Abstract
We augmented existing computationally predicted and experimentally determined interactions with evolutionarily conserved interactions between proteins of the malaria parasite, P. falciparum, and the human host. In a validation step, we found that conserved interacting host-parasite protein pairs were specifically expressed in host tissues where both the parasite and host proteins are known to be active. We compared host-parasite interactions with experimentally verified interactions between human host proteins and a very different pathogen, HIV-1. Both pathogens were found to use their protein repertoire in a combinatorial manner, providing a broad connection to host cellular processes. Specifically, the two biologically distinct pathogens predominately target central proteins to take control of a human host cell, effectively reaching into diversified cellular host cellular functions. Interacting signaling pathways and a small set of regulatory and signaling proteins were prime targets of both pathogens, suggesting remarkably similar patterns of host-pathogen interactions despite the vast biological differences of both pathogens. Such an identification of shared molecular strategies by the virus HIV-1 and the eukaryotic intracellular pathogen P. falciparum may allow us to illuminate new avenues of disease intervention.
The determination of protein interactions (1–4) and protein complexes (5–8) is being increasingly refined in many single and multicellular organisms as well as the human interactome (9–13). However, little is known about large-scale protein interactions between hosts and pathogens, although the identification of such maps will provide an essential foundation for the development of effective therapeutic and prevention strategies to combat diseases. Uetz et al. reported the first small map of computationally inferred physical protein interactions between the human host and the Kaposi's-Sarcoma associated Herpesvirus as well as the Varicella-Zoster-Virus (14). More recently, Calderwood et al. (15) empirically constructed a map of physical protein interactions between the Epstein-Barr-Virus and the human host. For HIV, subnetworks of virus-host protein interactions that are expressed at different stages of the infection have been identified (16) along with cofactors that enable HIV and the influenza virus to infect a host cell (17–19). Furthermore, Dyer et al. compared experimentally known interactions of different viruses with the human host (20). The first small map of experimentally determined protein interactions between the human host and the malaria parasite P. falciparum was released recently (21), and was preceded by computational predictions of host-parasite interactions (22, 23).
Using large-scale sets of intraspecies protein interactions between proteins of Homo sapiens and the malaria parasite Plasmodium falciparum, we inferred conserved host-parasite interactions using orthologous protein groups. Combining these interactions with experimentally determined (21) and previously predicted interactions based on protein structural features (22, 23), we pooled a large set of host-parasite interactions. Although completely different in their biology and evolution, we find that both HIV-1 and P. falciparum use their protein repertoires in a combinatorial manner. Because a similar strategy evolved independently in each case, it may highlight and potentially provide a broad and effective connection to host pathways and diversified cellular functions that may lead to a more stable intercalation of the host cell.
EXPERIMENTAL PROCEDURES
Intraspecies Protein Interaction and Pathway Data
We used human protein-protein interaction data from large-scale high-throughput screens (13, 24, 25) and several interaction databases (26–29) totaling 93,178 interactions among 11,691 human genes. For the parasite P. falciparum, we collected 2743 interactions between 1299 proteins (30). To examine human protein pathways, we utilized 184 annotated signaling pathways from the PID database (31).
Host-Parasite Protein Interaction Data
We used 444 experimentally obtained interactions (21) between proteins of the human host and the malaria parasite P. falciparum. In addition, we used 691 computationally determined protein interactions from protein structures of the human host and the parasite (23).
Human-HIV-1 Protein Interaction Data
We used a compilation of 702 experimentally obtained protein interactions between the human host and HIV-1. In particular, we accounted for interactions that have been found in vital cells in the human immune system such as helper T cells, macrophages, and dendritic cells (32).
Parasite Interacting Proteins
We compiled a list of 1014 proteins in P. falciparum that have molecular features, which potentially facilitate interactions with human host proteins. Specifically, we chose proteins carrying a host cell-targeting signal, allowing them to cross into the human host cell by passing several membranes (33–35). In addition, we picked proteins with trans-membrane domains, signaling peptides located in the red blood cell membrane (36). Finally, we augmented our set with proteins that have been implicated in host-pathogen interactions, pathogenesis, and defense response as well as those proteins that are located in the host cell as suggested by their corresponding GO annotation or PLasmoDraft prediction (37–39).
Host Protein/Gene Expression Data
From a mass-spectroscopic proteome analysis of the human red blood cell (40), we selected 1,578 human proteins. In addition, we pooled a set of 19,031 human proteins that were expressed in human liver tissue (41).
Parasite Protein Expression Data
We used a list of 838 proteins that are predominantly expressed in the merozoite stage, whereas we used 1038 parasite proteins that are detected in the sporozoite stage (36) of the parasites life cycle. In both studies the presence of proteins in the corresponding proteomes of the underlying cells was determined by mass-spectroscopy.
Orthologous Proteins
Using all-versus-all BLASTP searches determined by the InParanoid script (42) in protein sets of two species, sequence pairs with mutually best scores were selected as central orthologous pairs. To enhance quality, we only accepted BLAST matches with a score of >40 bits, covering at least 50% of the longer sequence. Proteins of both species that showed such an elevated degree of homology were clustered around these central pairs, forming orthologous groups. The quality of the clustering was further assessed by a standard bootstrap procedure. We only considered the central orthologous sequence pair that provided a confidence level of 100% as the real orthologous relationship, allowing us to obtain 1370 orthologous protein pairs in P. falciparum and H. sapiens.
