Abstract
Endoplasmic reticulum-mitochondrial contacts, known as mitochondria-associated membranes, regulate important cellular functions including calcium signaling, bioenergetics, and apoptosis. Human cytomegalovirus is a medically important herpesvirus whose growth increases energy demand and depends upon continued cell survival. To gain insight into how human cytomegalovirus infection affects endoplasmic reticulum-mitochondrial contacts, we undertook quantitative proteomics of mitochondria-associated membranes using differential stable isotope labeling by amino acids in cell culture strategy and liquid chromatography-tandem MS analysis. This is the first reported quantitative proteomic analyses of a suborganelle during permissive human cytomegalovirus infection. Human fibroblasts were uninfected or human cytomegalovirus-infected for 72 h. Heavy mitochondria-associated membranes were isolated from paired unlabeled, uninfected cells and stable isotope labeling by amino acids in cell culture-labeled, infected cells and analyzed by liquid chromatography-tandem MS analysis. The results were verified by a reverse labeling experiment. Human cytomegalovirus infection dramatically altered endoplasmic reticulum-mitochondrial contacts by late times. Notable is the increased abundance of several fundamental networks in the mitochondria-associated membrane fraction of human cytomegalovirus-infected fibroblasts. Chaperones, including HSP60 and BiP, which is required for human cytomegalovirus assembly, were prominently increased at endoplasmic reticulum-mitochondrial contacts after infection. Minimal translational and translocation machineries were also associated with endoplasmic reticulum-mitochondrial contacts and increased after human cytomegalovirus infection as were glucose regulated protein 75 and the voltage dependent anion channel, which can form an endoplasmic reticulum-mitochondrial calcium signaling complex. Surprisingly, mitochondrial metabolic enzymes and cytosolic glycolytic enzymes were confidently detected in the mitochondria-associated membrane fraction and increased therein after infection. Finally, proapoptotic regulatory proteins, including Bax, cytochrome c, and Opa1, were augmented in endoplasmic reticulum-mitochondrial contacts after infection, suggesting attenuation of proapoptotic signaling by their increased presence therein. Together, these results suggest that human cytomegalovirus infection restructures the proteome of endoplasmic reticulum-mitochondrial contacts to bolster protein translation at these junctions, calcium signaling to mitochondria, cell survival, and bioenergetics and, thereby, allow for enhanced progeny production.
Congenital human cytomegalovirus (HCMV)1 infection is the leading viral cause of birth defects, including sensorineuronal hearing loss and developmental delay, in developed countries (1–3). Most HCMV infections in immunocompetent persons are asymptomatic. Nonetheless, HCMV infection is a major cause of morbidity and mortality in immunocompromised individuals (1–3). Severe manifestations of HCMV infection in these patients include interstitial pneumonia, hepatitis, meningoencephalitis, gastrointestinal disease, myocarditis, bone marrow suppression, and retinitis. Because of its impact on human suffering and the health care system, the Institute of Medicine has assigned the highest priority to HCMV vaccine development (4).
Like that of most viruses, HCMV growth in permissive cells requires a large supply of energy and substrates from the infected cell (5–7). To obtain these, HCMV infection causes a global metabolic up-regulation of central carbon metabolic flux (5, 6). In particular, HCMV infection induces increased glucose and glutamine consumption during permissive infection of human foreskin fibroblasts (HFFs) (5–7). Increased aerobic glycolysis allows HCMV to use glucose biosynthetically, wherein most of the acetyl coenzyme A supports fatty acid synthesis needed for membrane formation for its progeny viruses. Thus, HCMV growth reprograms cellular mitochondrial metabolic flux and institutes its own metabolic program.
Because of its protracted growth cycle over several days, productive HCMV infection further requires antiapoptotic activity for efficient progeny production (8). To extend cell viability, HCMV encodes several products to undermine mitochondrial-mediated apoptosis. The HCMV UL36–38 locus encodes five proteins with antiapoptotic activities, including the predominant UL37 exon 1 protein (pUL37×1), also known as viral mitochondria localized inhibitor of apoptosis (vMIA), the lower abundance UL37Medium protein and the UL37 glycoprotein, the UL36 protein and the early UL38 protein (9–12). pUL37×1 blocks mitochondrial-mediated apoptosis by translocating proapoptotic Bax to the outer mitochondrial membrane (OMM) and suppressing its proapoptotic activity (13–15). In addition, pUL37×1/vMIA has been reported to alter mitochondrial ATP synthesis by decreasing the activity of the mitochondrial phosphate carrier in transfected cells (16). By early times, HCMV infection also encodes an untranslated beta 2.7 RNA that localizes to mitochondria (17, 18). Beta 2.7 RNA interacts with the GRIM-19 subunit of mitochondrial complex I and prevents its re-localization from the inner mitochondrial membrane to discrete perinuclear sites (19). Thus, HCMV infection redundantly blocks mitochondrial-mediated proapoptotic signaling pathways.
Mitochondrial activities, including its metabolic pathways and induction of apoptosis, are modulated by contacts with endoplasmic reticulum (ER) subdomains, known as mitochondria-associated membranes (MAM) (20, 21). Direct ER-mitochondrial contacts have been best visualized by electron tomography (22). The physical distance between the ER membrane and OMM is ∼10 nm. The distance between the organelles is larger (∼25 nm) when ribosomes are found between the juxtaposed membranes. Protein tethers can be seen between the ER and OMM. Mitofusin 2 (Mfn2) (23, 24), mitostatin (25), PACS-2 (26), and a calcium (Ca2+) signaling complex (27), have been implicated in stabilizing contacts between the ER and the OMM.
Our previous work demonstrated that HCMV infection targets ER-mitochondrial contacts by trafficking of its pUL37×1 and the N-terminal UL37 glycoprotein fragment (28–34). A highly conserved UL37×1 leader drives ER translocation, MAM association, and subsequent trafficking to the OMM (31, 34). pUL37×1 traffics from the ER, where it causes Ca2+ efflux (35), associates with internal MAM lipid rafts (36) in close proximity to sigma 1 receptor (Sig-1R) (32) and translocates to the OMM where it has potent antiapoptotic activity by recruiting Bax to the OMM and inhibiting its proapoptotic activities (10, 13, 14, 37, 38). In addition to targeting the MAM by pUL37×1, we found that HCMV infection increases the abundance of cytosolic glucose regulated protein 75 (GRP75), a component of the MAM Ca2+ macromolecular complex (29). Thus, HCMV positions itself to control ER-mitochondrial cross-talk and thereby modulate key mitochondrial functions.
By targeting the MAM, HCMV predictably acquires another level of control over mitochondrial metabolism and mitochondrial-mediated apoptosis. To gain insight into how HCMV infection alters the functionality of ER-mitochondrial contacts to its advantage, we undertook quantitative analyses of the MAM proteome using stable isotope labeling by amino acids in cell culture (SILAC) (39). A late time of infection was chosen for these studies as HCMV infection dramatically alters metabolic flux (5–7), the cell secretome (40), widespread changes in ER Ca2+ efflux (35), and a mitigated ER stress response (41) at these times; all events which are likely mediated by the MAM or to affect MAM domains. To date, no quantitative or nonquantitative proteomic analysis of the MAM fraction has yet been reported. Although proteomic analyses have identified HCMV virion components (42) and cellular proteins that interact with HCMV products (43, 44) as well as defined the HCMV-induced secretome (40), this is the first quantitative proteomic analyses of HCMV-infected HFFs and the effects of virus on ER-mitochondrial contacts. Using this quantitative proteomic approach, we observed dramatic changes in the MAM proteome by late times of HCMV infection, consistent with its global reprogramming of cellular metabolism. The association of chaperones, translation and ER-translocation machineries, Ca2+ signaling complexes, metabolic enzymes, and apoptotic regulatory proteins with the MAM fraction increased by late times of infection. These results suggest that HCMV infection induces a restructuring of ER-mitochondrial contacts by late times. This restructuring will predictably result in increased protein synthesis at the MAM, cell survival and bioenergetics. Further the increased abundance of mitochondrial inner mitochondrial membrane, intermembrane space and matrix proteins in the MAM fraction suggest that HCMV infection increases the stability of ER-mitochondrial contacts by late times of infection.
EXPERIMENTAL PROCEDURES
Cells and Viruses
HFFs were cultured in Dulbecco's Modified Eagle Medium containing 10% fetal calf serum (FCS), 100 U/ml of penicillin, 100 μg/ml of streptomycin (Invitrogen) as described (45). The bacterial artificial chromosome of HCMV strain AD169 (BAD) wild type (wt) was grown and titered as previously described (35, 46).
SILAC-Labeling of HFFs
Primary HFFs (starting at passage 5) were cultured for four cell doublings in custom-media containing 13C6-Arg and 15N2, 13C6-Lys (SILAC-labeled) or 12C6-Arg and 14N2, 12C6-Lys (unlabeled). HFFs were incubated with labeled medium or unlabeled medium for 3 days. Confluent cells were subcultured at ratios of 1:2 or 1:4 and fed unlabeled or labeled medium, respectively. Two roller bottles (769 cm2 each) of SILAC-labeled HFFs were uninfected or HCMV-infected (BADwt, multiplicity of infection of 3 plaque forming units/cell and maintained in DMEM containing 2% FCS until 72 h post infection (hpi). In parallel, two roller bottles of unlabeled HFFs were uninfected or infected with HCMV as above. At 72 hpi, infected and uninfected cells were washed 6 times with sterile PBS (Invitrogen), harvested, and paired for MAM fractionation as shown in Fig. 1. In the forward experiment, SILAC-labeled, HCMV-infected cells were mixed 1:1 with unlabeled uninfected cells. In the reverse experiment, the unlabeled HCMV-infected cells were mixed 1:1 with SILAC-labeled uninfected cells.
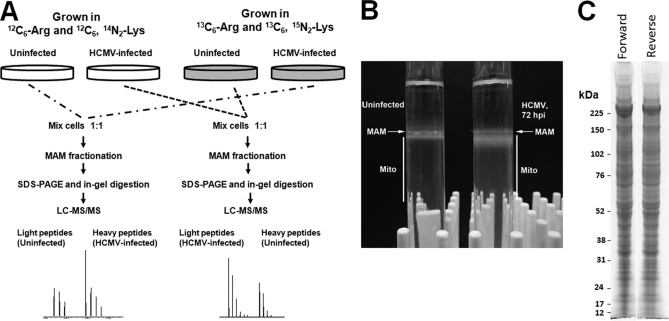
Fig. 1.
Experimental design for quantitative proteomics of enriched heavy MAM fraction from uninfected and HCMV-infected HFFs. A, Forward (left) and reverse (right) SILAC approaches. HFFs were grown in light (12C6-Arg and 12C6, 14N2-Lys) or heavy (13C6-Arg and 15N2, 13C6-Lys) medium for four cell doublings. HFFs, grown in heavy medium, were HCMV (strain BAD wt)-infected for 72 h; whereas, HFFs, grown in light medium, were uninfected for 72 h. Prior to subcellular fractionation, two roller bottles of SILAC-labeled, HCMV-infected HFFs were mixed (1:1) with two roller bottles of unlabeled, uninfected HFFs (forward experiment). In the reverse experiment, SILAC-labeled, uninfected HFFs were mixed (1:1) with unlabeled, HCMV-infected HFFs at 72 hpi. The mixed cells were fractionated to obtain heavy MAM as previously described (28, 47). Heavy MAM proteins (100 μg), obtained from mixed unlabeled and SILAC-labeled cells, were resolved by SDS-PAGE. Fifty sequential slices of each lane were in-gel digested with trypsin and analyzed by LC-MS/MS analysis. B, Percoll gradients of uninfected and HCMV infected HFFs. Uninfected HFFs and HCMV-infected HFFs (∼5 × 107 cells/each) were gently lysed by homogenization, pelleted by centrifugation at 10,300 × g for 10 min at 4 °C. The supernatant was removed and alternately processed, whereas the pellet was resuspended in 300 μl of ice-cold mannitol buffer A, briefly homogenized, layered over 10 ml of a 30% Percoll suspension in mannitol buffer B, and subjected to ultracentrifugation at 95,000 × g for 65 min at 4 °C. Banded MAM and mitochondria were recovered from the gradient by needle extraction from above (MAM, 200 μl) and below (Mito, 2 ml) the indicated points. C, SDS-PAGE of forward and reverse MAM samples. Heavy MAM from the paired forward or reverse paired HFFs were resolved by SDS-PAGE and stained by BioSafe Coomassie.
