Abstract
The study of chronic brain diseases including Alzheimer's disease in patients is typically limited to brain imaging or psychometric testing. Given the epidemic rise and insufficient knowledge about pathological pathways in sporadic Alzheimer's disease, new tools are required to identify the molecular changes underlying this disease. We hypothesize that levels of specific secreted cellular signaling proteins in cerebrospinal fluid or plasma correlate with pathological changes in the Alzheimer's disease brain and can thus be used to discover signaling pathways altered in the disease. Here we measured 91 proteins of this subset of the cellular communication proteome in plasma or cerebrospinal fluid in patients with Alzheimer's disease and cognitively normal controls to mathematically model disease-specific molecular traits. We found small numbers of signaling proteins that were able to model key pathological markers of Alzheimer's disease, including levels of cerebrospinal fluid β-amyloid and tau, and classify disease in independent samples. Several of these factors had previously been implicated in Alzheimer's disease supporting the validity of our approach. Our study also points to proteins which were previously unknown to be associated with Alzheimer's disease thereby implicating novel signaling pathways in this disorder.
The brain regulates key biological processes throughout the organism by releasing molecules into the blood and cerebrospinal fluid (CSF)1, and systemic factors in turn can induce cognitive and behavioral changes in animals and humans (1, 2). Thus, brain and periphery are interconnected not only on a cellular level but also in a complex molecular network of soluble communication factors.
Alzheimer's disease (AD) is the most common form of dementia affecting an estimated 5.4 million individuals in the US (3). The cause of AD is not known but genetic and pathological studies point to a key role for cerebral β-amyloid (Aβ) peptide and hyperphosphorylated tau in the disease. Levels of soluble Aβ42 in CSF decrease in AD in parallel to its deposition into amyloid plaques in the brain parenchyma, whereas tau increases as the disease progresses making these proteins good biomarkers of AD (4–6). Moreover, ratios between the concentrations of tau phosphorylated at threonine 181 (p-tau181) and total tau (t-tau) or Aβ42 are predictive of future development of AD in patients with mild cognitive impairment (MCI) (7–9) and cognitive decline (10–12) or brain atrophy in nondemented older adults (13). Despite the usefulness of these CSF markers in the diagnosis of AD patients (4, 5), they provide little information on biological mechanisms involved in this disorder.
Some members of the molecular network, which include cytokines, chemokines, or growth factors, have been reported to have different expression levels in patients with neurological diseases compared with those found in healthy controls [reviewed in (14)]. We and others have measured in plasma or CSF from AD patients and controls large numbers of communication factors, which we collectively called the communicome, and by statistically comparing levels in AD patients versus levels in controls we identified small groups of factors that could form a signature for the disease (15–19). Together, these findings implicate abnormal transcriptional regulation of peripheral leukocytes (20, 21) or neuroinflammatory pathways in AD (22). Technical problems and small sample sizes, however, have made it difficult to identify proteins, which are predictive of AD across various centers and studies (18) and the two-class analysis approach (AD patients versus a control group) is appealing for finding differences among groups but provides little insight into the role of these markers in AD.
Here we propose an alternative analytical approach in which levels of communication factors in CSF and plasma are used to mathematically model levels of Aβ42 and tau. We do this based on the hypothesis that the systemic network of the extracellular signaling proteome is linked with the levels of CSF markers of AD pathology and indicators of disease progression. The findings could help in identifying proteins and biological pathways hitherto unknown to be involved in AD.
EXPERIMENTAL PROCEDURES
Subjects and APOE Genotyping
AD patients and healthy nondemented controls (NDC) (supplemental Table S1) were recruited for multicenter studies that aim to identify molecular biomarkers for AD in blood and CSF. Subjects were chosen based on standardized inclusion and exclusion criteria (see Supplemental Methods). Written informed consent was obtained from all subjects, or assent from subjects and consent from caregivers in the case of subjects with significant impaired decisional capacity, in accordance with institutional ethics committee review boards at each participating institution. APOE genotyping was performed by the restriction digest method (23).
Collection of CSF and Plasma
CSF and plasma samples were generated by standard procedures as described (19, 24). Briefly, for the majority of patients lumbar puncture and blood draw was conducted between 9am and 11am after an overnight fast. After blood draw into K2-EDTA-coated lavender top vacutainers, blood was kept on ice, spun at 1000 × g in a refrigerated centrifuge within 1 h of draw, aliquoted, and frozen on dry ice. All samples were stored at −80 °C until use with no previous thaw cycle.
Measurement of Analytes in CSF and Plasma
We measured CSF levels of Aβ42, t-tau, and p-tau181 by Luminex xMAP method (AlzBio3, Innogenetics, Ghent, Belgium) a bead-based sandwich immunoassay. Plasma samples were shipped from Seattle to Palo Alto where they were thawed and aliquoted. For the multi-analyte profiling plasma aliquots were sent to Rules-Based Medicine (RBM, Austin, Texas) and measured in two big batches of samples from NDC and AD patients. CSF was sent directly to RBM and analyzed in one experiment. All samples were analyzed blinded by robotic sample handlers in a bead-based multiplex sandwich immunoassay (Luminex bead analyzer), according to Clinical Laboratory Standards Institute (CLSI) guidelines and rigorous assessment of fundamental assay parameters (http://www.rulesbasedmedicine.com/quality-policy/data-quality/). Out of 90 analytes measured by RBM IFN-γ, IGF-1, IL-1α, and lipoprotein-a were not detectable in any of the CSF and plasma samples. Additional analytes were excluded if they were detectable in less than 10% of samples. This resulted in 73 detectable proteins in each, plasma and CSF, with 60 proteins overlapping between the two biological fluids (supplemental Table S2). Soares and colleagues reported a similar sensitivity of this platform (18). As a quality control we remeasured 35 plasma samples at RBM 17 months after the initial plasma measurements. 86% of the detectable analytes correlated between the two measurements with a Pearson correlation coefficient R ≥ 0.7 (52% R ≥ 0.9, 34% r = 0.7–0.9) and 14% with R < 0.7. Lower R-values were because of changes in assay settings in the second experiment, which rendered most values for these analytes below least detectable dose or undetectable. Nevertheless, these data generated at two independent time points made us confident, that RBM produces highly reproducible data. Also, analytes with low correlation coefficient were deemed suitable for analysis because in the first experiment they were well above detection limit. Only data from the first experiment were subsequently used for the analysis. In addition, plasma levels of MCSF were quantified separately by Quantikine ELISA (R&D Systems, Minneapolis, MN) and were detectable in all plasma samples.
