Abstract
The nucleolus is a nuclear organelle that coordinates rRNA transcription and ribosome subunit biogenesis. Recent proteomic analyses have shown that the nucleolus contains proteins involved in cell cycle control, DNA processing and DNA damage response and repair, in addition to the many proteins connected with ribosome subunit production. Here we study the dynamics of nucleolar protein responses in cells exposed to stress and DNA damage caused by ionizing and ultraviolet (UV) radiation in diploid human fibroblasts. We show using a combination of imaging and quantitative proteomics methods that nucleolar substructure and the nucleolar proteome undergo selective reorganization in response to UV damage. The proteomic responses to UV include alterations of functional protein complexes such as the SSU processome and exosome, and paraspeckle proteins, involving both decreases and increases in steady state protein ratios, respectively. Several nonhomologous end-joining proteins (NHEJ), such as Ku70/80, display similar fast responses to UV. In contrast, nucleolar proteomic responses to IR are both temporally and spatially distinct from those caused by UV, and more limited in terms of magnitude. With the exception of the NHEJ and paraspeckle proteins, where IR induces rapid and transient changes within 15 min of the damage, IR does not alter the ratios of most other functional nucleolar protein complexes. The rapid transient decrease of NHEJ proteins in the nucleolus indicates that it may reflect a response to DNA damage. Our results underline that the nucleolus is a specific stress response organelle that responds to different damage and stress agents in a unique, damage-specific manner.
The nucleolus forms around hundreds of repeats of ribosomal DNA (rDNA)1 genes and comprises a complex set of proteins, ribosomal RNA (rRNA) and hundreds of small nucleolar RNA (snoRNA) species. Its key function is in ribosome subunit production. In higher eukaryotes, the nucleolus is organized in distinct substructures corresponding to fibrillar centers (FC), dense fibrillar component (DFC), and granular component (GC) (1–3), in which the processes of rRNA transcription (FC), maturation of pre-RNA transcripts (DFC), assembly of preribosomal particles, and late RNA processing (GC) take place (reviewed in 4–6). Following RNA polymerase I (pol I) mediated transcription, rRNA is extensively processed by cleavage and modification, and assembled with ribosomal proteins to form the separate large and small ribosome subunits. 40S and 60S ribosomal subunits are subsequently transported to the cytoplasm (7). The rate of ribosome subunit production depends on the rDNA epigenetic state, RNA pol I activity, and rate of processing (8). These are governed by cell cycle phase, cellular metabolism, external stimuli, and stresses. Most, if not all, signaling pathways affected by growth factor and nutrient availability regulate rRNA synthesis (9). The nucleolus is therefore crucial in proliferation control and metabolic activity of the cell (reviewed in 10).
Proteomic analyses of the nucleolus and classification of putative molecular functions of nucleolar proteins have broadened the perspective of nucleolar activities (11–17). Quantitative high-resolution mass spectrometry, combined with dynamic isotope-labeling of the cells with amino acids, has shown the extensive complexity of the nucleolar proteome with over 4000 identified proteins, and demonstrated extensive stress-specific responses of the proteome (13, 17). These, and other studies on specific pathways, have provided evidence of the relevance of the nucleolus, or nucleolus-mediated processes in the regulation of tumor suppressor and oncogene activities, cell-cycle regulation, signal recognition particle assembly, modification of small RNAs, control of aging and modulation of telomerase function (reviewed in 6, 10, 18, 19). Furthermore, interference of rDNA transcription by pol I, the general pol II transcription machinery, or cellular stress caused by various substances including cytotoxic drugs, DNA intercalating agents, proteasomal stress, or viruses, cause reorganization of the nucleolar proteins and structures, and nucleolar dysfunction (3, 20–22).
DNA damage response and repair is essential for the maintenance of genomic fidelity (23). Environmental carcinogens ultraviolet (UV) and ionizing radiation (IR) cause different types of DNA lesions and are sensed and repaired by distinct and evolutionarily highly conserved pathways. UV radiation causes DNA helix distorting bulges, cyclobutane pyrimidine dimer and 6–4-photoproduct formation, which are repaired by the nucleotide excision repair pathway (24–26), and causes selective repression of cellular transcription (27) and translation (28). IR causes DNA double-strand breaks that lead to the activation of non-homologous end-joining (NHEJ) or homologous recombination repair depending on the cell cycle stage (29). UV and IR activate damage sensors ATR (ataxia-telangiectasia mutated and RAD3-related) and ATM (ataxia-telangiectasia mutated) kinase cascades, respectively. ATR and ATM kinases evoke distinct signaling cascades leading to diverse cellular responses like cell cycle arrest, DNA damage repair and apoptosis (30). Although UV-caused DNA lesions inhibit pol I and halt pol II driven transcription elongation, IR is not considered to directly interfere with the transcription machineries. However, there is evidence that IR inhibits RNA pol I activity (31). Although UV leads to nucleoplasmic relocalization of several nucleolar proteins (NPM, Ki67, HRad17, WRN) (32–34), there are only few reports on nucleolar proteins relocalizing after IR. Daniely et al. (35) showed that nucleolin is relocalized to nucleoplasm after IR but not UV damage. Interestingly, the nucleolar proteome contains several key factors involved in sensing the DNA damage or DNA repair processes, including the ATM, ATR, BLM, MRE11, PARP1, TOPBP1, WRN, XRCC1 and Ku70/80 proteins. Whether they participate in the surveillance and repair of rDNA or are redistributed to chromatin upon DNA damage is currently not known.
Here, we provide a systematic analysis of the nucleolar responses to physiologically relevant DNA damaging agents, i.e. UV and IR, utilizing a combination of cellular imaging and quantitative proteomics. We demonstrate extensive damage specific responses of functionally-related groups of nucleolar proteins.
EXPERIMENTAL PROCEDURES
Cell Culture, Chemicals, Treatments and Transfections
WS1 skin fibroblasts (CRL-1502, ATCC) were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal calf serum (FCS), nonessential amino acids, and penicillin-streptomycin. U2OS osteosarcoma cells (HTB-96, ATCC) were maintained in DMEM supplemented with 15% FCS. Open reading frames, cloned into expression vectors generating fluorescent fusion proteins (FP) with cyan or yellow FP (CFP and YFP, respectively), were derived from the EMBL-DKFZ Protein Localization Project resource (http://www.dkfz.de/LIFEdb/). Stable U2OS cell lines were generated by cotransfecting FP-expression constructs and pCDNeo selection marker using Lipofectamine (Invitrogen). Following a 2-week selection in the presence of G418, stable cell colonies were isolated and verified for the expression of the CFP or YFP fusion proteins. Cells were maintained in DMEM supplemented with 15% FCS and 1 mg/ml of G418. All cells were kept at +37 °C in a humidified atmosphere containing 5% CO2. Chemicals used were actinomycin D (Sigma) and 5,6-dichloro-1-beta-d-ribofuranosylbenzimidazole (DRB) (Sigma). All other cell culture reagents were obtained from Invitrogen. Cells were treated with ultraviolet C radiation (UVC) (35 J/m2) (254 nm UVC light bulbs, Stratalinker) or IR (1 or 10 Gy) (137Cs γ-ray source, BioBeam 8000; STS, Braunschweig, Germany).
Plasmids
NPM-enhanced cyan green fluorescent protein (ECGFP) fusion protein was generated by excision of NPM1 cDNA from B23-GFP (a kind gift from Dr. M. Olson, University of Mississippi Medical Center, MS, ref. 36) and ligation to ECGFP-pRSETb (a kind gift from Dr. A. Miyawaki, Brain Science Institute, RIKEN, Saitama, Japan, ref. 37). The construct was further subcloned to pCDNA3.1+ (Invitrogen) to yield NPM-ECGFP.
Immunofluorescence
Cells were fixed with 3.5% paraformaldehyde followed by permeabilization with 0.5% Nonidet P-40. Following primary antibodies were used: mouse anti-nucleophosmin (NPM) (Zymed Laboratories Inc., South San Francisco, CA), rabbit anti-fibrillarin (FBL) (Abcam, Cambridge, MA), rabbit anti-UBF (H-300, Santa Cruz, Santa Cruz, CA), mouse anti-DDX56 (M03, Abnova), mouse anti-Ku70 (3C3.11, Santa Cruz), rabbit anti-AATF/Che-1 (Bethyl, Montgomery, TX), and rabbit anti-nucleostemin (H-270, Santa Cruz). Antibodies were detected with secondary antibodies conjugated to Alexa 488 or 594 (Molecular Probes, Eugene, OR) and nuclei were counterstained with Hoechst 33258. The fluorochromes were visualized with Zeiss Axioplan 2 Imaging MOT (Jena) equipped with 20x/0.5NA or 40x/0.75NA Plan-Neofluar objectives and Chroma 31000v2, Chroma 41001, and Chroma 41004 filters. Images were captured with Zeiss AxioCam HRm 14-bit grayscale CCD camera and AxioVision program version 4.6. Confocal imaging was performed with Zeiss LSM510 META microscope equipped with 63x/1.25NA Plan-Neofluar or 63x/1.4NA Plan-Apochromat objectives, diode, argon and HeNe lasers. Emissions were detected with the following filter settings: BP 420–480 for Hoechst and BP 505–530 for Alexa 488. HFT405/488/543 was used as dichroic beam splitter and NFT545 as emission splitter.