Subcellular Fractionation and MAM Isolation
Each paired mixture was separately fractioned to isolate enriched heavy MAM from bulk ER and mitochondria as previously described (28, 47). The Percoll method used for this analysis is one of the most rigorous procedures to obtain banded MAM and mitochondria. It relies on multiple fractionation steps, each used to separate organelles based upon their connectivity, size, shape, and density. Briefly, cells were homogenized and nuclei and debris were removed by pelleting. The supernatant was pelleted again to obtain crude mitochondria and interconnected MAM. The cytosol and microsomes remained soluble. The MAM and mitochondria were further separated by homogenization and resolved by banding in Percoll density gradients. Percoll gradients efficiently separate, based upon density and shape: the MAM is lighter; whereas, mitochondria are denser and band in the lower half of the gradient (see a representative separation in Fig. 1B). Banded MAM and mitochondria were obtained by side puncture of the ultracentrifuge tube from above and below the bands, respectively, and aspiration. Once extracted from the gradients, the MAM band was diluted and pelleted generating the heavy MAM, which pellets at lower centrifugal forces and is enriched in macromolecular complexes that join the ER and mitochondria (29). This banded fraction was used for the quantitative proteomic analyses. All of the fractions were resuspended in a minimal volume (50–200 μl) of sucrose homogenization medium and assayed for protein concentration using a BCA reagent kit (Pierce), according to the manufacturer's recommended protocol.
Protein Separation and Mass Spectrometry Analysis
One hundred micrograms of the heavy MAM fraction obtained from the mixture of unlabeled and SILAC-labels cells were further resolved by SDS-PAGE (Fig. 1C). The gel was stained with Bio-Safe Coomassie (Bio-Rad, Hercules, CA) and each lane was cut into 50 serial slices. Proteins in each gel slice were in-gel digested with trypsin. Concentrated peptides from each band were injected via an autosampler (6 μl) and loaded onto a Symmetry C18 trap column (5 μm, 300 μm i.d. × 23 mm, Waters) for 10 min at a flow rate of 10 μl/min, 100% A. The sample was subsequently separated by a C18 reverse-phase column (3.5 μm, 75 μm × 15 cm, LC Packings) at a flow rate of 250 nL/min using an Eksigent nano-hplc system (Dublin, CA). The mobile phases consisted of water with 0.1% formic acid (A) and 90% acetonitrile (B). A 65 min linear gradient from 5 to 60% B was employed. Eluted peptides were introduced into the mass spectrometer via a 10 μm silica tip (New Objective Inc., Ringoes, NJ) adapted to a nano-electrospray source (ThermoFisher Scientific). The spray voltage was set at 1.2 kV and the heated capillary at 200 °C. The LTQ-Orbitrap-XL (ThermoFisher Scientific) was operated in data-dependent mode with dynamic exclusion in which one cycle of experiments consisted of a full-MS in the Orbitrap (300–2000 m/z, resolution 30,000) survey scan and five subsequent MS/MS scans in the LTQ of the most intense peaks using collision-induced dissociation with the collision gas (helium) and normalized collision energy value set at 35%.
Database Search and SILAC Ratio Measurement
For protein identification and quantification we used Integrated Proteomics Pipeline (IP2) version 1.01 software developed by Integrated Proteomics Applications, Inc. (http://www.integratedproteomics.com/). Mass spectral data were uploaded into IP2 software. Files from each lane were searched against the forward and reverse Uniprot human database (UniProt release 15.4, June 2009, 22697 forward entries) for tryptic peptides allowing one missed cleavage, and possible modification of oxidized methionine (15.99492 Da), heavy arginine (6.0204 Da) and heavy lysine (8.0142 Da). IP2 uses the Sequest 2010 (06_10_13_1836) search engine. Mass tolerance was set at ± 50 ppm for MS and ± 1.5 Da for MS/MS. Data were filtered based on the following Xcorr values (Xcorr ≥ 1.9 for z = 1, ≥ 2.5 for z = 2, and ≥ 3.5 for z = 3). These criteria provided a 3% false discovery rate. Only proteins that were identified by at least two unique peptides were retained for further quantitative analysis. Proteins that were identified with one single peptide were excluded. The 50 bands from each lane were summed in the analysis. Census software version 1.77, built into the IP2 platform (48), was used to determine the ratios of unlabeled and labeled peptide pairs using an extracted chromatogram approach. The distribution of ratios was plotted and correction factors applied to adjust for error in sample mixing. Data were checked for validity by using regression correlation better than 0.98 for each peptide pair.
Western Blot Analysis
Total or fractionated proteins were resolved by SDS-PAGE in 4–12% Bis-Tris NuPage gels (Invitrogen) and analyzed by Western analyses as previously described (28, 49). Blots were probed with the following primary antibodies: rabbit anti-UL37×1 (amino acids 27 to 40) antiserum (DC35, 1:2,500), rabbit anti-erlin 2 (a kind gift from Dr. Stephen Robbins, 1:500), rabbit anti-calnexin (StressGen Biotechnologies, Victoria, BC, Canada; 1:500 or 1:5,000), rabbit anti-calreticulin (Affinity Bioreagents, Golden, CO; 1:1,000), goat anti-Cox 2 (Santa Cruz Biotechnology, Santa Cruz, CA; 1:200), rabbit anti-fatty acid coenzyme A ligase 4 (FACL4, Abgent, 1:100), goat anti-Golgin 97 (Molecular Probes, Eugene, OR; 1:100), mouse anti-GRP75/mHSP70 (StressGen,1:2,500 or 1:5,000), rabbit anti-GRP78/BiP (StressGen, 1:500), mouse anti-Hexokinase 1 (Santa Cruz, 1:100), rabbit anti-HSP60 (GeneTex, Irvine, CA; 1:2,000), mouse anti-Membrin (StressGen, 1:150), rabbit anti-Mfn1 (Santa Cruz, 1:100), mouse anti-Mfn2 (AbCam, Cambridge, MA; 1:200), rabbit anti-PACS-2 (GeneTex, 1:500), goat anti-phosphoglycerate mutase (PGM) 5 (Santa Cruz, 1:100), rabbit anti-Prohibitin (GeneTex, 1:500), mouse anti-Sig-1R (Santa Cruz, 1:200 or 1:250), mouse anti-voltage dependent anion channel (VDAC) (AbCam, 1:100 or 1:200), or mouse anti-α-tubulin (Sigma, 1:2,000) and with the corresponding horseradish peroxidase-conjugated secondary antibody (1:2,500 or 1:5,000).
Protein bands were detected using an ECL detection kit (Pierce). Each blot was stripped as previously described, and reprobed for the detection of other ER, MAM, mitochondrial markers (28, 49). Blots were exposed to film (Denville Scientific, Metuchen, NJ) and the exposures were scanned using a Bio-Rad GS-800 Calibrated Densitometer and analyzed using QuantityOne Software. Digital images were generated by using Scan Wizard Pro version 1.21 and processed in Adobe Photoshop version CS5 version 12.0.3.
Immunofluorescence Assays
HFFs were seeded onto sterile cover slips at 80% confluency and uninfected or HCMV (strain AD169) infected at a multiplicity of 1. Cells were harvested at 72 hpi by fixation with ice-cold methanol for 15 min. Fixed cells were stored at 4 °C until use. Cells were then probed with mouse anti-HSP60 IgG1 (Santa Cruz, 1:50), mouse anti-Hexokinase IgG1 (Santa Cruz, 1:50), or mouse anti-Sig-1R IgG1 (Santa Cruz; 1:50) and mouse anti-HCMV IE 1/2 IgG2A (Millipore, 1:50), and human anti-mitochondria IgG (Immunovision, 1:50) as described previously (28, 49, 50). Cells were then washed and probed simultaneously with 568Alexa Fluor conjugated goat anti-mouse IgG1 (1:1500), 488Alexa Fluor conjugated goat anti-mouse IgG2A (1:1500), and 647Alexa Fluor conjugated goat anti-human IgG (1:1500) secondary antibodies.
Confocal Microscopy
Images were acquired with a Zeiss LSM 510 confocal microscope (Intellectual and Developmental Disabilities Research Center) as described (28, 30). Excitation wavelengths for the Zeiss LSM 510 microscope were 488 nm, 561 nm, and 633 nm. Images were acquired by sequential excitation through 63x (NA = 1.4) objective. Post-acquisition processing was performed using Adobe Photoshop CS3.
RESULTS
Quantitative Proteomic Analyses of the MAM Proteome in Uninfected and HCMV-Infected HFFs
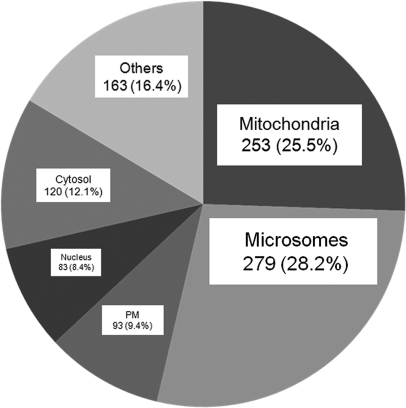
The enriched MAM fraction can be reproducibly isolated by banding in Percoll gradients (Fig. 1B) (47, 51–53). Using SILAC methodology, we initially identified 1719 unique proteins from proteomic analysis of the MAM fractions of forward (unlabeled uninfected cells and SILAC-labeled, HCMV-infected cells) and reverse (SILAC-labeled uninfected cells and unlabeled, HCMV-infected cells) experiments. Of these, 1254 unique proteins were identified in proteomic analyses of both forward and reverse experiments. Finally, 991 proteins were identified by at least two unique peptide hits in both proteomic sets, increasing the confidence of their authentic association with the MAM fraction. We focused our studies on these most confident 991 proteins. As expected by its identity as an ER subdomain, the MAM fraction predominantly contained proteins from the secretory apparatus (microsomes) and, to a lesser degree, nuclear proteins (nuclear), as the outer nuclear membrane is contiguous with the ER (Fig. 2). Further, consistent with its tight association with mitochondria, the next largest group of proteins was of mitochondrial origin. Surprisingly, a significant number of proteins known to be localized to the cytosol, plasma membrane, and other apparently unrelated subcellular compartments were also identified within the MAM fraction. This latter result suggests the MAM interacts with multiple cellular compartments in addition to mitochondria and may indicate that the identified proteins dually localize in the MAM as well. For example, GRP75 localizes in the mitochondrial matrix and has been recently found as the cytosolic component of the MAM macromolecular Ca2+ signaling complex (22, 29).
Fig. 2.
The subcellular distribution of cellular proteins identified in the heavy MAM fractions from uninfected and HCMV-infected HFFs. The known subcellular localization of 991 human proteins identified in heavy MAM proteome in both forward and reverse experiments (with identified peptide number cutoff of at least 2) using UniProt are represented by percent of total. Identified HCMV proteins were excluded from the analysis. The portion designated microsomes includes proteins, which localize in the ER, Golgi apparatus, endosomes, lysosomes, melanosomes, and vesicles using Uniprot.
