Abstract
The flagellum of African trypanosomes is an essential and multifunctional organelle that functions in motility, cell morphogenesis, and host-parasite interaction. Previous studies of the trypanosome flagellum have been limited by the inability to purify flagella without first removing the flagellar membrane. This limitation is particularly relevant in the context of studying flagellum signaling, as signaling requires surface-exposed proteins in the flagellar membrane and soluble signaling proteins in the flagellar matrix. Here we employ a combination of genetic and mechanical approaches to purify intact flagella from the African trypanosome, Trypanosoma brucei, in its mammalian-infectious stage. We combined flagellum purification with affinity-purification of surface-exposed proteins to conduct independent proteomic analyses of the flagellum surface and matrix fractions. The proteins identified encompass a broad range of molecular functionalities, including many predicted to function in signaling. Immunofluorescence and RNA interference studies demonstrate flagellum localization and function for proteins identified and provide insight into mechanisms of flagellum attachment and motility. The flagellum surface proteome includes many T. brucei-specific proteins and is enriched for proteins up-regulated in the mammalian-infectious stage of the parasite life-cycle. The combined results indicate that the flagellum surface presents a diverse and dynamic host-parasite interface that is well-suited for host-parasite signaling.
The eukaryotic flagellum (synonymous with cilium) is recognized as a major signaling center that acts as a cellular antenna to sense and transduce extracellular signals (1–4). A sensory function for the flagellum is broadly conserved across diverse taxa (5). In metazoans, receptor-guanylate cyclases, ion channels, and G protein-coupled receptors (GPCRs)1 in the flagellar membrane perceive chemical and mechanical cues that are necessary for normal development, physiology, and reproduction (6–9). Important examples include wingless (Wnt) and hedgehog signaling responses in vertebrates (1, 3, 10, 113). In protists, flagellum-localized ion channels, agglutinins, and receptor-kinases control motility, mating, and response to extracellular growth factors (11–13). Flagella are prominent among pathogenic protozoa, which cause tremendous human suffering worldwide and present a barrier to economic development in some of the poorest regions of the world (14–17). These include the etiological agents of African sleeping sickness, leishmaniasis, malaria, epidemic diarrhea, and trichomoniasis (14–16,18). In most cases these pathogens are obligate parasitic organisms whose survival depends upon their ability to sense and respond to extracellular cues in diverse host environments. The flagellum's motility function in protozoan parasites is self-evident, but its capacity for sensing and responding to external signals is largely unexplored.
African trypanosomes, e.g. Trypanosoma brucei, are uniflagellate protozoan parasites that cause African sleeping sickness in humans and related diseases in wild and domestic animals (19). T. brucei is transmitted to the bloodstream of a mammalian host through the bite of a tsetse fly vector. To be successful, T. brucei must integrate environmental signals that direct parasite movements and developmental transformations within specific host compartments (20–22). For example, entry into the mammalian bloodstream promotes cellular adaptations that define the bloodstream-form life cycle stage, including changes in metabolism, morphology, and surface protein composition (23). Prominent among these is differentiation of proliferative, “long-slender” forms into cell cycle-arrested, “short-stumpy” forms that are adapted for survival in the tsetse (23, 24). Parasite-host signaling is also reported to contribute to invasion of the central nervous system (25). In the tsetse, bloodstream-forms differentiate into procyclic-forms, which re-enter the cell cycle and establish an infection in the fly midgut. Procyclic-form parasites undergo a defined series of directional migrations and tissue-specific developmental transformations, culminating in flagellum attachment to epithelial cells in the tsetse salivary gland and differentiation into human-infectious forms (26, 27). Except for surface-exposed carboxylate transporters that participate in stumpy-to-procyclic differentiation (24), proteins that perceive signals for directing parasite navigation and tissue-specific development are mostly unknown.
The paradigm of the flagellum as a sensory organelle in other eukaryotes, together with the observation that the trypanosome flagellum interacts directly with host tissues (26, 28), has fueled the hypothesis that the parasite flagellum functions as a signaling organelle for integrating host-derived and parasite-derived signals (20, 22). In T. brucei, this idea is supported by the finding that specific proteins from cyclic nucleotide and Ca2+ signaling pathways are present in the flagellum (29–34). The T. brucei flagellum (Fig. 1) is built around a microtubule-based axoneme plus an extra-axonemal filament, termed the paraflagellar rod (PFR), which runs alongside and is attached to the axoneme (15, 35). The axoneme and PFR are ensheathed by a flagellar membrane whose protein and lipid composition are distinct from the cell surface membrane (36, 37). The lumen of the flagellum, termed the flagellar matrix, is contiguous with the cytoplasm, but selective filters at the base of the flagellum restrict access to the matrix, such that protein composition of the matrix is distinct from that of the cytoplasm (38). The flagellum emerges from the cytoplasm at the cell posterior and is laterally connected to the cell body by cytoskeletal filaments that connect the axoneme and PFR to subpellicular microtubules in the cell body and maintain tight apposition of the flagellar and cell surface membranes (39, 40). These connections form a “flagellum attachment zone” (FAZ) that runs along most of the length of the flagellum, with a small distal portion of the flagellum extending free of the cell body. A specialized membrane domain, termed the flagellar pocket, surrounds the flagellum at the site where it emerges from the cytoplasm at the cell posterior (39, 41, 42). As the sole site of surface protein turnover and macromolecular uptake in trypanosomes, the flagellar pocket is a key portal for host-parasite interaction (41, 42), yet little is known about its protein and lipid compositions.
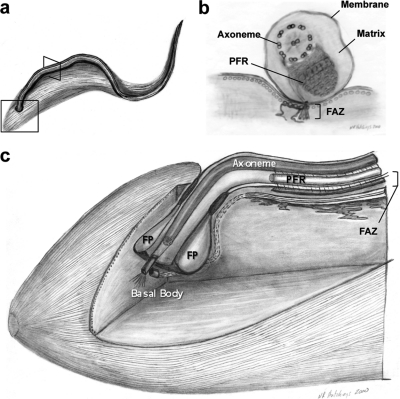
Fig. 1.
Flagellum architecture of T. brucei. Schematic diagrams showing the T. brucei cell and flagellum (A), together with cross sectional (B), and cut-away (C) views corresponding to the regions indicated by boxes in panel A. Relevant structural features are labeled, including the flagellum membrane, matrix, axoneme, and paraflagellar rod (PFR), as well as the flagellum attachment zone (FAZ), flagellar pocket (FP), and basal body. See text for details. Figure adapted from reference (110), with permission.
Lateral attachment of the flagellum to the cell body poses significant challenges for isolating intact flagella from T. brucei. Existing procedures employ detergent-extraction and salt-extraction to isolate the insoluble flagellum skeleton, which contains the axoneme and PFR, but lacks the membrane and matrix (43–45). Thus, although several hundred axonemal and PFR proteins have been identified (31, 43, 46), the protein compositions of the flagellar membrane and matrix in T. brucei are largely unknown. This poses a particular limitation for studying flagellum signaling, because signaling capacity is dictated by surface-exposed membrane proteins coupled to soluble components of signaling cascades in the matrix (2).
Here we employ a combined genetic and mechanical approach to isolate intact, membrane-enclosed flagella from T. brucei in its mammalian-infectious stage. We used flagellum purification, combined with affinity purification of surface-exposed proteins and multidimensional protein identification technology (MudPIT) to define the flagellum surface and flagellum matrix proteomes. Immunofluorescence and RNA interference (RNAi) studies demonstrate flagellum localization and function for proteins identified and provide insight into mechanisms of flagellum attachment and motility. Our combined studies indicate that the trypanosome flagellum presents a diverse and dynamic signaling platform adapted for host-pathogen interaction.
EXPERIMENTAL PROCEDURES
Cell Lines
Bloodstream-form trypanosomes, 221 single marker cell line (47), were used for all experiments and were cultivated in HMI-9 medium supplemented with 10–15% fetal bovine serum (Invitrogen) as described previously (48). The fla1 cell line was generated by transfection with the p2T7-Fla1 plasmid (49) using standard procedures (48). Selection for transformants was done using 5 μg/ml Phleomycin (InvivoGen, San Diego, CA).
Surface Biotinylation
Cells were washed twice in ice-cold phosphate-buffered saline (PBS) and resuspended in ice-cold PBS + 0.5 mg/ml sulfosuccinimidyl-2-[biotinamido]ethyl-1,3-dithiopropionate (Sulfo-NHS-SS-biotin; Thermo Scientific). After incubation on ice for 10–30 min with gentle agitation, unreacted Sulfo-NHS-SS-biotin was blocked by addition of Tris to 100 mm final concentration. Biotinylated cells were washed three times in ice-cold PBS + 100 mm Tris.
Purification of Flagella and Flagellar Surface Proteins
Cells harboring the fla1 tet-inducible RNAi cassette (density 5 × 105 cells/ml) were induced for 18 h with 1 μg/ml tetracycline. From this step onward all procedures were performed at 4 °C unless otherwise stated and all solutions were cooled on ice. Induced cells were surface biotinylated as described above. Flagella were removed from the cell bodies by repeated passage (five times) through a 28-G needle. Some (<1%) of the isolated flagella remained motile. The resulting flagella plus cell body mixture was loaded on a 30% sucrose bed and centrifuged for 10 min at 770 × g. The supernatant and interface containing isolated flagella were collected and sucrose sedimentation was repeated. Purified flagella were collected by a high-speed spin (15,000 × g, 1h) and resuspended in 200 μl PBS, containing 2.5 μg/ml Leupeptin and 0.5 μg/ml Pepstatin. Purified, biotinylated flagella were lysed by addition of Nonidet P-40 to 0.5% final concentration and incubation on ice for 10 min. Soluble proteins (flagellum matrix and membrane) were separated from insoluble proteins (flagellum skeleton) by centrifugation (15,000 × g, 30 min). The resulting soluble flagellar proteins (200 μl) were incubated with 50 μl streptavidin beads (Streptavidin Sepharose High Performance, GE Healthcare) for 30 min with agitation. Proteins bound to streptavidin beads were separated from the unbound fraction by centrifugation and washed at room temperature once in buffer A (8 m Urea, 200 mm NaCl, 2% SDS, 100 mm Tris, pH 8), once in buffer B (8 m Urea, 1.2 m NaCl, 0.2% SDS, 100 mm Tris, 10% Ethanol, 10% Isopropanol, pH 8), once in buffer C (8 m Urea, 200 mm NaCl, 0.2% SDS, 100 mm Tris, 10% Ethanol, 10% Isopropanol, pH 8), and twice in buffer D (8 m Urea, 100 mm Tris, pH 8). All fractions were analyzed by Western blotting using standard protocols (see Fig. 3). Antibodies used were anti-BiP 1:10,000 (50), anti-PFR 1:1,000 (45), anti-variable surface glycoprotein VSG 221 1:10,000 (51), anti-biotin 1:2,000 (Jackson Immunoresearch, West Grove, PA). Secondary antibodies were donkey anti-mouse or donkey anti-rabbit HRP coupled 1:2,500 (Bio-Rad, Hercules, CA).
Fig. 3.