Transmission Electron Microscopy (TEM)
WS1 cells were harvested by pelleting and fixed by 2.5% glutaraldehyde. The specimen was postfixed by 1% osmium tetroxide for 1 h in room temperature, dehydrated in graded ethanol and embedded in epoxy resin LX-112. Ultrathin sections were cut at 60 nm using Reichert-Jung ultra-microtome and stained by uranyl acetate and lead citrate in Leica EMstain automatic staining unit under standard protocols. Uranyl acetate and lead citrate increase resolution and contrast of cellular structures, such as nucleoli because of affinity to nucleic acids and protein. The sections were observed under Jeol JEM 1400 TEM at 80 KV. Electron micrographs were taken with Olympus-SIS Morada digital camera. All images are obtained at 5000 × magnification.
Live Cell Imaging and Data Analysis
U2OS cells were plated on 8-well Lab-Tek Chambered coverglass (Nunc). After reaching 50–80% confluency, cells were treated with UV, IR, or left untreated after which nuclei were stained with Hoechst 33342. Prior imaging, the culture medium was changed to DMEM without phenol red. Phase contrast was used for autofocusing. FP-proteins were detected using filter sets for DAPI (Semrock 5060B), CFP, and YFP (Chroma 52017; single excitation filters, double emission filter) or GFP and mRFP (Chroma 52022; single excitation filters, double emission filter) and images were captured every 10 min for 16 h using Zeiss/Intelligent Imaging Innovations (3i) - Stallion HIS live cell imaging system mounted on Zeiss Axiovert 100 with 20x/0.50NA Plan-Neofluar objective and equipped with humidified chambered heating stage and CO2 source. Cells in control experiments were viable throughout the incubation and divided at the expected rate. Raw data on each image capture were extracted and analyzed by creating an image analysis application called “CellGrain” (https://wiki.helsinki.fi/display/∼vhrantan@helsinki.fi/Cellgrain+Download+Page) in the analysis framework Anduril (38). Data on each image capture were analyzed by first identifying nuclei by Hoechst staining and to record nuclear intensities on GFP/CFP/YFP or red channels. Nuclear recognition was based on thresholding, watershedding, and removing objects smaller than 100 pixels. Nuclei were tracked to unambiguously identify changes in individual cells over time. Nucleoli were identified using a constant size local maxima finder, which finds small intensity areas brighter than immediate surroundings, and the nucleolar intensities were recorded. Background level for each image was set as 5% percentile intensity outside nuclei and all measured intensity values were subtracted with the background. Each video in the analysis contained 40 to 200 cells. Data from UV and IR treated cells were normalized to control experiments to exclude possible intensity changes of the FPs during imaging. Two-Way ANOVA analysis was applied to address statistical changes over time for each FP as compared with the control. Student's two-tailed t test was used for statistical analysis for fixed time point analyses.
Western Blotting
Nucleoli were isolated as previously described (13), lysed into Laemmli sample buffer supplemented with dithiotreitol (DTT) and sonicated briefly. Loading was normalized according to number of nucleoli in each sample. To obtain total cellular lysates, cells were scraped, solubilized in urea-Tris buffer (9 m urea, 75 mm Tris-HCl [pH 7.5] and 1 mm DTT) and sonicated. Protein concentration was determined using Bio-Rad Bradford protein assay (Bio-Rad, Hercules, CA). Equal amounts of protein were loaded into 9% SDS-PAGE and transferred into nitrocellulose membrane (Trans-Blot, Transfer Medium, Bio-Rad). Immunoblotting was carried out using anti-nucleophosmin (NPM) (Zymed Laboratories Inc.) and anti-fibrillarin (Abcam) antibodies followed by secondary antibodies conjugated to biotin and streptavidin conjugated to horseradish peroxidase (HRP), after which the signals were detected using enhanced chemiluminescence (ECL, Amersham Biosciences Life Sciences).
Stable Isotope Labeling With Amino Acids in Cell Culture (SILAC)
WS1 cells were cultivated for at least five passages in custom-made DMEM (Biowest) where arginine and lysine were replaced either by standard amino acids (Arg0, A8094; Lys0, L8662, Sigma; light) or by isotope-labeled amino acids (Arg6, CLM-2265 and Lys4, DLM-2640, Cambridge Isotope Laboratories; medium), or (Arg10, CNLM-539 and Lys8, CNLM-291 Cambridge Isotope Lab; heavy) and supplemented with 10% dialyzed FCS (Invitrogen) and penicillin-streptomycin. Cells were treated with UV, IR, or left untreated, and harvested at different times. Cells grown in light, medium, and heavy-isotope containing media treated at the indicated times were pooled and nucleoli were isolated as previously described (13). Two biological replicates were included for each treatment. Nucleoli were directly resuspended in Laemmli sample buffer supplemented with β-mercaptoethanol and boiled. Nucleolar proteins were separated by one-dimensional SDS-PAGE (4–12% Bis-Tris gel, BioRad) and visualized by colloidal Coomassie staining (Novex, Invitrogen). The entire protein gel lanes were excised and cut into six slices each. Proteins were reduced in 10 mm DTT and alkylated in 50 mm iodoacetamide. Every slice was subjected to in-gel digestion with trypsin and tryptic peptides were extracted by 1% formic acid, acetonitrile, lyophilized in a SpeedVac, and resuspended in 1% formic acid as previously described (17).
LC-MS/MS and Quantification and Analysis of Proteomic Data
Trypsin digested peptides were separated using an Ultimate U3000 (Dionex Corporation) nanoflow LC-system as in (17). Mass spectrometry was conducted essentially as in reference (17) using a LTQ Orbitrap XL (Thermo Fisher Scientific Inc) via a nano ES ion source (Proxeon Biosystems). Data were acquired using the Xcalibur software. Quantification was performed using MaxQuant version 1.0.13.13 (17, 39). The peak list generated by MaxQuant was searched using Mascot version 2.2.2 (Matrix Sciences, London, UK) as the database search engine for peptide identifications against the International Protein Index human protein database version 3.37 containing 69,290 proteins, to which 175 commonly observed contaminants and all the reversed sequences had been added (39). Mass tolerance was set to 7 ppm and MS/MS mass tolerance was 0.5 Da. Enzyme was set to trypsin with no proline restriction (trypsin/p) with two missed cleavages. Carbamidomethylation of cysteine was searched as a fixed modification, and N-acetyl protein and oxidation of methionine were searched as variable modifications. Identification of proteins was set to a false discovery rate of 1%. To achieve reliable identifications, all proteins were accepted based on the criteria that the number of forward hits in the database was at least 100-fold higher than the number of reverse database hits, thus resulting in a false discovery rate of 1%. A minimum of two peptides was quantified for each protein. Protein isoforms and proteins that cannot be distinguished based on the peptides identified are grouped.
Protein intensity values were converted to LOG2 scale to facilitate the comparison of the biological repeats. MaxQuant-indicated contaminations were excluded from the analysis. Only proteins that were identified in both biological repeats were included in further analysis. To minimize the effect of outliers, protein ratios were calculated as the means of the biological repeats. The variability of the biological repeats was defined as the standard deviation (S.D.). To minimize the variability, only proteins with S.D. less than average S.D. were considered for the downstream analysis. Pearson correlation analysis was performed on the biological repeats and was ≥0.63 for all. Student's two-tailed t test was used for statistical analysis to compare the data sets. All plotting and graphics were performed using Excel tools.
FUrd Labeling
U2OS cells plated on coverslips were treated with UV, IR, or left untreated. Cells were labeled with 5 mm 5-fluorouridine (FUrd, Sigma) for 30 min. Cells were fixed with 1% paraformaldehyde for 5 min and permeabilized with 0.1% Triton-X100. FUrd incorporation was detected using anti-BrdU antibody (Sigma) and Alexa488-conjugated secondary antibody.
Quantitative PCR
WS-1 cells were treated with UV, IR, or left untreated. Total RNAs were extracted using RNeasy kit (Qiagen) and quantified using Nanodrop. cDNA was constructed using SuperScript® III Reverse Transcriptase (Invitrogen) and Random hexamer according to the manufacture's instruction. qPCR was performed using SYBR Green (Atila Biosystem) and specific primer pairs for 5′ETS, ITS and GAPDH. Amplification was conducted for 40 cycles at 94 °C for 10 min each using ABI7900 (Applied Biosystems, Foster City, CA). The data were normalized against GAPDH. The primer sequences used were as follows. 5′ETS, forward: CCTGCTGTTCTCTCGCGCGTCCGAG, reverse: AACGCCTGACACGCACGGCACGGAG. ITS, forward: GTGGTGTGAAACCTTCCGAC, reverse: TACGAGGTCGATTTGGCG. GAPDH, forward: GGTGATGGCATCTGAATGAA, reverse: CCCTTGGCATCAGTTTCTGT. qPCR was performed using RNA from four biological repeats. Student's two-tailed t test was used for statistical analysis.