Statistical Analysis
Analysis was done in Prism 5 and R [http://www.r-project.org/, (25)]. The entire plasma sample set contained 78 AD patients and 118 NDC (supplemental Table S1) and we measured levels of tau, p-tau181, and Aβ42 in 72 AD patients and 112 controls. We standardized the values to z-scores by subtracting the mean and dividing by the standard deviation (26). Measurements below lowest detectable protein concentration were imputed conservatively with the lowest available value of an analyte. Values missing at random (e.g. analyte not measured because of insufficient amounts of sample) were imputed with 0 (which is the mean of the available observations) after standardization. The entire plasma set was randomly split into a training set (79 NDC, 52 AD) and a test set (39 NDC, 26 AD), whereby we insisted on the same AD versus NDC frequency distribution in the training and test as in the entire dataset and that all subjects in the training set have measurements for Aβ42 and tau. The CSF sample set was analyzed without further splitting. For the CSF sample set and the plasma training set high-dimensional linear regression models for continuous endpoints were computed with an elastic net penalty [http://cran.r-project.org/web/packages/elasticnet/index.html, (27)] using the add-on package elasticnet. We used the standardized levels of t-tau, p-tau181, Aβ42, or the ratios of Aβ42/t-tau, or Aβ42/p-tau181, respectively, as the continuous response variables in the linear regression equation and the standardized values of CSF or plasma protein levels together with sex and risk factors for AD APOE genotype and age as the explanatory variables. The penalty parameter was chosen via 10-fold cross validation minimizing prediction error. For fixed alpha, the regularization parameter lambda is varied from lambda_max, the smallest lambda such that all coefficients are estimated without constraint, down to a very small lambda_min; lambda = 0 renders the algorithm unstable (28). We use the built-in specification of lambdas that automatically chooses a suitable number of lambdas in the interval [lambda_min, lambda_max]. For alpha we used {0.01, 0.2, 0.4, 0.6, 0.8, 1}. Alpha = 1 specifies a pure Lasso penalty. For each response a separate model was calculated (Table I, Model C1–5 in CSF, P6–8 in plasma) and variables that fitted best into the corresponding model (selected predictors) are listed in order of decreasing association to the response variable (absolute size of regression coefficient, RC). To test the diagnostic utility of the selected predictors in both the training and the test set, we used the selected predictors to model the binary end point AD versus NDC (based on clinical diagnosis) and calculated a ROC curve where the linear predictor from the logistic regression was the continuous variable. This was compared with the ROC curve that is based on models with t-tau, p-tau181p, Aβ42, and their ratios Aβ42/t-tau, and Aβ42/p-tau181p, respectively. Comparison of the logistic regression analyses was done via ROC with 95% Wald-type confidence intervals for AUC on logit-scale were computed using the approach of Hanley and McNeil (29, 30). To assess the risk associated with APOE genotype we computed an ordinary logistic regression model with dependent variable AD versus non-AD and explanatory variable APOE4 status (ε4 versus non-ε4). In addition to the odds ratio we report a 95% profile likelihood confidence interval.
Table I. List of variables selected in the Elastic net models. All values were standardized and penalized linear regression models were calculated with the E-net method. Response variables: t-tau, p-tau181, Aβ42, or the ratios of Aβ42/t-tau, or Aβ42/p-tau181. Explanatory variables: CSF or plasma protein levels together with sex and risk factors for AD APOE genotype and age. For each response a separate model was calculated (Model C1–5 in CSF, P6–8 in plasma) and variables that fitted best into the corresponding model are listed in order of decreasing absolute size of the estimated regression coefficient (RC, indicated in parentheses). RC lies between −0.01 and 0 for variables without a coefficient. Since all variables had been standardized, the RC serves as a measure of the strength for the association between an explanatory variable and its corresponding response variable: the larger RC in absolute value, the higher the association to the response. A RC close to 0 points to a low degree of association between the corresponding variable and the response. For protein nomenclature see supplemental Table S2.
a Based on the CSF set with 43 subjects, AD n = 25, NDC n = 18 subjects.
b Based on the training set with 131 subjects, AD n = 52, NDC n = 79 subjects.
c In model C2 the E-net chose 26 predictors for modeling p-tau181p with CSF proteins and subjects' characteristics. To avoid overfitting for this model we limited the number of selected variables to eight. The additional 18 variables in order of how they were selected by the E-net here in an extended list: CRP, age, VEGF, SCF, CA19–9, IL-10, IL-5, C3, haptoglobin, TBG, calcitonin, GST, CCL2, α-fetoprotein, PAPP-A, ApoA1, CCL3, IgM.
Connectivity Network Diagrams
First, we calculated the Spearman rank correlation coefficients (RS) among all analytes in separate correlation matrices for AD patients and NDC, respectively. Confidence intervals for RS between analytes in plasma and CSF were calculated using Fisher's z-transformation and t-quantiles. The RS is an indicator of connectivity of two analytes and RS ranges [0.4,1] and [–1,–0.4] were used to draw connectivity network diagrams for selected analytes. To quantify the strength of integration of an analyte in the connectivity network we calculated for each analyte an interaction score, which is the sum of the squared values of all RS in the range of [–1,1] between this analyte and all analytes in the connectivity diagram.
RESULTS
Two Thirds of Measured Communication Factors are Detectable in CSF
We measured the concentrations of 91 secreted proteins in plasma from 78 AD patients and 118 cognitively normal elderly NDCs; we also measured 90 proteins in CSF of 25 AD patients and 18 NDC (supplemental Table S1 and S2). These proteins represent some of the best-studied cytokines, chemokines, growth factors, and acute phase response proteins and were selected based on availability in multiplex sandwich immunoassay. Seventy-four proteins were detectable in plasma and 73 in CSF, with 60 proteins overlapping between the two biological fluids. Protein concentrations were generally lower in CSF than in plasma (between 5- and 8000-fold) with a few interesting exceptions. β2-microglobulin, CD40, CCL11/eotaxin, tissue inhibitor of metalloproteinase (TIMP)-1, or tissue factor (TF) were measured at similar concentrations in both fluids and levels of fatty acid binding protein 3 (FABP3), CCL2/monocyte chemotactic protein-1 (MCP-1), CXCL8/interleukin-8, and vascular endothelial growth factor (VEGF) were 1.5- to 10-times higher in CSF than in plasma. Interestingly, systemic β2-microglobulin, CCL11, and CCL2 were recently identified in an unbiased screen as key aging factors with a possible role in adult neurogenesis in mice (31). These observations suggest that a majority of the communicome factors, many known classically for their role in peripheral immune function, developmental processes, or systemic diseases, may have a function in the CNS as well.