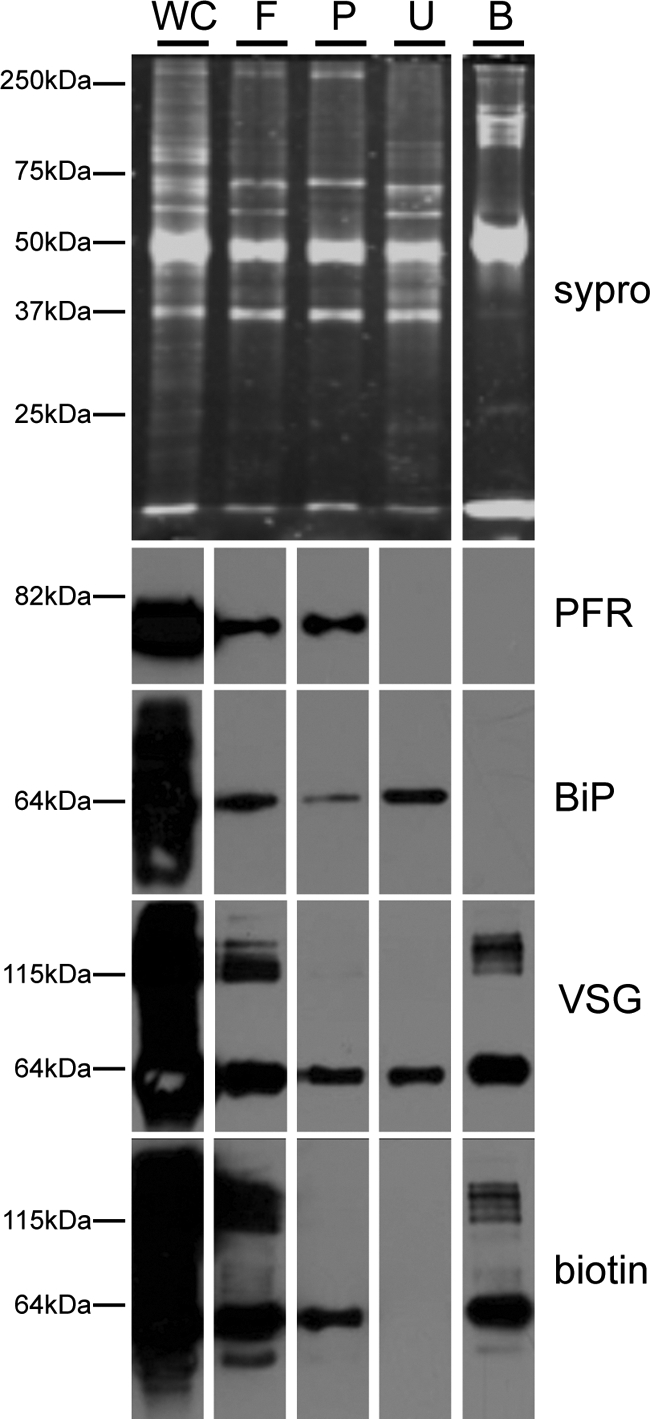
Analysis of flagellum purification fractions. Proteins from the indicated fractions of the purification were analyzed by SDS-PAGE and SYPRO-Ruby staining (top) or Western blotting (bottom) with antibodies against intracellular (BiP), flagellar skeleton (PFR) and cell surface (VSG) markers. Biotinylated proteins were detected with anti-biotin antibody. Whole cells (WC), purified flagella (F), detergent-insoluble flagellum pellets (P), and the streptavidin-unbound (U) and bound (B) fractions are as indicated in Fig. 2A. Flagellum enrichment is indicated by the increased ratio of the flagellar marker PFR to the intracellular marker BiP in the purified flagellum fraction (F) versus whole cells (WC). Both PFR and BiP are largely absent from the streptavidin bound fraction (B).
MudPIT Analysis
Streptavidin-bound proteins were digested directly on beads by the sequential addition of lys-C and trypsin proteases (52, 53). Peptide samples were fractionated online using multidimensional chromatography followed by tandem mass spectrometric analysis on a LTQ-Orbitrap mass spectrometer (ThermoFisher) as previously described (53, 54). RawXtract (version 1.8) was used to extract peaklist information from Xcalibur-generated RAW files. Database searching of the MS/MS spectra was performed using the ProLuCID algorithm (version 1.0) and a user assembled database consisting of all protein entries from the TriTrypDB for T. brucei strain 927 (version 2.3, 10533 entries) and seven sequences from T. brucei strain 427: 453391 (Tb-1.7g), 458439 (Tb-24), 18413545 (ESAG4.a from 221 expression site), 18413551 (ESAG4.b from 221 expression site), 189094632 (VSG-221), 18413541 (ESAG7 from 221 expression site), 18413542 (ESAG6 from 221 expression site) (55). Other database search parameters included: (1) precursor ion mass tolerance of ± 20 ppm, (2) fragment ion mass tolerance of ± 400 ppm, (3) only peptides with fully tryptic ends were considered candidate peptides in the search with no consideration for missed cleavages, and (4) static modification of +57.02156 on cysteine residues. Peptide identifications were organized and filtered using the DTASelect algorithm which uses a linear discriminant analysis to identify peptide scoring thresholds that yield a peptide-level false discovery rate of less than 5% as estimated using a decoy database approach. Proteins were considered present in the analysis if they were identified by two or more peptides using the 5% peptide-level false discovery rate (56–58). Details from mass spectrometric analysis are given in supplemental table S10.
Immunofluorescence Microscopy
Cells or isolated flagella were washed once in PBS and fixed by addition of paraformaldehyde to 0.1% for 5 min on ice. Fixed cells were washed once in PBS and air-dried onto coverslips. The coverslips were incubated for 10 min in −20 °C methanol and 10 min in −20 °C acetone. After a re-hydration step (10–30 min in PBS) the slides were blocked for 1.5 h in blocking solution (PBS + 5% bovine serum albumin (BSA) + 5% Normal donkey serum (Invitrogen)). Coverslips were incubated with primary antibodies diluted in blocking solution for 1.5 h. Antibodies used were anti-biotin 1:2000 (Jackson ImmunoResearch), anti-BiP 1:10,000 (50), anti-hemagglutinin HA.11 1:200–1:1,000 (Covance, Princeton, NJ), anti-PFR 1:500 (45), anti-tyrosinated tubulin YL1/2 1:500 (Chemicon International, Temecula, CA). After three washes in PBS + 0.05% Tween-20 for 10 min each, samples were stained with secondary antibodies diluted in blocking solution for 1.5 h (donkey anti-mouse Alexa Fluor 488, donkey anti-rabbit Alexa Fluor 594, goat anti-rat Alexa Fluor 594 1:500 (Molecular Probes, Eugene, OR)). Cells were washed three times in PBS + 0.05% Tween-20, once in PBS and mounted with Vectashield containing DAPI (Vector Laboratories). Flagellar pocket staining with biotinylated tomato lectin was performed as described (59). Biotinylated tomato lectin was visualized using streptavidin Alexa Fluor 594 (Molecular Probes). Images were taken using a 100x objective on a Zeiss Axioskop II compound microscope and processed using Axiovision (Zeiss, Inc., Jena, Germany) and Adobe Photoshop (Adobe Systems, Inc., Mountain View, CA).
Nonidet P-40 Fractionation of Whole Cells
Cells were washed once in PBS and lysed in ice-cold PBS + 0.5% Nonidet P-40 + protease inhibitors (complete mini, Roche) for 10 min on ice. Lysates were centrifuged for 30 min at 15,000 × g at 4 °C and supernatant and pellet fractions were analyzed by Western blotting using standard procedures. Primary antibodies used were anti-HA.11, anti-BiP (as described above) and anti-trypanin 1:1000 (60). Secondary antibodies were donkey anti-mouse or donkey anti-rabbit HRP coupled 1:2500 (Bio-Rad).
In situ Tagging
In situ tagging was carried out as previously described (61) to introduce epitope-tagged copies of each gene analyzed into the corresponding endogenous chromosomal locus. In brief, 500–800 bp DNA fragments homologous to the target gene open reading frame or 3′ UTR were PCR-amplified from genomic DNA and cloned upstream of the 3xHA or downstream of the puromycin resistance marker in pMOTag2H. pMOTag2H is an in situ tagging plasmid containing a 3xHA epitope tag and a puromycin resistance marker, adapted from (61). All sequences were verified by DNA sequencing at the UCLA Sequencing and Genotyping Core center. The tagging cassettes were excised from the pMOTag2H vector backbone by restriction digestion, then purified and transfected into 221 bloodstream-form T. brucei using standard methods (48). Stable transformants were selected using 0.1 μg/ml puromycin.
RNAi
The targets for RNAi against FS179 and RSU (FM458) were identified by the Trypanofan RNAit algorithm (62), then PCR-amplified from genomic DNA using primers listed below: (restriction enzyme cleavage sites are underlined):
FS179-RNAi-f: 5′ CATAAGCTTTCATTGCGTCATTTTGCCTA 3′
FS179-RNAi-r: 5′ CATCTAGAAAGGCTGACGAGATCTTGGA 3′
RSU-RNAi-f: 5′ TGTGAAGCAACCCAACACAT 3′
RSU-RNAi-r: 5′ GTAATGCGAGAGCGGAGTTC 3′
The resulting DNA fragments for RNAi were ligated into the p2T7-Ti/B-RNAi vector, which is a tetracycline-controlled expression vector with opposing T7 promoters (49). Inserts were verified by sequencing at the UCLA genomics center. The p2T7-Ti/B-RNAi vector containing the FS179 or FM458 target sequence was linearized with NotI, ethanol precipitated and transfected into 221 bloodstream form T. brucei as described above. Transformants were selected using 5 μg/ml phleomycin.
Imaging and Motility Traces
FS179 RNAi imaging (see Fig. 7A–C): Cells were induced for 48 h with 1 μg/ml tetracycline, fixed using 4% PFA and mounted with Vectashield containing DAPI (Vector Laboratories). Images were taken and processed as described above. FM458 RNAi motility traces (see Fig. 7D, 7E): Cells were induced for 48h with 1 μg/ml tetracycline and motility traces were obtained for induced and uninduced control cells as described previously (48). The movie of the isolated flagellum was recorded and played at 30 frames per second as described previously (48).
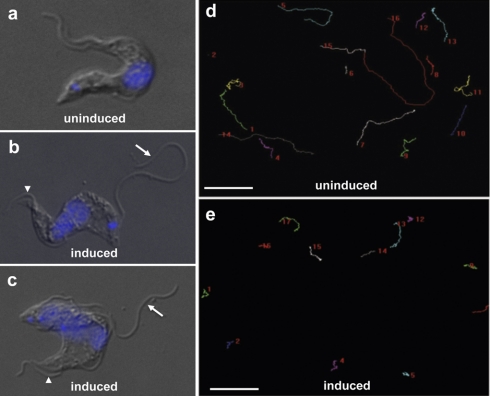
Fig. 7.
RNAi knockdown indicates flagellum function for FS179 and FM458. A–C, Phase contrast images of tetracycline-inducible FS179 RNAi cells grown in the absence (A) or presence (B and C) of tetracycline to induce RNAi. FS179 knockdown causes the daughter flagellum to become detached (arrows in B and C), while the parental flagellum remains connected to the cell body (arrowheads in b and c). DNA is visualized with DAPI. (d and e) Motility trace analysis of FM458 RNAi cells grown in the absence (D) or presence (E) of tetracycline to induce RNAi. Each line traces the path of a single cell for 2 min (Scale bar 50 μm).
