e. it does not require complex setup procedures or local bioinformatics support. First, MIAPE reports will be automatically created as long as users have the appropriate input PSI XML files, such as mzIdentML, which can be exported directly from Mascot. Second, it provides permanent storage within the MIAPE repository (regardless of the source format from which it was generated) and allows the users to read the documents in a user-friendly manner (unlike the XML files) on the web interface. Third, the automated connection between MIAPE guidelines and PSI XML standards allows, for the first time, a useful aid for reporting proteomics experiments following journals requirements. Both the MIAPE reports and the data exchange formats of the same experiment will be automatically filled out, validated and connected, providing a valid syntax and an appropriate annotation of experimental data.
The example enclosed in this manuscript describes in detail the complete pipeline from a peak list to the automatic generation of MIAPE reports and the PRIDE XML, which is amenable to submission to the PRIDE central repository as recommended by the main proteomic journals. In addition, the connection of both XML schemas for protein and peptide identification (mzIdentML and PRIDE) provides a utility for converting mzIdentML to PRIDE XML which, to our knowledge, has not previously been demonstrated by any other tool. It thus provides users with a more comprehensive set of utilities to report their experiments, connecting some of the most accepted data standards and we hope will thus improve capabilities for data sharing in proteomics.
Additional Information: Database access
The software described in this article is accessible from http://www.proteored.org/MIAPE/
URLs to link direct to source data files from http://www.proteored.org/MIAPE/Data/)
Peaklists
CID peaklist (peaklistA.mgf): http://www.proteored.org/MIAPE/Data/PeakListA.mgf
ETD peaklist (peaklistB.mgf): http://www.proteored.org/MIAPE/Data/PeakListB.mgf
Identification files (mascot mzIdentML files)
CID id. (identificationA.mzid):http://www.proteored.org/MIAPE/Data/IdentificationA.mzid
ETD id. (identificationB.mzid): http://www.proteored.org/MIAPE/Data/IdentificationB.mzid
URLs to link direct to these records in the MIAPE Generator database
MIAPE MS from PeaklistA (CID): http://estrellapolar.cnb.csic.es/proteored/MIAPE/MIAPE_MS.asp?pmCodigoAcceso=706d3d17d13d344d5938b2c46dedd4262453f8abd579d5b42a1c269779c0b073&pmIDUsuario=70&pmId=2805
MIAPE MS from PeaklistB (ETD): http://estrellapolar.cnb.csic.es/proteored/MIAPE/MIAPE_MS.asp?pmCodigoAcceso=7846ae7be2a181d4eaf79fc57ffdca27d8548368574c2ca0d848cd3c553c3767&pmIDUsuario=70&pmId=2806
MIAPE MSI from IdentificationA.mzid (CID): http://estrellapolar.cnb.csic.es/proteored/MIAPE/MIAPE_MSI.asp?pmCodigoAcceso=fcba3b7b55bb7a3f909142b57f2e882706bce8d0a2039ffa2f4054f1f8ce5722&pmIDUsuario=70&pmId=2643
MIAPE MSI from IdentificationB.mzid (ETD): http://estrellapolar.cnb.csic.es/proteored/MIAPE/MIAPE_MSI.asp?pmCodigoAcceso=55ad9b2516dbc6b2e3aa3d74139a87d7276b2121e1745b66a0fd5afa04fbd97d&pmIDUsuario=70&pmId=2644
PRIDE database: http://www.ebi.ac.uk/pride
PRIDE accession numbers 16437 and 16438; login: user: review75163, password YM#T7sTQ
Footnotes
* This work was supported by the Spanish National Research Council (CSIC), the Spanish National Proteomics Institute (ProteoRed-ISC III) (grant number 2005X747_3) and by the EU FP7 ProteomeXchange (grant number 260558) grants.
The ProteoRed MIAPE web toolkit.
1 The abbreviations used are:
- PRIDE
- proteomics identifications database
- PSI
- proteomics standards initiative
- MIAPE
- minimum information about a proteomics experiment
- XML
- eXtensible Markup Language
- EMBL-EBI
- The European Molecular Biology Laboratory - The European Bioinformatics Institute
- CV
- controlled vocabulary
- ABRF
- The Association of Biomolecular Resource Facilities
- ETD
- electron transfer dissociation.