CSF Communication Factors Can Model CSF Aβ and Tau And Hence Discriminate Between AD and NDC
Consistent with published reports (9, 11, 32) CSF levels of Aβ42, t-tau, p-tau181 alone or in combination were strong classifiers of AD in our data set (area under the curve (AUC) 0.75–0.90 for various models; Table II). If levels of communication factors are indeed associated with the disease process (discussed in (6)) they should be able to model the pathological changes characteristic of AD (Fig. 1). We thus modeled levels of CSF Aβ42, t-tau, p-tau181, or ratios Aβ42/t-tau or Aβ42/p-tau181 with all measurements of communication factors in CSF (73 detectable factors) using the linear regression-modeling tool Elastic net (E-net, (27)). We adjusted for sex and AD risk factors age and APOE4 status by including them as variables in the analysis. In E-net models C1–C5 a total of 12 CSF proteins were selected to be associated with the 5 pathological markers of AD. Interestingly, FABP3 and creatine kinase-muscle/brain (CK-MB), showed a consistently strong association in all models involving tau; (indicated by shades of green in Table I) endothelin-1, and interleukin-13 (IL-13) were also required to model p-tau181. IL-7 was the only factor selected to model Aβ42 but with a regression coefficient of nearly zero, implying that its influence on the response variable in the regression model is small.
Table II. Classification of AD and nondemented controls. Compare AUC calculated using variables selected in elastic net models (Table I) with AUC calculated using established markers for pathology. See also ROC curves in Figures 2 and 3.
| Established biomarker model | CSF sample set (n = 43) |
||
|---|---|---|---|
| AUC calculated based on established biomarkers in CSFa | E-net model | AUC calculated based on CSF communicome, age, and APOE4 statusb | |
| t-tau | 0.86 (0.71–0.94) | C1 | 0.75 (0.56–0.88) |
| p-tau181 | 0.90 (0.74–0.97) | C2 | 0.88 (0.70–0.96) |
| Aβ1–42 | 0.75 (0.58–0.87) | C3 | n.a. |
| Aβ1–42/t-tau | 0.84 (0.69–0.93) | C4 | 0.91 (0.77–0.97) |
| Aβ1–42/p-tau181 | 0.86 (0.70–0.94) | C5 | 0.93 (0.78–0.98) |
| Established biomarker model | Training set (n = 131) |
||
|---|---|---|---|
| AUC calculated based on established biomarkers in CSFa | E-net model | AUC calculated based on plasma communicome, age, and APOE4 statusb | |
| t-tau | 0.80 (0.71–0.87) | P6 | 0.86 (0.79–0.92) |
| p-tau181 | 0.82 (0.73–0.89) | P7 | 0.86 (0.78–0.92) |
| Aβ1–42 | 0.81 (0.72–0.87) | P8 | 0.85 (0.76–0.90) |
| Aβ1–42/t-tau | 0.85 (0.78–0.91) | P9 | 0.86 (0.78–0.92) |
| Aβ1–42/p-tau181 | 0.84 (0.76–0.90) | P10 | 0.84 (0.76–0.90) |
| Established biomarker model | Test set (n = 65) |
||
|---|---|---|---|
| AUC calculated based on established biomarkers in CSFc | Validation of variables selected in E-net model | AUC calculated based on plasma communicome, age, and APOE4 status selected in the training setd | |
| t-tau | 0.80 (0.630–0.907) | P6 | 0.77 (0.63–0.87) |
| p-tau181 | 0.81 (0.648–0.909) | P7 | 0.76 (0.61–0.86) |
| Aβ1–42 | 0.84 (0.704–0.925) | P8 | 0.75 (0.60–0.85) |
| Aβ1–42/t-tau | 0.86 (0.715–0.938) | P9 | 0.77 (0.63–0.87) |
| Aβ1–42/p-tau181 | 0.86 (0.725–0.934) | P10 | 0.74 (0.60–0.85) |
a AUC calculated based on the levels of established pathological CSF biomarkers t-tau, p-tau181, Aβ1–42 and their ratios in subjects used for CSF or plasma models, respectively.
b AUC (in bold) calculated based on communicome, age and APOE4 status and their corresponding regression coefficients as selected in Elastic net (E-net) models C1–5 (CSF) and P6–10 (plasma) in subjects used for CSF or plasma models, respectively. Note that for Aβ1–42 only IL-7 with a regression coefficient close to 0 was selected in the E-net model C3. Thus, no ROC/AUC could be calculated (n.a., not applicable).
c These AUC results are based on 53 subjects because 6 controls and 6 AD patients had no measurement of Aβ and tau in CSF.
d Variables selected by the E-net models P6–10 in the training set were validated in an independent test set and AUC was calculated for all 65 subjects (italicized and bold).
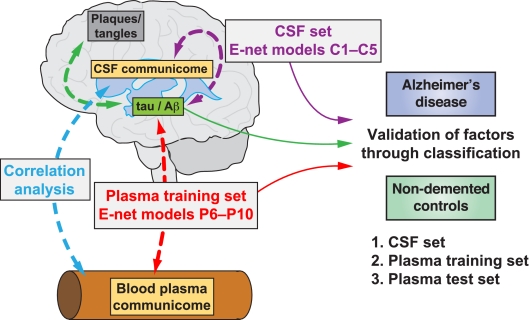
Fig. 1.
Modeling of pathological markers in CSF and classification of AD based on communication factors in CSF and plasma. Accumulation of plaques and tangles in the AD brain is associated with decreased levels of Aβ and increased levels of tau in CSF, respectively (green dashed double arrow). Relative levels of these proteins alone or their ratios can be used to classify AD and NDC (green solid arrow). In Elastic net (E-net) regression models C1–C5 soluble communication factors in CSF (CSF communicome; see text for details) are used to model tau and Aβ levels or their ratios (dashed purple double arrow) and subsequently to classify AD and NDC (solid purple line). In E-net models P6-P10 soluble communication factors in plasma (plasma communicome) derived from the training set are used to model tau and Aβ levels or their ratios (dashed red double arrow) and subsequently to classify AD and NDC (solid red line). These variables were validated by classifying AD and NDC in an independent test set. In a separate analysis correlations are studied between CSF communicome proteins and corresponding plasma communicome proteins in AD patients and NDCs (dashed blue double arrow).