Bioinformatic and Protein Comparisons
Protein domains were assessed using the SMART database (63). For homology searches, we employed eight ciliated (Leishmania major, Trypanasoma cruzi, Plasmodium falciparum, Chlamydomonas reinhardtii, Monosiga brevicollis, Caenorhabditis elegans, Drosophila melanogaster, and Homo sapiens) and four nonciliated (Dictyostelium discoideum, Saccharomyces cerevisiae, Cyanidioschyzon merolae, and Arabidopsis thaliana) eukaryotic species, including both unicellular and multicellular representatives. Homology searches were performed using NCBI BLAST with default parameters. Proteins with an expect-value of ≤1 × 10−10 were considered as homologs and identical criteria were applied across all data sets and comparisons. For T. brucei flagellum skeleton proteomes, we used the combined, nonoverlapping set of proteins identified in three proteomic analyses of extracted flagellum skeletons from procyclic-form parasites (31, 43, 46). Comparison to the C. reinhardtii flagellum proteome was done locally with proteins downloaded from (64). For analysis of molecular function (see Table I), the DAVID Bioinformatics Resource, version 6.7 (65, 66) was used for functional annotation of proteins in TbFSP and TbFMP. Proteins were categorized using annotated Gene Ontology (GO) terms for molecular function (67), using the GOTERM_MF_2 category with a threshold count of 2 and an EASE of 0.1. Expression analysis (see Fig.10) was done using RNA sequencing data from (68).
Table I. Predicted molecular function of proteins identified in TbFSP and TbFMP. Proteins in the flagellum surface proteome (TbFSP) and flagellum matrix proteome (TbFMP) were analyzed for predicted molecular function using the DAVID database for gene ontology and protein domain analysis.
| Predicted molecular function | TbFSP (158) | TbFMP (666) |
|---|---|---|
| Cyclase activity | 17 | 0 |
| Lyase activity | 18 | 0 |
| Substrate-specific transporter activity | 12 | 0 |
| Transmembrane transporter activity | 15 | 0 |
| Nucleotide binding | 31 | 117 |
| Hydrolase activity | 0 | 117 |
| Protein binding | 0 | 54 |
| Oxidoreductase activity | 0 | 52 |
| Cofactor binding | 0 | 20 |
| Translation factor activity, nucleic acid binding | 0 | 12 |
| Carboxylic acid binding | 0 | 5 |
| Peroxidase activity | 0 | 4 |
| Carbohydrate binding | 0 | 4 |
| No Annotated Function | 83 | 350 |
| Not in DAVID | 22 | 46 |
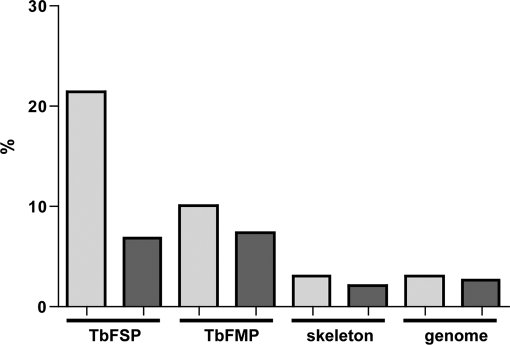
Fig. 10.
Flagellum surface proteins are up-regulated in bloodstream-form cells. Chart shows the percentage of proteins in each proteomic dataset that are up-regulated in bloodstream-form cells (light gray bars) or procyclic-form cells (dark gray bars) by twofold or more based on RNA sequencing analysis (68). Proteomic data sets are for the flagellum surface (TbFSP), the flagellum matrix (TbFMP), and the flagellum skeleton (TbFP) proteomes (31, 43, 46).
RESULTS
Purification of Intact, Membrane-enclosed Flagella from T. brucei in its Mammalian-infectious Form
The T. brucei flagellum is laterally connected to the cell body along most of its length by filamentous connectors and tightly apposed membrane-membrane contacts (40). These connections present significant challenges for isolating intact flagella. Current approaches employ detergents to remove membranes, together with salt-extraction to disrupt filamentous connectors, followed by centrifugation to isolate flagellum skeletons from solubilized material (43–45). Resultant preparations contain the axoneme and PFR, but are stripped of flagellar membrane and soluble matrix proteins. To obviate the need for detergent and salt-extraction, and thus retain the flagellar membrane, we employed tetracycline-inducible RNAi against fla1 to disrupt lateral flagellum attachment to the cell body (49). This allowed intact flagella to be separated from cell bodies by shearing, without using detergent (Fig. 2). Importantly, some isolated flagella remained motile without addition of exogenous ATP (Fig. 2B, supplemental Movie S1), indicating that they retained an intact flagellum membrane, as well as internal proteins and small molecules necessary for motility. Flagella were purified from cell bodies by sucrose density sedimentation (Fig. 2). To enable purification of surface-exposed proteins, cells were surface biotinylated before flagellum purification (Fig. 2D, supplemental Fig. S1). Surface-biotinylation was retained in purified flagella (Fig. 2E). The bulbous enlargement at one end of purified flagella contains material from the basal body, kinetoplast, and flagellar pocket (Fig. 2E). Thus, purified flagella retain an intact membrane that includes flagellar pocket membrane.
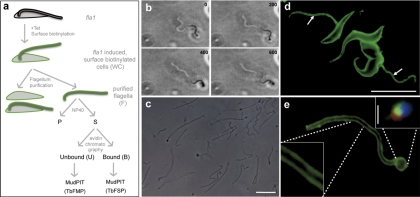
Fig. 2.
Purification of flagellum surface and matrix proteins from bloodstream-form T. brucei. A, Schematic diagram illustrating the strategy for flagellum purification and separation of flagellum surface and matrix proteins. Lateral connections between the flagellum and cell body were removed by tetracycline-induced (+Tet) RNAi against Fla1 (49). Induced fla1 cells were surface biotinylated (fla1-induced, surface biotinylated cells) and flagella were removed from the cell body by mechanical shearing, then purified by sucrose sedimentation (purified flagella). Purified, surface-biotinylated flagella were lysed with nonionic detergent (Nonidet P-40) and soluble proteins (S) were separated from insoluble flagellar skeletons (P) by centrifugation. Soluble proteins were applied to streptavidin beads (avidin chromatography). Proteins in the streptavidin-bound (B) and streptavidin-unbound (U) fractions were identified using multidimensional protein identification technology (MudPIT). B, Time-lapsed image sequence (from supplemental movie 1, recorded and played at 30 fps), showing that some flagella continued to beat after detachment from cell bodies (timestamps are given in ms). C, Phase contrast image of purified flagella (Scale bar 5 μm). D, Antibiotin immunofluorescence (green) of surface-biotinylated RNAi-induced fla1 cells showing detached flagella (arrows, scale bar 25 μm). E, Anti-biotin immunofluorescence (green) of purified flagella shows surface labeling (inset, bottom left). The bulbous structure at one end of the flagellum (inset, upper right) includes material from the flagellar pocket (tomato lectin staining, red), the basal body (tyrosinated tubulin staining, green) and kinetoplast DNA (DAPI staining, blue). (scale bar 0.5 μm)
Purified, surface-biotinylated flagella were lysed with nonionic detergent (Nonidet P-40) and lysates were centrifuged to separate the insoluble fraction, containing flagellum skeleton proteins, from the soluble fraction, containing matrix plus membrane proteins. Biotinylated proteins in the soluble fraction were purified by adsorption to streptavidin beads and fractions from the purification were analyzed for total protein, biotin, and specific marker proteins (Fig. 3). Purified flagella were enriched for flagellum markers (PFR) relative to intracellular markers (BiP). Streptavidin-binding quantitatively purified biotinylated proteins and SYPRO-Ruby staining revealed unique protein profiles for the streptavidin-bound and unbound fractions. The bound fraction contained surface proteins (variable surface glycoprotein, VSG (69)), but not abundant intracellular proteins (Binding Protein, BiP (50)), or intraflagellar proteins (PFR (45)) (Fig. 3). These results support immunofluorescence data (Fig. 2D and 2E, supplemental Fig. S1) indicating that biotinylation was specific for surface proteins.
Identification of Flagellum Surface and Matrix Proteins by Mass Spectrometry
Proteins in the streptavidin-bound and unbound fractions were identified by MudPIT proteomic analysis. In some cases peptide coverage allowed unambiguous identification of proteins. In other cases, peptides did not distinguish between proteins with very similar sequences. As such, each proteomic data set represents a maximum number and minimum number of total proteins, with the minimum number reflecting groups of proteins whose sequences could not be unambiguously distinguished. From here on, we refer to the minimum number of proteins identified in each fraction. This analysis identified 158 proteins in the bound fraction, termed the T. brucei flagellum surface proteome (“TbFSP,” supplemental Table S1) and 666 proteins in the unbound fraction, termed the T. brucei flagellum matrix proteome (“TbFMP,” supplemental Table S3). Proteins in the two data sets encompassed a broad range of predicted functionalities and both data sets included a large number of proteins with no annotated molecular function (Table 1). We next employed several independent analyses to validate the proteomic data sets. Each dataset included suspected contaminants, e.g. ribosomal proteins. However, to avoid user bias, no subjective filters were applied to remove these suspected contaminants and analyses were performed on the unfiltered datasets, i.e. 158 proteins in TbFSP and 666 proteins in TbFMP. We focused on the flagellum surface proteome because the primary interest was surface-exposed proteins.
Validation of the Flagellum Surface Proteome
As a first step in assessing the quality of the proteomic data sets, we probed each dataset for proteins known to localize to specific flagellum compartments (Fig. 4). The flagellum surface proteome contained most known flagellum membrane proteins and was generally devoid of intraflagellar proteins, i.e. flagellum matrix and skeleton proteins (Fig. 4A, supplemental Tables S2, S5, and S8). By comparison, the flagellum matrix proteome contained seven of 21 known matrix proteins, nine of 10 known membrane proteins and 62 of 532 flagellum skeleton proteins (Fig. 4B, supplemental Tables S4, S8). We also examined each data set for predicted transmembrane (TM) domains, as these are expected to be enriched in the surface proteome. As expected, proteins with predicted TM domains were enriched in the surface proteome (51%) compared with the genome as a whole (19%) (70, 71), the flagellum matrix (20%) or the flagellum skeleton (2%) (Fig. 5). Proteins with a predicted signal peptide were also enriched in the flagellum surface proteome (38%) versus the genome (24%) and the matrix (20%) (supplemental Tables S2, S4). By using the minimal number of proteins identified, our analysis is suspected to underestimate the enrichment of TM domains and signal peptides in the flagellum surface proteome, because the genome numbers include redundant and closely related sequences.
Fig. 4.