RESULTS
UV but Not IR Leads to Reorganization of Nucleolar Structures and Represses RNA Pol I Transcription
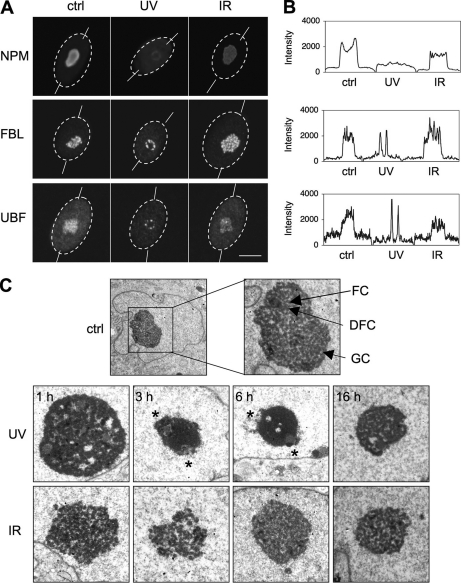
We have shown previously that nucleophosmin (NPM, B23) is a sensitive marker for UV damage stress (40) that undergoes nucleoplasmic translocation in response to cytotoxic drugs (41). To address whether different types of DNA damage have the propensity to affect nucleolar protein localization we compared changes in nucleolar protein localization following exposure to UV and ionizing radiation (IR). We irradiated normal human diploid WS1 fibroblasts either with UVC (35 J/m2) or IR (10 Gy) and stained the cells for nucleophosmin (NPM), fibrillarin (FBL) and upstream binding factor (UBF) marking GC, DFC, and FC, respectively. The changes in the cellular intensity we recorded as line plots (supplemental Fig. S1). After exposure to UV, the intensity of nucleolar NPM decreased whereas FBL and UBF accumulated in structures corresponding to nucleolar caps previously described to form consequent to transcription stress (Fig. 1A and B, ref. 42). These changes were consistent with nucleolar reorganization, and did not take place following IR (Fig. 1A and 1B). To address whether nucleoli undergo temporal structural changes following DNA damage we irradiated WS1 cells with either UV or IR and detected the nucleolar structures with transmission EM (TEM). The nucleolus was reorganized 3 to 6 h after UV treatment (Fig. 1C). This was evident by extensive condensation of GC and relocalization of FCs to the nucleolar periphery (Fig. 1C, asterisks). These features appeared to be partially reversed by 16 h. In contrast, IR did not elicit gross alterations in the nucleolar substructures, suggesting that the nucleolar response to UV was more pronounced than to IR-induced damage (Fig. 1C).
Fig. 1.
UV causes reorganization of nucleolar compartments. A, WS1 cells were treated with UVC (35 J/m2) or ionizing radiation (IR) (10 Gy) and incubated for 6 h. Cells were fixed and stained for NPM, FBL, and UBF. Confocal images are shown. Nuclear borders and line plots used in B are outlined. Full line plots are provided in supplemental Fig. S1. Scale bar, 10 μm. B, Line plots of the cellular intensities were analyzed using Fiji image processing software. C, Transmission EM images of WS1 cells left untreated or treated with UV (35 J/m2) or IR (10 Gy) for the indicated times. Nucleolar compartments, fibrillar center (FC) surrounded by dense fibrillar center (DFC) and granular component (GC) are indicated in control nucleoli. Asterisks indicate nucleolar caps.
As UV damage causes bulky DNA lesions inhibiting transcription (43), and IR has been implicated to transiently repress RNA pol I (31), we assessed RNA pol I activity in UV and IR-treated cells over time. Cells were labeled with 5-fluorouridine (FUrd) to detect nascent RNA transcripts. As shown in supplemental Fig. S2A, UV strongly and rapidly (within 15 min) repressed nascent RNA synthesis, whereas IR was without effect. Concordantly, UV but not IR caused significant and sustained inhibition of ribosomal gene transcription as measured by qPCR of ITS1 rRNA transcript (p = 0.0095) (supplemental Fig. S2B). This suggests that the cellular responses to IR at the level of ribosomal gene transcription are limited. We conclude that UV but not IR causes nucleolar protein relocalization, reorganization of nucleolar substructures, and down-regulation of RNA pol I transcription.
UV Radiation Causes Selective Changes in the Distribution of FP-tagged Nucleolar Proteins
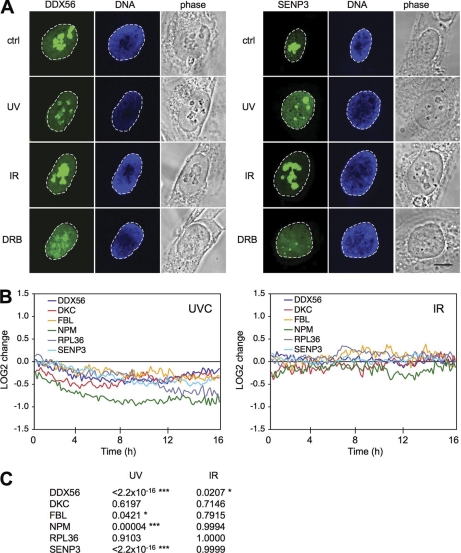
To address dynamic changes of a larger set of nucleolar proteins to DNA damage and to compare the effects of known nucleolar stressors to UV and IR, we transiently expressed in U2OS cells, separately, 24 YFP- and CFP-tagged nucleolar human ORFs from the GFP-cDNA resource (EMBL Heidelberg and DKFZ Heidelberg, http://www.dkfz.de/LIFEdb/), encoding both known and putative novel nucleolar proteins (supplemental Table S1). Additionally, we used six FP-tagged validated nucleolar proteins, i.e. UBF (44), FBL, RPAC1, RL27 (45), RRN3/TIFIA (46), and NPM-ECGFP (47). Of the initial FP-ORFs tested, only proteins that displayed nucleolar localization when expressed with both N-and C-terminal tags were included in the study. We treated the transfected cells separately with either UV or IR, and for comparison, also treated cells with the RNA pol I and II inhibitors actinomycin D and DRB. Protein phenotypes were classified, based on their nucleolar localization, as either “no change,” “increased nucleoplasmic intensity ” (NoE, indicating partial or complete nucleolar exit), and “nucleolar reorganization ” (RE, retention in nucleolar remnants, like caps or necklace-like structures), respectively. DDX56-GFP and SENP3-YFP are shown as examples for the reorganization and nucleolar exit phenotypes, respectively, after UV radiation (Fig. 2A). UV caused a noticeable change in the localization of 24 out of 30 proteins, whereas IR caused little or no changes in localization of any of the proteins tested. DRB and actinomycin D both caused extensive changes in protein localization, similar to UV, although the extent of changes varied somewhat in a manner that may depend on either the drug concentrations and/or duration of the treatments (supplemental Table S1). However, as we used fixed times for these experiments, we considered it possible that certain proteins could undergo rapid dynamic changes not detected in these assays. We therefore generated stable U2OS cell lines expressing FP-tagged nucleolar proteins for live cell image analysis and developed an algorithm for single-cell tracking and quantitative image analysis for nucleolar and nucleoplasmic intensity changes. Cells were treated with either UV or IR, or were left untreated and were followed by time-lapse imaging for 16 h. UV radiation caused a progressive and significant decrease in the nucleolar intensity in three out of six nucleolar FP-proteins, consistent with their relocalization from the nucleolus to the nucleoplasm (Fig. 2B and C). IR treatment elicited only minor changes in the localization of the nucleolar FP-proteins (Fig. 2B). In conclusion, UV but not IR induced dynamic changes in the localization of several nucleolar proteins.
Fig. 2.
Fluorescent-tagged nucleolar proteins relocalize in a damage-specific manner. A, U2OS cells stably expressing DDX56-GFP or SENP3-YFP fusion proteins were treated with UV (35 J/m2), DRB (100 μm), or IR (10 Gy) for 6 h or left untreated and fixed. Nuclei were counterstained with Hoechst 33258. Confocal images are shown. Scale bar, 10 μm. B, Live-cell image analyses of U2OS cells stably expressing YFP/CFP-tagged proteins. Cells were treated with UV (35 J/m2) or IR (10 Gy) or left untreated and stained with Hoechst 33342 (1 μg/ml). Cells were imaged with Stallion HSI wide-field microscope for 16 h and images were captured every 10 mins. Individual cells were tracked, and nucleolar and nucleoplasmic intensities were recorded. Intensity difference between nucleoli and nucleoplasm for individual cells (n = 40–200) was analyzed, normalized to untreated cells and plotted as LOG2 values. C, p values were calculated by Two-Way ANOVA analysis.