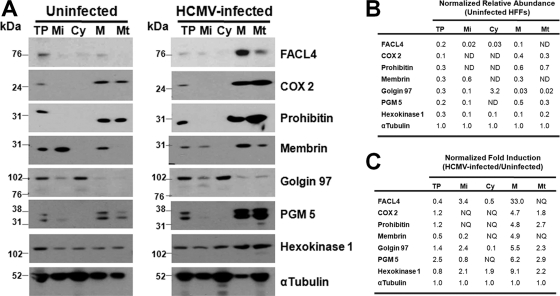
To verify the identity of our subcellular fractions, we examined the relative abundances of known ER-mitochondrial, secretory apparatus, and glycolytic enzymes in the subcellular fractions from uninfected and from HCMV-infected HFFs (Fig. 3A). FACL4, a known MAM marker (26) was faintly detected in the MAM fraction (0.1), but dramatically and specifically increased in the MAM (33.0-fold) despite decreasing in the total cell fraction (0.4-fold) after HCMV infection (Fig. 3B and 3C). Analogously, mitochondrial markers, COX2 and Prohibitin, were detected in the enriched MAM and mitochondria fractions. The abundance of COX2 increased in the MAM (4.7-fold) and in mitochondria (1.8-fold) above the induction observed in total protein (1.2-fold) during HCMV infection. Similarly, Prohibitin increased 4.8-fold and 2.7-fold in MAM and mitochondria, respectively, during HCMV infection despite being marginally induced in total extracts (1.2-fold).
Fig. 3.
Validation of subcellular fractionation using Western analyses and induction of MAM components during HCMV infection. A, Total protein (TP, 30 μg) or fractions were isolated from uninfected or HCMV (BADwt) HFFs at 72 hpi. Fractionated microsomes (Mi), cytosol (Cy), heavy MAM (M) and mitochondria (Mt) (10 μg) were resolved by SDS-PAGE, blotted, and probed with antibodies against markers of ER-mitochondria (FACL4, COX2, and Prohibitin), the secretory apparatus (Membrin and Golgin 97), and glycolytic enzymes (PGM 5 and Hexokinase 1). B, The relative abundances of cellular markers in uninfected HFFs. The relative abundance of each marker tested in Panel A was determined in each fraction by densitometry of the Western blots. The values were normalized to the relative abundance of α-tubulin in the same fraction. ND, not detected. C, Fold induction of cellular markers during HCMV infection of HFFs. The induction of each protein tested in Panel A was determined by comparing the normalized values in HCMV-infected cell fractions to the normalized abundances of the corresponding fractions from uninfected cells. NQ, not quantifiable.
Membrin, an ER-Golgi SNARE protein (54), was detected in the MAM (0.3) but to levels less than in microsomes (0.6). Membrin abundance increased in the MAM (4.9-fold), despite being reduced in total protein lysates (0.5-fold) during HCMV infection. Intriguingly, the abundance of Membrin decreased fivefold in the microsomal fraction (0.2) commensurate with the ∼fivefold increase in the MAM fraction, suggesting a potential relocalization during HCMV infection.
Conversely, the trans-Golgi protein, Golgin 97, was very weakly detected in MAM (0.03) and mitochondria (0.02) at levels ∼100-fold less than in the cytosol (3.2). Nonetheless, its abundance increased in the MAM (5.5-fold) or mitochondria (2.3-fold) after HCMV infection even though the increase in the total extract was only modestly increased (1.4-fold).
Glycolytic enzymes (PGM 5 and Hexokinase 1) were weakly detectable in the MAM of uninfected HFFs. Nonetheless, after HCMV infection, the relative abundances of both PGM 5 (6.2-fold) and Hexokinase 1 (9.1-fold) increased markedly in the MAM above the values in total extracts (2.5-fold and 0.8-fold, respectively). Together, our results establish the presence of known MAM (FACL4) and mitochondrial (COX2, Prohibitin) markers in the enriched MAM fraction and their strong induction in the MAM after HCMV infection, above that observed with their corresponding changes in total protein. Further, these results show the very low abundance of nonresident proteins (such as the trans-Golgi protein, Golgin 97) in the enriched MAM. Moreover, HCMV infection specifically alters the abundance of some proteins in the enriched MAM fraction (FACL4, COX2, Prohibitin, Membrin, PGM 5, and Hexokinase 1) even though their total abundances are not proportionately increased in the HCMV-infected cell, suggesting their relocalization from other subcellular compartments.
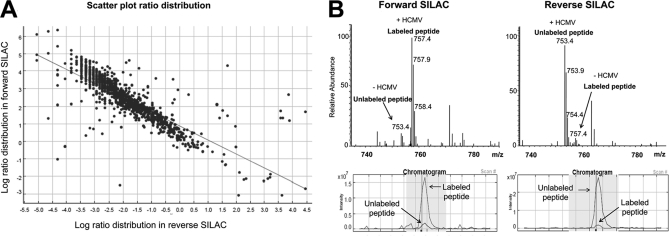
To comprehensively and independently quantify proteins that were altered in the cellular MAM fraction after HCMV infection, we performed differential SILAC strategy where one set of cells for example cells that will be HCMV-infected are metabolically labeled with heavy 13C6-Arg and 13C6, 15N2-Lys whereas cells that will be uninfected are metabolically labeled with light 12C6-Arg and 12C6, 14N2-Lys. HCMV-infected and uninfected cells are then mixed at 1:1 ratio for subsequent MAM fractionation and proteome profiling. Because trypsin cuts proteins after Lys and Arg residues, each peptide from each protein is detected as pairs of light and heavy ion corresponding to 12C6-Arg/12C6, 15N2-Lys labeled peptide and 13C6-Arg and 13C6, 15N2-Lys, respectively. The intensity ratio of the heavy to light peptide reflects the amount of the corresponding protein in the SILAC labeled versus unlabeled cells. To further validate our data, we have performed SILAC switch labeling experiment (see Fig. 1 for detailed experimental design). MAM proteome profiling of HCMV-infected versus uninfected cells, derived from two independent experiments using forward and reverse SILAC strategies, exhibited close symmetry, as shown by scatter plot comparison (Fig. 4A). Comparing labeled and unlabeled peptide pairs of a MAM-specific marker protein Ero1α (55) between sample sets, we observed a consistent trend of Ero1α up-regulation in MAM domains in HCMV-infected cells (Fig. 4B). Fold changes are presented as a ratio of light unlabeled peptides over heavy labeled peptides. Because labeling media, heavy and light, were reversed in the two experiments, changes because of HCMV infection appear inversely related. In the case of Ero1α, for example, 13- and ninefold increases in MAM abundance of the protein were detected after HCMV infection.
Fig. 4.
A, Scatter Plot showing correlation in ratio distribution of proteins detected in forward and reverse SILAC experiments. Log ratio distribution of all proteins detected in the MAM fraction of uninfected and unlabeled HFFs versus HCMV-infected and SILAC-labeled HFFs were plotted versus log ratios of the same set of proteins detected in HCMV-infected and unlabeled HFF cells versus uninfected and SILAC-labeled HFF cells. This plot shows strong correlation between the two experiments with an R2 = -0.8. B, Example of MS spectra of labeled and unlabeled peptide pairs detected for an MAM marker protein, Ero1 α, in both forward and reverse experiments. The peptide was detected as doubly charged species at m/z of 753.4 and 757.4 corresponding to the unlabeled and labeled peptide, respectively. The peptide intensity was consistently increased in HCMV-infected versus uninfected cells.
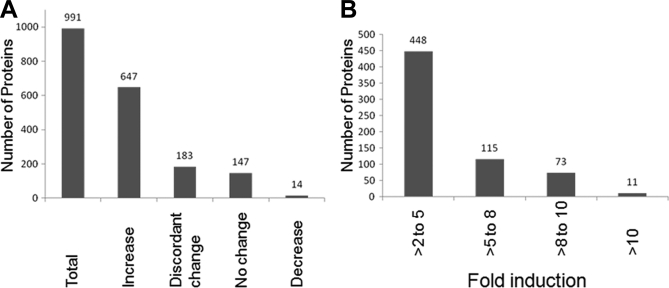
An overall trend of MAM proteome enrichment was observed at late times of HCMV infection (Fig. 5A). Over 65.3% (647/991) of identified MAM proteins increased in abundance by 72 hpi. Most of these were induced between two- and fivefold, although a small subset of proteins increased more than 10-fold at late times of HCMV infection (Fig. 5B). Another 33.3% of identified MAM proteins were not altered by HCMV infection, either staying at the same level in the presence or absence of HCMV infection (14.8%, 147/991) or changing in discordant ways between forward and reverse experiments (18.5%, 183/991). Surprisingly, only 1.4% (14/991) of the identified MAM proteins were consistently decreased at late times of HCMV infection, suggesting the importance of the MAM proteome for HCMV infection and progeny production.
Fig. 5.
A, Alteration of the MAM proteome by HCMV at late times of infection. Total cellular proteins (991 proteins) associated with the MAM fraction in uninfected and in HCMV-infected HFFs at 72 hpi were identified using quantitative proteomics. The number of proteins (at least two identified unique peptides) whose expression was increased (>twofold), showed discordant change (inconsistent values in forward and reverse experiments), had no change (0.5 to twofold), or was decreased (<0.5) after HCMV infection is shown. B, The fold induction levels of MAM-associated cellular proteins after HCMV infection. The number of MAM-associated cellular proteins showing low (>two- to fivefold), intermediate (>five- to eightfold), intermediate high (>eight- to 10-fold), or high (>10-fold) induction is shown.
We first mined the heavy MAM proteomic data for known MAM proteins (Table I). Multiple MAM proteins including lipid synthetic enzymes, Ca2+ signaling proteins, ER-mitochondrial membrane tethering complexes were detected. Markers with nearly exclusive MAM localization under normal cell conditions, such as ACAT1, ERp57, ERp44, Ero1α, erlin 1, and erlin 2 were detected in the MAM proteome by SILAC technology. MAM-enriched chaperones, notably BiP/GRP78, calnexin, and calreticulin, were readily detected in both proteomic sets. MAM junction proteins, which physically associate the MAM and mitochondria membranes, were also observed. These include components of the MAM Ca2+ macromolecular bridge, GRP75 and VDAC, as well as Mfn1, which can form heterodimers with Mfn2. VDAC-interacting proteins and proteins involved in ER-to-mitochondria Ca2+ signaling were detected as well as cell stress chaperones involved in regulating Ca2+ homeostasis. Nonetheless, Sig-1R, an MAM lipid raft marker (56), was detected only in one of the replicate proteomic experiments. Intriguingly, some MAM markers such as PACS-2, Autocrine motility factor receptor, inositol 1,4,5-triphosphate receptor type 3 (IP3R), phosphatidylserine synthases 1 and 2, Mfn2, ceramide synthase, and FACL4 were not detected by proteomic analysis. This may reflect their relatively low abundance within the MAM proteome, their low efficiency of retention during MAM fractionation, or a masking of their detection by other proteins. After HCMV infection, chaperones associated with the MAM (20) increased dramatically (Table I). Particularly notable is the increased abundance (10-fold) of GRP78/BiP, known to be required for permissive HCMV infection (57). The induction levels of the MAM proteins were independently verified by the reverse approach.