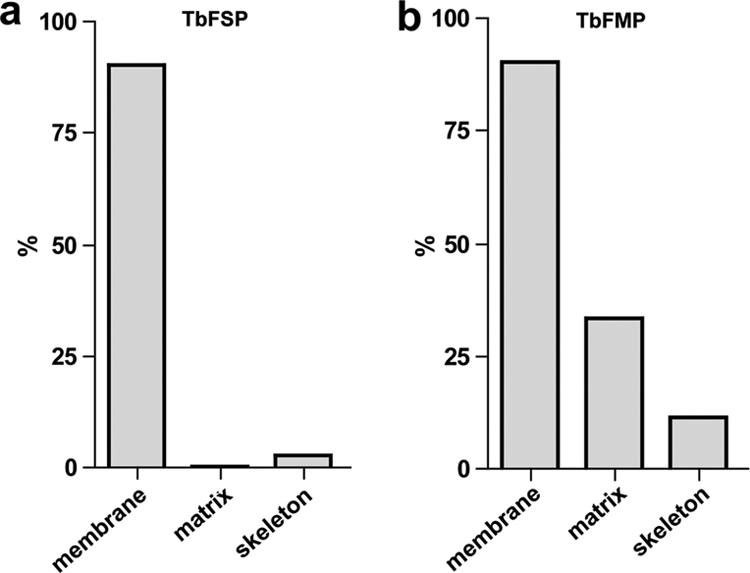
The flagellum surface proteome contains known flagellum surface proteins but is generally devoid of intraflagellar proteins. Chart shows the percentage of known flagellum membrane, matrix, and skeleton proteins that were identified in the flagellum surface proteome (TbFSP) and flagellum matrix proteome (TbFMP). Flagellum skeleton proteins are from (31, 43, 46). Known flagellum membrane and matrix proteins are given in supplemental Tables S5 and table S8, respectively. TbFSP included 9 of 10 (90%) membrane, none of 21 (0%) matrix and 14 of 532 (2.6%) skeleton proteins. TbFMP included 9 of 10 (90%) membrane, 7 of 21 (33%) matrix and 62 of 532 (11.6%) skeleton proteins.
Fig. 5.
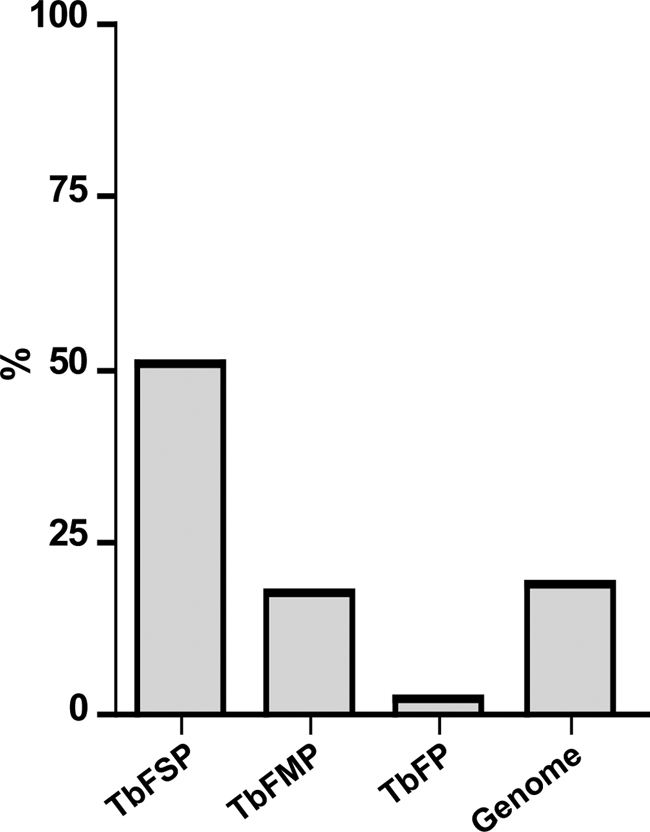
The flagellum surface proteome is enriched for transmembrane proteins. Chart shows the percentage of proteins in the flagellum surface (TbFSP), matrix (TbFMP), and skeleton (TbFP) (31, 43, 46) proteomes that are predicted to contain transmembrane domains. For comparison, the relative number of predicted transmembrane-containing proteins in the T. brucei genome (71) is shown. Numbers are 80/158, 118/666, 12/532, and 2133/11425 for TbFSP, TbFMP, TbFP, and genome, respectively.
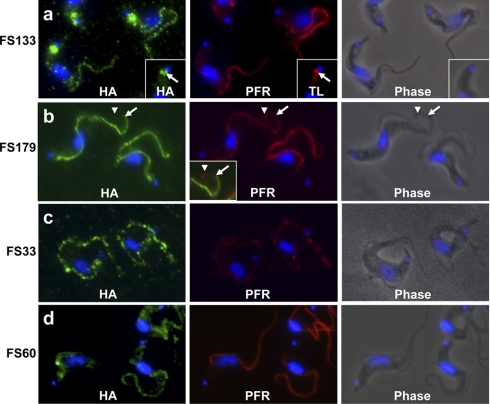
We next used epitope-tagging and immunofluorescence microscopy to determine the subcellular location of ten proteins from the flagellum surface proteome. All tagged proteins were detergent-soluble (supplemental Fig. S2 and not shown), consistent with membrane association. Eight of ten tagged proteins localized to one or more subdomains of the flagellum membrane, e.g. along the entire length of the flagellum, the flagellar pocket or the flagellum attachment zone (Fig. 6, supplemental Fig. S3 and data not shown). Four of these eight proteins were exclusively located in the flagellum and/or flagellar pocket, one was distributed throughout the whole cell surface and three were also observed in intracellular compartments (summarized in supplemental Table S6). In sum, the flagellum surface proteome includes most experimentally characterized flagellum membrane proteins, is largely devoid of intraflagellar proteins and immunolocalization demonstrates flagellar location for several proteins identified. Thus, the results indicate that the flagellum surface proteome is enriched for flagellum membrane proteins.
Fig. 6.
Immunofluorescence shows flagellum localization for proteins identified in the flagellum surface proteome. (A–D) Immunofluorescence analysis of bloodstream-form trypanosomes expressing the indicated HA-tagged flagellum surface proteins (FS133, FS179, FS33, or FS60). Cells were co-stained for the HA epitope (green), the paraflagellar rod (PFR, red) or the flagellar pocket marker tomato lectin (TL, red) as indicated. DNA was visualized with DAPI. A, FS133 localizes along the flagellum and in the flagellar pocket (inset, arrows). B, FS179 localizes along the flagellum, but does not extend to the flagellum distal tip, as evidenced by the extension of PFR staining (arrowhead) beyond the end of FS179 staining (arrow). See inset for merged images. C, D, FS33 and FS60 localize along the entire length of the flagellum. FS60 was also observed in cytoplasmic puncta (not shown).
RNAi Knockdown Reveals Flagellar Functions for Proteins Identified
We used tetracycline-inducible RNAi to determine whether loss of a particular protein impacted flagellum function. In most cases, RNAi directed against proteins in the flagellum surface proteome did not reveal an obvious phenotype in culture (not shown). One notable exception was FS179, which encodes a putative calcium channel. Immunofluorescence showed that FS179 was localized to the flagellum attachment zone (Fig. 6B), suggesting a potential role in flagellum attachment. RNAi knockdown of FS179 caused the flagellum to become detached from the cell body and the daughter flagellum was preferentially affected (Fig. 7A–C). This result supports FS179 localization to the flagellum attachment zone and is consistent with previous studies suggesting a requirement for Ca2+ in flagellum attachment (40). The flagellar matrix protein FM458 corresponds to the protein kinase A regulatory subunit (RSU) (72) and was of interest based on the importance of protein kinase A and cyclic nucleotide signaling to flagellum function in other organisms (13, 73, 74). RNAi knockdown of FM458 inhibited forward motility, consistent with a role in flagellum function (Fig. 7D,7E). Interestingly, despite motility defects, bloodstream-form FM458 knockdowns were viable, which contrasts to the lethal phenotype caused by knockdown of flagellum skeleton proteins (29, 43, 60).
Host-Parasite Signaling Capacity of the Flagellum Surface
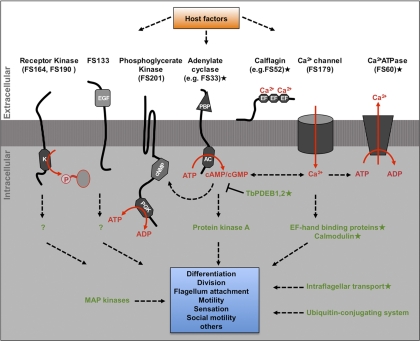
Prominent within the flagellum surface proteome were proteins known or suspected to function in host-parasite interaction and/or signaling (Fig. 8). For example, VSG, MSP-A, and GPI-PLC, are characterized surface proteins that function in antigenic variation and VSG clearance (69, 75, 76). Likewise, transferrin receptor and glucose transporter function in host nutrient uptake (77, 78), whereas adenylate cyclases and calflagins are predicted to function in host-parasite signaling (30, 33, 34, 79). In addition, many uncharacterized flagellum surface (“FS”) proteins have domain architectures that indicate receptor function, with a large extracellular domain suitable for binding host ligands connected to an intracellular signaling module, e.g. kinase domains in FS164, FS190, and FS201. In some cases, extracellular domains exhibit homology to characterized ligand-binding motifs, such as the periplasmic binding protein superfamily (receptor-type adenylate cyclases) or EGF-like domain superfamily involved in protein-protein interaction (FS133) (80, 81). Also well-represented among uncharacterized proteins are several classes of ion and metabolite transporters, including ABC-transporters (FS166), P-type ATPase pumps (FS60, FS80, FS163, FS188, FS193, and FS198), ion channels (FS179), amino acid transporters (FS group 108 and 121), glucose transporters (FS192) and a group of major facilitator proteins (FS group 126) related to the PAD surface receptors that perceive extracellular signals for bloodstream-form to procyclic-form differentiation (24). In sum, the data indicate a diverse flagellar surface protein repertoire that is suitable for mediating a broad range of host-parasite interactions.
Fig. 8.
The trypanosome flagellum provides a diverse signaling platform. Diagram shows representative components of signaling pathways identified in the flagellum surface proteome (black text) and flagellum matrix proteome (green text). These include receptors and transporters for initiating signal transduction (black text) and downstream signaling proteins (green text). Predicted domain structures are indicated for flagellum surface proteins (K, kinase; EGF, epidermal growth factor domain superfamily; cNMP, cyclic nucleotide binding domain; PGK, phosphoglycerate kinase domain; PBP, periplasmic binding protein domain; AC, adenylate cyclase domain; EF, EF hand calcium-binding domain). Predicted interactions are indicated with dashed lines. Unknown host factors (orange box) are predicted to engage signaling pathways, which mediate downstream trypanosome processes (blue box). [star] Indicates proteins that have been demonstrated to be flagellar by independent studies (29–33, 93, 96, 97, 111).