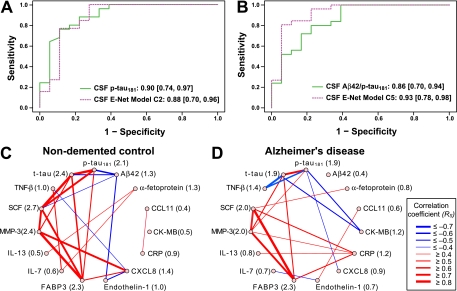
The communication factors selected in E-net Models C2 and C5 (the latter in conjunction with APOE4 status) were capable of independently classifying AD and NDC similar to or slightly better than p-tau181 or Aβ42/p-tau181 levels measured in these patients (Fig 2A, B and Table II). This validates the chosen communication proteins and suggests that they are biologically related to the abundance of p-tau181 and Aβ42 in CSF, the pathological hallmarks of AD.
Fig. 2.
Pathology associated CSF communicome models can classify AD and NDC and the connectivity network of the proteins in these models is altered in AD patients. A, B, CSF pathological markers or CSF Elastic net (E-net) models C2 and C5 were used to calculate ROC curves. The color-coding is based on Fig. 1 and the numbers indicate the AUC values and 95% C.I. (listed also in Table II). A, CSF samples from AD patients and NDC were classified based on levels of CSF p-tau181 (solid green line) or levels of CSF communicome proteins (FABP3, CCL11, endothelin-1, CK-MB, IL-13, CXCL8) and APOE4 status selected in model C2 (dashed purple line). B, CSF samples from AD patients and NDC were classified based on levels of CSF Aβ42/p-tau181 (solid green line) or levels of CSF communicome proteins (FABP3, IL-13, SCF, CK-MB, α-fetoprotein, endothelin-1, CRP) and APOE4 status selected in model C5 (dashed purple line). C, D, Network diagrams illustrating connectivity between CSF pathological markers and CSF communication factors selected in models C1–C5. The connectivity between two proteins is expressed as Spearman rank correlation coefficient RS calculated from all measurements for these two proteins in CSF samples of nondemented controls (C) or AD patients (D), respectively. The correlations are visualized in a network with different strokes and colors depending on strength and type of connectivity among proteins. Red strokes RS ≥ 0.4, blue strokes RS ≤ –0.4. In parentheses next to the name of the analyte is the interaction score, which is the sum of the squared values of all RS among all analytes in the connectivity diagram. Note the considerable change of the interaction scores between communication factors in AD patients (D) compared with NDC (C).
Consistent with the E-net analysis, we observed in the connectivity network diagram of the overall 12 selected CSF communication factors strong, positive Spearman rank correlations (RS) between t-tau or p-tau181 and several communication factors in healthy individuals but Aβ42 was only weakly integrated into this network (Fig 2C). Other factors including CXCL8, FABP3, matrix metalloproteinase 3 (MMP-3), and stem cell factor (SCF) show strong correlations with each other in the network (RS ≥ 0.8). Although some of the relationships are maintained in AD (FABP3, MMP-3, SCF), the strong correlations with tau are missing; instead, negative relationships among tau, CK-MB, and TNF-β appear, and C-reactive protein (CRP) becomes a network hub with several positive interactions (Fig 2D). This change in connectivity between pathological and communicome factors in AD versus NDC is reflected by differences in the interaction scores.
Plasma Communication Factors Can Model CSF Tau and Aβ and Hence Discriminate Between AD and NDC
We next asked whether blood-derived proteins have a relationship to CSF pathological markers of AD and may thus be related to the disease itself (Fig. 1). We again used E-net to model Aβ42, t-tau, p-tau181, or ratios Aβ42/t-tau or Aβ42/p-tau181 but this time based on values for 74 detectable plasma communication proteins (supplemental Table S2) in a training set of 52 AD patients and 79 NDC (supplemental Table S1), and we adjusted for sex, age and APOE4 status. A total of 22 plasma communication factors were selected to best model pathological CSF markers in E-net Models P6-P10 (Table I). Notably, CK-MB and tumor necrosis factor-β (TNF-β) overlapped with proteins selected in the CSF models. Among the 22 proteins, macrophage-colony stimulating factor (MCSF) showed the strongest and most consistent association with all 5 pathological markers in CSF although APOE4 status and age were key drivers in two of these models (P8, P9). Other prominent factors were selected in four models (CK-MB and granulocyte-colony stimulating factor (G-CSF)) or in three models (TNF-RII, IL-3, and PAI-1) (indicated by shades of orange in Table I).
To assess the potential diagnostic utility of the selected plasma proteins we calculated ROC curves for the training set. Several models outperformed the pathological CSF markers in classifying NDC and AD, and this effect was not simply explained by APOE4 status and age (Table II and Fig 3A, C). Moreover, plasma communication factors in combination with APOE4 status and age were as effective in classifying AD as CSF measurements of Aβ42/tau and Aβ42/p-tau181 (Table II). We validated the results of these models in an independent test set with 65 subjects (supplemental Table S1), producing a classification (AUC 0.74–0.77), which was clearly stronger than with APOE4 status and age alone (AUC 0.63, Fig 3B, D).
Fig. 3.
Pathology associated plasma communicome models that efficiently classify AD and NDC are validated in an independent test set. ROC curves for the plasma training and a test set were calculated with CSF pathological markers (solid green lines), AD risk factors APOE4 status and age (solid black lines), or plasma Elastic net (E-net) models P2 and P5 (dashed red lines), respectively. The color-coding is based on Fig. 1 and the numbers indicate the AUC values and 95% C.I. (listed also in Table II). A, AD patients and NDC in the training set were classified based on levels of CSF p-tau181 or the variables selected to be associated with p-tau181 in e-net model P2: plasma communicome proteins (MCSF, CD40L, G-CSF, adiponectin, CCL2, CK-MB, TNF-β, β2-microglobulin, TNF RII, IL-4, IL-3, IL-18, PAI-1, Apo CIII, TBG), APOE4 status, and age. B, The variables selected in the training set E-net model P2 are validated in an independent test set of AD and NDC and compared with the ROC curves that were calculated based either on levels of CSF p-tau181 or APOE4 status and age of 53 of the 65 subjects in the test set; measurements of Aβ42 and tau were not done for six NDC and six AD in the test set. C, AD patients and NDC in the training set were classified based on levels of CSF Aβ42/p-tau181 or the variables selected to be associated with p-tau181 in E-net model P10: plasma communicome proteins (MCSF, G-CSF, GH, TPO), APOE4 status, and age. D, The variables selected in the training set E-net model P10 are validated in an independent test set of AD and NDC and compared with the ROC curves that were calculated based either on levels of CSF Aβ42/p-tau181 or APOE4 status and age of the subjects in the test set. See connectivity network in Fig. 4 for illustration of the changes in interactions among communication factors in AD patients compared with controls.