Table I. Known MAM proteins induced at late times of HCMV infection as detected by quantitative proteomics.
| UniProt ID | Proteina | Descriptionb | Forward |
Reverse |
|---|---|---|---|---|
| SILAC-HCMV-Infected/Uninfected average±S.D. (P)c | SILAC-Uninfected/HCMV-Infected average±S.D. (P)c | |||
| Q99720 | Sigma 1 Receptor | MAM lipid raft protein | ND | 0.23 ± 0 (2) |
| Q96HE7 | Ero1-alpha | MAM protein | 13.41 ± 7.12 (6) | 0.11 ± 0.01 (6) |
| P30101 | Erp57 (PDIA3) | MAM enriched, | 11.54 ± 0.93 (79) | 0.12 ± 0.02 (110) |
| interacts with Ero1-alpha | ||||
| Q9BS26 | Erp44 | interacts with Ero1-alpha | 12.57 ± 3.67 (9) | 0.13 ± 0.03 (6) |
| P11021 | BiP (GRP78) | MAM enriched | 10.27 ± 1.76 (88) | 0.16 ± 0.03 (111) |
| P24752 | ACAT1 | MAM protein | 13.32 ± 1.73 (17) | 0.12 ± 0.02 (17) |
| P27824 | calnexin | MAM enriched | 6.23 ± 0.53 (81) | 0.22 ± 0.02 (73) |
| P27797 | calreticulin | MAM enriched | 10.04 ± 1.82 (56) | 0.13 ± 0.03 (66) |
| P21796 | VDAC1 | MAM Ca2+ signaling complex | 7.12 ± 0.78 (19) | 0.22 ± 0.02 (19) |
| P45880 | VDAC2 | MAM Ca2+ signaling complex | 10.66 ± 1.66 (16) | 0.17 ± 0.03 (40) |
| Q9Y277 | VDAC3 | MAM Ca2+ signaling complex | 9.97 ± 1.92 (8) | 0.15 ± 0.02 (8) |
| P38646 | Grp75 (HspA9; mortalin; mHsp70) | MAM Ca2+ signaling complex | 23.91 ± 5.4 (55) | 0.11 ± 0.11 (108) |
| Q8IWA4 | Mfn 1 | MAM tethering complex | 11.18 ± 2.03 (3) | 0.12 ± 0.03 (2) |
| O75477 | Erlin 1 | MAM lipid raft protein | 3.7 ± 0.26 (17) | 0.31 ± 0.04 (20) |
| O94905 | Erlin 2 | MAM lipid raft protein | 3.9 ± 0.53 (20) | 0.3 ± 0.03 (20) |
a Name given by UniProt database.
b Relevant descriptions from the literature.
c Average ratio ± standard deviation (number of identified unique peptides).
In addition, HCMV proteins, including pUL37×1, which is known to traffic to the MAM, were detected exclusively after HCMV infection (supplementary Table S1). Further, pUL37×1 interacting proteins, including ATPase inhibitory factor 1, Bax, adenine nucleotide translocator (ANT), lactate dehydrogenase A, nonmetastatic cells 2, protein expressed, and ribosomal phosphoprotein, large, P1 (58) and pUL37×1/vMIA regulated HtrA2 (59) were also detected (data not shown).
Electron tomography imaging has visualized ribosomes at ER-mitochondrial contact sites (22). To determine the feasibility of translation and translocation occurring at or near MAM domains, we examined the detection of minimal translational and translocation machineries in the MAM fraction (Table II). Signal recognition particle-dependent translation in the ER minimally requires the signal recognition particle receptor, the SEC63 translocon, and an oligosaccharyltransferase protein. All the minimal components were identified in our proteomics analysis, along with other facilitator proteins known to aid protein translation and translocation at the ER (60). This finding suggests for the first time the presence of minimal translation/translocation machinery in the MAM, and that protein synthesis and translocation can occur at these sites. Similarly to the induction of MAM chaperones and Ca2+ signaling and homeostasis proteins, HCMV infection increased the translational and translocation machineries associated with the MAM (Table II).
Table II. Minimal required translation and translocation machinery is MAM associated and induced by HCMV infection at late times.
| UniProt ID | Proteina | Descriptionb | Forward |
Reverse |
|---|---|---|---|---|
| SILAC-HCMV-Infected/Uninfected average±S.D. (P)c | SILAC-Uninfected/HCMV-Infected average±S.D. (P)c | |||
| Translation/Translocation Machinery (Required Minimal Components) | ||||
| P61619 | SEC61A1 | Translocon protein | 4.61 ± 0.49 (18) | 0.3 ± 0.02 (22) |
| P60468 | SEC61B | Translocon protein | 5.59 ± 0.34 (3) | 0.28 ± 0.02 (6) |
| P04843 | RPN1 | OST protein | 4.51 ± 0.41 (51) | 0.3 ± 0.02 (66) |
| P04844 | RPN2 | OST protein | 3.93 ± 0.27 (57) | 0.33 ± 0.02 (41) |
| P61803 | DAD1 | OST protein | 4.07 ± 0.13 (6) | 0.33 ± 0.02 (7) |
| P39656 | DDOST | OST protein | 3.86 ± 0.3 (22) | 0.33 ± 0.03 (26) |
| P61011 | SRP54 | Subunit of signal recognition particle | 3.98 ± 0.96 (3) | 0.41 ± 0.02 (2) |
| O76094 | SRP72 | Subunit of signal recognition particle | 3.71 ± 0.54 (2) | ND |
| Q9UHB9 | SRP68 | Subunit of signal recognition particle | ND | 0.4 (1) |
| P08240 | SRPRA | Subunit of signal recognition particle receptor | 7.4 ± 0.97 (13) | 0.19 ± 0.05 (14) |
| Q9Y5M8 | SRPRB | subunit of signal recognition particle receptor | 6.13 ± 0.44 (10) | 0.26 ± 0.02 (16) |
| Translation/Translocation Machinery (Accessory Components) | ||||
| Q99442 | SEC62 | Translocon protein | 5.98 ± 1.11 (6) | 0.19 ± 0.03 (5) |
| Q9UGP8 | SEC63 | Translocon protein | 3.23 ± 0.41 (13) | 0.33 ± 0.08 (12) |
| P43307 | SSR1 (TRAPA) | Translocon associated protein | 4.67 ± 0.54 (7) | 0.33 ± 0.03 (10) |
| Q9UNL2 | SSR3 (TRAPG) | Translocon associated protein | 5.41 ± 0.21 (3) | 0.26 ± 0.02 (3) |
| P51571 | SSR4 (TRAPD) | Translocon associated protein | 5.1 ± 0.35 (15) | 0.27 ± 0.01 (11) |
a Name given by UniProt database.
b Relevant descriptions from the literature.
c Average ratio ± standard deviation (number of identified unique peptides).
The twelve host cell proteins with the highest increases in MAM abundance during late HCMV infection are listed in Table III. This list of proteins represents all unique peptide hits of host cell MAM proteins detected to be induced by at least 10-fold during HCMV infection in both forward and reverse experiments. Tellingly, the most induced proteins are cellular chaperones including heat shock proteins, HSP10, HSP60, and GRP75, which are known to participate in various signaling cascades and affect lipid bilayer fluidity when they become membrane-associated (61). In contrast, the MAM proteins decreased the most are listed in Table IV. Notably, collagen and other secreted proteins were dramatically decreased in the MAM fraction of HCMV-infected HFFs at late times of infection.
Table III. MAM-associated proteins with highest induction at late times of HCMV infection.
| UniProt ID | Proteina | Descriptionb | Forward |
Reverse |
|---|---|---|---|---|
| SILAC-HCMV-Infected/Uninfected average±S.D. (Peptide #)c | SILAC-Uninfected/HCMV-Infected average±S.D. (Peptide #)c | |||
| P61604 | HSP10 | Mitochondrial chaperone | 27.57 ± 1.75 (11) | 0.08 ± 0.02 (15) |
| P10809 | HSP60 | Mitochondrial chaperone | 24.08 ± 3.74 (51) | 0.09 ± 0.09 (106)(H) |
| P38646 | GRP75/mHSP70 | Mitochondrial chaperone, Component of the MAM Ca2+ signaling complex | 23.91 ± 5.4 (55) | 0.11 ± 0.01 (108)(H) |
| P05091 | Aldehyde dehydrogenase | Oxidation reduction, mitochondrion matrix | 23.84 ± 4.73 (11) | 0.09 ± 0.02 (18) |
| Q969H8 | Stromal cell-derived growth factor SF20 | Secreted protein | 22.35 ± 1.74 (3) | 0.07 ± 0.03 (6) |
| Q14696 | LDLR chaperone MESD | Chaperone of low-density lipoprotein receptors | 20.62 ± 2.94 (4) | 0.07 ± 0.02 (7) |
| Q9UJS0 | Ca2+-binding mitochondrial carrier protein Aralar2 | Mitochondrial Ca2+-dependent aspartate and glutamate carrier | 20.49 ± 6.09 (16) | 0.08 ± 0.03 (19) |
| Q56VL3 | Ovarian carcinoma immunoreactive antigen domain-containing protein 2 | Endosomal protein | 20.19 ± 2.12 (5) | 0.08 ± 0.01 (4) |
| P55145 | Mesencephalic astrocyte-derived neurotrophic factor | Secreted protein, Inhibits ER stress-induced cell death | 20.17 ± 2.91 (3) | 0.08 ± 0.02 (6) |
| Q99714 | 3-hydroxyacyl-CoA dehydrogenase type-2 | Mitochondrial tRNA maturation | 19.82 ± 5.36 (8) | 0.09 ± 0.03 (11) |
| O43615 | TIM44 | Mitochondrial IMM translocase | 19.1 ± 4.77 (17) | 0.09 ± 0.03 (17) |
| Q9HAV7 | GrpE protein homolog 1 | Mitochondrial Hsp70 Chaperone | 16.68 ± 2.48 (2) | 0.07 ± 0.03 (7) |
a Name given by UniProt database.
b Relevant descriptions from the literature.
c Average ratio ± standard deviation (number of identified unique peptides).
Table IV. MAM-associated proteins decreased at late times of HCMV infection.
| UniProt ID | Proteina | Descriptionb | Forward |
Reverse |
|---|---|---|---|---|
| SILAC-HCMV-Infected/Uninfected average±S.D. (P)c | SILAC-Uninfected/HCMV-Infected average±S.D. (P)c | |||
| P02452 | Collagen alpha-1 (I) chain | Secreted | 0.20 ± 0.21 (54) | 7.21 ± 6.72 (39) |
| P12109 | Collagen alpha-1 (VI) chain | Secreted | 0.27 ± 0.03 (31) | 4.35 ± 1.51 (35) |
| Q99715 | Collagen alpha-1 (XII) chain | Secreted | 0.12 ± 0.06 (220) | 12.29 ± 4.1 (245) |
| P08123 | Collagen alpha-2 (I) chain | Secreted | 0.20 ± 0.12 (33) | 4.15 ± 1.74 (28) |
| P12110 | Collagen alpha-2 (VI) chain | Secreted | 0.39 ± 0.04 (20) | 3.19 ± 0.42 (24) |
| P12111 | Collagen alpha-3 (VI) chain | Secreted | 0.25 ± 0.23 (168) | 7.82 ± 5.54 (133) |
| Q05682 | Caldesmon | Cytoplasm | 0.48 ± 0.20 (18) | 2.82 ± 0.96 (18) |
| P14209 | CD99 antigen (T-cell surface glycoprotein E2) | Secreted | 0.11 ± 0.03 (2) | 3.74 ± 4.92 (2) |
| P13646 | Keratin, type I cytoskeletal 13 | Cytoplasm | 0.76 ± 0.58 (2) | 4.39 ± 3.97 (3) |
| P02751 | Fibronectin | Secreted | 0.26 ± 0.13 (6) | 8.73 ± 4.76 (6) |
| P07996 | Thrombospondin-1 | External surface protein | 0.23 ± 0.04 (10) | 5.27 ± 2.81 (10) |
| Q01995 | Transgelin | Cytoplasm | 0.37 ± 0.04 (9) | 3.01 ± 0.11 (8) |
a Name given by UniProt database.
b Relevant descriptions from the literature.
c Average ratio ± standard deviation (number of identified unique peptides).