The Flagellum Surface is Enriched for T. brucei-specific Proteins
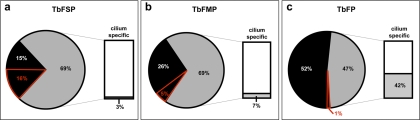
The flagellum surface is postulated to function in detection of extracellular signals and is therefore anticipated to include cell-specific proteins that accommodate environments uniquely encountered by T. brucei. The availability of an independent flagellum surface proteome allowed us to test this idea directly by examining the phylogenetic distribution of proteins in the flagellum surface and matrix proteomes (Fig. 9, supplemental table S1 and S3). As reference organisms, we chose eight ciliated (L. major, T. cruzi, P. falciparum, C. reinhardtii, M. brevicollis, C. elegans, D. melanogaster, H. sapiens) and four nonciliated (D. discoideum, S. cerevisiae, C. merolae, A. thaliana) eukaryotic species, including both unicellular and multicellular representatives. Among the 158 flagellum surface proteins, 16% were specific to T. brucei. By comparison, 5% of matrix proteins and 1% of flagellum skeleton proteins were T. brucei-specific.
Fig. 9.
The flagellum surface proteome contains many T. brucei-specific proteins. (A–C) Pie charts show phylogenetic distribution of proteins in the flagellum surface (TbFSP), flagellum matrix (TbFPM), or flagellum skeleton (TbFP) proteomes. Percentages are shown for proteins that are conserved in at least one nonkinetoplastid organism (gray), or are kinetoplastid-specific (black), or T. brucei-specific (red outline). Bar charts show the percentage of conserved proteins, i.e. not kinetoplastid-specific, that are restricted to ciliated organisms.
Conserved Flagellum Surface Proteins are not Biased Toward Flagellated Organisms
Proteins required for flagellum structure and motility are broadly conserved among organisms with flagella, but are generally absent in organisms that lack flagella (82–84). To determine if the same bias applied to flagellum surface and matrix proteins, we examined the phylogenetic distribution of “conserved” proteins in each data set, i.e. proteins that were encoded in the genome of at least one non-kinetoplastid organism (Fig. 9). As expected (82–84), a large fraction (42%) of conserved flagellum skeleton proteins were restricted to organisms with flagella. In contrast, only 3% of conserved flagellum surface proteins and 7% of conserved flagellum matrix proteins were restricted to organisms with flagella.
DISCUSSION
Flagellum Purification
The T. brucei flagellum is an essential and multifunctional organelle that is required for cell motility and cell morphogenesis and is postulated to function in immune evasion and host-parasite signaling (15, 20, 35, 85–87). Previous studies of the T. brucei flagellum have been limited by the inability to purify intact, membrane-enclosed flagella. Here we combine genetic manipulation and mechanical force to isolate intact flagella from mammalian-infectious T. brucei. The quality of the preparation is evidenced by motility of flagella separated from cell bodies, combined with biochemical and microscopic analysis of purified flagella. The flagellum preparation is further validated by independent analyses showing flagellum localization and/or function for several proteins identified in the purified sample. The availability of intact flagellum preparations opens new opportunities for studying unique and conserved features of T. brucei flagella. In other organisms, the ability to purify intact flagella has been critical for advancing studies of flagellum biology, including the recent resurgence in awareness of the important roles played by flagella in human health and disease (7, 64, 88). Development of a method for flagellum isolation from T. brucei is anticipated to advance efforts to understand flagellum biology in these pathogens and will expand the utility of trypanosomes as an experimental system for studying flagellum biology.
Identification of Flagellum Surface and Flagellum Matrix Proteins
We took advantage of our flagellum purification to conduct a proteomic analysis of surface-exposed flagellum membrane proteins and soluble flagellum matrix proteins. We identified 158 and 666 proteins in the flagellum surface and matrix fractions, respectively, with 87 proteins common between the two datasets. The combined surface and matrix proteomes include 165 proteins for which homologs were identified in a proteomic analysis of intact flagella from C. reinhardtii (64) (supplemental Table S7). Gene ontology annotation indicates that proteins identified in the T. brucei surface and matrix fractions encompass a wide range of functionalities (Table I). A notable feature of each dataset was the large number of proteins, 52% of the surface and 52% of the matrix proteome, for which gene ontology analyses did not reveal predicted functionality. Previously, pioneering proteomic studies of detergent-extracted flagellum skeletons from procyclic T. brucei provided important insights into flagellum biology (31, 43, 46). Novelty of the membrane and matrix proteomes identified in the current study is illustrated by comparison with these previous analyses. There is minimal overlap among the proteomes, with 91% of proteins identified in the surface and matrix proteomes being unique to the current analysis. The combined studies make T. brucei one of few organisms for which analysis of the entire flagellum, i.e. membrane, matrix, and skeleton, is available and, to our knowledge, provides the first example where an independent analysis of surface-exposed flagellar proteins is available.
Analysis of the surface data set is particularly interesting, as it shows enrichment for transmembrane proteins, including several predicted transporters and proteins predicted to initiate host-parasite signaling (see below). The number and predicted functionalities of proteins identified in the flagellum surface fraction indicate that T. brucei surface protein diversity is greater than might be inferred from the relatively few previously characterized surface proteins (89). The surface data set has relevance from a therapeutic standpoint because the identified proteins are accessible to small molecules added to live, mammalian-infectious parasites.
Signaling Capacity of the Trypanosome Flagellum
The trypanosome flagellum has been hypothesized to participate in host-parasite signaling, but proteins responsible for detection and transduction of extracellular signals are mostly unknown. Previously, only a handful of flagellar membrane (including the flagellar pocket) and matrix proteins have been identified in T. brucei (24, 30, 33, 34, 75, 90–97). In other organisms, flagellum-dependent signaling is dominated by three signal transduction pathways: cyclic nucleotide signaling, Ca2+ signaling and phosphorylation cascades initiated by receptor-kinases on the flagellar membrane (2, 8). The T. brucei flagellum surface proteome is replete with proteins capable of initiating signal transduction in each of these signaling pathways (Fig. 8), including receptor kinases, receptor adenylate cyclases (30) and proteins predicted to function in Ca2+ signaling (33, 36). The surface proteome also includes many proteins predicted to function in transport of solutes across the cell membrane (Table I). The combined data indicate a diverse flagellar surface protein repertoire suitable for mediating a broad range of signaling functions, thus providing molecular support for the hypothesis that the flagellum membrane is an important host-parasite signaling interface.
A role for the T. brucei flagellum as a signaling platform is supported by previous work that identified downstream targets of cyclic nucleotide, Ca2+ and phospho-signaling pathways anchored to the T. brucei flagellum skeleton (Fig. 8) (29, 31, 32, 43). The flagellar matrix fraction included many of these effectors, as well as several other candidate signaling proteins (supplemental Table S3) (Fig. 8). A major effector of cyclic nucleotide signaling that was not identified in previous studies of the T. brucei flagellum is the protein kinase A RSU, which binds cyclic nucleotide (72). RSU mediates cyclic nucleotide signaling in diverse organisms (98, 99), although a function has not been defined for this protein in T. brucei. We identified RSU in the flagellum matrix fraction (supplemental Table S3). Absence of RSU from detergent-extracted flagellum preparations (31, 43, 46) indicates it is not stably associated with the axoneme or PFR. To test for a requirement of RSU in flagellum function, we used RNAi. RSU knockdown inhibited motility of bloodstream-form cells (Fig. 7D,7E). The motility defect caused by RSU knockdown supports the identification of RSU in the flagellum matrix and provides experimental evidence for cyclic nucleotide-dependent signaling in regulating flagellum function.
Functional Analysis of Flagellum Surface Proteins Provides Insight into Mechanisms of Flagellum Attachment
One of the most distinctive features of the T. brucei flagellum is lateral attachment to the cell body, via regularly-spaced, desmosome-like adhesions between the flagellar and cell surface membranes (40). Immunofluorescence revealed that some proteins in the flagellum surface fraction, e.g. FS179 (Fig. 6B) and FS105 (supplemental Fig. S3), are restricted to the flagellum-cell body interface and do not extend to the flagellum tip, suggesting a potential role in flagellum attachment. RNAi knockdown of FS179, which encodes a putative Ca2+ channel, caused the flagellum to become detached from the cell body, with the daughter flagellum being preferentially affected (Fig. 7A–7C). The phenotype was ultimately lethal (supplemental Fig. S4). FS179 knockdown phenocopies flagellum detachment caused by treatment of bloodstream trypanosomes with Ca2+ chelators, as reported by Vickerman forty years ago (40). These results indicate a requirement for Ca2+ in establishment of flagellum-cell body adhesion in T. brucei (40) and suggest that FS179 is critical for maintaining Ca2+ homeostasis at the flagellum-cell body interface. In other organisms Ca2+ is required for homophilic cell-cell adhesion, including fibrous linkages between the connecting cilium and the periciliary inner segment collar of mammalian photoreceptors (100). Defects in these periciliary attachments cause Usher syndrome in humans (101). Previous analogies have been drawn between mammalian Ca2+-dependent cell-cell adhesions and trypanosome flagellum-cell body adhesions (40). Our findings support this view at the molecular level. Flagellum attachment is essential in T. brucei, suggesting that Ca2+ channels might be exploited as targets for therapeutic intervention in trypanosomiases. Indeed, Ca2+ channels are major targets of the pharmaceutical industry for treating of a variety of human diseases (102) and Ca2+ channel blockers have antiprotozoal activity in vitro (103).
Matrix-Specific Proteins Include IFT Proteins and Components of the Ubiquitin Conjugating System
Many proteins identified in the flagellum surface fraction were also identified in the flagellum matrix fraction. This result is expected because biotinylation and avidin purification are not 100% efficient. On the other hand, the vast majority (87%) of proteins in the flagellum matrix were exclusive to this fraction. Matrix-specific proteins include components of the intraflagellar transport (IFT) system that are required for flagellum assembly and signaling (104, 105). We identified six IFT complex B proteins, and the IFT anterograde kinesin motor, KIF3A (supplemental Table S8). Notably, none of the IFT proteins were identified in the surface fraction, or in previous proteomic analyses of the flagellum skeleton (31, 43, 46).
Another interesting group of proteins specifically identified in the matrix fraction includes components of the ubiquitin conjugating system (supplemental Table S9). Ubiquitination is a post-translational protein modification that has essential roles in many cellular processes, including cell-cycle control, protein quality control and signaling (106–108). A recent study identified a functional ubiquitination system in the flagellum of C. reinhardtii, where it is postulated to control flagellar resorption and cAMP-dependent signaling in the flagellum during mating (109). Several ubiquitinated proteins were identified, including the polycystin 2 cation channel (CrPKD2) and a cyclic GMP-dependent protein kinase (CrPKG) that both participate in flagellum signaling during mating (109). Polyubiquitin was identified in all three T. brucei flagellum fractions (supplemental Tables S9 (31, 43, 46)), indicating ubiquitination of T. brucei flagellar proteins. The identification of an ubiquitin-activating enzyme and three ubiquitin conjugating enzymes in the flagellum matrix fraction suggests that ubiquitination might be carried out within the flagellum itself. Although the function of ubiquitination in the T. brucei flagellum remains to be determined, we postulate that this pathway participates in regulation of flagellum function as described for C. reinhardtii (109).