Consistent with the E-net analysis, connectivity network diagrams illustrated a considerable complexity of relationships among all 22 plasma communication factors selected by E-net models P6-P10 and tau and Aβ in NDC (Fig 4A). Interestingly, this network becomes seemingly more complex in AD subjects where immune and inflammatory factors become important network hubs (Fig 4B). For example, β2-microglobulin shows strong correlations with inflammatory factors TIMP-1, PAI-1 and members of the TNF superfamily (CD40L, TNF RII, and TNF-β) in the AD network.
Fig. 4.
Plasma predictors of pathological markers point to changes in the communicome network in plasma of AD patients. A, B, Connectivity diagrams of pathological markers in CSF and communication factors in plasma, which were chosen in Elastic net regression models for these markers. The relationship between two proteins is expressed as Spearman rank correlation coefficient RS calculated from all measurements for these two proteins in nondemented controls (A) or AD patients (B), respectively. The correlations are visualized in a network with different strokes and colors depending on strength and type of a relationship among proteins. Red strokes RS ≥ 0.4, blue strokes RS ≤ −0.4. In parentheses next to the name of the analyte is the interaction score, which is the sum of the squared values of all RS among all analytes in the connectivity diagram. Note the considerable difference in connectivity among communication factors in AD patients compared with NDC.
Correlations and Connectivity Between Communication Factors in CSF and Plasma are Altered in AD
To assess whether levels of a given protein factor in the CSF are related to its levels in plasma we compared CSF and plasma concentrations in 43 subjects using Spearman rank correlation. In healthy individuals, levels of 10 out of the 60 proteins detectable in CSF and plasma (supplemental Table S2) correlated considerably between the two fluids (Rs ≥ 0.5; Fig 5A). Strikingly, 6 of these 10 correlations were much weaker in AD (FABP3, IL-10, TIMP-1, CCL4 (macrophage inflammatory protein-1β, MIP-1β), myoglobin, and TSH. Conversely, levels of intercellular adhesion molecule-1 (ICAM-1) and CCL5 did not correlate between CSF and plasma in NDCs but correlated highly in AD patients (Fig 5A and supplemental Fig. S1). Similarly, it becomes apparent that the connectivity network of factors selected in models C1–5 and P6–10 (Fig 5B, C) are overall very different in AD and NDC, and this is supported by the change in interaction scores for each factor. Many of the factors appearing as well-connected network hubs in healthy individuals (e.g. insulin, α-fetoprotein, FABP3, or SCF) have very low connectivity in AD patients. In turn, MCSF and CRP have high positive correlations in AD but negative or fewer correlations in NDC. Together, these analyses across the BBB support the notion that the disease process in AD is accompanied by changes in secreted signaling proteins in CSF and plasma.
Fig. 5.
Correlations and connectivity between communication factors in CSF and plasma are different in AD patients compared with NDC. A, Spearman rank correlations (correlation coefficient RS) between CSF and plasma in nondemented controls (green bars) and AD patients (blue bars). Proteins with RS ≥ 0.5 in at least one group are shown. RS ≤ –0.5 were not observed. Note that levels of CCL5 and ICAM-1 correlate much stronger in AD than in NDC across the BBB and that ApoH and CCL2 reach RS = 0.5 in AD but not in NDC. B, C, Connectivity diagrams illustrating correlations between CSF communication factors selected in models C1–C5 and plasma communication factors selected in models P6–P10. In parentheses are the interaction scores. See legend to Fig. 2 for more details on calculations. Note the considerable lack and difference in connectivity between communication factors in AD patients (C) compared with NDC (B). See also supplemental Fig. S1. BBB, blood-brain barrier.
DISCUSSION
The study of genetic forms of AD has greatly advanced our knowledge about disease processes linked to the disease but the cause of sporadic forms of AD affecting >95% of all patients is unknown. One reason may be that studying this disease is typically limited to imaging or psychometric testing of patients. Also, existing in vitro or transgenic animal models based on genetic forms of AD provide limited unbiased information on molecular or cellular networks involved in the pathogenesis of sporadic AD. Here we analyzed AD in a more continuous and complex fashion based on the hypothesis that the systemic network of the extracellular signaling proteome is linked with the pathological traits in AD. To test this hypothesis we modeled known and accepted pathological indicators of the disease (Aβ42, t-tau, p-tau181, or ratios Aβ42/t-tau or Aβ42/p-tau181) (4, 5) with other CSF or plasma proteins (Fig. 1 and supplemental Table S2) using the penalized linear regression method E-net (27). We also correlated levels of proteins measured in CSF and plasma with each other and analyzed how the connectivity or “network” of correlations among these factors changes with disease (Fig. 1). The current study attempts therefore to provide methods for discovery of potentially diagnostic or biologically relevant proteins in AD.
We discovered several small groups of proteins in CSF which could be used to model or predict levels of pathological markers and classify AD and non-demented controls with similar or better diagnostic utility than the CSF pathological markers themselves (Fig 2A, B and Table II). These proteins alone or in combination may therefore be associated with the disease process either in a direct or indirect way [see also discussion on fluid biomarkers in relation to AD pathology in (6)], and future experimental studies will need to test this. Consistent with their ability to model CSF tau and Aβ, several of the selected CSF communicome proteins correlate with these markers and it is striking that most correlations are prominently changed in AD patients (Fig 2C, D). This is particularly obvious for correlations between FABP3 or SCF, and pathological markers. FABP3, which shows many strong correlations with other proteins and functions as a network hub, is a cytosolic protein involved in cellular fatty acid uptake, transport, and metabolism (33). It is highly expressed in the adult human brain (34) and has previously been suggested to be a peripheral marker for mild traumatic brain injury and stroke (34–36). In support of a possible change in the half-life of FABP3 in brain and the systemic environment in disease, we observed a moderate positive correlation between CSF and blood FABP3 levels in NDC but a negative correlation in AD patients (Fig 5A and supplemental Fig. S1). FABP3 was also independently identified as a possible predictor of AD in a recent ELISA study (37) and two studies of CSF communicome proteins using the same platform that was used in our study (15, 38). Other proteins overlapping between our analysis and the latter two studies include IL-7, SCF, and CCL11/eotaxin (with Hu et al. identifying its close relative Eotaxin-3/CCL26).