Validation Studies of HCMV Induction of Key MAM Proteins
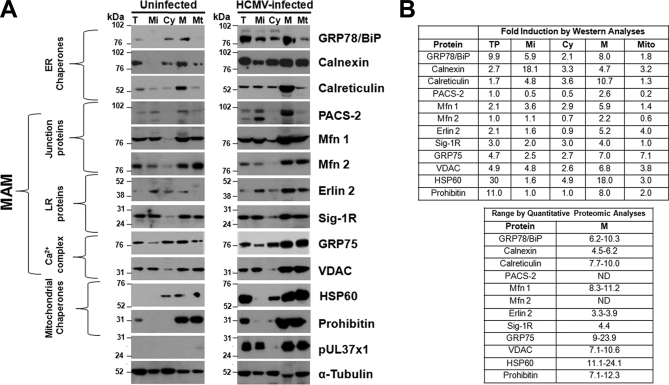
Proteomic data of key MAM proteins at late times of HCMV infection were validated using Western analyses of subcellular fractions. HCMV induced substantial increases in ER chaperones, BiP/GRP78 (eightfold), calnexin (4.7-fold), and calreticulin (10.7-fold) in the heavy MAM (Fig. 6). More moderate increases of MAM junction proteins, PACS-2 (2.6-fold), Mfn1 (5.9-fold), and Mfn2 (2.2-fold), were documented by Western analyses as were increased MAM lipid raft proteins, erlin 2 (5.2-fold) and Sig-1R (fourfold). Components of the MAM Ca2+ signaling complex, VDAC (6.8-fold) and GRP75 (sevenfold) and mitochondrial chaperones, HSP60 (18-fold) and prohibitin (8-fold), were more dramatically increased. In contrast, minor changes were observed for α-tubulin, which was used to normalize protein levels in each fraction. HCMV pUL37×1 was detected in the microsomal, MAM and mitochondrial fractions of HCMV-infected HFFs, as we have previously found (28, 29, 31). The values of induction observed for key MAM components by Western analyses mostly fell within the induction value ranges determined by quantitative proteomics (Fig. 6B). Together, these studies verified the induction of key, known MAM proteins at late times of HCMV infection. These results further suggest that HCMV infection induces MAM restructuring to benefit its growth.
Fig. 6.
A, Verification of quantitative MAM proteomic results using Western analyses. HFFs were uninfected (left) or HCMV-infected (right). At 72 hpi, microsomal (Mi), cytosol (Cy), heavy MAM (M), mitochondria (Mt) were isolated as previously described (28, 47). Proteins in total (T) lysates (30 μg) or indicated fractions (10 μg) were resolved by SDS-PAGE and analyzed by Western analyses using ER chaperones (anti-GRP78/BiP, -calnexin, -calreticulin), MAM junction (anti-PACS-2, -Mfn 1, -Mfn 2), MAM lipid rafts (anti-erlin 2, Sig-1R), MAM Ca2+ signaling complex (anti-GRP75, -VDAC) and mitochondrial chaperones (anti-HSP60, -Prohibitin). The presence of pUL37×1 was verified by anti-UL37×1 (DC35) and α-tubulin was used as a loading control. B, Comparison of HCMV induction of key proteins associated with the MAM calculated by Western analyses (top) or quantitative proteomic analyses (bottom). Top right: The HCMV induction for each protein was calculated for each fraction in the Western analyses (Panel A) by comparing the indicated protein levels in HCMV-infected cells normalized to α-tubulin levels and then divided by the protein levels in uninfected cells normalized to α-tubulin levels. Bottom right: The range of HCMV induction of each protein was determined by quantitative proteomics as in Tables I and III. ND: Proteins were not detected by MS/MS analyses. Prohibitin data from quantitative proteomics were not shown.
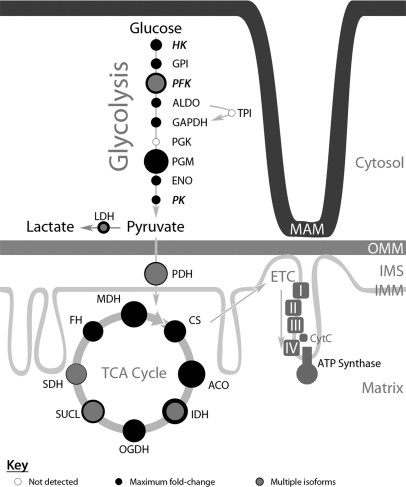
Fig. 7 pictorially represents the observed association of glycolytic enzymes to the MAM fraction at late times of HCMV infection. The quantitative proteomics demonstrated increases in glycolytic enzymes (supplementary Table S2) of most glycolysis reactions with values from 1.9-fold (pyruvate kinase) to 14.4-fold (phosphoglycerate mutase). These values were verified by the reverse experiment. Nonetheless, two glycolytic enzymes, triose phosphate isomerase and phosphoglycerolkinase, were not detected with confidence in the MAM fraction but were detected in banded mitochondria (data not shown).
Fig. 7.
Increased levels of the glycolytic, TCA cycle, and ETC pathways associated with ER-mitochondrial contacts at late times of HCMV infection. The relative fold-inductions for proteins involved in glycolysis, TCA cycle, and ETC detected in association with the MAM fraction from HCMV-infected HFFs above uninfected cells (numeric values are shown in supplementary Table S2) are represented by the areas of corresponding filled black circles. The open circles represent glycolytic enzymes not detected by this quantitative proteomic analysis. Proteins with multiple isoforms are also represented by a smaller, gray-filled circle (isoform with lowest induction) and surrounded by a darker outline (isoform with the largest induction). Constituent members of the ETC and ATP synthase complexes are represented only by solid blocks, as these proteins were observed to have increased expression levels of at least sevenfold, but typically over 10-fold higher than in uninfected cells. Glycolysis hexokinase (HK), glucose-6-phosphate isomerase (GPI), phosphofructokinase (PFK), fructose-biphosphate aldolase (ALDO), glyceraldehyde-3-phosphate dehydrogenase (GAPDH), phosphoglycerate mutase (PGM), alpha-enolase (ENO), pyruvate kinase (PK), lactate dehydrogenase (LDH), pyruvate dehydrogenase (PDH), citrate synthase (CS), aconitase hydratase (ACO), isocitrate dehydrogenase (IDH), 2-oxoglutarate dehydrogenase (OGDH), succinyl-CoA ligase (SUCL), succinate dehydrogenase (SDH), fumarate hydratase (FH), malate dehydrogenase (MDH).
Furthermore, the glycolytic pyruvate kinase enzyme detected in our proteomics showed peptides specific for the M2 isoform. Pyruvate kinase M2 has been demonstrated to promote glycolytic flux as well as allow the buildup of intermediate metabolites to support anabolic metabolism, a process linked to rapidly dividing cancer cells (62, 63) and also critical for productive HCMV infection (5, 6).
In addition to the glycolytic enzymes, all the tricarboxylic acid (TCA) cycle enzymes, except for malic enzyme, were increasingly associated with the MAM after HCMV infection. Notably the induction of enzymes ranged from 7.4-fold (isocitrate dehydrogenase) to 17.6-fold (aconitase). In addition, the electron transport chain (ETC) complexes I, II, III, and IV, and the ATP synthase complex localized in the inner mitochondrial membrane were detected and increased (>7.9- to 15.5-fold) in the MAM fraction after HCMV infection.
Furthermore, we observed up-regulation of proteins involved in glutaminolytic pathways and anaplerosis. We detected inductions of malate shuttle proteins (oxoglutarate/malate carrier protein, malic enzyme, and malate dehydrogenase) and glutamate intake proteins (Aralar, aspartate aminotransferase, glutamate/OH carrier, and glutamate dehydrogenase), but did not detect inhibitors of glutamate intake (such as aminooxyacetate or ribulose-5-phosphate). Enzymes needed to convert glutamine to α-ketoglutarate to enter the TCA cycle (glutaminase and glutamate dehydrogenase) were also detected. Finally, the quantitative proteomic analyses detected an increase in the abundance of lipid synthetic or degradative enzymes listed in supplementary Table S3. Together, these studies place critical metabolic and synthetic pathways needed for HCMV infection at the MAM, where they can be coordinated by ER-mitochondrial crosstalk.
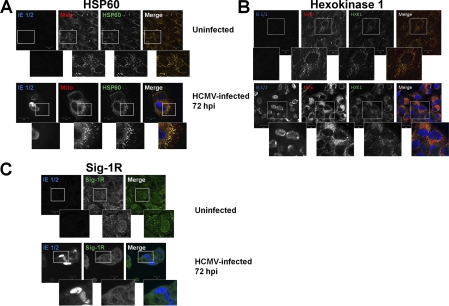
To determine whether HCMV infection induced relocalization of selected MAM proteins, we compared the localization of HSP60, Hexokinase 1, and Sig-1R in uninfected HFFs and HCMV-infected HFFs by immunofluorescence assay (Fig. 8). Although HSP60 and Hexokinase 1 increased in abundance in the HCMV-infected cell, they did not appear to markedly relocalize during HCMV infection. In stark contrast, Sig-1R relocalized from a primarily nuclear localization in uninfected cells to a cytosolic membrane localization during late times of HCMV infection. Together with the results above, these findings show that HCMV selectively induces relocalization of some MAM proteins such as Sig-1R.
Fig. 8.
Relocalization of Sig-1R during HCMV infection of HFFs. HFFs were uninfected or HCMV infected at a multiplicity of 1 and harvested at 72 hpi. A, Induction of HSP60 during HCMV infection. Uninfected and HCMV-infected HFFs were probed with anti-HSP60 (green), anti-HCMV IE 1/2 (blue), and human anti mitochondria (red) antibodies and the corresponding secondary antibodies. The stained cells were imaged by confocal microscopy. The three leftmost panels are greyscale and the right panel shows the overlay of confocal sections. The insets show enlarged regions of interest in the cells. B, Induction of Hexokinase 1 at late times of HCMV infection. Cells were probed with anti-Hexokinase (green), anti-HCMV IE 1/2 (blue), and human anti mitochondria (red) and the corresponding secondary antibodies and imaged as above. The three leftmost panels are greyscale and the right panel shows the overlay of confocal sections. The insets show enlarged regions of interest in the cells. C, Relocalization of Sig-1R during HCMV infection of HFFs. Cells were probed with anti-Sig 1R (green) and anti HCMV IE 1/2 (blue) antibodies. The left and middle panels are greyscale and the right shows the overlay of confocal sections. The insets show enlarged regions of interest in the cells.
DISCUSSION
Currently, the only licensed antiviral drugs for HCMV target its DNA replication. However, other regulated cellular metabolic reactions occurring during infection have emerged as important to HCMV progeny production and maturation, which may serve as future therapeutic targets. These events include the uncoupling of respiration from glucose metabolism, the concomitant use of glutamine for anaplerosis to maintain cycling of oxidative phosphorylation reactions in the TCA cycle, and the biosynthesis of lipids and membranes. The current studies analyzed HCMV alterations of the MAM, which regulates Ca2+-responsive mitochondrial enzymes and is enriched in lipid synthetic enzymes. Importantly for these studies, the MAM can be physically fractionated by exploiting their tight association to mitochondria, and distinguishable banding patterns in Percoll gradients (29, 47). Using SILAC methodology we probed the MAM compositional changes induced by HCMV infection during late stages of infection, when the cell has been reprogrammed into a virus factory.
The MAM is increasingly being appreciated as a key control of mitochondrial mediated apoptotic signaling, calcium signaling from the ER to mitochondria and consequent regulation of cell bioenergetics (20). Proteomic analyses of enriched MAM allowed for increased sensitivity in detecting MAM components in primary human fibroblasts and the changes induced in the MAM by HCMV infection. Although some MAM markers have been identified, a more comprehensive, unbiased analysis of the MAM proteome has not yet been reported. To our knowledge, this is the first proteomic analysis of enriched MAM, and uses some of the most rigorous methodology currently available to fractionate the MAM and quantify the changes induced by HCMV infection. The detected changes during infection provide insight into which MAM proteins could be playing important roles during HCMV infection. These studies have provided valuable insight into the MAM and alteration during HCMV infection and will serve as a foundation to test the role of individual components during HCMV infection.