REFERENCES
- 1. Kaiser J. (2002) Proteomics. Public-private group maps out initiatives. Science 296, 827. [DOI] [PubMed] [Google Scholar]
- 2. Orchard S., Hermjakob H., Apweiler R. (2003) The proteomics standards initiative. Proteomics 3, 1374–1376 [DOI] [PubMed] [Google Scholar]
- 3. Orchard S., Kersey P., Hermjakob H., Apweiler R. (2003) The HUPO proteomics standards initiative meeting: towards common standards for exchanging proteomics data. Comp. Funct. Genomics 4, 16–19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Orchard S., Kersey P., Zhu W., Montecchi-Palazzi L., Hermjakob H., Apweiler R. (2003) Progress in establishing common standards for exchanging proteomics data: the second meeting of the HUPO proteomics standards initiative. Comp. Funct. Genomics 4, 203–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Orchard S., Hermjakob H., Binz P. A., Hoogland C., Taylor C. F., Zhu W., Julian R. K., Jr., Apweiler R. (2005) Further steps towards data standardisation: The Proteomic Standards Initiative HUPO 3(rd) annual congress. Beijing 25–27th October. Proteomics 5, 337–339 [DOI] [PubMed] [Google Scholar]
- 6. Orchard S., Hermjakob H., Taylor C. F., Potthast F., Jones P., Zhu W., Julian R. K., Jr., Apweiler R. (2005) Further steps in standardisation. Report of the second annual Proteomics Standards Initiative Spring Workshop (Siena, Italy 17–20th April 2005). Proteomics 5, 3552–3555 [DOI] [PubMed] [Google Scholar]
- 7. Orchard S., Hermjakob H., Taylor C., Binz P. A., Hoogland C., Julian R., Garavelli J. S., Aebersold R., Apweiler R. (2006) Workshop of the Human Proteome Organisation Proteomics Standards Initiative (HUPO-PSI) Geneva, September, 4–6, 2005 Proteomics 6, 738–741 [DOI] [PubMed] [Google Scholar]
- 8. Orchard S., Apweiler R., Barkovich R., Field D., Garavelli J. S., Horn D., Jones A., Jones P., Julian R., McNally R., Nerothin J., Paton N., Pizarro A., Seymour S., Taylor C., Wiemann S., Hermjakob H. (2006) Proteomics and Beyond: a report on the 3rd Annual Spring Workshop of the HUPO-PSI 21–23 April 2006, San Francisco, CA, U.S.A. Proteomics 6, 4439–4443 [DOI] [PubMed] [Google Scholar]
- 9. Orchard S., Taylor C. F., Jones P., Montechi-Palazzo L., Binz P. A., Jones A. R., Pizarro A., Julian R. K., Jr., Hermjakob H. (2007) Entering the implementation era: a report on the HUPO-PSI Fall workshop 25–27 September 2006, Washington DC, U.S.A. Proteomics 7, 337–339 [DOI] [PubMed] [Google Scholar]
- 10. Orchard S., Montechi-Palazzi L., Deutsch E. W., Binz P. A., Jones A. R., Paton N., Pizarro A., Creasy D. M., Wojcik J., Hermjakob H. (2007) Five years of progress in the Standardization of Proteomics Data 4th Annual Spring Workshop of the HUPO-Proteomics Standards Initiative April 23–25, 2007 Ecole Nationale Supérieure (ENS), Lyon, France. Proteomics 7, 3436–3440 [DOI] [PubMed] [Google Scholar]
- 11. Orchard S., Martens L., Tasman J., Binz P. A., Albar J. P., Hermjakob H. (2008) 6th HUPO Annual World Congress - Proteomics Standards Initiative Workshop 6–10 October 2007, Seoul, Korea Proteomics 8, 1331–1333 [DOI] [PubMed] [Google Scholar]
- 12. Orchard S., Albar J. P., Deutsch E. W., Binz P. A., Jones A. R., Creasy D., Hermjakob H. (2008) Annual spring meeting of the Proteomics Standards Initiative 23–25 April 2008, Toledo, Spain Proteomics 8, 4168–4172 [DOI] [PubMed] [Google Scholar]