Enrichment
To estimate whether nodes shared a certain feature of possible biological relevance (e.g. being a parasite target), we calculated the corresponding fraction of such nodes by fa,≥k = |Na,≥k|/|N≥k|, where each node in N≥k had at least a certain number of neighbors k in an underlying network of interactions. As a null hypothesis, we assumed a random distribution, fa,≥kr, being the random fraction of feature a among all nodes that had at least k neighbors and defined Ea,≥k = fa,≥k/fa,≥kr as the enrichment of feature a. Averaging E over 10,000 randomizations we considered sets enriched with feature a if E >1 and vice versa.
Kernel Density Function
A simple way to analyze a series of values x = x1,. ., xn would be a histogram. However, if the number of observations is low the significance of a histogram is rather limited. Therefore, we defined the kernel density approximation, a smoothing operation that allowed the estimation of a putative probability density function of data points around a certain point x as . K(y) is the kernel function, satisfying , and h is a smoothing parameter. In particular, we chose the Gaussian as kernel function .
RESULTS
To predict protein-protein interactions between human host and malaria parasite proteins, we assembled a network of 93,178 interactions between human proteins (13, 24–29) and a web of 2743 interactions between parasite proteins (30). Using InParanoid database (42), we compiled 1370 orthologous pairs of human and parasite proteins. We identified a candidate host-parasite interaction in the web of human interactions if a protein had a parasite homolog. Analogously, we determined a potential interaction in the parasite interaction network if a parasite protein had a human ortholog, revealing a total of 98,224 conserved host-parasite interactions.
Although this set of interactions included 1137 parasite proteins we assumed that only a subset of these parasite proteins indeed interact with the human host. Specifically, malaria parasites export remodeling and virulence proteins to the erythrocyte surface after invasion of a red blood cell. Recent studies independently uncovered a host-cell targeting signal that participates in the process of moving proteins across membranes to the host cell membrane (33–35). We further augmented this set with proteins that have transmembrane domains, signaling peptides and/or are located in the red blood cell membrane (36). Furthermore, we added proteins that have been implicated in host-pathogen interactions, pathogenesis, defense responses, or are located in the host cell as suggested by their corresponding GO annotation or PlasmoDraft prediction (37–39). In total, we compiled a list of 1014 parasite proteins that potentially interact with human host proteins. If host-pathogen interactions were conserved, we expected many interacting parasite proteins would have molecular characteristics that make them accessible to host proteins. Indeed, we found a small but significant overlap (Fig. 1A, p < 10−10, hypergeometric test) between the set of accessible parasite proteins and those identified in our network, suggesting that the set of evolutionarily conserved interactions were a result of selective pressure.
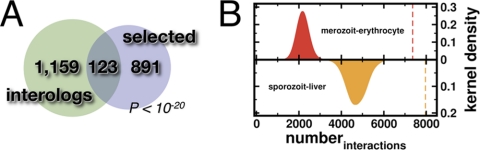
Fig. 1.
A, Using orthologous protein pairs between the human host and the parasite P. falciparum we derived a total of 98,224 conserved protein interactions, involving 1,282 parasite proteins. Compiling a list of 1014 parasite proteins that potentially can interact with human host proteins, we find a small but significant overlap (p < 10−20, hypergeometric test), suggesting that the set of evolutionary conserved interactions are enriched with potentially interacting parasite proteins. In the upper panel of (B) we assumed that host-parasite interactions predominantly appeared between proteins that are expressed in human liver tissue and the sporozoite stage of the parasite, allowing us to find 7366 interactions (dotted line). Randomizing the sets of expressed genes we confirmed the significance of this observation (p < 10−3). In the lower panel we determined the number of interactions between proteins that are expressed in the human red blood cell and the parasitic merozoite stage. Analogously, we found that 7967 interactions (dotted line) were statistically significant, randomizing the sets of expressed genes (p < 10−3).
Given that the malaria parasite infects both human liver cells and erythrocytes, we hypothesized that conserved interactions might be enriched with interactions between proteins that are expressed in the liver tissue (41) and the sporozoite stage of the parasite (36) as well as the erythrocytes and parasite blood stages. We found that the presence of 7366 interactions between simultaneously expressed host and parasite proteins was highly significant (p < 10−3). In the lower panel of Fig. 1B, we determined the number of interactions between proteins that were simultaneously expressed in the red blood cell (40) and the merozoite stage (36). Analogously, we found 7967 significantly placed interactions (dotted line, p < 10−3), confirming that the co-expression of proteins in conserved interactions is a highly nonrandom event.
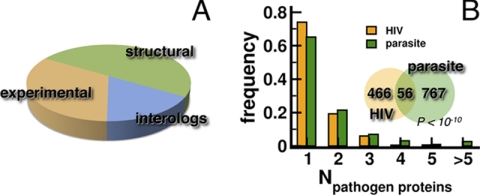
To establish potential evolutionary conserved interactions, we selected all 250 interactions that involved parasite proteins, carrying molecular features that make them accessible to interactions with human host proteins. In addition, we demanded that the interacting parasite proteins were either expressed in the sporozoite or merozoite stage, whereas their human interaction partners had to be present in human liver or the red blood cell, respectively. Randomizing such sets of expressed genes and accessible parasite proteins, we found that the emergence of such a set of conserved interactions is a highly nonrandom event as well (p < 10−3). Finally, we augmented our set of conserved interactions with experimentally determined host-parasite interactions (21) and interactions that were computationally inferred from protein structure data (23). In total, we assembled 1385 interactions between 379 parasite and 823 host proteins, where the majority of interactions were provided by computationally predicted interactions, followed by experimental observations and conserved interactions (Fig. 2A, supplemental Table S1).