Using levels of plasma communicome proteins to model AD pathological markers we identified between 4 and 15 proteins (out of 74 detectable proteins; supplemental Table S2) for the various models P6-P10. The selected proteins were slightly weaker in their diagnostic utility than the ones selected in CSF models C2, C4, and C5 but outperformed models C1 and C3 (Table II). All plasma models required age and APOE4 status as parameters to achieve sufficient classification accuracy (Fig. 3 and Table II). Further validation of the models in an independent test set resulted in reduced AUC values but a clear improvement over the classification accuracy with APOE4 status and age. Because each of the 5 plasma models was developed independently with the E-net tool, it is reassuring that several proteins were selected in 3, 4 or even all models (shades of orange in Table I). Of these, MCSF and G-CSF may be particularly relevant in AD, as they are reduced in AD plasma (17) and systemic administration of either MCSF (39) or G-CSF (40) ameliorated memory deficits in mouse models of the disease. Furthermore, MCSF functions as a network hub and shows strong positive correlations with FABP3, SCF, and MMP-3 in the CSF-plasma connectivity diagram in AD patients but not in NDC (Fig 5B, C).
The comparison of individual correlations of communicome proteins between CSF and plasma in AD and NDC (Fig 5A) resulted in an additional set of proteins which overlapped in part with those identified in models C1-C5 (FABP3, CRP) and P6-P10 (CCL2) as well as proteins reported in the literature as having a role in AD (CCL5, ICAM-1, leptin). Notably, many of these correlations are different in AD compared with NDC. Whether the correlations between CSF and plasma are the result of exchange between the compartments or co-regulation of these proteins and why the correlations change with disease needs to be explored mechanistically. It is reassuring that out of a 18 protein signature we previously described to classify AD or model progression (17) all six proteins that were detectable on the two distinct platforms were selected in at least one model here (CCL5, CXCL8, GCSF, ICAM-1, IL-3, MCSF). Likewise, in a recent study that used the multiplex sandwich immunoassay platform to measure 108 serum proteins (including the ones measured here) (16), several predictors of AD overlapped with the ones identified here (e.g. CK-MB, G-CSF).
In the future, the analytical methods described here could be improved by increasing the number of communication factors measured in the plasma or CSF and modeling not only pathological CSF markers but clinical or imaging parameters as well. This could strengthen the value of biological information obtained with such an approach. At the same time, the analytical method used here could go beyond the communicome as we know it today by combining it with the analysis of cellular networks (41) or other “-omic” approaches (42) to discover associations with disease patterns in tissue. Naturally, these methods are not limited to AD but could be used for other diseases where direct access to the diseased organ is difficult. Ultimately, the proteins and biological pathways identified by analytical approaches need to be validated using appropriate disease models and ultimately, human studies.
In summary, we used a targeted proteomic multiplex assay to measure soluble proteins involved in intercellular communication in CSF and plasma of AD patients and cognitively normal elderly controls. We found small numbers of systemic extracellular signaling proteins that were able to model key pathological markers of Alzheimer's disease, including levels of CSF β-amyloid and tau, and classify disease in independent samples. We propose that the network-based analytical approaches employed here can help identify proteins or signaling pathways involved in AD or other chronic neurodegenerative disorders with higher confidence than traditional two-class, disease and control modeling approaches. Future studies need to replicate our findings in large multi-center cohorts and test the various proteins for a biological role in AD.
Acknowledgments
We thank the individuals who participated in this study. We also thank H. Johns for technical assistance and S. D. Edland for support in accessing the database and acknowledge numerous unnamed staff at our institutions for their effort in patient recruitment, clinical assessment, and sample preparation.
Footnotes
* This study was supported by the Stanford University School of Medicine Med Scholars Program Grant (SLBH), Anonymous (TWC), the Department of Veterans Affairs (TWC, ERP, GL), and the US National Institute on Aging (AG27505, TWC; AG10491, AG05136, ERP; AG08017, JFQ and JAK; AG05131 and AGO23185, DRG).
 This article contains supplemental Fig. S1 and Tables S1 and S2.
This article contains supplemental Fig. S1 and Tables S1 and S2.
Abbreviations of all measured proteins are listed separately in supplemental Table S2.
AUTHORS' CONTRIBUTION: MB and TWC designed the study and wrote the paper. MB, KR, and TWC prepared the statistical analysis plan, KR did the statistical analysis, and MB and TWC did the network analysis and the further interpretation of the data. SLBH contributed to data analysis. CMC, DRG, JAK, GL, ERP, and JFQ provided plasma and CSF samples, did measurements of Aβ42, total tau, and p-tau181, identified patients and controls, and were responsible for collecting data from them.
CONFLICT OF INTEREST: The authors do not report a conflict of interest.
1 The abbreviations used are:
- Aβ
- β-amyloid
- AD
- Alzheimer's disease
- AUC
- area under the curve
- BBB
- blood-brain barrier
- CSF
- cerebrospinal fluid
- E-net
- Elastic net
- MCI
- mild cognitive impairment
- NDC
- nondemented controls
- p-tau181
- tau phosphorylated at threonine 181
- RC
- regression coefficient
- Rs
- Spearman Rank correlation coefficient
- t-tau
- total tau
- CLSI
- Clinical Laboratory Standards Institute.