Quantitative Proteomics Reveal Extensive Dynamic Nucleolar Protein Changes Consequent to UV
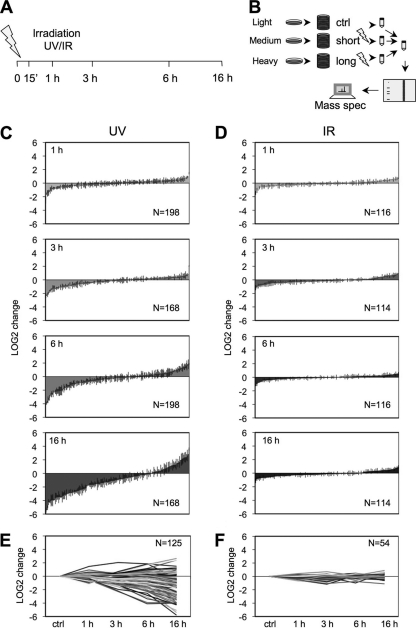
The extensive localization changes of a subset of nucleolar proteins prompted us to conduct a systematic study for dynamic nucleolar responses to UV and IR using quantitative mass spectrometry-based proteomics. This also allows the extent of changes in protein ratios to be assessed using a sensitive, unbiased and complementary technology to fluorescence imaging and to clarify the apparent absence of responses to IR. We utilized SILAC (48) of fibroblasts treated with either UV, or IR, followed by isolation of the nucleoli to measure changes in the nucleolar proteome. For this purpose we labeled WS1 cells with light, medium, and heavy isotope-containing arginine and lysine. Because every label-containing tryptic peptide occurs in three isotopic forms, its intensity, as analyzed by mass spectrometry, reveals the relative ratio of the corresponding protein over time (13, 17, 48). We hence cultivated WS1 cells with media containing the three different isotopes, treated with either UV (35 J/m2), or IR (10 Gy), and either incubated the cells for 1, 3, 6, or 16 h or left the cells untreated (control) (Fig. 3A and B). We separately verified that the cells displayed appropriate UV and IR-related damage responses, such as phosphorylation of H2AX, evident in both UV and IR-treated cells as diffuse and focal nuclear staining representing replicative and double-strand break damage, respectively (supplemental Fig. S3). Cells were harvested, pooled, and nucleoli were isolated as previously described, proteins were separated by SDS-PAGE, trypsin-digested, and analyzed by mass spectrometry (13). The resulting peptides were analyzed and quantified using MaxQuant (39), resulting in differences between ratios in isotopes for each peptide (see supplemental Data set S1 for an example of raw data). Two biological replicates were analyzed for each condition. To increase the robustness of the analysis, only proteins quantifiable in both biological experiments and with LOG2 values less than the average standard deviation of each experiment were included in the final data set. By using these stringent inclusion criteria, 100–200 proteins were included for each treatment condition (see supplemental Tables S2–S5 for complete proteomic responses of the biological replicates). Following UV radiation, the number of proteins showing decreased nucleolar accumulation increased over time (Fig. 3C and 3E). At the same time, a subset of proteins increased their accumulation in the nucleolus. The ratios of proteomic changes following UV consistently increased up to 16 h (Fig. 3C and 3E). However, the IR treated cells displayed only minor changes in the steady state content of the nucleolar proteome (Fig. 3D and 3F). The UV provoked changes could reflect gross nucleolar reorganization and altered functional activities of the nucleolus.
Fig. 3.
Quantitative proteomics of nucleolar UV and IR responses. A, WS1 cells were treated with UV (35 J/m2) or IR (10 Gy) or left untreated and harvested at the indicated times. B, Stable isotope labeling of amino acids in cell culture (SILAC). WS1 cells were passaged at least five times in media containing different isotopes of carbon and nitrogen. Cells were then treated with UV or IR followed by harvesting and pooling of cells, and nucleoli were isolated. Nucleolar extracts were resolved by gradient SDS-PAGE, the gel was cut into pieces and proteins were trypsin digested. The resulting peptides were analyzed by LTQ Orbitrap XL mass spectrometer and quantified using MaxQuant software. Two biological repeats were performed for each treatment and only proteins identifiable in both experiments with standard deviations under the average standard deviation of the experiment were included in the analyses. All data was plotted as normalized LOG2 values against the control. Negative LOG2 values indicate reduced nucleolar abundance whereas positive values indicate increased nucleolar abundance as compared with the untreated sample. Numbers (N) of proteins are indicated. Error bars, S.D. C, LOG2 values of proteins following UV. D, LOG2 values of proteins following IR. E, LOG2 changes of proteins following UV over time. F, LOG2 changes of proteins following IR over time.
UV and IR Radiation Lead to Markedly Divergent Responses
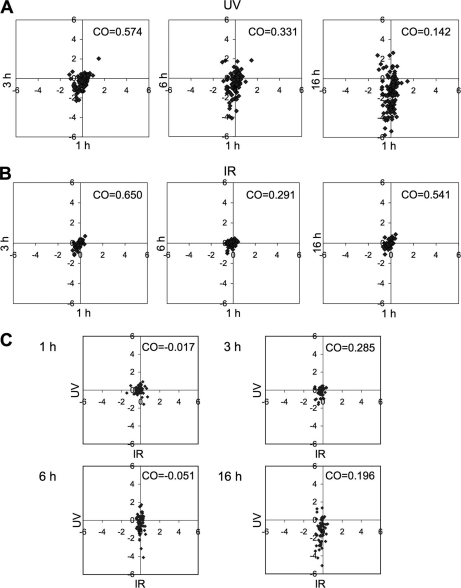
The dynamic nucleolar protein responses to the different stresses were evaluated by Pearson correlation (CO) analysis by comparing the changes in nucleolar protein abundances over time. UV caused a progressive change in the nucleolar protein content as estimated by CO analysis (Fig. 4A). This was evident as a marked decrease of CO (from 0.574 to 0.142) in the UV treated cells when 3, 6, and 16 h proteomes were compared with the 1 h proteome, indicating an increasing difference in the UV response over time (Fig. 4A). Interestingly, similar comparison of IR treated cells indicated a marked decrease in the CO between the 1 and 6 h proteomes (0.291), whereas 3 and 16 h proteomes differed only slightly (0.650 and 0.541, respectively) (Fig. 4B). This suggested that the IR-induced nucleolar proteomic changes are subtler and temporally restricted. Lastly, we compared the changes in nucleolar proteins caused by UV and IR. As shown by the CO analysis, the UV and IR responses were distinct (CO –0.017, 0.285, –0.051 and 0.196 at 1, 3, 6 and 16 h, respectively) (Fig. 4C).
Fig. 4.
Comparative analyses of nucleolar proteomes in UV and IR treated cells. A, Kinetic changes in the nucleolar proteome following UV treatment. LOG2 values at 1 h (x axis) were compared with those at 3, 6 and 16 h (y axis). n = 125 proteins. B, Kinetic changes in the nucleolar proteome following IR treatment. LOG2 values at 1 h (x axis) were compared with those at 3, 6, and 16 h (y axis). n = 53 proteins. C, Comparison of LOG2 values of UV and IR nucleolar proteomes. IR and UV data are plotted on x- and y axis, respectively. n = 53 proteins. Correlation coefficiencies (CO) were measured for each set.
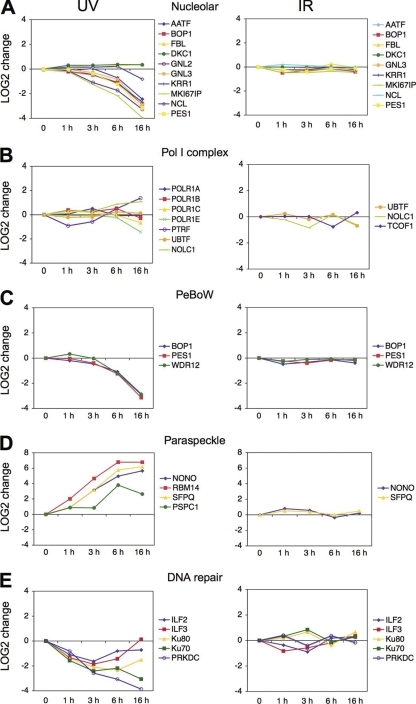
Changes in Functionally Related Groups of Nucleolar Proteins by UV
Analysis of ten accredited nucleolar proteins showed examples of proteins that underwent a marked change over time and proteins that did not (Fig. 5A). Proteins showing the most extensive change in response to UV included Ki67, GNL2 and GNL3 (nucleostemin), PES1, a representative of PeBoW RNA maturation complex, and AATF/Che-1, a RNA polymerase II binding protein (Fig. 5A). However, FBL and DKC1 were retained within the nucleolar structures following UV (Fig. 5A). Unexpectedly, there was no change in nucleolar ratio of NPM, which was predicted to relocalize to the nucleoplasm both by immunofluorescence and live cell imaging. This possibly results from the saturation of NPM signals because of the highly abundant nature of the protein and low dynamic range at intensity levels > 1 × 109, which were observed for NPM (not shown). Following IR treatment none of the above nucleolar proteins displayed marked alterations suggesting that they have no discernible responses after IR damage, at least with respect to their nucleolar accumulation.
Fig. 5.
Dynamic profiling of functional protein groups. Profiles of the proteins are plotted as normalized LOG2 values against the control over time. A, Selected nucleolar proteins. B, RNA polymerase I complex proteins. C, PeBoW complex proteins. D, Paraspeckle proteins. E, NHEJ-affiliated proteins.