Fig. 2.
In (A) we augmented our set of evolutionarily conserved set with experimentally determined and protein structure inferred host-parasite interactions. In a total of 1385 interactions between 379 parasite and 823 host proteins, most interactions were inferred from structural protein data, followed by sets of experimentally and evolutionarily determined interactions. B, Comparing the characteristics of interactions between the human host, the virus HIV-1 and the parasite P. falciparum we calculated the number of pathogen proteins that targeted a host protein. We generally found that the majority of host proteins interacted with a low number of pathogen proteins and vice versa. In the inset, we counted the number of human proteins that were targeted by both parasite and virus proteins and found a small but significant overlap (p < 10−10, hypergeometric test).
We were interested in exploring features of host-pathogen interactions shared between pathogens as diverse as a virus and a eukaryotic parasite. As a counterpart to the parasite P. falciparum, we chose the human virus HIV-1 and collected 702 physical interactions between 17 HIV-1 and 522 human proteins (32). Considering topological integrity, we found that the bipartite network of interactions between parasite and host proteins was composed of 104 disconnected subnetworks. Notably, the largest subnetwork contained the vast majority of parasite and host proteins. By comparison, a bipartite network of interactions between viral and human host proteins was composed of one such subnetwork. If the placement of host-pathogen interactions were a random process, we would expect that randomly assembled bipartite networks would break into many disconnected parts. By randomly connecting parasite and human host proteins, we indeed observed that randomized networks on average broke into more than observed connected components (p < 10−4). Analogously, we rewired HIV and human proteins and found a similar result (p < 10−4), confirming that the integrity of a host-pathogen interaction network is the result of a significantly nonrandom process.
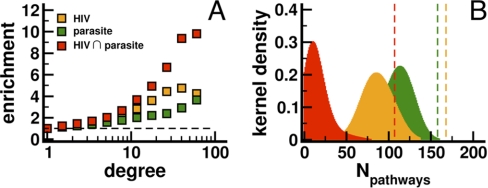
Comparing the characteristics of interactions with the human host, we calculated the number of pathogen proteins that target a host protein according to our data. We generally found that the majority of host proteins interacted with a low number of pathogen proteins and vice versa (Fig. 2B). In the inset, we counted the number of human proteins that interacted with the parasite and the virus and found a small but significant set of 56 proteins (Fig. 2B, p < 10−10, hypergeometric test). Assuming that important proteins strongly interact we expected that hubs might predominantly be targets of pathogen proteins. Using a network of human interactions, we determined the enrichment of all human proteins that were targeted by P. falciparum. We found that especially highly connected proteins are indeed affected by the parasite, a result that was confirmed by interactions between human host and HIV proteins (Fig. 3A). Such trends were reinforced when we considered the enrichment of simultaneously targeted 56 proteins, suggesting the presence of core that is exploited by pathogens to seize control of the host cells.
Fig. 3.
A, Determining the enrichment of all human proteins that were targeted by P. falciparum, we found that especially highly connected proteins were predominantly affected by pathogens. Focusing on proteins that were targeted by HIV we found a similar result. Considering the set of proteins that both pathogens target, we observed an amplification of the pathogens specific enrichment signal. In (B) we counted the number of pathways that pathogens reached by targeting corresponding proteins (dashed lines). Randomizing the set of targeted proteins, we found that the choice of attacked proteins is highly nonrandom (p < 10−5), a result we confirmed by focusing on proteins that interacted with both pathogens.
Protein pathways provide an additional level of systems information to leverage in assessing patterns that show the ways pathogens exploit the host cell. Our approach relied on the strength of 184 manually curated signaling protein pathways from the Pathway Interaction Database (31). We hypothesized that the comparably small number of targeted human proteins would allow pathogens to interact with a larger number of pathways than would appear by chance. Out of 184 pathways, human proteins targeted by the virus were part of 168 pathways, whereas the malaria parasite touched 158 pathways. Randomizing sets of attacked human proteins, we found significantly fewer pathways than expected by chance alone (Fig. 3B, p < 10−5). In turn, interactions involving 56 human proteins that were attacked by both pathogens targeted human host proteins in 107 pathways. By randomizing the human sets, we determined that this observation was statistically significant (p < 10−5). Given the nonrandom points of interaction, the intersection of attacked human proteins potentially allows the broadest possible reach into host pathways, suggesting that the parasite may diversify its contact at the host-parasite interface.