REFERENCES
- 1. Steinman L. (2008) Nuanced roles of cytokines in three major human brain disorders. J. Clin. Invest. 118, 3557–3563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Perry V. H., Cunningham C., Holmes C. (2007) Systemic infections and inflammation affect chronic neurodegeneration. Nat. Rev. Immunol. 7, 161–167 [DOI] [PubMed] [Google Scholar]
- 3. Alzheimer's Association National Office (2011) 2011 Alzheimer's Disease Facts and Figures. Alzheimer's Association National Office, IL: [DOI] [PubMed] [Google Scholar]
- 4. Dubois B., Feldman H. H., Jacova C., Cummings J. L., Dekosky S. T., Barberger-Gateau P., Delacourte A., Frisoni G., Fox N. C., Galasko D., Gauthier S., Hampel H., Jicha G. A., Meguro K., O'Brien J., Pasquier F., Robert P., Rossor M., Salloway S., Sarazin M., de Souza L. C., Stern Y., Visser P. J., Scheltens P. (2010) Revising the definition of Alzheimer's disease: a new lexicon. Lancet Neurol. 9, 1118–1127 [DOI] [PubMed] [Google Scholar]
- 5. Dubois B., Feldman H. H., Jacova C., Dekosky S. T., Barberger-Gateau P., Cummings J., Delacourte A., Galasko D., Gauthier S., Jicha G., Meguro K., O'brien J., Pasquier F., Robert P., Rossor M., Salloway S., Stern Y., Visser P. J., Scheltens P. (2007) Research criteria for the diagnosis of Alzheimer's disease: revising the NINCDS-ADRDA criteria. Lancet Neurol. 6, 734–746 [DOI] [PubMed] [Google Scholar]
- 6. Perrin R. J., Fagan A. M., Holtzman D. M. (2009) Multimodal techniques for diagnosis and prognosis of Alzheimer's disease. Nature 461, 916–922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. De Meyer G., Shapiro F., Vanderstichele H., Vanmechelen E., Engelborghs S., De Deyn P. P., Coart E., Hansson O., Minthon L., Zetterberg H., Blennow K., Shaw L., Trojanowski J. Q. (2010) Diagnosis-independent Alzheimer disease biomarker signature in cognitively normal elderly people. Arch. Neurol. 67, 949–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Shaw L. M., Vanderstichele H., Knapik-Czajka M., Clark C. M., Aisen P. S., Petersen R. C., Blennow K., Soares H., Simon A., Lewczuk P., Dean R., Siemers E., Potter W., Lee V. M., Trojanowski J. Q. (2009) Cerebrospinal fluid biomarker signature in Alzheimer's disease neuroimaging initiative subjects. Ann. Neurol. 65, 403–413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Visser P. J., Verhey F., Knol D. L., Scheltens P., Wahlund L. O., Freund-Levi Y., Tsolaki M., Minthon L., Wallin A. K., Hampel H., Bürger K., Pirttila T., Soininen H., Rikkert M. O., Verbeek M. M., Spiru L., Blennow K. (2009) Prevalence and prognostic value of CSF markers of Alzheimer's disease pathology in patients with subjective cognitive impairment or mild cognitive impairment in the DESCRIPA study: a prospective cohort study. Lancet Neurol. 8, 619–627 [DOI] [PubMed] [Google Scholar]
- 10. Fagan A. M., Roe C. M., Xiong C., Mintun M. A., Morris J. C., Holtzman D. M. (2007) Cerebrospinal fluid tau/beta-amyloid(42) ratio as a prediction of cognitive decline in nondemented older adults. Arch Neurol. 64, 343–349 [DOI] [PubMed] [Google Scholar]
- 11. Li G., Sokal I., Quinn J. F., Leverenz J. B., Brodey M., Schellenberg G. D., Kaye J. A., Raskind M. A., Zhang J., Peskind E. R., Montine T. J. (2007) CSF tau/Abeta42 ratio for increased risk of mild cognitive impairment: a follow-up study. Neurology 69, 631–639 [DOI] [PubMed] [Google Scholar]
- 12. Okonkwo O. C., Alosco M. L., Griffith H. R., Mielke M. M., Shaw L. M., Trojanowski J. Q., Tremont G. (2010) Cerebrospinal fluid abnormalities and rate of decline in everyday function across the dementia spectrum: normal aging, mild cognitive impairment, and Alzheimer disease. Arch. Neurol. 67, 688–696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Fagan A. M., Head D., Shah A. R., Marcus D., Mintun M., Morris J. C., Holtzman D. M. (2009) Decreased cerebrospinal fluid Abeta(42) correlates with brain atrophy in cognitively normal elderly. Ann. Neurol. 65, 176–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Britschgi M., Wyss-Coray T. (2007) Systemic and acquired immune responses in Alzheimer's disease. Int. Rev. Neurobiol. 82, 205–233 [DOI] [PubMed] [Google Scholar]
- 15. Hu W. T., Chen-Plotkin A., Arnold S. E., Grossman M., Clark C. M., Shaw L. M., Pickering E., Kuhn M., Chen Y., McCluskey L., Elman L., Karlawish J., Hurtig H. I., Siderowf A., Lee V. M., Soares H., Trojanowski J. Q. (2010) Novel CSF biomarkers for Alzheimer's disease and mild cognitive impairment. Acta Neuropathol. 119, 669–678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. O'Bryant S. E., Xiao G., Barber R., Reisch J., Doody R., Fairchild T., Adams P., Waring S., Diaz-Arrastia R. (2010) A serum protein-based algorithm for the detection of Alzheimer disease. Arch. Neurol. 67, 1077–1081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ray S., Britschgi M., Herbert C., Takeda-Uchimura Y., Boxer A., Blennow K., Friedman L. F., Galasko D. R., Jutel M., Karydas A., Kaye J. A., Leszek J., Miller B. L., Minthon L., Quinn J. F., Rabinovici G. D., Robinson W. H., Sabbagh M. N., So Y. T., Sparks D. L., Tabaton M., Tinklenberg J., Yesavage J. A., Tibshirani R., Wyss-Coray T. (2007) Classification and prediction of clinical Alzheimer's diagnosis based on plasma signaling proteins. Nat. Med. 13, 1359–1362 [DOI] [PubMed] [Google Scholar]
- 18. Soares H. D., Chen Y., Sabbagh M., Roher A., Rohrer A, Schrijvers E., Breteler M. (2009) Identifying early markers of Alzheimer's disease using quantitative multiplex proteomic immunoassay panels. Ann. N.Y. Acad. Sci. 1180, 56–67 [DOI] [PubMed] [Google Scholar]