To address whether the dynamic proteomic changes reflect changes in functionally related groups of nucleolar proteins, we then classified individual proteins using public and nucleolar proteomic databases and identified functional protein groups (13, 14). Considering that UV led to inhibition of RNA pol I activity, we analyzed for changes in RNA pol I protein subunits and associated factors. In contrast to changes observed in the majority of nucleolar proteins, the changes of RNA pol I subunits (A, B, C, E), UBF, and NOLC1 were less pronounced (Fig. 5B). PTRF, which causes dissociation of pol I transcription complex, underwent transient nucleolar decrease. IR did not cause consistent discernible changes in three RNA pol I associated proteins (Fig. 5B). These findings suggest that although UV inhibits RNA pol I transcription, proportions of RNA pol I components do not decrease in the nucleolus, although they may be reorganized either to nucleolar caps or necklace-like structures (Fig. 1) (41, 49).
Given that small ribosomal subunit proteins are extensively exported from nucleoli following actinomycin D treatment (13), we compared ribosomal protein responses to UV and IR. As shown in Supplemental Fig. S4A and B, UV caused initial accumulation of large ribosomal proteins within 1 h, whereas the proportions of small ribosomal proteins in the nucleolus decreased. Over time these changes became more varied. IR did not cause major changes in the ratios of either large or small subunit ribosomal proteins. Similarly, we analyzed changes in the small subunit (SSU) processome. These contain both ribosomal and nonribosomal proteins that bind to U3 snoRNA and are required for 18S biogenesis (50, 51). UV caused a marked decrease in the nucleolar levels of the SSU processome proteins, whereas IR had only a marginal effect (supplemental Fig. S4C).
RNA helicases remodel RNA, RNA-protein complexes, or unwind RNA (reviewed in ref. 52). We identified DEAD box proteins and detected their changes in response to stresses. Although IR did not elicit any marked effect, UV caused predominantly a decrease in the DEAD box proteins, with the exception of DDX5 and DDX17, which have previously been identified as proteins accumulating in the nucleolus also in actinomycin d-treated cells (supplemental Fig. S4D) (12). Based on these results we conclude that despite domain similarities RNA helicases do not respond as a protein group to UV damage.
We then addressed responses of exosome complex proteins that function in RNA processing in eukaryotes, and whose nucleolar presence is prominently reduced in actinomycin d-treated cells (13). Proportions of all exosome proteins identified in this study were decreased in nucleoli after UV treatment, with similar kinetics of decrease indicating their close connection (supplemental Fig. S4E). IR responses of the exosome proteins did not display any consistent pattern (supplemental Fig. S4E). However, EXOSC1, 2 and 5 displayed rapid and transient decreases, the significance of which is not presently clear.
PeBoW complex proteins (PES1, BOP1 and WDR12) function in maturation of 28S and 5.8S rRNAs and formation of the 60S complex. We observed that upon UV their association with the nucleolus decreased with similar kinetics, suggesting complex formation and a possible functional response to UV (Fig. 5C).
Paraspeckle proteins form distinct subnuclear bodies together with an essential large noncoding RNA, NEAT (53, 54). The core paraspeckle proteins (PSF/SFPQ, P54NRB/NONO, PSPC1, RBM14) accumulated prominently in the nucleolus after UV (Fig. 5D). This finding is consistent with paraspeckle protein movement to the nucleolar periphery after inhibition of RNA polymerases (53). IR increased the nucleolar accumulation of paraspeckle proteins at early times after the damage. Interestingly, a group of RNA-interacting proteins present in nuclear speckles (ASF/SFRS1, RBMX, RBM4, SLTM) displayed similar marked nucleolar accumulation after UV, suggesting they could either be affiliated with paraspeckle proteins or share functional activities (data not shown).
Several DNA damage sensing and repair pathway proteins localize to nucleoli (13). We were therefore interested whether the DNA stresses also change their nucleolar occupancy. As shown in Fig. 5E, nonhomologous end-joining (NHEJ) pathway proteins Ku80 (XRCC5) and Ku70 (XRCC6) and the catalytic subunit of DNA-PK (PRKDC) were present in the nucleoli and detectable throughout the time course studied. Moreover, heterodimeric ILF2 and ILF3, which are dsRNA binding proteins and interact with Ku70/80 and PRKDC, displayed similar kinetic responses to UV (Fig. 5E). The proportions of these proteins decreased in the nucleolus by 3 h after UV, and although Ku70/80 and PRKDC demonstrated sustained nucleolar depletion, nucleolar ILF2/3 were recovered 6 and 16 h after the damage. However, the responses of the NHEJ-like proteins to IR were more diverse. Ku70/80 were accumulated selectively 3 h after IR, whereas ILF2/3 were decreased over the first 3 h, and PRKDC fluctuated (Fig. 5E).
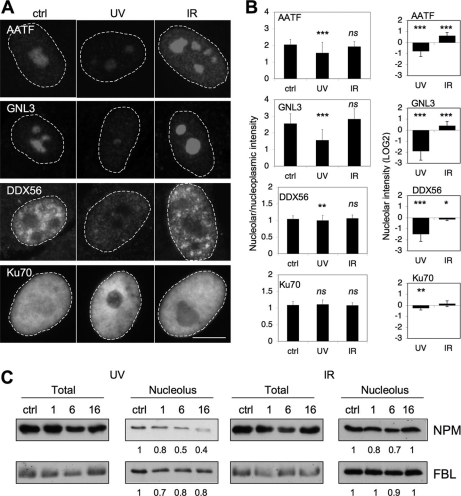
To corroborate the proteomic data we used immunofluorescence and Western blotting to detect changes in a subset of proteins which underwent prominent changes. UV or IR-treated WS1 cells were stained with antibodies against AATF, GNL3, DDX56, and Ku70 proteins, and the intensities were quantified. As shown in Fig. 6A and B, all proteins showed decreased nucleolar prominence after UV, which was consistent with the proteomic data. None of the proteins showed altered localizations 16 h following IR (Fig. 6A). Subsequent analysis of nucleolar extracts isolated from UV-treated cells by immunoblotting for NPM and FBL indicated substantial depletion of NPM whereas FBL was retained in the nucleolar fraction (Fig. 6C).
Fig. 6.
Immunofluorescence and immunoblotting analysis of selected nucleolar proteins. A, WS1 cells were treated with UV (35 J/m2) and IR (10 Gy) and fixed after 16 h. Cells were stained for AATF, GNL3 (nucleostemin), DDX56, and Ku70 proteins and imaged using widefield fluorescence microscopy. Scale bar, 10 μm. B, Ratios between nucleolar and nucleoplasmic intensities and LOG2 changes in nucleolar intensity as compared with control are shown. Error bars, S.D. n = 25–158 cells. **p < 0.01; ***p < 0.001; ns, nonsignificant. C, WS1 cells were treated as above and harvested at 1, 6 and 16 h postdamage. Cells were extracted with urea buffer and isolated nucleoli were resuspended directly in Laemmli sample buffer and sonicated briefly. Equal numbers of nucleoli or total protein were separated by SDS-PAGE and immunoblotted for FBL and NPM.
IR Induces Fast and Transient Changes of Nucleolar Proteins
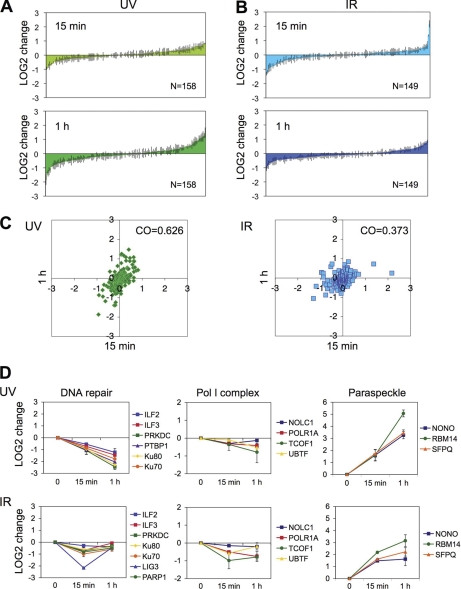
Given that IR has been reported to cause rapid changes in nucleolar activity (31) and that the UV and IR responses differed markedly 1 h after the damage, we analyzed the nucleolar proteomic changes 15 min and 1 h after the insults by quantitative proteomics. IR caused rapid changes of the nucleolar response, which was more extensive in the 15 min than 1 h proteome (LOG2 change 15 min compared with 1 h, 0.266 and 0.255, respectively, p = 0.000000298). UV-induced proteomic changes, however, were more marked at 1 h than at 15 min (LOG2 change 15 min compared with 1 h, 0.255 and 0.380, respectively p = 0.000000631) (Fig. 7A and 7B). These changes were further corroborated by Pearson correlation analyses that supported the presence of rapid proteomic changes, especially in the IR-treated cells (Fig. 7C). The proteomic analysis was therefore suggestive of rapid response of a subset of nucleolar proteome consequent to IR, and progressive changes in response to UV.