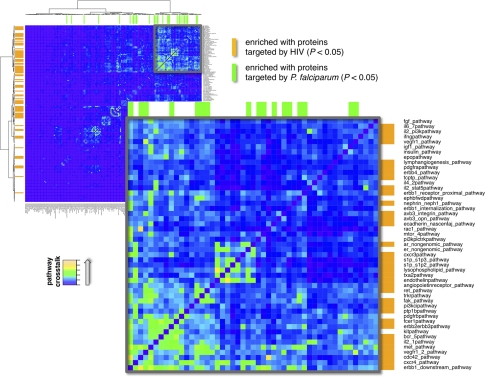
In Fig. 4, we illustrate the level of pathway crosstalk between 184 signaling pathways. Specifically, we counted the number of proteins that a pair of pathways shares by constructing a symmetric matrix. Applying Ward clustering, we notably observed a cluster of 50 prominent signaling pathways that strongly share proteins. Assuming that targeted human proteins allow the pathogens the broadest possible reach into host functions, we expected that this group of pathways would be specifically attacked. Applying Fisher's exact test we found 99 pathways enriched with proteins that were targeted by HIV-1 (p < 0.05) whereas 49 pathways significantly harbored parasite targeted proteins (p < 0.05). Qualitatively, we indeed observed that interacting pathways were predominantly enriched with targeted proteins, suggesting that strongly cross-talking pathways are prime targets of pathogens.
Fig. 4.
Using 184 signaling pathways, we determined the number of overlapping proteins in all pairs of pathways. Applying Ward clustering, we observed a cluster of prominent signaling pathways that strongly shared proteins (boxes). Determining pathways with proteins that were enriched with proteins targeted by the virus and parasite, respectively (Fisher's exact test, p < 0.05), we found that groups of strongly cross-talking pathways are predominately enriched with targeted proteins.
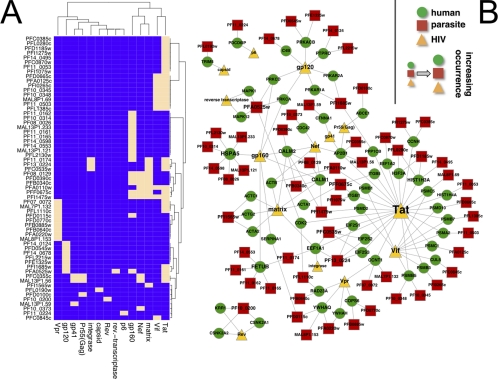
Focusing in detail on 56 human host proteins that were targeted by both pathogens we constructed a bipartite matrix that indicated if two pathogen proteins interact with at least one common host protein (Fig. 5A). In addition, we mapped all interactions between pathogen and host proteins if a host protein shared interactions with both the virus and the parasite. Qualitatively, human proteins that were targeted by both pathogens were involved in interactions with prominent signaling proteins (Fig. 5B). Moreover, we observed that key HIV-1 proteins Vif and Tat as well as several P. falciparum proteins interacted with several components of the human proteasome, a protein assembly that is important in both the red blood cell (44) and T-cells (45). For example, the proteasome component PSMB1, interacting with both Vif and the malaria parasite protein (PFA0110w) is up-regulated in sickle cell disease (43), a condition known to influence malaria parasite infection. In human T-cells, the proteasome is part of an innate anti-HIV defense that is circumvented by the viral proteins Vif and Tat (44).
Fig. 5.
A, Using 56 proteins that were targeted by both the virus HIV-1 and the parasite P. falciparum we constructed a bipartite matrix. Specifically, we indicated a link if a pair of a virus and parasite proteins shared at least one common host protein. Specifically, we observed that Vif, Tat, matrix, gp160 and Nef shared the majority of targeted proteins with the parasite. B, On a qualitative basis, we mapped interactions between pathogen and host proteins if a host protein shared interactions with both the virus and the parasite. We observed a large assembly of interactions with proteasomal proteins and viral proteins Tat and Vif. Although the parasite interacted with proteasomal subunits as well interactions were distributed among many different parasite proteins. In addition, we found numerous signaling and regulation proteins that interacted with a variety of parasite and viral proteins.
DISCUSSION
Detecting sequence homology of interacting proteins is a widely used approach to identify evolutionary conserved interactions. Although sequence homology is a powerful technique, large inserts might obscure homology signals in gene and protein sequences of P. falciparum that might hamper the detection of homologs in different organisms. To account for such constraints, we utilized orthologous groups of proteins that were determined in very conservative ways. Although we imposed a strict threshold of significance for each match, we also demanded that the potential match covers more than half of the underlying sequences. After checking their nonrandom nature, we only chose bona fide orthologous pairs that allowed us to infer evolutionary conserved interactions.
Investigating the power of evolutionary conserved interactions, we checked if they carry any relevant signals, indicating the presence of evolutionary pressure. Indeed, we found that conserved interactions enriched parasite proteins with molecular characteristic that make them accessible to interact with a human host protein. In addition, such interactions showed strong nonrandom co-expression of parasite and host proteins in the respective cell-cycle stages and invaded host tissues, strongly suggesting that interologs reflect strong evolutionary pressure.