The Flagellum Surface Presents a Dynamic and Cell-specific Host-Parasite Interface
Phylogenetic analysis revealed that 16% of flagellar surface proteins are T. brucei-specific. By comparison, only 5% of proteins in the flagellar matrix and 1% in the flagellum skeleton are T. brucei-specific. These results suggest that flagellum surface proteins must accommodate cell-specific functions, such as perception of signals specific to the parasite's extracellular environment. In contrast, composition of the flagellar matrix and skeleton fractions reflect more broadly conserved functions, e.g. downstream signal transduction and motility capacity of the organelle. Trypanosomes infect a broad range of organisms and survival in diverse host environments necessitates specialized surface proteins for signaling, nutrient acquisition and protection against host defenses. A cell-specific flagellum surface proteome would provide an interface that is tailored to meet the demands specifically imposed on T. brucei in each host. In support of this idea, we found that 21% of flagellum surface proteins are up-regulated by twofold or more in the bloodstream life cycle stage (Fig. 10) (68, 71). This is higher than seen for matrix proteins (10%), flagellum skeleton proteins (3%) or for the genome as a whole (3%) (Fig. 10) (68, 71). Thus, the flagellum surface proteome is a dynamic host-parasite interface that is particularly subject to life cycle stage-specific regulation.
Conserved Flagellum Surface Proteins are not Biased Toward Flagellated Organisms
Comparing the phylogenetic distribution of conserved proteins, i.e. those that are not kinetoplastid-specific, in each flagellum sub-fraction revealed an interesting feature of surface and matrix proteins. Namely, conservation of surface and matrix proteins is not biased toward flagellated organisms. This result contrasts markedly from what is observed for conserved flagellum skeleton proteins, which are generally more restricted to flagellated organisms (Fig. 9). The bias toward flagellated organisms reflects the requirement of flagellum skeleton proteins for axoneme structure and motility, which are unique to organisms with flagella (82–84). Flagellum surface and matrix proteins on the other hand, are predicted to be involved in perception and transduction of extracellular signals, i.e. processes that are not unique to flagellated organisms. An exception is the group of IFT proteins in the matrix, which are involved in flagellum assembly but represent a small fraction of matrix proteins. The combined phylogenetic analyses indicate that protein composition of the eukaryotic flagellum surface is shaped by selective forces that are a combination of cell-specific demands, imposed by unique extracellular environments, and more broadly conserved signaling needs. This finding has practical implications, as it means comparative genomic approaches, which are useful for identifying flagellum assembly and motility genes (82–84), are not well-suited for identification of genes involved in flagellum signaling.
Acknowledgments
We thank Dr. Zakayi Pius Kabututu for assistance and input for flagellum purification. Prof. G. Cross for the pMOTag2H vector and the 221 cell line; Prof. T. Seebeck and Prof. J. Bangs for antibodies. We also thank colleagues for critical reading of the manuscript.
Footnotes
* This work was supported by the Swiss National Science Foundation (MO), National Institutes of Health (NIH)-AI052348 (KLH), the Burroughs Wellcome Fund (KLH), NIH-NRSA-GM07185 (HTN), the A. G. Leventis Foundation (GL), NIH-NRSA-GM GM07104 (EAS), URC-CARE-GM55052 and UC-LEADS (SMN), Jonsson Cancer Center at UCLA and NIH-GM089778 (JAW).
 This article contains supplemental Figs. S1 to S4, Tables, S1 to S10 and Movie S1.
This article contains supplemental Figs. S1 to S4, Tables, S1 to S10 and Movie S1.
1 The abbreviations used are:
- PFR
- paraflagellar rod
- MudPIT
- multidimensional protein identification technology
- RNAi
- RNA interference
- VSG
- variable surface glycoprotein
- FS
- flagellum surface
- TM
- transmembrane
- RSU
- protein kinase A regulatory subunit
- IFT
- intraflagellar transport
- HA
- hemagglutinin
- PBS
- phosphate-buffered saline
- GRCR
- G protein coupled receptor
- BIP
- intracellular binding protein.
REFERENCES
- 1. Berbari N. F., O'Connor A. K., Haycraft C. J., Yoder B. K. (2009) The primary cilium as a complex signaling center. Curr. Biol. 19, R526–535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Christensen S. T., Ott C. M. (2007) Cell signaling. A ciliary signaling switch. Science 317, 330–331 [DOI] [PubMed] [Google Scholar]
- 3. Corbit K. C., Aanstad P., Singla V., Norman A. R., Stainier D. Y., Reiter J. F. (2005) Vertebrate Smoothened functions at the primary cilium. Nature 437, 1018–1021 [DOI] [PubMed] [Google Scholar]
- 4. Bloodgood R. A. Sensory reception is an attribute of both primary cilia and motile cilia. J. Cell Sci. 123, 505–509 [DOI] [PubMed] [Google Scholar]
- 5. Satir P., Mitchell D. R., Jékely G. (2008) How did the cilium evolve? Curr. Top. Dev. Biol. 85, 63–82 [DOI] [PubMed] [Google Scholar]
- 6. Badano J. L., Mitsuma N., Beales P. L., Katsanis N. (2006) The ciliopathies: an emerging class of human genetic disorders. Annu. Rev. Genomics Hum. Genet. 7, 125–148 [DOI] [PubMed] [Google Scholar]
- 7. Fliegauf M., Benzing T., Omran H. (2007) When cilia go bad: cilia defects and ciliopathies. Nat. Rev. Mol. Cell Biol. 8, 880–893 [DOI] [PubMed] [Google Scholar]
- 8. Kaupp U. B., Kashikar N. D., Weyand I. (2008) Mechanisms of sperm chemotaxis. Annu. Rev. Physiol. 70, 93–117 [DOI] [PubMed] [Google Scholar]
- 9. Pan J., Wang Q., Snell W. J. (2005) Cilium-generated signaling and cilia-related disorders. Lab. Invest. 85, 452–463 [DOI] [PubMed] [Google Scholar]
- 10. Rohatgi R., Milenkovic L., Scott M. P. (2007) Patched1 regulates hedgehog signaling at the primary cilium. Science 317, 372–376 [DOI] [PubMed] [Google Scholar]
- 11. Christensen S. T., Guerra C. F., Awan A., Wheatley D. N., Satir P. (2003) Insulin receptor-like proteins in Tetrahymena thermophila ciliary membranes. Curr. Biol. 13, R50–R52 [DOI] [PubMed] [Google Scholar]
- 12. Ogura A., Takahashi K. (1976) Artificial deciliation causes loss of calcium-dependent responses in Paramecium. Nature 264, 170–172 [DOI] [PubMed] [Google Scholar]
- 13. Pan J., Snell W. J. (2000) Signal transduction during fertilization in the unicellular green alga, Chlamydomonas. Curr. Opin. Microbiol. 3, 596–602 [DOI] [PubMed] [Google Scholar]
- 14. Dawson S. C., House S. A. (2010) Life with eight flagella: flagellar assembly and division in Giardia. Curr. Opin. Microbiol. 13, 480–490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ralston K. S., Kabututu Z. P., Melehani J. H., Oberholzer M., Hill K. L. (2009) The Trypanosoma brucei flagellum: moving parasites in new directions. Annu. Rev. Microbiol. 63, 335–362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Sinden R. E., Talman A., Marques S. R., Wass M. N., Sternberg M. J. (2010) The flagellum in malarial parasites. Curr. Opin. Microbiol. 13, 491–500 [DOI] [PubMed] [Google Scholar]
- 17. WHO (2004) Available at http://www.who.int/evidence/bod
- 18. Bastin P. The peculiarities of flagella in parasitic protozoa. Curr. Opin. Microbiol. 13, 450–452 [DOI] [PubMed] [Google Scholar]
- 19. Brun R., Blum J., Chappuis F., Burri C. (2010) Human African trypanosomiasis. Lancet 375, 148–159 [DOI] [PubMed] [Google Scholar]
- 20. Maric D., Epting C. L., Engman D. M. (2010) Composition and sensory function of the trypanosome flagellar membrane. Curr. Opin. Microbiol. 13, 466–472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Parsons M., Ruben L. (2000) Pathways involved in environmental sensing in trypanosomatids. Parasitol. Today 16, 56–62 [DOI] [PubMed] [Google Scholar]
- 22. Rotureau B., Morales M. A., Bastin P., Spath G. F. (2009) The flagellum-MAP kinase connection in Trypanosomatids: a key sensory role in parasite signaling and development? Cell Microbiol. 11, 710–718 [DOI] [PubMed] [Google Scholar]
- 23. Fenn K., Matthews K. R. (2007) The cell biology of Trypanosoma brucei differentiation. Curr. Opin. Microbiol. 10, 539–546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Dean S., Marchetti R., Kirk K., Matthews K. R. (2009) A surface transporter family conveys the trypanosome differentiation signal. Nature 459, 213–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Nikolskaia O. V., de A, Lima A. P., Kim Y. V., Lonsdale-Eccles J. D., Fukuma T., Scharfstein J., Grab D. J. (2006) Blood-brain barrier traversal by African trypanosomes requires calcium signaling induced by parasite cysteine protease. J. Clin. Invest. 116, 2739–2747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Tetley L., Vickerman K. (1985) Differentiation in Trypanosoma brucei: host-parasite cell junctions and their persistence during acquisition of the variable antigen coat. J. Cell Sci. 74, 1–19 [DOI] [PubMed] [Google Scholar]
- 27. Van Den Abbeele J., Claes Y., van Bockstaele D., Le Ray D., Coosemans M. (1999) Trypanosoma brucei spp. development in the tsetse fly: characterization of the post-mesocyclic stages in the foregut and proboscis. Parasitology 118, 469–478 [DOI] [PubMed] [Google Scholar]
- 28. Gluenz E., Höög J. L., Smith A. E., Dawe H. R., Shaw M. K., Gull K. (2010) Beyond 9+0: noncanonical axoneme structures characterize sensory cilia from protists to humans. Faseb J. 24, 3117–3121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Oberholzer M., Marti G., Baresic M., Kunz S., Hemphill A., Seebeck T. (2007) The Trypanosoma brucei cAMP phosphodiesterases TbrPDEB1 and TbrPDEB2: flagellar enzymes that are essential for parasite virulence. Faseb J. 21, 720–731 [DOI] [PubMed] [Google Scholar]
- 30. Paindavoine P., Rolin S., Van Assel S., Geuskens M., Jauniaux J. C., Dinsart C., Huet G., Pays E. (1992) A gene from the variant surface glycoprotein expression site encodes one of several transmembrane adenylate cyclases located on the flagellum of Trypanosoma brucei. Mol. Cell. Biol. 12, 1218–1225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Portman N., Lacomble S., Thomas B., McKean P. G., Gull K. (2009) Combining RNA interference mutants and comparative proteomics to identify protein components and dependences in a eukaryotic flagellum. J. Biol. Chem. 284, 5610–5619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Ridgley E., Webster P., Patton C., Ruben L. (2000) Calmodulin-binding properties of the paraflagellar rod complex from Trypanosoma brucei. Mol. Biochem. Parasitol. 109, 195–201 [DOI] [PubMed] [Google Scholar]