Fig. 7.
Rapid IR damage responses of the nucleolar proteome. A, Changes in LOG2 values of proteins in UV-treated cells (35 J/m2) at 15 min and 1 h postdamage as compared with control. Error bars, S.D. B, Changes in LOG2 values of proteins in IR-treated cells (10 Gy) at 15 min and 1 h postdamage as compared with control. Error bars, S.D. C, Rapid kinetic changes in the nucleolar proteome following UV and IR treatments. LOG2 values at 15 min (x axis) were compared with those at 1 h (y axis). Correlation coefficiencies (CO) were measured for each set. D, Profiles of DNA repair proteins, RNA polymerase I (Pol I) complex proteins and paraspeckle proteins are plotted as normalized LOG2 values as compared with control over time. UV and IR treatments are indicated. Error bars, S.D.
To assess the nature of the IR-induced rapid changes, we looked at changes in the functionally related protein groups. The UV-induced decrease in nucleolar association of the NHEJ-like repair complex was already evident within 15 min (Fig. 7D). IR caused a transient decrease in several NHEJ-complex proteins, including PARP1. The decrease in DNA ligase III (LIG3), which is an essential component of single-strand break repair and base excision repair, was most pronounced (4-fold) and was fully recovered by 1 h (Fig. 7D). Likewise, Treacher Collins syndrome gene product (TCOF1), which interacts with UBF and regulates rDNA transcription (55) was rapidly decreased together with UBF and pol I catalytic subunit (POLR1A) (Fig. 7D). Paraspeckle proteins, which accumulated strongly in response to UV and modestly in response to IR, demonstrated also rapid accumulation in nucleoli after IR (Fig. 7D).
Taken together, these results suggest that multiple classes of nucleolar proteins, including DNA damage pathway proteins, show temporally distinct and damage-specific responses possibly reflecting alterations in the nucleolar function or their involvement in the cellular stress responses.
DISCUSSION
We present here extensive mapping of kinetics of human nucleolar proteins in response to two physiologically and therapeutically relevant DNA damaging agents, i.e. UV and IR. By utilizing cellular imaging methods and quantitative proteomics we show extensive alterations in nucleolar proteins in response to UV radiation. The UV induced change is largely progressive and reflects extensive nucleolar reorganization. In contrast, we show that IR does not substantially alter the nucleolar morphology and provokes highly selective proteomic responses that are very rapid and fluctuate over time. By grouping proteins based on their annotated functions, we identify proteins with similar dynamics. Highly coordinated dynamics were detected for PeBoW complex, paraspeckle proteins, a subset of NHEJ-like proteins and exosome components in response to UV, which represented both decreases and increases of nucleolar proteins. IR altered the proportions of several NHEJ-like proteins and RNA pol I components, and increased the accumulation of paraspeckle proteins with the nucleolar structures.
The nucleolus has been suggested to act as a cellular stress sensor (56). We and others have previously shown that the nucleolus responds to different cellular stresses, like UV radiation and cytotoxic drugs, by causing relocalization of NPM to the nucleoplasm (17, 20, 40). These relocalizations likely reflect a meaningful cellular response and active engagement of the translocated proteins in the alternative compartments. For example, the translocation of NPM promotes its interaction with p53 and MDM2, binding to chromatin both in a damage-specific and possibly in a RNA-specific manner, and is responsible for the transport of other nucleolar proteins, like RPL5 and ARF, and 40S and 60S ribosomal subunits (19, 40, 57–59). NPM also controls APE1/Ref-1 base excision repair protein localization and activity thus impacting rRNA quality control (60). In addition, many RNA virus proteins localize to the nucleolus and scavenge the nucleolar proteome for virus production (22). Furthermore, quantitative proteomic analyses have indicated that the nucleolus undergoes a constant bidirectional flux of dynamically moving proteins in cells exposed to RNA pol I stress, whereas RNA pol II stress mediated changes are more limited (13). These findings underscore that the nucleolar responses are distinct in face of different physiological and pathophysiological states, and that the nucleolar proteins may possess also extranucleolar activities.
We show here by live cell imaging, TEM and quantitative proteomics that the nucleolus undergoes massive reorganization following UV but not IR. The UV-induced morphological changes, condensation of GC, and disengagement of FCs to form caplike and nucleolar necklace-like structures are consistent with inhibition of RNA pol I and pol II (3, 41, 49, 61) (Fig. 1). Analysis of UV responses of thirty FP-tagged nucleolar proteins and comparisons to localization changes caused by actinomycin D and DRB, under conditions inhibiting RNA pol I and RNA pol II, was compatible with a complex response befitting inactivation of both RNA polymerases. However, IR did not cause either nucleolar segregation, or detectable changes in the representative nucleolar proteins analyzed. These findings were further confirmed with live-cell fluorescence microscopy analysis of localization changes in FP-tagged nucleolar proteins. Consistent with the morphological changes, we find that ribosomal gene transcription, as measured by FUrd incorporation and qPCR, is significantly down-regulated by UV, but not by IR, even at the high dose (10 Gy) used here. This is in contrast to results in a prior study by Kruhlak et al. (31), who reported transient ATM-pathway dependent decrease of FUrd incorporation in murine embryonic fibroblasts. It is possible that either transient decreases in RNA pol I activity by IR are not sufficiently captured by the assays used here, or that the discrepancies result from cell type-specific variation. It is also of interest that the prominent repression of RNA pol I activity by UV is not associated with extensive changes in RNA pol I components in the nucleoli. These findings are consistent with the imaging data and previous functional and proteomic analyses showing RNA pol I complex reorganization to nucleolar caps or necklaces consequent to inhibition of RNA pol I and RNA pol II (3, 13, 41). In contrast, IR caused a rapid and transient reduction in nucleolar expression of UBF, TCOF1, and POLR1A in IR-treated cells, which however appear to be compensated at the level of rRNA transcription.
The UV-induced reorganization is accompanied by changes in numerous nucleolar proteins. Most of these changes are progressive and lead to extensive decrease in the amount of bona fide nucleolar proteins, like SSU processome proteins, several DEAD box helicases, and PeBoW complex proteins. Although many ribosomal proteins are decreased, the kinetics of large and small subunit proteins are distinct. UV causes a biphasic response of small ribosomal proteins as visualized by a group with rapid decrease at 1 h and recovery at later times (RPS3, RPS10, RPS20, RPS25), whereas others undergo a slow and sustained reduction. However, the proportion of large ribosomal proteins was initially increased, with varying degrees of increase and decrease over time. The extensive reductions of the SSU processome and PeBoW proteins are indicative that rRNA processing is markedly affected. Overall, these findings are consistent with severe repression of ribosome subunit production and processing, and that individual ribosomal proteins, like RPS3, implicated in DNA damage response (62), may have more specific functions in repair or transcriptional stress responses.
The paraspeckle protein (PSF/SFPQ, p54NRB/NONO, PSPC1, RBM14) ratios were strongly increased in response to UV in a manner that resembled the RNA pol I mediated stress (13). Furthermore, a group of RNA-binding proteins, present in nucleoplasmic speckles (ASF/SFRS1, RBMX, RBM4, SLTM) showed highly similar dynamics suggesting that they may represent novel paraspeckle proteins or that the nucleolar caps may retain also other RNA splicing and processing proteins. In addition, we find an extensive decrease in several NHEJ-repair complex proteins in response to UV. These include DNA-PK, Ku70, and Ku80 proteins, ILF2 and ILF3, which interact with the DNA-PK/Ku-complex. Interestingly, DNA-PK activity may not only be relevant to damage repair, but may also mediate selective repression of protein translation in response to UV damage (63).
We detected fast and transient changes in nucleolar proteome rapidly (15 min) after IR. The amplitude of these changes was not substantial but the imminent 15 min IR response differed in a highly significant manner from the 1 h response (Fig. 7). These IR responses were exemplified by rapid decrease of DNA repair proteins, including PARP1 and DNA ligase III (LIG3), and their nucleolar recovery by 1 h. Therefore it seems possible that these alterations reflect a biologically relevant response to DNA double-strand break damage. Unexpectedly, we find that IR leads to a transient nucleolar increase in paraspeckle proteins. Whether this reflects inhibition of RNA pol I function or engagement of the paraspeckle proteins in other nucleolus-associated activities in the nucleolar domain is not presently known. p54NRB/NONO has also recently been associated with DNA double-strand break repair and damage recovery (64, 65).
Here we have studied nucleolar responses to UV and IR induced cellular stress and DNA damage. This is the first time that nucleolar responses of diploid human cells to physiologically relevant cellular damage caused by UV and IR have been mapped to this extent. This study reveals detailed responses of a large number of individual proteins in a damage- and time-dependent manner. By utilizing quantitative proteomics, live cell imaging and biochemical analysis, we demonstrate changes of nucleolar proteins responding specifically to different DNA damaging and cellular stress agents UV and IR. These data provides a framework for understanding dynamics of nucleolar responses to DNA damage in normal human cells.