Pooling conserved interactions with other sets of experimentally and structurally determined interactions, we established a set of interactions between selected parasite and human host proteins. Subsequently, we compared such host-parasite interactions to an experimentally determined set of interactions between the human host and the virus HIV-1. Although profoundly different in their basic biology, we found striking similarities between interaction patterns of the virus and the malaria parasite. Both HIV-1 and P. falciparum invoked intricate, yet surprisingly similar processes of interaction with a remarkably low number of proteins to exploit the human host cell. In particular, the subtle structure of the human interactome revealed sites that were not only topologically important on their own, but also were significant pathogen targets. The functional and topological role of central proteins allows the host to maintain a complex system with relatively fewer proteins. Because highly interacting proteins are prime targets of a single pathogen, they may be key players in the subtle molecular strategies a pathogen employs in invading and establishing an infection in a host cell. This trend was reinforced by host proteins targeted by both pathogens suggesting a core of functions that allow a pathogen to stably intercalate with a host cell. Tapping such a topological feature with a limited proteomic repertoire in an economic, yet effective way, the malaria parasite and the virus use combinations of pathogen proteins to interact with a variety of different functions. Therefore, untangling the intricate web of host-pathogen interactions is essential to thoroughly understand their pathogenesis. Furthermore, the analysis of host-pathogen interfaces might present a molecular and functional Achilles heel that could be exploited for new interventions to limit the pathogens in a systematic way (45).
We identified malaria parasite specific counterparts to the key players of viral infection, a surprising observation that implied an evolutionary convergent nature of infection by vastly distinct pathogens. On a different note, we found parasite proteins and targets that could contribute to the pathogenicity of the parasite and suggest testable hypotheses about P. falciparum biology for which future experimentation could reveal possible exploitable features. In this way, webs of well-defined host-pathogen interactions can serve as evolutionary blueprints of pathogenic interference that suggests avenues guiding future efforts to eradicate pathogens that plague human kind.
Acknowledgments
This work was partly supported by NIH Grants AI055035 and AI071121 to MTF.
Footnotes
 This article contains supplemental Table S1.
This article contains supplemental Table S1.
REFERENCES
- 1. Giot L., Bader J. S., Brouwer C., Chaudhuri A., Kuang B., Li Y., Hao Y. L., Ooi C. E., Godwin B., Vitols E., Vijayadamodar G., Pochart P., Machineni H., Welsh M., Kong Y., Zerhusen B., Malcolm R., Varrone Z., Collis A., Minto M., Burgess S., McDaniel L., Stimpson E., Spriggs F., Williams J., Neurath K., Ioime N., Agee M., Voss E., Furtak K., Renzulli R., Aanensen N., Carrolla S., Bickelhaupt E., Lazovatsky Y., DaSilva A., Zhong J., Stanyon C. A., Finley R. L., Jr., White K. P., Braverman M., Jarvie T., Gold S., Leach M., Knight J., Shimkets R. A., McKenna M. P., Chant J., Rothberg J. M. (2003) A protein interaction map of Drosophila melanogaster. Science 302, 1727–1736 [DOI] [PubMed] [Google Scholar]
- 2. Reguly T., Breitkreutz A., Boucher L., Breitkreutz B. J., Hon G. C., Myers C. L., Parsons A., Friesen H., Oughtred R., Tong A., Stark C., Ho Y., Botstein D., Andrews B., Boone C., Troyanskya O. G., Ideker T., Dolinski K., Batada N. N., Tyers M. (2006) Comprehensive curation and analysis of global interaction networks in Saccharomyces cerevisiae. J. Biol. 5, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Li S., Armstrong C. M., Bertin N., Ge H., Milstein S., Boxem M., Vidalain P. O., Han J. D., Chesneau A., Hao T., Goldberg D. S., Li N., Martinez M., Rual J. F., Lamesch P., Xu L., Tewari M., Wong S. L., Zhang L. V., Berriz G. F., Jacotot L., Vaglio P., Reboul J., Hirozane-Kishikawa T., Li Q., Gabel H. W., Elewa A., Baumgartner B., Rose D. J., Yu H., Bosak S., Sequerra R., Fraser A., Mango S. E., Saxton W. M., Strome S., Van Den Heuvel S., Piano F., Vandenhaute J., Sardet C., Gerstein M., Doucette-Stamm L., Gunsalus K. C., Harper J. W., Cusick M. E., Roth F. P., Hill D. E., Vidal M. (2004) A map of the interactome network of the metazoan C. elegans. Science 303, 540–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Rain J. C., Selig L., De Reuse H., Battaglia V., Reverdy C., Simon S., Lenzen G., Petel F., Wojcik J., Schächter V., Chemama Y., Labigne A., Legrain P. (2001) The protein-protein interaction map of Helicobacter pylori. Nature 409, 211–215 [DOI] [PubMed] [Google Scholar]