- 13. Orchard S., Hoogland C., Bairoch A., Eisenacher M., Kraus H. J., Binz P. A. (2009) Managing the data explosion. A report on the HUPO-PSI Workshop. August 2008, Amsterdam, The Netherlands. Proteomics 9, 499–501 [DOI] [PubMed] [Google Scholar]
- 14. Orchard S., Deutsch E. W., Binz P. A., Jones A. R., Creasy D., Montechi-Palazzi L., Corthals G., Hermjakob H. (2009) Annual spring meeting of the Proteomics Standards Initiative. Proteomics 9, 4429–4432 [DOI] [PubMed] [Google Scholar]
- 15. Orchard S., Albar J. P., Deutsch E. W., Eisenacher M., Binz P. A., Hermjakob H. (2010) Implementing data standards. a report on the HUPOPSI workshop September 2009, Toronto, Canada. Proteomics 10, 1895–1898 [DOI] [PubMed] [Google Scholar]
- 16. Taylor C. F., Paton N. W., Lilley K. S., Binz P. A., Julian R. K., Jr., Jones A. R., Zhu W., Apweiler R., Aebersold R., Deutsch E. W., Dunn M. J., Heck A. J., Leitner A., Macht M., Mann M., Martens L., Neubert T. A., Patterson S. D., Ping P., Seymour S. L., Souda P., Tsugita A., Vandekerckhove J., Vondriska T. M., Whitelegge J. P., Wilkins M. R., Xenarios I., Yates J. R., 3rd, Hermjakob H. (2007) The minimum information about a proteomics experiment (MIAPE). Nat. Biotechnol. 25, 887–893 [DOI] [PubMed] [Google Scholar]
- 17. Gibson F., Anderson L., Babnigg G., Baker M., Berth M., Binz P. A., Borthwick A., Cash P., Day B. W., Friedman D. B., Garland D., Gutstein H. B., Hoogland C., Jones N. A., Khan A., Klose J., Lamond A. I., Lemkin P. F., Lilley K. S., Minden J., Morris N. J., Paton N. W., Pisano M. R., Prime J. E., Rabilloud T., Stead D. A., Taylor C. F., Voshol H., Wipat A., Jones A. R. (2008) Guidelines for reporting the use of gel electrophoresis in proteomics. Nat. Biotechnol. 26, 863–864 [DOI] [PubMed] [Google Scholar]
- 18. Hoogland C., O'Gorman M., Bogard P., Gibson F., Berth M., Cockell S. J., Ekefjärd A., Forsstrom-Olsson O., Kapferer A., Nilsson M., Martínez-Bartolomé S., Albar J. P., Echevarría-Zomeño S., Martínez-Gomariz M., Joets J., Binz P. A., Taylor C. F., Dowsey A., Jones A. R. (2010) Guidelines for reporting the use of gel image informatics in proteomics. Nat. Biotechnol. 28, 655–656 [DOI] [PubMed] [Google Scholar]
- 19. Taylor C. F., Binz P. A., Aebersold R., Affolter M., Barkovich R., Deutsch E. W., Horn D. M., Hühmer A., Kussmann M., Lilley K., Macht M., Mann M., Müller D., Neubert T. A., Nickson J., Patterson S. D., Raso R., Resing K., Seymour S. L., Tsugita A., Xenarios I., Zeng R., Julian R. K., Jr. (2008) Guidelines for reporting the use of mass spectrometry in proteomics. Nat. Biotechnol. 26, 860–861 [DOI] [PubMed] [Google Scholar]
- 20. Binz P. A., Barkovich R., Beavis R. C., Creasy D., Horn D. M., Julian R. K., Jr., Seymour S. L., Taylor C. F., Vandenbrouck Y. (2008) Guidelines for reporting the use of mass spectrometry informatics in proteomics. Nat. Biotechnol. 26, 862. [DOI] [PubMed] [Google Scholar]
- 21. Jones A. R., Carroll K., Knight D., Maclellan K., Domann P. J., Legido-Quigley C., Huang L., Smallshaw L., Mirzaei H., Shofstahl J., Paton N. W. (2010) Guidelines for reporting the use of column chromatography in proteomics. Nat. Biotechnol. 28, 654. [DOI] [PubMed] [Google Scholar]