Acknowledgments
We thank Maija Salo, Hester Liu, Anni-Helena Sukupolvi and Kaisa Penttilä for excellent technical assistance, Dr. Haiping Hao for help with statistical analyses, Dr. Atsushi Miyawaki for ECGFP-vector, Dr. Mark Olson for B23-GFP construct, Molecular Imaging Unit (University of Helsinki) for image analysis, Fang Zhao (Advanced Microscopy Unit, University of Helsinki) for help with TEM, Douglas Lamont and the University of Dundee CLS Proteomics Facility for mass spectrometry analyses and Laiho lab members for helpful discussions.
Footnotes
* This work was supported by Academy of Finland (M.L. grant no 129699, L.L. grant no. 108828) and Biocentrum Helsinki. AIL is a Wellcome Trust Principal Research Fellow. HMM was supported by the Helsinki Biomedical Graduate School. FMB is supported by a fellowship from the Caledonian Research Foundation.
 This article contains supplemental Figs. S1 to S4, Tables S1 to S5 and Data set S1.
This article contains supplemental Figs. S1 to S4, Tables S1 to S5 and Data set S1.
1 The abbreviations used are:
- rDNA
- ribosomal DNA
- CFP
- cyan fluorescent protein
- CO
- correlation coefficiency
- DFC
- dense fibrillar center
- DRB
- 5,6-dichloro-1-beta-D-ribofuranosylbenzimidazole
- dsRNA
- double-strand RNA
- ECGFP
- enhanced cyan green fluorescent protein
- FBL
- fibrillarin
- FC
- fibrillar center
- FP
- fluorescent protein
- GC
- granular component
- IR
- ionizing radiation
- NHEJ
- nonhomologous end-joining
- NPM
- nucleophosmin
- RNA pol I
- RNA polymerase I
- RNA pol II
- RNA polymerase II
- rRNA
- ribosomal RNA
- SSU
- small subunit
- TEM
- transmission electron microscopy
- UBF
- upstream binding factor
- YFP
- yellow fluorescent protein.
REFERENCES
- 1. Olson M. O., Dundr M., Szebeni A. (2000) The nucleolus: An old factory with unexpected capabilities. Trends Cell Biol. 10, 189–196 [DOI] [PubMed] [Google Scholar]
- 2. Hernandez-Verdun D. (2006) Nucleolus: From structure to dynamics. Histochem. Cell Biol. 125, 127–137 [DOI] [PubMed] [Google Scholar]
- 3. Sirri V., Urcuqui-Inchima S., Roussel P., Hernandez-Verdun D. (2008) Nucleolus: The fascinating nuclear body. Histochem. Cell Biol. 129, 13–31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Leary D. J., Huang S. (2001) Regulation of ribosome biogenesis within the nucleolus. FEBS Lett. 509, 145–150 [DOI] [PubMed] [Google Scholar]
- 5. Fatica A., Tollervey D. (2002) Making ribosomes. Curr. Opin. Cell Biol. 14, 313–318 [DOI] [PubMed] [Google Scholar]
- 6. Olson M. O., Dundr M. (2005) The moving parts of the nucleolus. Histochem. Cell Biol. 123, 203–216 [DOI] [PubMed] [Google Scholar]
- 7. Zemp I., Kutay U. (2007) Nuclear export and cytoplasmic maturation of ribosomal subunits. FEBS Lett. 581, 2783–2793 [DOI] [PubMed] [Google Scholar]
- 8. McStay B., Grummt I. (2008) The epigenetics of rRNA genes: From molecular to chromosome biology. Annu. Rev. Cell Dev. Biol. 24, 131–157 [DOI] [PubMed] [Google Scholar]
- 9. Mayer C., Grummt I. (2006) Ribosome biogenesis and cell growth: MTOR coordinates transcription by all three classes of nuclear RNA polymerases. Oncogene 25, 6384–6391 [DOI] [PubMed] [Google Scholar]
- 10. Boisvert F. M., van Koningsbruggen S., Navascués J., Lamond A. I. (2007) The multifunctional nucleolus. Nat. Rev. Mol. Cell Biol. 8, 574–585 [DOI] [PubMed] [Google Scholar]
- 11. Scherl A., Couté Y., Déon C., Callé A., Kindbeiter K., Sanchez J. C., Greco A., Hochstrasser D., Diaz J. J. (2002) Functional proteomic analysis of human nucleolus. Mol. Biol. Cell 13, 4100–4109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Andersen J. S., Lyon C. E., Fox A. H., Leung A. K., Lam Y. W., Steen H., Mann M., Lamond A. I. (2002) Directed proteomic analysis of the human nucleolus. Curr. Biol. 12, 1–11 [DOI] [PubMed] [Google Scholar]
- 13. Andersen J. S., Lam Y. W., Leung A. K., Ong S. E., Lyon C. E., Lamond A. I., Mann M. (2005) Nucleolar proteome dynamics. Nature 433, 77–83 [DOI] [PubMed] [Google Scholar]
- 14. Hinsby A. M., Kiemer L., Karlberg E. O., Lage K., Fausbøll A., Juncker A. S., Andersen J. S., Mann M., Brunak S. (2006) A wiring of the human nucleolus. Mol. Cell 22, 285–295 [DOI] [PubMed] [Google Scholar]
- 15. Ahmad Y., Boisvert F. M., Gregor P., Cobley A., Lamond A. I. (2009) NOPdb: Nucleolar proteome database–2008 update. Nucleic Acids Res. 37, D181–4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Pederson T., Tsai R. Y. (2009) In search of nonribosomal nucleolar protein function and regulation. J. Cell Biol. 184, 771–776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Boisvert F. M., Lam Y. W., Lamont D., Lamond A. I. (2010) A quantitative proteomics analysis of subcellular proteome localization and changes induced by DNA damage. Mol. Cell. Proteomics 9, 457–470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ruggero D., Pandolfi P. P. (2003) Does the ribosome translate cancer? Nat. Rev. Cancer 3, 179–192 [DOI] [PubMed] [Google Scholar]
- 19. Grisendi S., Mecucci C., Falini B., Pandolfi P. P. (2006) Nucleophosmin and cancer. Nat. Rev. Cancer 6, 493–505 [DOI] [PubMed] [Google Scholar]
- 20. Rubbi C. P., Milner J. (2003) Disruption of the nucleolus mediates stabilization of p53 in response to DNA damage and other stresses. EMBO J. 22, 6068–6077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Drygin D., Rice W. G., Grummt I. (2010) The RNA polymerase I transcription machinery: An emerging target for the treatment of cancer. Annu. Rev. Pharmacol. Toxicol. 50, 131–156 [DOI] [PubMed] [Google Scholar]
- 22. Hiscox J. A. (2007) RNA viruses: Hijacking the dynamic nucleolus. Nat. Rev. Microbiol. 5, 119–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Jackson S. P., Bartek J. (2009) The DNA-damage response in human biology and disease. Nature 461, 1071–1078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ravanat J. L., Douki T., Cadet J. (2001) Direct and indirect effects of UV radiation on DNA and its components. J. Photochem. Photobiol. B. 63, 88–102 [DOI] [PubMed] [Google Scholar]
- 25. Friedberg E. C. (2001) How nucleotide excision repair protects against cancer. Nat. Rev. Cancer 1, 22–33 [DOI] [PubMed] [Google Scholar]
- 26. Herrlich P., Karin M., Weiss C. (2008) Supreme EnLIGHTenment: Damage recognition and signaling in the mammalian UV response. Mol. Cell 29, 279–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Latonen L., Laiho M. (2005) Cellular UV damage responses–functions of tumor suppressor p53. Biochim. Biophys. Acta 1755, 71–89 [DOI] [PubMed] [Google Scholar]
- 28. Deng J., Harding H. P., Raught B., Gingras A. C., Berlanga J. J., Scheuner D., Kaufman R. J., Ron D., Sonenberg N. (2002) Activation of GCN2 in UV-irradiated cells inhibits translation. Curr. Biol. 12, 1279–1286 [DOI] [PubMed] [Google Scholar]
- 29. Wyman C., Kanaar R. (2006) DNA double-strand break repair: All's well that ends well. Annu. Rev. Genet. 40, 363–383 [DOI] [PubMed] [Google Scholar]
- 30. Durocher D., Jackson S. P. (2001) DNA-PK, ATM and ATR as sensors of DNA damage: Variations on a theme? Curr. Opin. Cell Biol. 13, 225–231 [DOI] [PubMed] [Google Scholar]
- 31. Kruhlak M., Crouch E. E., Orlov M., Montaño C., Gorski S. A., Nussenzweig A., Misteli T., Phair R. D., Casellas R. (2007) The ATM repair pathway inhibits RNA polymerase I transcription in response to chromosome breaks. Nature 447, 730–734 [DOI] [PubMed] [Google Scholar]
- 32. Chang M. S., Sasaki H., Campbell M. S., Kraeft S. K., Sutherland R., Yang C. Y., Liu Y., Auclair D., Hao L., Sonoda H., Ferland L. H., Chen L. B. (1999) HRad17 colocalizes with NHP2L1 in the nucleolus and redistributes after UV irradiation. J. Biol. Chem. 274, 36544–36549 [DOI] [PubMed] [Google Scholar]
- 33. Al-Baker E. A., Boyle J., Harry R., Kill I. R. (2004) A p53-independent pathway regulates nucleolar segregation and antigen translocation in response to DNA damage induced by UV irradiation. Exp. Cell Res. 292, 179–186 [DOI] [PubMed] [Google Scholar]