Functionally, the physical association of the ER and mitochondria regulates important cellular processes such as lipid biosynthesis and nonvesicular transfer of lipids to mitochondria (52, 53, 64, 65). Transfer of ceramide synthesized in the MAM to mitochondria can induce channel formation and apoptosis (66). The MAM accommodates internal lipid rafts, detergent-resistant membranes, which are enriched in Sig-1R (56, 67, 68). Erlin-1 and erlin-2, associated with IP3R degradation, are also present in internal lipid rafts (69–71). Importantly, the ER can transmit Ca2+ signals to mitochondria without increasing bulk cytosolic Ca2+ concentrations above physiological thresholds. An MAM macromolecular complex composed of IP3R type 3 (IP3R3)-GRP75-VDAC generates high Ca2+ microdomains at ER-mitochondrial contact sites and thereby enables the function of the mitochondrial uniporter (72). The MAM is enriched in Ca2+ binding chaperones, calreticulin, Sig-1R, and ERp57 (20, 53, 56, 65, 73). Sig-1R, a ligand-dependent MAM chaperone, increases ER Ca2+ efflux by stabilizing IP3R3 (74). IP3R-mediated Ca2+ release is essential in maintaining cellular bioenergetics (21). Nonetheless, if unchecked, Ca2+ release can induce mitochondrial-mediated apoptosis (20). IP3R-mediated Ca2+ signaling activates apoptosis by inducing cytochrome c release from mitochondria (75). Increased mitochondrial Ca2+ has also been associated with increasing electron transport, increased reactive oxygen species production, and opening of the mitochondrial permeability transition pore (76).
We observed a dramatic alteration of the MAM proteome at late times of HCMV infection. This conclusion is supported by the augmented congregation of proteins at the MAM, which regulate Ca2+ cross-talk between ER and mitochondria, glutamate uptake and utilization for anaplerosis, metabolic flux of both glycolysis and oxidative phosphorylation, along with an impressive concentration of heat shock proteins. The broad induction of MAM constituents during infection indicates an important role for ER-mitochondrial cross-talk at late times of infection, predictably for the maintenance of cell viability and increased energy production for progeny production. The altered MAM proteome during HCMV infection could be because of the recruitment of crucial proteins to ER-mitochondria junctions, which were either absent or under-represented in MAM of uninfected cells. The induction of membrane association of soluble cytosolic proteins such as the glycolytic enzymes suggests that protein scaffolding at MAM domains also helps mediate the critical functional changes in ER and mitochondria observed during HCMV infection.
Of the proteins increased in the MAM, heat shock proteins had the highest fold induction caused by HCMV infection. Cellular chaperones are known to be induced by HCMV infection, as well as by ER stress, however their dramatic accumulation at MAM sites was unexpected. Previous evidence supports unique functions for membrane-tethered chaperone proteins, beyond assisting protein-folding (61). HSP10, HSP60, and GRP75, which plays a role in Ca2+ signaling when tethered at the MAM membrane, may likely engage in unique cell signaling functions at MAM domains. Membrane fluidity and microdomain organization are decisive in the transduction of stresses into signals for activation of HSPs (61). Association of certain HSPs with membranes can reestablish fluidity and bilayer stability and restore membrane functionality during stress and modulate thereby cellular responses to stress.
ER stress is known to increase MAM junctions twofold (26). HCMV induces specific components of ER stress at late times of infection, but truncates the full unfolded protein response (41, 77, 78). This allows the virus to utilize the increased levels of chaperones for the production and assembly of virion components, without a widespread block on nascent polypeptide translation. We detected a prominent trend of induction of MAM proteins around two- to fivefold during late times of HCMV infection, consistent with an increase in MAM junctions because of HCMV-induced ER stress. Importantly, however, we also detect MAM components increased well beyond this modest level of induction, suggesting functionally directed restructuring of MAM subdomains for the functional benefits to HCMV growth, not merely an increase in the number of ER-mitochondria contacts or of the tightness of the ER-mitochondrial junctions.
HCMV infection alters the localizations, shapes, and sizes of secretory organelles at late times of infection during its assembly (79). Viral proteins were also detected in the MAM of HCMV-infected cells but not in uninfected cells. This finding may indicate trafficking or recruitment of other HCMV proteins, in addition to pUL37×1, to the MAM. Alternatively, the identified HCMV proteins may result from virions exiting the nucleus through the outer nuclear membrane, which is contiguous with the ER membrane whose subdomains were used for these studies. Importantly to the enrichment of the MAM compartment, the trans-Golgi marker was very low in abundance in the MAM (1/100th) of the levels in the cytosol. Furthermore, we did not detect the highly abundant HCMV proteins that traffic to other membrane-bound compartments including the nucleus (pp150/pUL32 and UL53), secretory apparatus (gB, gH, gL, and gO), or endosomes (US27, UL33, and US28) as well as the assembly compartment (pp28/UL99). Because the analyses were performed at late times of infection, the undetected HCMV virion proteins, localized to the secretory apparatus, are of much greater abundance than pUL37×1, which we reliably detect in the MAM fraction. Consistent with the proteomic results, pUL37×1 has been detected in the ER, MAM, and mitochondria but not in the nucleus, Golgi or plasma membrane (29, 31, 50). Together, these findings argue for the enrichment of the MAM in the banded fraction and against a simple mixing of cellular membranes during late times of HCMV infection.
There appear to be multiple potential mechanisms underlying the increased abundance of proteins in the MAM. The most common mechanism, notable for the heat shock proteins, appears to be increased total expression during HCMV infection and a proportional increase in the MAM abundances. Examples of these include GRP78, GRP75, and HSP60. HCMV infection is known to induce a modified ER stress response (77, 78, 80). The ability of HCMV to modulate the cellular ER stress response is evident by the finding that Sig-1R increases in the MAM despite the fact that HCMV causes ER stress. Sig-1R is a sensor that relocalizes from the MAM to bulk ER when cells undergo chronic ER stress responses (74). Sig-1R relocalizes from the periphery of the nucleus (observed in uninfected cells) to the MAM in about 50% of HCMV-infected HFFs (Fig. 8), suggesting a finely tuned regulation of its location in the infected cell. The abundance of GRP78 is increased during HCMV infection and is essential for HCMV assembly (57). Herein, we found that a significant fraction of the increased GRP78 in HCMV-infected cells localizes in the MAM, suggesting the functional importance of the MAM during HCMV infection. Our finding of GRP78 enrichment in the MAM of infected cells is consistent with its known enrichment in the MAM of uninfected cells (74). GRP75 is also documented to increase in abundance in the heavy MAM fraction of HCMV-infected cells (29) and our current results solidify that finding using quantitative proteomics and Western analyses. Nevertheless, the increase in GRP75 was greater in the MAM than in total abundance suggesting that in addition to a corresponding increase with total abundance, GRP75 may also be relocalized during HCMV infection.
In contrast, the abundances of other cellular proteins selectively increase in the MAM fraction even though there is little to no change in their abundances in total cellular extracts. These changes in their relative abundance in the MAM suggest a targeted relocalization of these proteins from other subcellular compartments to the MAM during HCMV infection. Examples of these include calreticulin whose marginal increase in total fraction is considerably less than the increases in microsomes or heavy MAM fractions. Similarly, the increases in calnexin and PACS-2 abundances in the MAM fraction are greater than in their corresponding increases in total extracts. Consistent with this potential mechanism is the finding that calnexin is a known cargo for PACS-2 (81) and its relocalization to the MAM may be mediated by PACS-2. Finally, relocalization of glycolytic enzymes from the cytosol to ER-mitochondrial contacts appears to underlie at least in part their increased abundance in the MAM. Intriguingly, membrane association of glycolytic enzymes has been found in pancreatic cancer cells (82). Thus, HCMV may selectively retarget key cellular proteins to the MAM and thereby affect its functions during infection.
Mining our proteomic results for the MAM Ca2+ signaling complex components, we gain some understanding of the structural organization of MAM domains during late times of HCMV infection. The OMM porin, VDAC, is known to be involved in more than one macromolecular complex at the MAM. In one of these complexes, it associates with cytosolic GRP75 and IP3R of the ER membrane to form a Ca2+ signaling complex bridging the ER and mitochondria (27). Here, we observed a dramatic increase in GRP75 abundance in the MAM, which we had previously shown with proteinase K digestion to be cytosolic GRP75 (29). VDAC can also form complexes with ANT proteins of the inner mitochondrial membrane to regulate mitochondrial membrane permeability, Bax association, and cytochrome c release. Importantly, VDAC can directly bind ANT of the inner mitochondrial membrane when it is in a low conductance, cationically selective, tensed conformation. This tensed pore structure allows transfer of extramitochondrial Ca2+ into the intermembrane space of the mitochondria. HCMV pUL37×1/vMIA causes ER Ca2+ efflux (35), interacts with Bax and ANT (10, 13, 14) but blocks the proapoptotic activity of Bax at the OMM (13, 37, 38). Hexokinase, which was up-regulated in the MAM of HCMV-infected cells, can bind to this complex to promote mitochondrial oxidative phosphorylation, and malate shuttling through VDAC to the cytosol (83). As cytochrome c stably binds ANT in this macromolecular complex configuration, we expected to detect cytochrome c, which resides within the intermembrane space of the mitochondria, by proteomic analyses of the MAM. Consistently, the HCMV-specific induction of cytochrome c levels directly matched induced ANT levels in our experiments (supplementary Table S3). However, we did not detect other components of lipid raft-associated apoptosomes in our MAM fractions, which should have been detected if the cytochrome c in the MAM fraction induced apoptosis. Together, these data suggest that the majority of MAM associated VDAC is engaged in activities allowing Ca2+ signaling to mitochondria, and increased metabolic flux and blocking of proapoptotic signaling.
One important finding of these studies was the increased MAM association of several metabolic enzymes during HCMV infection. Strikingly, we detected HCMV-specific induction of MAM association with cytosolic glycolytic enzymes. Because the cytosol is not pelleted with the MAM and mitochondria during early fractionation steps, but remains in the supernatant, it is unlikely that significant carryover to the Percoll gradient and subsequent contamination of the banded MAM, unless these components were membrane tethered. A similar phenomenon of membrane-tethering of these enzymes has been characterized in rapidly dividing cancer cells (82). Revealingly, a Ca2+ responsive aspartate-glutamate carrier, Aralar, which supplies mitochondria with glutamate for oxidative phosphorylation reactions, emerges as one of the most enriched proteins at MAM domains at 72 hpi. We found a large (12- to 20-fold) increase in MAM-associated Aralar 2 during HCMV infection, which when compared with the mean 20-fold induction seen for HCMV viral proteins in MAM (which are absent in uninfected cells), exposes a significant induction of this protein during late times of HCMV infection, corresponding with the requirement of glutamate for anaplerosis at late times of infection (7).
Our current studies revealed exceptionally few decreased MAM proteins. All of the downregulated MAM proteins were secretory proteins, destined for the extracellular matrix. It appears that HCMV infection specifically reduces the known ability of MAM to support transport of secretory proteins to the Golgi (53). In contrast, global protein secretion from peripheral ER to the cell surface is not diminished during HCMV infection. Previous studies on the HCMV secretome demonstrate an increase in other secreted proteins including cytokines, as well as angiogenic and wound healing proteins from HCMV-infected cells at 72 hpi (40). Taken together, these results argue that HCMV infection selectively diminishes specific MAM functional activities whereas augmenting others.
Acknowledgments
We are grateful to Dr. Stephen Robbins for the kind gift of anti-erlin 2 antiserum.
Footnotes
* This work was funded in part by National Institutes of Health Grant R21 AI081957 (to ACP) and by Funds from the Children's Research Institute (to ACP). Additional support was provided by NIH NCMRR/NINDS 2R24HD050846–06 (NCMRR-DC Core Molecular and Functional Outcome Measures in Rehabilitation Medicine), by NIH NCRR UL1RR031988 (GWU-CNMC CTSI), and by NIH/NICHD 5P30HD040677-10 (Intellectual and Developmental Disabilities Research Centers) to ACP, KB and YH.