- 33. Wu Y., Deford J., Benjamin R., Lee M. G., Ruben L. (1994) The gene family of EF-hand calcium-binding proteins from the flagellum of Trypanosoma brucei. Biochem. J. 304, 833–841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wu Y., Haghighat N. G., Ruben L. (1992) The predominant calcimedins from Trypanosoma brucei comprise a family of flagellar EF-hand calcium-binding proteins. Biochem. J. 287, 187–193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kohl L., Bastin P. (2005) The flagellum of trypanosomes. Int. Rev. Cytol. 244, 227–285 [DOI] [PubMed] [Google Scholar]
- 36. Balber A. E. (1990) The pellicle and the membrane of the flagellum, flagellar adhesion zone, and flagellar pocket: functionally discrete surface domains of the bloodstream form of African trypanosomes. Crit. Rev. Immunol. 10, 177–201 [PubMed] [Google Scholar]
- 37. Tyler K. M., Fridberg A., Toriello K. M., Olson C. L., Cieslak J. A., Hazlett T. L., Engman D. M. (2009) Flagellar membrane localization via association with lipid rafts. J. Cell Sci. 122, 859–866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hu Q., Milenkovic L., Jin H., Scott M. P., Nachury M. V., Spiliotis E. T., Nelson W. J. (2010) A septin diffusion barrier at the base of the primary cilium maintains ciliary membrane protein distribution. Science 329, 436–439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lacomble S., Vaughan S., Gadelha C., Morphew M. K., Shaw M. K., McIntosh J. R., Gull K. (2009) Three-dimensional cellular architecture of the flagellar pocket and associated cytoskeleton in trypanosomes revealed by electron microscope tomography. J. Cell Sci. 122, 1081–1090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Vickerman K. (1969) On the surface coat and flagellar adhesion in trypanosomes. J. Cell Sci. 5, 163–193 [DOI] [PubMed] [Google Scholar]
- 41. Field M. C., Carrington M. (2009) The trypanosome flagellar pocket. Nat. Rev. Microbiol. 7, 775–786 [DOI] [PubMed] [Google Scholar]
- 42. Webster P., Russell D. G. (1993) The flagellar pocket of trypanosomatids. Parasitol. Today 9, 201–206 [DOI] [PubMed] [Google Scholar]
- 43. Broadhead R., Dawe H. R., Farr H., Griffiths S., Hart S. R., Portman N., Shaw M. K., Ginger M. L., Gaskell S. J., McKean P. G., Gull K. (2006) Flagellar motility is required for the viability of the bloodstream trypanosome. Nature 440, 224–227 [DOI] [PubMed] [Google Scholar]
- 44. Robinson D. R., Gull K. (1991) Basal body movements as a mechanism for mitochondrial genome segregation in the trypanosome cell cycle. Nature 352, 731–733 [DOI] [PubMed] [Google Scholar]
- 45. Schlaeppi K., Deflorin J., Seebeck T. (1989) The major component of the paraflagellar rod of Trypanosoma brucei is a helical protein that is encoded by two identical, tandemly linked genes. J. Cell Biol. 109, 1695–1709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Hart S. R., Lau K. W., Hao Z., Broadhead R., Portman N., Hühmer A., Gull K., McKean P. G., Hubbard S. J., Gaskell S. J. (2009) Analysis of the trypanosome flagellar proteome using a combined electron transfer/collisionally activated dissociation strategy. J. Am. Soc. Mass Spectrom. 20, 167–175 [DOI] [PubMed] [Google Scholar]
- 47. Wirtz E., Leal S., Ochatt C., Cross G. A. (1999) A tightly regulated inducible expression system for conditional gene knock-outs and dominant-negative genetics in Trypanosoma brucei. Mol. Biochem. Parasitol. 99, 89–101 [DOI] [PubMed] [Google Scholar]
- 48. Oberholzer M., Lopez M. A., Ralston K. S., Hill K. L. (2009) Approaches for functional analysis of flagellar proteins in African trypanosomes. Methods Cell Biol. 93, 21–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. LaCount D. J., Barrett B., Donelson J. E. (2002) Trypanosoma brucei FLA1 is required for flagellum attachment and cytokinesis. J. Biol. Chem. 277, 17580–17588 [DOI] [PubMed] [Google Scholar]
- 50. Bangs J. D., Uyetake L., Brickman M. J., Balber A. E., Boothroyd J. C. (1993) Molecular cloning and cellular localization of a BiP homologue in Trypanosoma brucei. Divergent ER retention signals in a lower eukaryote. J. Cell Sci. 105, 1101–1113 [DOI] [PubMed] [Google Scholar]
- 51. McDowell M. A., Ransom D. M., Bangs J. D. (1998) Glycosylphosphatidylinositol-dependent secretory transport in Trypanosoma brucei. Biochem. J. 335, 681–689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Tagwerker C., Flick K., Cui M., Guerrero C., Dou Y., Auer B., Baldi P., Huang L., Kaiser P. (2006) A tandem affinity tag for two-step purification under fully denaturing conditions: application in ubiquitin profiling and protein complex identification combined with in vivocross-linking. Mol. Cell Proteomics 5, 737–748 [DOI] [PubMed] [Google Scholar]
- 53. Wohlschlegel J. A. (2009) Identification of SUMO-conjugated proteins and their SUMO attachment sites using proteomic mass spectrometry. Methods Mol. Biol. 497, 33–49 [DOI] [PubMed] [Google Scholar]
- 54. Florens L., Carozza M. J., Swanson S. K., Fournier M., Coleman M. K., Workman J. L., Washburn M. P. (2006) Analyzing chromatin remodeling complexes using shotgun proteomics and normalized spectral abundance factors. Methods 40, 303–311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Xu T. V., Venable J. D., Cociorva D., Lu B., Liao L., Wohlschlegel J., Hewel J., Yates J. R., 3rd (2006) ProLuCID, a fast and sensitive tandem mass spectra-based protein identification program. Mol. Cellular Proteomics 5 [Google Scholar]
- 56. Eng J. K., McCormack A. L. YJI. (1994) An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J. Am. Soc. Mass Spectrom. 5, 976–989 [DOI] [PubMed] [Google Scholar]
- 57. Tabb D. L., McDonald W. H., Yates J. R., 3rd (2002) DTASelect and Contrast: tools for assembling and comparing protein identifications from shotgun proteomics. J. Proteome Res. 1, 21–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Cociorva D. L. T. D., Yates J. R. (2007) Validation of tandem mass spectrometry database search results using DTASelect. Curr. Protoc. Bioinformatics 13 [DOI] [PubMed] [Google Scholar]
- 59. Tazeh N. N., Silverman J. S., Schwartz K. J., Sevova E. S., Sutterwala S. S., Bangs J. D. (2009) Role of AP-1 in developmentally regulated lysosomal trafficking in Trypanosoma brucei. Eukaryot. Cell 8, 1352–1361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Ralston K. S., Hill K. L. (2006) Trypanin, a component of the flagellar Dynein regulatory complex, is essential in bloodstream form African trypanosomes. PLoS Pathog. 2, e101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Oberholzer M., Morand S., Kunz S., Seebeck T. (2006) A vector series for rapid PCR-mediated C-terminal in situ tagging of Trypanosoma brucei genes. Mol. Biochem. Parasitol. 145, 117–120 [DOI] [PubMed] [Google Scholar]
- 62. Redmond S., Vadivelu J., Field M. C. (2003) RNAit: an automated web-based tool for the selection of RNAi targets in Trypanosoma brucei. Mol. Biochem. Parasitol 128, 115–118 [DOI] [PubMed] [Google Scholar]
- 63. Schultz J., Milpetz F., Bork P., Ponting C. P. (1998) SMART, a simple modular architecture research tool: identification of signaling domains. Proc. Natl. Acad. Sci. U.S.A. 95, 5857–5864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Pazour G. J., Agrin N., Leszyk J., Witman G. B. (2005) Proteomic analysis of a eukaryotic cilium. J. Cell Biol. 170, 103–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Huang da W., Sherman B. T., Lempicki R. A. (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57 [DOI] [PubMed] [Google Scholar]
- 66. Huang da W., Sherman B. T., Lempicki R. A. (2009) Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 37, 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Ashburner M., Ball C. A., Blake J. A., Botstein D., Butler H., Cherry J. M., Davis A. P., Dolinski K., Dwight S. S., Eppig J. T., Harris M. A., Hill D. P., Issel-Tarver L., Kasarskis A., Lewis S., Matese J. C., Richardson J. E., Ringwald M., Rubin G. M., Sherlock G. (2000) Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 25, 25–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Siegel T. N., Hekstra D. R., Wang X., Dewell S., Cross G. A. (2010) Genome-wide analysis of mRNA abundance in two life-cycle stages of Trypanosoma brucei and identification of splicing and polyadenylation sites. Nucleic Acids Res. 38, 4946–4957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Vanhamme L., Pays E., McCulloch R., Barry J. D. (2001) An update on antigenic variation in African trypanosomes. Trends Parasitol. 17, 338–343 [DOI] [PubMed] [Google Scholar]
- 70. Berriman M., Ghedin E., Hertz-Fowler C., Blandin G., Renauld H., Bartholomeu D. C., Lennard N. J., Caler E., Hamlin N. E., Haas B., Böhme U., Hannick L., Aslett M. A., Shallom J., Marcello L., Hou L., Wickstead B., Alsmark U. C., Arrowsmith C., Atkin R. J., Barron A. J., Bringaud F., Brooks K., Carrington M., Cherevach I., Chillingworth T. J., Churcher C., Clark L. N., Corton C. H., Cronin A., Davies R. M., Doggett J., Djikeng A., Feldblyum T., Field M. C., Fraser A., Goodhead I., Hance Z., Harper D., Harris B. R., Hauser H., Hostetler J., Ivens A., Jagels K., Johnson D., Johnson J., Jones K., Kerhornou A. X., Koo H., Larke N., Landfear S., Larkin C., Leech V., Line A., Lord A., Macleod A., Mooney P. J., Moule S., Martin D. M., Morgan G. W., Mungall K., Norbertczak H., Ormond D., Pai G., Peacock C. S., Peterson J., Quail M. A., Rabbinowitsch E., Rajandream M. A., Reitter C., Salzberg S. L., Sanders M., Schobel S., Sharp S., Simmonds M., Simpson A. J., Tallon L., Turner C. M., Tait A., Tivey A. R., Van Aken S., Walker D., Wanless D., Wang S., White B., White O., Whitehead S., Woodward J., Wortman J., Adams M. D., Embley T. M., Gull K., Ullu E., Barry J. D., Fairlamb A. H., Opperdoes F., Barrell B. G., Donelson J. E., Hall N., Fraser C. M., Melville S. E., El-Sayed N. M. (2005) The genome of the African trypanosome Trypanosoma brucei. Science 309, 416–422 [DOI] [PubMed] [Google Scholar]