- 5. Gavin A. C., Aloy P., Grandi P., Krause R., Boesche M., Marzioch M., Rau C., Jensen L. J., Bastuck S., Dümpelfeld B., Edelmann A., Heurtier M. A., Hoffman V., Hoefert C., Klein K., Hudak M., Michon A. M., Schelder M., Schirle M., Remor M., Rudi T., Hooper S., Bauer A., Bouwmeester T., Casari G., Drewes G., Neubauer G., Rick J. M., Kuster B., Bork P., Russell R. B., Superti-Furga G. (2006) Proteome survey reveals modularity of the yeast cell machinery. Nature 440, 631–636 [DOI] [PubMed] [Google Scholar]
- 6. Gavin A. C., Bösche M., Krause R., Grandi P., Marzioch M., Bauer A., Schultz J., Rick J. M., Michon A. M., Cruciat C. M., Remor M., Höfert C., Schelder M., Brajenovic M., Ruffner H., Merino A., Klein K., Hudak M., Dickson D., Rudi T., Gnau V., Bauch A., Bastuck S., Huhse B., Leutwein C., Heurtier M. A., Copley R. R., Edelmann A., Querfurth E., Rybin V., Drewes G., Raida M., Bouwmeester T., Bork P., Seraphin B., Kuster B., Neubauer G., Superti-Furga G. (2002) Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature 415, 141–147 [DOI] [PubMed] [Google Scholar]
- 7. Krogan N. J., Cagney G., Yu H., Zhong G., Guo X., Ignatchenko A., Li J., Pu S., Datta N., Tikuisis A. P., Punna T., Peregrin-Alvarez J. M., Shales M., Zhang X., Davey M., Robinson M. D., Paccanaro A., Bray J. E., Sheung A., Beattie B., Richards D. P., Canadien V., Lalev A., Mena F., Wong P., Starostine A., Canete M. M., Vlasblom J., Wu S., Orsi C., Collins S. R., Chandran S., Haw R., Rilstone J. J., Gandi K., Thompson N. J., Musso G., St Onge P., Ghanny S., Lam M. H., Butland G., Altaf-Ul A. M., Kanaya S., Shilatifard A., O'Shea E., Weissman J. S., Ingles C. J., Hughes T. R., Parkinson J., Gerstein M., Wodak S. J., Emili A., Greenblatt J. F. (2006) Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature 440, 637–643 [DOI] [PubMed] [Google Scholar]
- 8. Sprinzak E., Altuvia Y., Margalit H. (2006) Characterization and prediction of protein-protein interactions within and between complexes. Proc. Natl. Acad. Sci. U.S.A. 103, 14718–14723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lehner B., Fraser A. G. (2004) A first-draft human protein-interaction map. Genome Biol. 5, R63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ramani A. K., Bunescu R. C., Mooney R. J., Marcotte E. M. (2005) Consolidating the set of known human protein-protein interactions in preparation for large-scale mapping of the human interactome. Genome Biol. 6, R40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Gandhi T. K., Zhong J., Mathivanan S., Karthick L., Chandrika K. N., Mohan S. S., Sharma S., Pinkert S., Nagaraju S., Periaswamy B., Mishra G., Nandakumar K., Shen B., Deshpande N., Nayak R., Sarker M., Boeke J. D., Parmigiani G., Schultz J., Bader J. S., Pandey A. (2006) Analysis of the human protein interactome and comparison with yeast, worm and fly interaction datasets. Nat. Genet. 38, 285–293 [DOI] [PubMed] [Google Scholar]
- 12. Rhodes D. R., Tomlins S. A., Varambally S., Mahavisno V., Barrette T., Kalyana-Sundaram S., Ghosh D., Pandey A., Chinnaiyan A. M. (2005) Probabilistic model of the human protein-protein interaction network. Nat. Biotechnol. 23, 951–959 [DOI] [PubMed] [Google Scholar]
- 13. Stelzl U., Worm U., Lalowski M., Haenig C., Brembeck F. H., Goehler H., Stroedicke M., Zenkner M., Schoenherr A., Koeppen S., Timm J., Mintzlaff S., Abraham C., Bock N., Kietzmann S., Goedde A., Toksöz E., Droege A., Krobitsch S., Korn B., Birchmeier W., Lehrach H., Wanker E. E. (2005) A human protein-protein interaction network: a resource for annotating the proteome. Cell 122, 957–968 [DOI] [PubMed] [Google Scholar]
- 14. Uetz P., Dong Y. A., Zeretzke C., Atzler C., Baiker A., Berger B., Rajagopala S. V., Roupelieva M., Rose D., Fossum E., Haas J. (2006) Herpesviral protein networks and their interaction with the human proteome. Science 311, 239–242 [DOI] [PubMed] [Google Scholar]
- 15. Calderwood M. A., Venkatesan K., Xing L., Chase M. R., Vazquez A., Holthaus A. M., Ewence A. E., Li N., Hirozane-Kishikawa T., Hill D. E., Vidal M., Kieff E., Johannsen E. (2007) Epstein-Barr virus and virus human protein interaction maps. Proc. Natl. Acad. Sci. U.S.A. 104, 7606–7611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bandyopadhyay S., Kelley R., Ideker T. (2006) Discovering regulated networks during HIV-1 latency and reactivation. Pac. Symp. Biocomput. 354–366 [PMC free article] [PubMed] [Google Scholar]