- 19. Zhang J., Sokal I., Peskind E. R., Quinn J. F., Jankovic J., Kenney C., Chung K. A., Millard S. P., Nutt J. G., Montine T. J. (2008) CSF multianalyte profile distinguishes Alzheimer and Parkinson diseases. Am. J. Clin. Pathol. 129, 526–529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Booij B. B., Lindahl T., Wetterberg P., Skaane N. V., Sæbo S., Feten G., Rye P. D., Kristiansen L. I., Hagen N., Jensen M., Bårdsen K., Winblad B., Sharma P., Lönneborg A. (2011) A gene expression pattern in blood for the early detection of Alzheimer's disease. J. Alzheimers Dis. 23, 109–119 [DOI] [PubMed] [Google Scholar]
- 21. Rye P. D., Booij B. B., Grave G., Lindahl T., Kristiansen L., Andersen H. M., Horndalsveen P. O., Nygaard H. A., Naik M., Hoprekstad D., Wetterberg P., Nilsson C., Aarsland D., Sharma P., Lönneborg A. (2011) A novel blood test for the early detection of Alzheimer's disease. J. Alzheimers Dis. 23, 121–129 [DOI] [PubMed] [Google Scholar]
- 22. Lucin K. M., Wyss-Coray T. (2009) Immune activation in brain aging and neurodegeneration: too much or too little? Neuron 64, 110–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hixson J. E., Vernier D. T. (1990) Restriction isotyping of human apolipoprotein E by gene amplification and cleavage with HhaI. J. Lipid Res. 31, 545–548 [PubMed] [Google Scholar]
- 24. Li G., Peskind E. R., Millard S. P., Chi P., Sokal I., Yu C. E., Bekris L. M., Raskind M. A., Galasko D. R., Montine T. J. (2009) Cerebrospinal fluid concentration of brain-derived neurotrophic factor and cognitive function in non-demented subjects. PLoS One 4, e5424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. R Development Core Team (2009) R: A language and environment for statistical computing., R Foundation for Statistical Computing, Vienna, Austria [Google Scholar]
- 26. van den Berg R. A., Hoefsloot H. C., Westerhuis J. A., Smilde A. K., van der Werf M. J. (2006) Centering, scaling, and transformations: improving the biological information content of metabolomics data. BMC Genomics 7, 142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zou H., Hastie T. (2005) Regularization and variable selection via the elastic net. J. Roy. Statistical Soc. B 67, 301–320 [Google Scholar]
- 28. Friedman J., Hastie T., Tibshirani R. (2010) Regularization paths for generalized linear models via coordinate descent. J. Stat. Software 33, 1–22 [PMC free article] [PubMed] [Google Scholar]
- 29. Hanley J. A., McNeil B. J. (1982) The meaning and use of the area under a receiver operating characteristic (ROC) curve. Radiology 143, 29–36 [DOI] [PubMed] [Google Scholar]
- 30. Pepe M. (2003) The statistical evaluation of medical tests for classification and prediction, Oxford University Press [Google Scholar]
- 31. Villeda S. A., Luo J., Mosher K. I., Zou B., Britschgi M., Bieri G., Stan T. M., Fainberg N., Ding Z., Eggel A., Lucin K. M., Czirr E., Park J.-S., Couillard-Després S., Aigner L., Li G., Peskind E. R., Kaye J. A., Quinn J. F., Galasko D. R., Xie X. S., Rando T. A., Wyss-Coray T. (2011) The aging systemic milieu negatively regulates neurogenesis and cognitive function. Nature 477, 90–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Blennow K., Hampel H. (2003) CSF markers for incipient Alzheimer's disease. Lancet Neurol. 2, 605–613 [DOI] [PubMed] [Google Scholar]
- 33. Veerkamp J. H., Zimmerman A. W. (2001) Fatty acid-binding proteins of nervous tissue. J. Mol. Neurosci. 16, 133–142; discussion 151–157 [DOI] [PubMed] [Google Scholar]
- 34. Pelsers M. M., Hanhoff T., Van der Voort D., Arts B., Peters M., Ponds R., Honig A., Rudzinski W., Spener F., de Kruijk J. R., Twijnstra A., Hermens W. T., Menheere P. P., Glatz J. F. (2004) Brain- and heart-type fatty acid-binding proteins in the brain: tissue distribution and clinical utility. Clin. Chem. 50, 1568–1575 [DOI] [PubMed] [Google Scholar]
- 35. Wunderlich M. T., Hanhoff T., Goertler M., Spener F., Glatz J. F., Wallesch C. W., Pelsers M. M. (2005) Release of brain-type and heart-type fatty acid-binding proteins in serum after acute ischaemic stroke. J. Neurol. 252, 718–724 [DOI] [PubMed] [Google Scholar]
- 36. Zimmermann-Ivol C. G., Burkhard P. R., Le Floch-Rohr J., Allard L., Hochstrasser D. F., Sanchez J. C. (2004) Fatty acid binding protein as a serum marker for the early diagnosis of stroke: a pilot study. Mol. Cell. Proteomics 3, 66–72 [DOI] [PubMed] [Google Scholar]
- 37. Chiasserini D., Parnetti L., Andreasson U., Zetterberg H., Giannandrea D., Calabresi P., Blennow K. (2010) CSF levels of heart fatty acid binding protein are altered during early phases of Alzheimer's disease. J. Alzheimers Dis. 22, 1281–1288 [DOI] [PubMed] [Google Scholar]
- 38. Hu W. T., Chen-Plotkin A., Arnold S. E., Grossman M., Clark C. M., Shaw L. M., McCluskey L., Elman L., Karlawish J., Hurtig H. I., Siderowf A., Lee V. M., Soares H., Trojanowski J. Q. (2010) Biomarker discovery for Alzheimer's disease, frontotemporal lobar degeneration, and Parkinson's disease. Acta Neuropathol. 120, 385–399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Boissonneault V., Filali M., Lessard M., Relton J., Wong G., Rivest S. (2009) Powerful beneficial effects of macrophage colony-stimulating factor on beta-amyloid deposition and cognitive impairment in Alzheimer's disease. Brain 132, 1078–1092 [DOI] [PubMed] [Google Scholar]
- 40. Tsai K. J., Tsai Y. C., Shen C. K. (2007) G-CSF rescues the memory impairment of animal models of Alzheimer's disease. J. Exp. Med. 204, 1273–1280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Park J., Lee D. S., Christakis N. A., Barabási A. L. (2009) The impact of cellular networks on disease comorbidity. Mol. Syst. Biol. 5, 262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Inouye M., Kettunen J., Soininen P., Silander K., Ripatti S., Kumpula L. S., Hämäläinen E., Jousilahti P., Kangas A. J., Männistö S., Savolainen M. J., Jula A., Leiviskä J., Palotie A., Salomaa V., Perola M., Ala-Korpela M., Peltonen L. (2010) Metabonomic, transcriptomic, and genomic variation of a population cohort. Mol. Syst. Biol. 6, 441. [DOI] [PMC free article] [PubMed] [Google Scholar]