- 34. Blander G., Zalle N., Daniely Y., Taplick J., Gray M. D., Oren M. (2002) DNA damage-induced translocation of the werner helicase is regulated by acetylation. J. Biol. Chem. 277, 50934–50940 [DOI] [PubMed] [Google Scholar]
- 35. Daniely Y., Dimitrova D. D., Borowiec J. A. (2002) Stress-dependent nucleolin mobilization mediated by p53-nucleolin complex formation. Mol. Cell. Biol. 22, 6014–6022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Dundr M., Misteli T., Olson M. O. (2000) The dynamics of postmitotic reassembly of the nucleolus. J. Cell Biol. 150, 433–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Sawano A., Miyawaki A. (2000) Directed evolution of green fluorescent protein by a new versatile PCR strategy for site-directed and semi-random mutagenesis. Nucleic Acids Res. 28, E78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Ovaska K., Laakso M., Haapa-Paananen S., Louhimo R., Chen P., Aittomäki V., Valo E., Núñez-Fontarnau J., Rantanen V., Karinen S., Nousiainen K., Lahesmaa-Korpinen A. M., Miettinen M., Saarinen L., Kohonen P., Wu J., Westermarck J., Hautaniemi S. (2010) Large-scale data integration framework provides a comprehensive view on glioblastoma multiforme. Genome Med. 2, 65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Cox J., Mann M. (2008) MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372 [DOI] [PubMed] [Google Scholar]
- 40. Kurki S., Peltonen K., Latonen L., Kiviharju T. M., Ojala P. M., Meek D., Laiho M. (2004) Nucleolar protein NPM interacts with HDM2 and protects tumor suppressor protein p53 from HDM2-mediated degradation. Cancer Cell 5, 465–475 [DOI] [PubMed] [Google Scholar]
- 41. Chan P. K., Qi Y., Amley J., Koller C. A. (1996) Quantitation of the nucleophosmin/B23-translocation using imaging analysis. Cancer Lett. 100, 191–197 [DOI] [PubMed] [Google Scholar]
- 42. Shav-Tal Y., Blechman J., Darzacq X., Montagna C., Dye B. T., Patton J. G., Singer R. H., Zipori D. (2005) Dynamic sorting of nuclear components into distinct nucleolar caps during transcriptional inhibition. Mol. Biol. Cell 16, 2395–2413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. de Laat W. L., Jaspers N. G., Hoeijmakers J. H. (1999) Molecular mechanism of nucleotide excision repair. Genes Dev. 13, 768–785 [DOI] [PubMed] [Google Scholar]
- 44. Chen D., Huang S. (2001) Nucleolar components involved in ribosome biogenesis cycle between the nucleolus and nucleoplasm in interphase cells. J. Cell Biol. 153, 169–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Leung A. K., Gerlich D., Miller G., Lyon C., Lam Y. W., Lleres D., Daigle N., Zomerdijk J., Ellenberg J., Lamond A. I. (2004) Quantitative kinetic analysis of nucleolar breakdown and reassembly during mitosis in live human cells. J. Cell Biol. 166, 787–800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Dundr M., Hoffmann-Rohrer U., Hu Q., Grummt I., Rothblum L. I., Phair R. D., Misteli T. (2002) A kinetic framework for a mammalian RNA polymerase in vivo. Science 298, 1623–1626 [DOI] [PubMed] [Google Scholar]
- 47. Latonen L., Moore H. M., Bai B., Jäämaa S., Laiho M. (2011) Proteasome inhibitors induce nucleolar aggregation of proteasome target proteins and polyadenylated RNA by altering ubiquitin availability. Oncogene 30, 790–805 [DOI] [PubMed] [Google Scholar]
- 48. Ong S. E., Blagoev B., Kratchmarova I., Kristensen D. B., Steen H., Pandey A., Mann M. (2002) Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol. Cell. Proteomics 1, 376–386 [DOI] [PubMed] [Google Scholar]
- 49. Haaf T., Ward D. C. (1996) Inhibition of RNA polymerase II transcription causes chromatin decondensation, loss of nucleolar structure, and dispersion of chromosomal domains. Exp. Cell Res. 224, 163–173 [DOI] [PubMed] [Google Scholar]
- 50. Dragon F., Gallagher J. E., Compagnone-Post P. A., Mitchell B. M., Porwancher K. A., Wehner K. A., Wormsley S., Settlage R. E., Shabanowitz J., Osheim Y., Beyer A. L., Hunt D. F., Baserga S. J. (2002) A large nucleolar U3 ribonucleoprotein required for 18S ribosomal RNA biogenesis. Nature 417, 967–970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Bernstein K. A., Gallagher J. E., Mitchell B. M., Granneman S., Baserga S. J. (2004) The small-subunit processome is a ribosome assembly intermediate. Eukaryot. Cell 3, 1619–1626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Bleichert F., Baserga S. J. (2007) The long unwinding road of RNA helicases. Mol. Cell 27, 339–352 [DOI] [PubMed] [Google Scholar]
- 53. Fox A. H., Lam Y. W., Leung A. K., Lyon C. E., Andersen J., Mann M., Lamond A. I. (2002) Paraspeckles: A novel nuclear domain. Curr. Biol. 12, 13–25 [DOI] [PubMed] [Google Scholar]
- 54. Bond C. S., Fox A. H. (2009) Paraspeckles: Nuclear bodies built on long noncoding RNA. J. Cell Biol. 186, 637–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Valdez B. C., Henning D., So R. B., Dixon J., Dixon M. J. (2004) The treacher collins syndrome (TCOF1) gene product is involved in ribosomal DNA gene transcription by interacting with upstream binding factor. Proc. Natl. Acad. Sci. U.S.A. 101, 10709–10714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Olson M. O. (2004) Sensing cellular stress: Another new function for the nucleolus? Sci. STKE 224, pe10. [DOI] [PubMed] [Google Scholar]
- 57. Yang C., Maiguel D. A., Carrier F. (2002) Identification of nucleolin and nucleophosmin as genotoxic stress-responsive RNA-binding proteins. Nucleic Acids Res. 30, 2251–2260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Yu Y., Maggi L. B., Jr., Brady S. N., Apicelli A. J., Dai M. S., Lu H., Weber J. D. (2006) Nucleophosmin is essential for ribosomal protein L5 nuclear export. Mol. Cell. Biol. 26, 3798–3809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Maggi L. B., Jr., Kuchenruether M., Dadey D. Y., Schwope R. M., Grisendi S., Townsend R. R., Pandolfi P. P., Weber J. D. (2008) Nucleophosmin serves as a rate-limiting nuclear export chaperone for the mammalian ribosome. Mol. Cell. Biol. 28, 7050–7065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Vascotto C., Fantini D., Romanello M., Cesaratto L., Deganuto M., Leonardi A., Radicella J. P., Kelley M. R., D'Ambrosio C., Scaloni A., Quadrifoglio F., Tell G. (2009) APE1/Ref-1 interacts with NPM1 within nucleoli and plays a role in the rRNA quality control process. Mol. Cell. Biol. 29, 1834–1854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Panse S. L., Masson C., Héliot L., Chassery J. M., Junéra H. R., Hernandez-Verdun D. (1999) 3-D organization of ribosomal transcription units after DRB inhibition of RNA polymerase II transcription. J. Cell Sci. 112, 2145–2154 [DOI] [PubMed] [Google Scholar]
- 62. Yadavilli S., Mayo L. D., Higgins M., Lain S., Hegde V., Deutsch W. A. (2009) Ribosomal protein S3: A multi-functional protein that interacts with both p53 and MDM2 through its KH domain. DNA Repair 8, 1215–1224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Powley I. R., Kondrashov A., Young L. A., Dobbyn H. C., Hill K., Cannell I. G., Stoneley M., Kong Y. W., Cotes J. A., Smith G. C., Wek R., Hayes C., Gant T. W., Spriggs K. A., Bushell M., Willis A. E. (2009) Translational reprogramming following UVB irradiation is mediated by DNA-PKcs and allows selective recruitment to the polysomes of mRNAs encoding DNA repair enzymes. Genes Dev. 23, 1207–1220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Li S., Kuhne W. W., Kulharya A., Hudson F. Z., Ha K., Cao Z., Dynan W. S. (2009) Involvement of p54(nrb), a PSF partner protein, in DNA double-strand break repair and radioresistance. Nucleic Acids Res. 37, 6746–6753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Salton M., Lerenthal Y., Wang S. Y., Chen D. J., Shiloh Y. (2010) Involvement of matrin 3 and SFPQ/NONO in the DNA damage response. Cell Cycle 9, 1568–1576 [DOI] [PubMed] [Google Scholar]