- 17. Bushman F. D., Malani N., Fernandes J., D'Orso I., Cagney G., Diamond T. L., Zhou H., Hazuda D. J., Espeseth A. S., König R., Bandyopadhyay S., Ideker T., Goff S. P., Krogan N. J., Frankel A. D., Young J. A., Chanda S. K. (2009) Host cell factors in HIV replication: meta-analysis of genome-wide studies. PLoS Pathog. 5, e1000437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. König R., Stertz S., Zhou Y., Inoue A., Hoffmann H. H., Bhattacharyya S., Alamares J. G., Tscherne D. M., Ortigoza M. B., Liang Y., Gao Q., Andrews S. E., Bandyopadhyay S., De Jesus P., Tu B. P., Pache L., Shih C., Orth A., Bonamy G., Miraglia L., Ideker T., Garcia-Sastre A., Young J. A., Palese P., Shaw M. L., Chanda S. K. (2010) Human host factors required for influenza virus replication. Nature 463, 813–817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Brass A. L., Dykxhoorn D. M., Benita Y., Yan N., Engelman A., Xavier R. J., Lieberman J., Elledge S. J. (2008) Identification of host proteins required for HIV infection through a functional genomic screen. Science 319, 921–926 [DOI] [PubMed] [Google Scholar]
- 20. Dyer M. D., Murali T. M., Sobral B. W. (2008) The landscape of human proteins interacting with viruses and other pathogens. PLoS Pathog. 4, e32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Vignali M., McKinlay A., LaCount D. J., Chettier R., Bell R., Sahasrabudhe S., Hughes R. E., Fields S. (2008) Interaction of an atypical Plasmodium falciparum ETRAMP with human apolipoproteins. Malar J 7, 211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Dyer M. D., Murali T. M., Sobral B. W. (2007) Computational prediction of host-pathogen protein-protein interactions. Bioinformatics 23, i159–166 [DOI] [PubMed] [Google Scholar]
- 23. Davis F. P., Barkan D. T., Eswar N., McKerrow J. H., Sali A. (2007) Host pathogen protein interactions predicted by comparative modeling. Protein Sci. 16, 2585–2596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ewing R. M., Chu P., Elisma F., Li H., Taylor P., Climie S., McBroom-Cerajewski L., Robinson M. D., O'Connor L., Li M., Taylor R., Dharsee M., Ho Y., Heilbut A., Moore L., Zhang S., Ornatsky O., Bukhman Y. V., Ethier M., Sheng Y., Vasilescu J., Abu-Farha M., Lambert J. P., Duewel H. S., Stewart II, Kuehl B., Hogue K., Colwill K., Gladwish K., Muskat B., Kinach R., Adams S. L., Moran M. F., Morin G. B., Topaloglou T., Figeys D. (2007) Large-scale mapping of human protein-protein interactions by mass spectrometry. Mol. Syst. Biol. 3, 89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Rual J. F., Venkatesan K., Hao T., Hirozane-Kishikawa T., Dricot A., Li N., Berriz G. F., Gibbons F. D., Dreze M., Ayivi-Guedehoussou N., Klitgord N., Simon C., Boxem M., Milstein S., Rosenberg J., Goldberg D. S., Zhang L. V., Wong S. L., Franklin G., Li S., Albala J. S., Lim J., Fraughton C., Llamosas E., Cevik S., Bex C., Lamesch P., Sikorski R. S., Vandenhaute J., Zoghbi H. Y., Smolyar A., Bosak S., Sequerra R., Doucette-Stamm L., Cusick M. E., Hill D. E., Roth F. P., Vidal M. (2005) Towards a proteome-scale map of the human protein-protein interaction network. Nature 437, 1173–1178 [DOI] [PubMed] [Google Scholar]
- 26. Chatr-aryamontri A., Ceol A., Palazzi L. M., Nardelli G., Schneider M. V., Castagnoli L., Cesareni G. (2007) MINT: the Molecular INTeraction database. Nucleic Acids Res. 35, D572–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kerrien S., Alam-Faruque Y., Aranda B., Bancarz I., Bridge A., Derow C., Dimmer E., Feuermann M., Friedrichsen A., Huntley R., Kohler C., Khadake J., Leroy C., Liban A., Lieftink C., Montecchi-Palazzi L., Orchard S., Risse J., Robbe K., Roechert B., Thorneycroft D., Zhang Y., Apweiler R., Hermjakob H. (2007) IntAct–open source resource for molecular interaction data. Nucleic Acids Res. 35, D561–565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Matthews L., Gopinath G., Gillespie M., Caudy M., Croft D., de Bono B., Garapati P., Hemish J., Hermjakob H., Jassal B., Kanapin A., Lewis S., Mahajan S., May B., Schmidt E., Vastrik I., Wu G., Birney E., Stein L., D'Eustachio P. (2008) Reactome knowledgebase of human biological pathways and processes. Nucleic Acids Res. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Peri S., Navarro J. D., Kristiansen T. Z., Amanchy R., Surendranath V., Muthusamy B., Gandhi T. K., Chandrika K. N., Deshpande N., Suresh S., Rashmi B. P., Shanker K., Padma N., Niranjan V., Harsha H. C., Talreja N., Vrushabendra B. M., Ramya M. A., Yatish A. J., Joy M., Shivashankar H. N., Kavitha M. P., Menezes M., Choudhury D. R., Ghosh N., Saravana R., Chandran S., Mohan S., Jonnalagadda C. K., Prasad C. K., Kumar-Sinha C., Deshpande K. S., Pandey A. (2004) Human protein reference database as a discovery resource for proteomics. Nucleic Acids Res. 32, D497–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. LaCount D. J., Vignali M., Chettier R., Phansalkar A., Bell R., Hesselberth J. R., Schoenfeld L. W., Ota I., Sahasrabudhe S., Kurschner C., Fields S., Hughes R. E. (2005) A protein interaction network of the malaria parasite Plasmodium falciparum. Nature 438, 103–107 [DOI] [PubMed] [Google Scholar]