 This article contains supplemental Tables S1, S2, and S3, as well as a supplemental database containing peptide results from proteomic analyses (Database S4).
This article contains supplemental Tables S1, S2, and S3, as well as a supplemental database containing peptide results from proteomic analyses (Database S4).
1 The abbreviations used are:
- ANT
- adenine nucleotide translocator
- Arg
- Arginine
- BAD
- bacterial artificial chromosome of human cytomegalovirus strain AD169
- Ca2+
- calcium
- DMEM
- Dulbecco's Modified Eagle Medium
- ER
- endoplasmic reticulum
- ETC
- electron transport chain
- GRP
- glucose regulated protein
- HCMV
- human cytomegalovirus
- HFFs
- human foreskin fibroblasts
- hpi
- hours post infection
- HSP
- heat shock protein
- IP3R
- inositol 1,4,5-triphosphate receptor
- LTQ
- linear trap quadrupole
- Lys
- Lysine
- MAM
- mitochondria-associated membranes
- Mfn
- mitofusin
- min
- minutes
- OMM
- outer mitochondrial membrane
- PBS
- phosphate buffered saline
- PGM
- phosphoglycerate mutase
- pUL37x1
- UL37 exon 1 protein
- Sig-1R
- sigma 1 receptor
- SILAC
- stable isotope labeling by amino acids in cell culture
- SR
- signal recognition particle receptor
- SRP
- signal recognition particle
- TCA
- tricarboxylic acid
- VDAC
- voltage dependent anion channel
- vMIA
- viral mitochondria localized inhibitor of apoptosis
- wt
- wild type.
REFERENCES
- 1. Mocarski E. S., Shenk T., Pass R. F. (2007) Cytomegaloviruses. In: Knipe D. M., Howley P. M. eds. Fields Virology, 5th Ed., pp. 2701–2772, Wolters Kluwer Health, Lippincott Williams & Wilkins, Philadelphia [Google Scholar]
- 2. Gerna G., Baldanti F., Revello M. G. (2004) Pathogenesis of human cytomegalovirus infection and cellular targets. Hum. Immunol. 65, 381–386 [DOI] [PubMed] [Google Scholar]
- 3. Boeckh M., Nichols W. G. (2004) The impact of cytomegalovirus serostatus of donor and recipient before hematopoietic stem cell transplantation in the era of antiviral prophylaxis and preemptive therapy. Blood. 103, 2003–2008 [DOI] [PubMed] [Google Scholar]
- 4. Khanna R., Diamond D. J. (2006) Human cytomegalovirus vaccine: time to look for alternative options. Trends Mol. Med. 12, 26–33 [DOI] [PubMed] [Google Scholar]
- 5. Munger J., Bajad S. U., Coller H. A., Shenk T., Rabinowitz J. D. (2006) Dynamics of the cellular metabolome during human cytomegalovirus infection. PLoS Pathog. 2, e132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Munger J., Bennett B. D., Parikh A., Feng X. J., McArdle J., Rabitz H. A., Shenk T., Rabinowitz J. D. (2008) Systems-level metabolic flux profiling identifies fatty acid synthesis as a target for antiviral therapy. Nat. Biotechnol. 26, 1179–1186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chambers J. W., Maguire T. G., Alwine J. C. (2010) Glutamine metabolism is essential for human cytomegalovirus infection. J. Virol. 84, 1867–1873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Reboredo M., Greaves R. F., Hahn G. (2004) Human cytomegalovirus proteins encoded by UL37 exon 1 protect infected fibroblasts against virus-induced apoptosis and are required for efficient virus replication. J. Gen. Virol. 85, 3555–3567 [DOI] [PubMed] [Google Scholar]
- 9. Kouzarides T., Bankier A. T., Satchwell S. C., Preddy E., Barrell B. G. (1988) An immediate early gene of human cytomegalovirus encodes a potential membrane glycoprotein. Virology 165, 151–164 [DOI] [PubMed] [Google Scholar]
- 10. Goldmacher V. S., Bartle L. M., Skaletskaya A., Dionne C. A., Kedersha N. L., Vater C. A., Han J. W., Lutz R. J., Watanabe S., Cahir McFarland E. D., Kieff E. D., Mocarski E. S., Chittenden T. (1999) A cytomegalovirus-encoded mitochondria-localized inhibitor of apoptosis structurally unrelated to Bcl-2. Proc. Natl. Acad. Sci. U.S.A. 96, 12536–12541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Terhune S., Torigoi E., Moorman N., Silva M., Qian Z., Shenk T., Yu D. (2007) Human cytomegalovirus UL38 protein blocks apoptosis. J. Virol. 81, 3109–3123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. McCormick A. L., Roback L., Livingston-Rosanoff D., St Clair C. (2010) The human cytomegalovirus UL36 gene controls caspase-dependent and -independent cell death programs activated by infection of monocytes differentiating to macrophages. J. Virol. 84, 5108–5123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Arnoult D., Bartle L. M., Skaletskaya A., Poncet D., Zamzami N., Park P. U., Sharpe J., Youle R. J., Goldmacher V. S. (2004) Cytomegalovirus cell death suppressor vMIA blocks Bax- but not Bak-mediated apoptosis by binding and sequestering Bax at mitochondria. Proc. Natl. Acad. Sci. U.S.A. 101, 7988–7993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Poncet D., Larochette N., Pauleau A. L., Boya P., Jalil A. A., Cartron P. F., Vallette F., Schnebelen C., Bartle L. M., Skaletskaya A., Boutolleau D., Martinou J. C., Goldmacher V. S., Kroemer G., Zamzami N. (2004) An anti-apoptotic viral protein that recruits Bax to mitochondria. J. Biol. Chem. 279, 22605–22614 [DOI] [PubMed] [Google Scholar]
- 15. McCormick A. L., Meiering C. D., Smith G. B., Mocarski E. S. (2005) Mitochondrial cell death suppressors carried by human and murine cytomegalovirus confer resistance to proteasome inhibitor-induced apoptosis. J. Virol. 79, 12205–12217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Poncet D., Pauleau A. L., Szabadkai G., Vozza A., Scholz S. R., Le Bras M., Brière J. J., Jalil A., Le Moigne R., Brenner C., Hahn G., Wittig I., Schägger H., Lemaire C., Bianchi K., Souquere S., Pierron G., Rustin P., Goldmacher V. S., Rizzuto R., Palmieri F., Kroemer G. (2006) Cytopathic effects of the cytomegalovirus-encoded apoptosis inhibitory protein vMIA. J. Cell Biol. 174, 985–996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Spector D. H. (1996) Activation and regulation of human cytomegalovirus early genes. Intervirology 39, 361–377 [DOI] [PubMed] [Google Scholar]
- 18. Greenaway P. J., Wilkinson G. W. (1987) Nucleotide sequence of the most abundantly transcribed early gene of human cytomegalovirus strain AD169. Virus Res. 7, 17–31 [DOI] [PubMed] [Google Scholar]
- 19. Reeves M. B., Davies A. A., McSharry B. P., Wilkinson G. W., Sinclair J. H. (2007) Complex I binding by a virally encoded RNA regulates mitochondria-induced cell death. Science 316, 1345–1348 [DOI] [PubMed] [Google Scholar]
- 20. Hayashi T., Rizzuto R., Hajnoczky G., Su T. P. (2009) MAM: more than just a housekeeper. Trends Cell Biol. 19, 81–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Cárdenas C., Miller R. A., Smith I., Bui T., Molgó J., Müller M., Vais H., Cheung K. H., Yang J., Parker I., Thompson C. B., Birnbaum M. J., Hallows K. R., Foskett J. K. (2010) Essential regulation of cell bioenergetics by constitutive InsP3 receptor Ca2+ transfer to mitochondria. Cell 142, 270–283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Csordás G., Renken C., Várnai P., Walter L., Weaver D., Buttle K. F., Balla T., Mannella C. A., Hajnóczky G. (2006) Structural and functional features and significance of the physical linkage between ER and mitochondria. J. Cell Biol. 174, 915–921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. de Brito O. M., Scorrano L. (2008) Mitofusin 2 tethers endoplasmic reticulum to mitochondria. Nature 456, 605–610 [DOI] [PubMed] [Google Scholar]
- 24. de Brito O. M., Scorrano L. (2009) Mitofusin-2 regulates mitochondrial and endoplasmic reticulum morphology and tethering: the role of Ras. Mitochondrion. 9, 222–226 [DOI] [PubMed] [Google Scholar]
- 25. Cerqua C., Anesti V., Pyakurel A., Liu D., Naon D., Wiche G., Baffa R., Dimmer K. S., Scorrano L. (2010) Trichoplein/mitostatin regulates endoplasmic reticulum-mitochondria juxtaposition. EMBO Rep. 11, 854–860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Simmen T., Aslan J. E., Blagoveshchenskaya A. D., Thomas L., Wan L., Xiang Y., Feliciangeli S. F., Hung C. H., Crump C. M., Thomas G. (2005) PACS-2 controls endoplasmic reticulum-mitochondria communication and Bid-mediated apoptosis. EMBO J. 24, 717–729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Szabadkai G., Bianchi K., Várnai P., De Stefani D., Wieckowski M. R., Cavagna D., Nagy A. I., Balla T., Rizzuto R. (2006) Chaperone-mediated coupling of endoplasmic reticulum and mitochondrial Ca2+ channels. J. Cell Biol. 175, 901–911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bozidis P., Williamson C. D., Colberg-Poley A. M. (2008) Mitochondrial and secretory human cytomegalovirus UL37 proteins traffic into mitochondrion-associated membranes of human cells. J. Virol. 82, 2715–2726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Bozidis P., Williamson C. D., Wong D. S., Colberg-Poley A. M. (2010) Trafficking of UL37 proteins into mitochondrion-associated membranes during permissive human cytomegalovirus infection. J. Virol. 84, 7898–7903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Williamson C. D., Colberg-Poley A. M. (2009) Access of viral proteins to mitochondria via mitochondria-associated membranes. Rev. Med. Virol. 19, 147–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Williamson C. D., Colberg-Poley A. M. (2010) Intracellular Sorting Signals for Sequential Trafficking of Human Cytomegalovirus UL37 Proteins to the Endoplasmic Reticulum and Mitochondria. J. Virol. 84, 6400–6409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Williamson C. D., Zhang A., Colberg-Poley A. M. (2011) The human cytomegalovirus UL37 exon 1 protein associates with internal lipid rafts. J. Virol. 85, 2100–2111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Mavinakere M. S., Colberg-Poley A. M. (2004) Internal cleavage of the human cytomegalovirus UL37 immediate-early glycoprotein and divergent trafficking of its proteolytic fragments. J. Gen. Virol. 85, 1989–1994 [DOI] [PubMed] [Google Scholar]
- 34. Mavinakere M. S., Colberg-Poley A. M. (2004) Dual targeting of the human cytomegalovirus UL37 exon 1 protein during permissive infection. J. Gen. Virol. 85, 323–329 [DOI] [PubMed] [Google Scholar]
- 35. Sharon-Friling R., Goodhouse J., Colberg-Poley A. M., Shenk T. (2006) Human cytomegalovirus pUL37x1 induces the release of endoplasmic reticulum calcium stores. Proc. Natl. Acad. Sci. U.S.A. 103, 19117–19122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Williamson C. D., Zhang A., Colberg-Poley A. M. (2011) The human cytomegalovirus protein UL37 exon 1 associates with internal lipid rafts. J. Virol. 85, 2100–2111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Pauleau A. L., Larochette N., Giordanetto F., Scholz S. R., Poncet D., Zamzami N., Golmacher V. S., Kroemer G. (2007) Structure-function analysis of the interaction between Bax and the cytomegalovirus-encoded protein vMIA. Oncogene 26, 7067–7080 [DOI] [PubMed] [Google Scholar]
- 38. Norris K. L., Youle R. J. (2008) Cytomegalovirus proteins vMIA and m38.5 link mitochondrial morphogenesis to Bcl-2 family proteins. J. Virol. 82, 6232–6243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ong S. E., Blagoev B., Kratchmarova I., Kristensen D. B., Steen H., Pandey A., Mann M. (2002) Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol. Cell. Proteomics 1, 376–386 [DOI] [PubMed] [Google Scholar]