- 22. Domann P. J., Akashi S., Barbas C., Huang L., Lau W., Legido-Quigley C., McClean S., Neususs C., Perrett D., Quaglia M., Rapp E., Smallshaw L., Smith N. W., Smyth W. F., Taylor C. F. (2010) Guidelines for reporting the use of capillary electrophoresis in proteomics. Nat. Biotechnol. 28, 654–655 [DOI] [PubMed] [Google Scholar]
- 23. Orchard S., Ping P. (2009) HUPO World Congress Publication Committee meeting. August 2008, Amsterdam, The Netherlands. Proteomics, 9, 502–503 [DOI] [PubMed] [Google Scholar]
- 24. Orchard S., Binz P. A., Hermjakob H. (2009) Second Joint HUPO publication and Proteomics Standards Initiative workshop. Proteomics 9, 4426–4428 [DOI] [PubMed] [Google Scholar]
- 25. Martens L., Hermjakob H., Jones P., Adamski M., Taylor C., States D., Gevaert K., Vandekerckhove J., Apweiler R. (2005) PRIDE: The PRoteomics IDEntifications database. Proteomics 5, 3537–3545 [DOI] [PubMed] [Google Scholar]
- 26. Jones P., Côté R. G., Martens L., Quinn A. F., Taylor C. F., Derache W., Hermjakob H., Apweiler R. (2006) PRIDE: a public repository of protein and peptide identifications for the proteomics community. Nucleic Acids Res. 34, D659–D663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Vizcaíno J. A., Côté R., Reisinger F., Foster J. M., Mueller M., Rameseder J., Hermjakob H., Martens L. (2009) A guide to the Proteomics Identifications Database proteomics data repository. Proteomics 9, 4276–4283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Paradela A., Escuredo P. R., Albar J. P. (2006) Geographical focus. Proteomics initiatives in Spain: ProteoRed. Proteomics 6, 73–76 [DOI] [PubMed] [Google Scholar]
- 29. Martinez-Bartolome S., Blanco F., Albar J. P. (2010) Relevance of proteomics standards for the ProteoRed Spanish organization. J Proteomics 73, 1061–1066 [DOI] [PubMed] [Google Scholar]
- 30. Martínez-Bartolomé S., Medina-Aunon J. A., Jones A. R., Albar J. P. (2010) Semi-automatic tool to describe, store and compare proteomics experiments based on MIAPE compliant reports. Proteomics 10, 1256–1260 [DOI] [PubMed] [Google Scholar]
- 31. Gibson F., Hoogland C., Martinez-Bartolomé S., Medina-Aunon J. A., Albar J. P., Babnigg G., Wipat A., Hermjakob H., Almeida J. S., Stanislaus R., Paton N. W., Jones A. R. (2010) The gel electrophoresis markup language (GelML) from the Proteomics Standards Initiative. Proteomics 10, 3073–3081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Martens L., Chambers M., Sturm M., Kessner D., Levander F., Shofstahl J., Tang W. H., Rompp A., Neumann S., Pizarro A. D., Montecchi-Palazzi L., Tasman N., Coleman M., Reisinger F., Souda P., Hermjakob H., Binz P. A., Deutsch E. W. (2011) mzML–a community standard for mass spectrometry data. Mol. Cell. Proteomics 10, R110.000133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Eisenacher M. (2011) mzIdentML: An Open Community-Built Standard Format for the Results of Proteomics Spectrum Identification Algorithms. Methods Mol Biol. 696, 161–177 [DOI] [PubMed] [Google Scholar]
- 34. Montecchi-Palazzi L., Kerrien S., Reisinger F., Aranda B., Jones A. R., Martens L., Hermjakob H. (2009) The PSI semantic validator: A framework to check MIAPE compliance of proteomics data. Proteomics 9, 5112–5119 [DOI] [PubMed] [Google Scholar]
- 35. Medina-Aunon J. A., Carazo J. M., Albar J. P. (2011) PRIDEViewer: A novel user-friendly interface to visualize PRIDE XML files. Proteomics 11, 334–337 [DOI] [PubMed] [Google Scholar]