- 31. Schaefer C. F., Anthony K., Krupa S., Buchoff J., Day M., Hannay T., Buetow K. H. (2009) PID: The Pathway Interaction Database. Nucleic Acids Res. 37, D674–679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Fu W., Sanders-Beer B. E., Katz K. S., Maglott D. R., Pruitt K. D., Ptak R. G. (2009) Human immunodeficiency virus type 1, human protein interaction database at NCBI. Nucleic Acids Res. 37, D417–422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hiller N. L., Bhattacharjee S., van Ooij C., Liolios K., Harrison T., Lopez-Estraño C., Haldar K. (2004) A host-targeting signal in virulence proteins reveals a secretome in malarial infection. Science 306, 1934–1937 [DOI] [PubMed] [Google Scholar]
- 34. Marti M., Good R. T., Rug M., Knuepfer E., Cowman A. F. (2004) Targeting malaria virulence and remodeling proteins to the host erythrocyte. Science 306, 1930–1933 [DOI] [PubMed] [Google Scholar]
- 35. Sargeant T. J., Marti M., Caler E., Carlton J. M., Simpson K., Speed T. P., Cowman A. F. (2006) Lineage-specific expansion of proteins exported to erythrocytes in malaria parasites. Genome Biol. 7, R12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Aurrecoechea C., Brestelli J., Brunk B. P., Dommer J., Fischer S., Gajria B., Gao X., Gingle A., Grant G., Harb O. S., Heiges M., Innamorato F., Iodice J., Kissinger J. C., Kraemer E., Li W., Miller J. A., Nayak V., Pennington C., Pinney D. F., Roos D. S., Ross C., Stoeckert C. J., Jr., Treatman C., Wang H. (2009) PlasmoDB: a functional genomic database for malaria parasites. Nucleic Acids Res. 37, D539–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. de Koning-Ward T. F., Gilson P. R., Boddey J. A., Rug M., Smith B. J., Papenfuss A. T., Sanders P. R., Lundie R. J., Maier A. G., Cowman A. F., Crabb B. S. (2009) A newly discovered protein export machine in malaria parasites. Nature 459, 945–949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Harris M. A., Clark J., Ireland A., Lomax J., Ashburner M., Foulger R., Eilbeck K., Lewis S., Marshall B., Mungall C., Richter J., Rubin G. M., Blake J. A., Bult C., Dolan M., Drabkin H., Eppig J. T., Hill D. P., Ni L., Ringwald M., Balakrishnan R., Cherry J. M., Christie K. R., Costanzo M. C., Dwight S. S., Engel S., Fisk D. G., Hirschman J. E., Hong E. L., Nash R. S., Sethuraman A., Theesfeld C. L., Botstein D., Dolinski K., Feierbach B., Berardini T., Mundodi S., Rhee S. Y., Apweiler R., Barrell D., Camon E., Dimmer E., Lee V., Chisholm R., Gaudet P., Kibbe W., Kishore R., Schwarz E. M., Sternberg P., Gwinn M., Hannick L., Wortman J., Berriman M., Wood V., de la Cruz N., Tonellato P., Jaiswal P., Seigfried T., White R. (2004) The Gene Ontology (GO) database and informatics resource. Nucleic Acids Res. 32, D258–261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Brèhèlin L., Dufayard J. F., Gascuel O. (2008) PlasmoDraft: a database of Plasmodium falciparum gene function predictions based on postgenomic data. BMC Bioinformatics 9, 440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Roux-Dalvai F., Gonzalez de Peredo A., Simo C., Guerrier L., Bouyssié D., Zanella A., Citterio A., Burlet-Schiltz O., Boschetti E., Righetti P. G., Monsarrat B. (2008) Extensive analysis of the cytoplasmic proteome of human erythrocytes using the peptide ligand library technology and advanced mass spectrometry. Mol. Cell Proteomics 7, 2254–2269 [DOI] [PubMed] [Google Scholar]
- 41. Durinck S., Moreau Y., Kasprzyk A., Davis S., De Moor B., Brazma A., Huber W. (2005) BioMart and Bioconductor: a powerful link between biological databases and microarray data analysis. Bioinformatics 21, 3439–3440 [DOI] [PubMed] [Google Scholar]
- 42. Remm M., Storm C. E., Sonnhammer E. L. (2001) Automatic clustering of orthologs and in-paralogs from pairwise species comparisons. J. Mol. Biol. 314, 1041–1052 [DOI] [PubMed] [Google Scholar]
- 43. Goodman S. R., Kurdia A., Ammann L., Kakhniashvili D., Daescu O. (2007) The human red blood cell proteome and interactome. Exp. Biol. Med. (Maywood) 232, 1391–1408 [DOI] [PubMed] [Google Scholar]
- 44. Marin M., Rose K. M., Kozak S. L., Kabat D. (2003) HIV-1 Vif protein binds the editing enzyme APOBEC3G and induces its degradation. Nat. Med. 9, 1398–1403 [DOI] [PubMed] [Google Scholar]
- 45. Tan S. L., Ganji G., Paeper B., Proll S., Katze M. G. (2007) Systems biology and the host response to viral infection. Nat. Biotechnol. 25, 1383–1389 [DOI] [PMC free article] [PubMed] [Google Scholar]