- 40. Dumortier J., Streblow D. N., Moses A. V., Jacobs J. M., Kreklywich C. N., Camp D., Smith R. D., Orloff S. L., Nelson J. A. (2008) Human cytomegalovirus secretome contains factors that induce angiogenesis and wound healing. J. Virol. 82, 6524–6535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Isler J. A., Skalet A. H., Alwine J. C. (2005) Human cytomegalovirus infection activates and regulates the unfolded protein response. J. Virol. 79, 6890–6899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Varnum S. M., Streblow D. N., Monroe M. E., Smith P., Auberry K. J., Pasa-Tolic L., Wang D., Camp D. G., 2nd, Rodland K., Wiley S., Britt W., Shenk T., Smith R. D., Nelson J. A. (2004) Identification of proteins in human cytomegalovirus (HCMV) particles: the HCMV proteome. J. Virol. 78, 10960–10966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Moorman N. J., Sharon-Friling R., Shenk T., Cristea I. M. (2010) A targeted spatial-temporal proteomics approach implicates multiple cellular trafficking pathways in human cytomegalovirus virion maturation. Mol. Cell. Proteomics 9, 851–860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Gao Y., Colletti K., Pari G. S. (2008) Identification of human cytomegalovirus UL84 virus- and cell-encoded binding partners by using proteomics analysis. J. Virol. 82, 96–104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Adair R., Liebisch G. W., Su Y., Colberg-Poley A. M. (2004) Alteration of cellular RNA splicing and polyadenylation machineries during productive human cytomegalovirus infection. J. Gen. Virol. 85, 3541–3553 [DOI] [PubMed] [Google Scholar]
- 46. Yu D., Smith G. A., Enquist L. W., Shenk T. (2002) Construction of a self-excisable bacterial artificial chromosome containing the human cytomegalovirus genome and mutagenesis of the diploid TRL/IRL13 gene. J. Virol. 76, 2316–2328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Bozidis P., Williamson C. D., Colberg-Poley A. M. (2007) Isolation of endoplasmic reticulum, mitochondria, and mitochondria-associated membrane fractions from transfected cells and from human cytomegalovirus-infected primary fibroblasts. Current Protocols Cell Biol. Chapter 3, Unit 3, 27, 1–23 [DOI] [PubMed] [Google Scholar]
- 48. Park S. K., Venable J. D., Xu T., Yates J. R., 3rd (2008) A quantitative analysis software tool for mass spectrometry-based proteomics. Nat. Methods 5, 319–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Mavinakere M. S., Williamson C. D., Goldmacher V. S., Colberg-Poley A. M. (2006) Processing of human cytomegalovirus UL37 mutant glycoproteins in the endoplasmic reticulum lumen prior to mitochondrial importation. J. Virol. 80, 6771–6783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Colberg-Poley A. M., Patel M. B., Erezo D. P., Slater J. E. (2000) Human cytomegalovirus UL37 immediate-early regulatory proteins traffic through the secretory apparatus and to mitochondria. J. Gen. Virol. 81, 1779–1789 [DOI] [PubMed] [Google Scholar]
- 51. Stone S. J., Vance J. E. (2000) Phosphatidylserine synthase-1 and -2 are localized to mitochondria-associated membranes. J. Biol. Chem. 275, 34534–34540 [DOI] [PubMed] [Google Scholar]
- 52. Vance J. E. (1990) Phospholipid synthesis in a membrane fraction associated with mitochondria. J. Biol. Chem. 265, 7248–7256 [PubMed] [Google Scholar]
- 53. Rusiñol A. E., Cui Z., Chen M. H., Vance J. E. (1994) A unique mitochondria-associated membrane fraction from rat liver has a high capacity for lipid synthesis and contains pre-Golgi secretory proteins including nascent lipoproteins. J. Biol. Chem. 269, 27494–27502 [PubMed] [Google Scholar]
- 54. Honda A., Al-Awar O. S., Hay J. C., Donaldson J. G. (2005) Targeting of Arf-1 to the early Golgi by membrin, an ER-Golgi SNARE. J. Cell Biol. 168, 1039–1051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Gilady S. Y., Bui M., Lynes E. M., Benson M. D., Watts R., Vance J. E., Simmen T. (2010) Ero1alpha requires oxidizing and normoxic conditions to localize to the mitochondria-associated membrane (MAM). Cell Stress Chaperones 15, 619–629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Hayashi T., Fujimoto M. (2010) Detergent-resistant microdomains determine the localization of sigma-1 receptors to the endoplasmic reticulum-mitochondria junction. Mol Pharmacol. 77, 517–528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Buchkovich N. J., Maguire T. G., Yu Y., Paton A. W., Paton J. C., Alwine J. C. (2008) Human cytomegalovirus specifically controls the levels of the endoplasmic reticulum chaperone BiP/GRP78, which is required for virion assembly. J. Virol. 82, 31–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Smith G. B., Mocarski E. S. (2005) Contribution of GADD45 family members to cell death suppression by cellular bcl-xL and cytomegalovirus vMIA. J. Virol. 79, 14923–14932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. McCormick A. L., Roback L., Mocarski E. S. (2008) HtrA2/Omi terminates cytomegalovirus infection and is controlled by the viral mitochondrial inhibitor of apoptosis (vMIA). PLoS Pathog. 4, e1000063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Corsi A. K., Schekman R. (1996) Mechanism of polypeptide translocation into the endoplasmic reticulum. J. Biol. Chem. 271, 30299–30302 [DOI] [PubMed] [Google Scholar]
- 61. Horváth I., Multhoff G., Sonnleitner A., Vigh L. (2008) Membrane-associated stress proteins: more than simply chaperones. Biochim. Biophys. Acta 1778, 1653–1664 [DOI] [PubMed] [Google Scholar]
- 62. Vander Heiden M. G., Locasale J. W., Swanson K. D., Sharfi H., Heffron G. J., Amador-Noguez D., Christofk H. R., Wagner G., Rabinowitz J. D., Asara J. M., Cantley L. C. (2010) Evidence for an alternative glycolytic pathway in rapidly proliferating cells. Science 329, 1492–1499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Mazurek S., Eigenbrodt E., Failing K., Steinberg P. (1999) Alterations in the glycolytic and glutaminolytic pathways after malignant transformation of rat liver oval cells. J. Cell. Physiol. 181, 136–146 [DOI] [PubMed] [Google Scholar]
- 64. Voelker D. R. (1989) Reconstitution of phosphatidylserine import into rat liver mitochondria. J. Biol. Chem. 264, 8019–8025 [PubMed] [Google Scholar]
- 65. Bionda C., Portoukalian J., Schmitt D., Rodriguez-Lafrasse C., Ardail D. (2004) Subcellular compartmentalization of ceramide metabolism: MAM (mitochondria-associated membrane) and/or mitochondria? Biochem. J. 382, 527–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Stiban J., Caputo L., Colombini M. (2008) Ceramide synthesis in the endoplasmic reticulum can permeabilize mitochondria to proapoptotic proteins. J. Lipid Res. 49, 625–634 [DOI] [PubMed] [Google Scholar]
- 67. Hayashi T., Su T. P. (2003) Sigma-1 receptors (sigma(1) binding sites) form raft-like microdomains and target lipid droplets on the endoplasmic reticulum: roles in endoplasmic reticulum lipid compartmentalization and export. J Pharmacol Exp Ther. 306, 718–725 [DOI] [PubMed] [Google Scholar]
- 68. Hayashi T., Su T. P. (2004) Sigma-1 receptors at galactosylceramide-enriched lipid microdomains regulate oligodendrocyte differentiation. Proc. Natl. Acad. Sci. U.S.A. 101, 14949–14954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Browman D. T., Resek M. E., Zajchowski L. D., Robbins S. M. (2006) Erlin-1 and erlin-2 are novel members of the prohibitin family of proteins that define lipid-raft-like domains of the ER. J. Cell Sci. 119, 3149–3160 [DOI] [PubMed] [Google Scholar]
- 70. Hoegg M. B., Browman D. T., Resek M. E., Robbins S. M. (2009) Distinct regions within the erlins are required for oligomerization and association with high molecular weight complexes. J. Biol. Chem. 284, 7766–7776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Pearce M. M., Wang Y., Kelley G. G., Wojcikiewicz R. J. (2007) SPFH2 mediates the endoplasmic reticulum-associated degradation of inositol 1,4,5-trisphosphate receptors and other substrates in mammalian cells. J. Biol. Chem. 282, 20104–20115 [DOI] [PubMed] [Google Scholar]
- 72. Rizzuto R., Pozzan T. (2006) Microdomains of intracellular Ca2+: molecular determinants and functional consequences. Physiol. Rev. 86, 369–408 [DOI] [PubMed] [Google Scholar]
- 73. Ardail D., Popa I., Bodennec J., Louisot P., Schmitt D., Portoukalian J. (2003) The mitochondria-associated endoplasmic-reticulum subcompartment (MAM fraction) of rat liver contains highly active sphingolipid-specific glycosyltransferases. Biochem. J. 371, 1013–1019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Hayashi T., Su T. P. (2007) Sigma-1 receptor chaperones at the ER-mitochondrion interface regulate Ca(2+) signaling and cell survival. Cell 131, 596–610 [DOI] [PubMed] [Google Scholar]
- 75. Joseph S. K., Hajnóczky G. (2007) IP3 receptors in cell survival and apoptosis: Ca2+ release and beyond. Apoptosis 12, 951–968 [DOI] [PubMed] [Google Scholar]
- 76. Camello-Almaraz C., Gomez-Pinilla P. J., Pozo M. J., Camello P. J. (2006) Mitochondrial reactive oxygen species and Ca2+ signaling. Am. J. Physiol. Cell Physiol. 291, C1082–1088 [DOI] [PubMed] [Google Scholar]
- 77. Tirosh B., Iwakoshi N. N., Lilley B. N., Lee A. H., Glimcher L. H., Ploegh H. L. (2005) Human cytomegalovirus protein US11 provokes an unfolded protein response that may facilitate the degradation of class I major histocompatibility complex products. J. Virol. 79, 2768–2779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Alwine J. C. (2008) Modulation of host cell stress responses by human cytomegalovirus. Curr. Top. Microbiol. Immunol. 325, 263–279 [DOI] [PubMed] [Google Scholar]
- 79. Das S., Pellett P. E. (2011) Spatial Relationships between Markers for Secretory and Endosomal Machinery in Human Cytomegalovirus-Infected Cells versus Those in Uninfected Cells. J. Virol. 85, 5864–5879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Isler J. A., Maguire T. G., Alwine J. C. (2005) Production of infectious human cytomegalovirus virions is inhibited by drugs that disrupt calcium homeostasis in the endoplasmic reticulum. J. Virol. 79, 15388–15397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Myhill N., Lynes E. M., Nanji J. A., Blagoveshchenskaya A. D., Fei H., Carmine Simmen K., Cooper T. J., Thomas G., Simmen T. (2008) The subcellular distribution of calnexin is mediated by PACS-2. Mol. Biol. Cell. 19, 2777–2788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Liu X., Zhang M., Go V. L., Hu S. (2010) Membrane proteomic analysis of pancreatic cancer cells. J. Biomed. Sci. 17, 74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Vyssokikh M. Y., Brdiczka D. (2003) The function of complexes between the outer mitochondrial membrane pore (VDAC) and the adenine nucleotide translocase in regulation of energy metabolism and apoptosis. Acta Biochim. Pol. 50, 389–404 [PubMed] [Google Scholar]