- 71. Aslett M., et al. TriTrypDB: a functional genomic resource for the Trypanosomatidae. Nucleic Acids Res. 38: D457–462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Shalaby T., Liniger M., Seebeck T. (2001) The regulatory subunit of a cGMP-regulated protein kinase A of Trypanosoma brucei. Eur. J. Biochem. 268, 6197–6206 [DOI] [PubMed] [Google Scholar]
- 73. Gaillard A. R., Diener D. R., Rosenbaum J. L., Sale W. S. (2001) Flagellar radial spoke protein 3 is an A-kinase anchoring protein (AKAP). J. Cell Biol. 153, 443–448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Eisenbach M., Giojalas L. C. (2006) Sperm guidance in mammals - an unpaved road to the egg. Nat. Rev. Mol. Cell Biol. 7, 276–285 [DOI] [PubMed] [Google Scholar]
- 75. Hanrahan O., Webb H., O'Byrne R., Brabazon E., Treumann A., Sunter J. D., Carrington M., Voorheis H. P. (2009) The glycosylphosphatidylinositol-PLC in Trypanosoma brucei forms a linear array on the exterior of the flagellar membrane before and after activation. PLoS Pathog. 5, e1000468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. LaCount D. J., Gruszynski A. E., Grandgenett P. M., Bangs J. D., Donelson J. E. (2003) Expression and function of the Trypanosoma brucei major surface protease (GP63) genes. J. Biol. Chem. 278, 24658–24664 [DOI] [PubMed] [Google Scholar]
- 77. Salmon D., Geuskens M., Hanocq F., Hanocq-Quertier J., Nolan D., Ruben L., Pays E. (1994) A novel heterodimeric transferrin receptor encoded by a pair of VSG expression site-associated genes in T. brucei. Cell 78, 75–86 [DOI] [PubMed] [Google Scholar]
- 78. Tetaud E., Barrett M. P., Bringaud F., Baltz T. (1997) Kinetoplastid glucose transporters. Biochem. J. 325, 569–580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Emmer B. T., Daniels M. D., Taylor J. M., Epting C. L., Engman D. M. (2010) Calflagin inhibition prolongs host survival and suppresses parasitemia in Trypanosoma brucei infection. Eukaryot. Cell 9, 934–942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Felder C. B., Graul R. C., Lee A. Y., Merkle H. P., Sadee W. (1999) The Venus flytrap of periplasmic binding proteins: an ancient protein module present in multiple drug receptors. AAPS PharmSci. 1, E2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Campbell I. D., Baron M., Cooke R. M., Dudgeon T. J., Fallon A., Harvey T. S., Tappin M. J. (1990) Structure-function relationships in epidermal growth factor (EGF) and transforming growth factor-alpha (TGF-alpha). Biochem. Pharmacol 40, 35–40 [DOI] [PubMed] [Google Scholar]
- 82. Li J. B., Gerdes J. M., Haycraft C. J., Fan Y., Teslovich T. M., May-Simera H., Li H., Blacque O. E., Li L., Leitch C. C., Lewis R. A., Green J. S., Parfrey P. S., Leroux M. R., Davidson W. S., Beales P. L., Guay-Woodford L. M., Yoder B. K., Stormo G. D., Katsanis N., Dutcher S. (2004) Comparative genomics identifies a flagellar and basal body proteome that includes the BBS5 human disease gene. Cell 117, 541–552 [DOI] [PubMed] [Google Scholar]
- 83. Avidor-Reiss T., Maer A. M., Koundakjian E., Polyanovsky A., Keil T., Subramaniam S., Zuker C. S. (2004) Decoding cilia function: defining specialized genes required for compartmentalized cilia biogenesis. Cell 117, 527–539 [DOI] [PubMed] [Google Scholar]
- 84. Baron D. M., Ralston K. S., Kabututu Z. P., Hill K. L. (2007) Functional genomics in Trypanosoma brucei identifies evolutionarily conserved components of motile flagella. J. Cell Sci. 120, 478–491 [DOI] [PubMed] [Google Scholar]
- 85. Bastin P. (2010) The peculiarities of flagella in parasitic protozoa. Curr. Opin. Microbiol. 13, 450–452 [DOI] [PubMed] [Google Scholar]
- 86. Rodriguez J. A., Lopez M. A., Thayer M. C., Zhao Y., Oberholzer M., Chang D. D., Kisalu N. K., Penichet M. L., Helguera G., Bruinsma R., Hill K. L., Miao J. (2009) Propulsion of African trypanosomes is driven by bihelical waves with alternating chirality separated by kinks. Proc. Natl. Acad. Sci. U.S.A. 106, 19322–19327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Engstler M., Pfohl T., Herminghaus S., Boshart M., Wiegertjes G., Heddergott N., Overath P. (2007) Hydrodynamic flow-mediated protein sorting on the cell surface of trypanosomes. Cell 131, 505–515 [DOI] [PubMed] [Google Scholar]
- 88. Smith J. C., Northey J. G., Garg J., Pearlman R. E., Siu K. W. (2005) Robust method for proteome analysis by MS/MS using an entire translated genome: demonstration on the ciliome of Tetrahymena thermophila. J. Proteome Res. 4, 909–919 [DOI] [PubMed] [Google Scholar]
- 89. Borst P., Fairlamb A. H. (1998) Surface receptors and transporters of Trypanosoma brucei. Annu. Rev. Microbiol. 52, 745–778 [DOI] [PubMed] [Google Scholar]
- 90. Mussmann R., Engstler M., Gerrits H., Kieft R., Toaldo C. B., Onderwater J., Koerten H., van Luenen H. G., Borst P. (2004) Factors affecting the level and localization of the transferrin receptor in Trypanosoma brucei. J. Biol. Chem. 279, 40690–40698 [DOI] [PubMed] [Google Scholar]
- 91. Vanhollebeke B., De Muylder G., Nielsen M. J., Pays A., Tebabi P., Dieu M., Raes M., Moestrup S. K., Pays E. (2008) A haptoglobin-hemoglobin receptor conveys innate immunity to Trypanosoma brucei in humans. Science 320, 677–681 [DOI] [PubMed] [Google Scholar]
- 92. Engstler M., Weise F., Bopp K., Grünfelder C. G., Günzel M., Heddergott N., Overath P. (2005) The membrane-bound histidine acid phosphatase TbMBAP1 is essential for endocytosis and membrane recycling in Trypanosoma brucei. J. Cell Sci. 118, 2105–2118 [DOI] [PubMed] [Google Scholar]
- 93. Luo S., Rohloff P., Cox J., Uyemura S. A., Docampo R. (2004) Trypanosoma brucei plasma membrane-type Ca(2+)-ATPase 1 (TbPMC1) and 2 (TbPMC2) genes encode functional Ca(2+)-ATPases localized to the acidocalcisomes and plasma membrane, and essential for Ca(2+) homeostasis and growth. J. Biol. Chem. 279, 14427–14439 [DOI] [PubMed] [Google Scholar]
- 94. Ziegelbauer K., Multhaup G., Overath P. (1992) Molecular characterization of two invariant surface glycoproteins specific for the bloodstream stage of Trypanosoma brucei. J. Biol. Chem. 267, 10797–10803 [PubMed] [Google Scholar]
- 95. Vanhamme L., Paturiaux-Hanocq F., Poelvoorde P., Nolan D. P., Lins L., Van Den Abbeele J., Pays A., Tebabi P., Van Xong H., Jacquet A., Moguilevsky N., Dieu M., Kane J. P., De Baetselier P., Brasseur R., Pays E. (2003) Apolipoprotein L-I is the trypanosome lytic factor of human serum. Nature 422, 83–87 [DOI] [PubMed] [Google Scholar]
- 96. Absalon S., Blisnick T., Kohl L., Toutirais G., Doré G., Julkowska D., Tavenet A., Bastin P. (2008) Intraflagellar transport and functional analysis of genes required for flagellum formation in trypanosomes. Mol. Biol. Cell 19, 929–944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Franklin J. B., Ullu E. Biochemical analysis of PIFTC3, the Trypanosoma brucei orthologue of nematode DYF-13, reveals interactions with established and putative intraflagellar transport components. Mol. Microbiol. 78, 173–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Howard D. R., Habermacher G., Glass D. B., Smith E. F., Sale W. S. (1994) Regulation of Chlamydomonas flagellar dynein by an axonemal protein kinase. J. Cell Biol. 127, 1683–1692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Francis S. H., Corbin J. D. (1999) Cyclic nucleotide-dependent protein kinases: intracellular receptors for cAMP and cGMP action. Crit. Rev. Clin. Lab. Sci. 36, 275–328 [DOI] [PubMed] [Google Scholar]
- 100. Maerker T., van Wijk E., Overlack N., Kersten F. F., McGee J., Goldmann T., Sehn E., Roepman R., Walsh E. J., Kremer H., Wolfrum U. (2008) A novel Usher protein network at the periciliary reloading point between molecular transport machineries in vertebrate photoreceptor cells. Hum. Mol. Genet. 17, 71–86 [DOI] [PubMed] [Google Scholar]
- 101. Williams D. S. (2008) Usher syndrome: animal models, retinal function of Usher proteins, and prospects for gene therapy. Vision Res. 48, 433–441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Abernethy D. R., Schwartz J. B. (1999) Calcium-antagonist drugs. N. Engl. J. Med. 341, 1447–1457 [DOI] [PubMed] [Google Scholar]
- 103. Reimao J. Q., Scotti M. T., Tempone A. G. (2010) Anti-leishmanial and anti-trypanosomal activities of 1,4-dihydropyridines: In vitro evaluation and structure-activity relationship study. Bioorg. Med. Chem. 18, 8044–8053 [DOI] [PubMed] [Google Scholar]
- 104. Cole D. G. (2003) The intraflagellar transport machinery of Chlamydomonas reinhardtii. Traffic 4, 435–442 [DOI] [PubMed] [Google Scholar]
- 105. Rosenbaum J. L., Witman G. B. (2002) Intraflagellar transport. Nat. Rev. Mol. Cell Biol. 3, 813–825 [DOI] [PubMed] [Google Scholar]
- 106. Okiyoneda T., Barriere H., Bagdany M., Rabeh W. M., Du K., Hohfeld J., Young J. C., Lukacs G. L. (2010) Peripheral protein quality control removes unfolded CFTR from the plasma membrane. Science 329, 805–810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Hershko A. (2005) The ubiquitin system for protein degradation and some of its roles in the control of the cell division cycle. Cell Death Differ. 12, 1191–1197 [DOI] [PubMed] [Google Scholar]
- 108. Wilkinson K. D. (1999) Ubiquitin-dependent signaling: the role of ubiquitination in the response of cells to their environment. J. Nutr. 129, 1933–1936 [DOI] [PubMed] [Google Scholar]
- 109. Huang K., Diener D. R., Rosenbaum J. L. (2009) The ubiquitin conjugation system is involved in the disassembly of cilia and flagella. J. Cell Biol. 186, 601–613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Hill K. L., Hutchings N. R., Grandgenett P. M., Donelson J. E. (2000) T lymphocyte-triggering factor of african trypanosomes is associated with the flagellar fraction of the cytoskeleton and represents a new family of proteins that are present in several divergent eukaryotes. J. Biol. Chem. 275, 39369–39378 [DOI] [PubMed] [Google Scholar]
- 111. Buchanan K. T., Ames J. B., Asfaw S. H., Wingard J. N., Olson C. L., Campana P. T., Araújo A. P., Engman D. M. (2005) A flagellum-specific calcium sensor. J. Biol. Chem. 280, 40104–40111 [DOI] [PubMed] [Google Scholar]
- 112. Deleted in proof.
- 113. Lancaster M. A., Schroth J., Gleeson J. G. (2011) Subcellular spatial regulation of canonical Wnt signaling at the primary cillum. Nat. Cell Biol. 13, 700–707, e pub, May 22 [DOI] [PMC free article] [PubMed] [Google Scholar]