Abstract
Despite their importance in many biological processes, membrane proteins are underrepresented in proteomic analysis because of their poor solubility (hydrophobicity) and often low abundance. We describe a novel approach for the identification of plasma membrane proteins and intracellular microsomal proteins that combines membrane fractionation, a centrifugal proteomic reactor for streamlined protein extraction, protein digestion and fractionation by centrifugation, and high performance liquid chromatography-electrospray ionization-tandem MS. The performance of this approach was illustrated for the study of the proteome of ER and Golgi microsomal membranes in rat hepatic cells. The centrifugal proteomic reactor identified 945 plasma membrane proteins and 955 microsomal membrane proteins, of which 63 and 47% were predicted as bona fide membrane proteins, respectively. Among these proteins, >800 proteins were undetectable by the conventional in-gel digestion approach. The majority of the membrane proteins only identified by the centrifugal proteomic reactor were proteins with ≥2 transmembrane segments or proteins with high molecular mass (e.g. >150 kDa) and hydrophobicity. The improved proteomic reactor allowed the detection of a group of endocytic and/or signaling receptor proteins on the plasma membrane, as well as apolipoproteins and glycerolipid synthesis enzymes that play a role in the assembly and secretion of apolipoprotein B100-containing very low density lipoproteins. Thus, the centrifugal proteomic reactor offers a new analytical tool for structure and function studies of membrane proteins involved in lipid and lipoprotein metabolism.
Membrane proteins play an important role in a variety of cellular functions, ranging from signal transduction, subcellular compartmentalization, membrane trafficking, and protein secretion, in addition to their function in providing and maintaining the structural integrity of membranes. Not surprisingly, the membrane associated proteins account for nearly 60% of pharmaceutical drug targets (1–3). Depending upon the physical nature of interactions between proteins and membranes, membrane proteins are broadly classified as integral membrane proteins (proteins that contain hydrophobic transmembrane segments in their sequences) and peripheral membrane proteins. Although the peripheral membrane proteins are in general soluble and relatively amiable for identification using proteomic approaches, integral membrane proteins pose greater challenges owing to low abundance and hydrophobicity (4, 5).
Many analytical approaches and strategies have been developed to facilitate membrane protein characterization. The low abundance issue has been addressed by enriching samples using subcellular fractionation, (e.g. sucrose or sorbitol density ultracentrifugation) (6–9), cationic colloidal silica absorption (10), aqueous-polymer two-phase system (11), and triton X-114 phase separation (12). In addition, high abundant nonmembrane proteins can be separated from membrane proteins by using high-salt and high-pH buffers (13, 14). The hydrophobicity issue of membrane proteins has been circumvented by the use of mass spectrometry compatible surfactants (15), organic solvents (60% methanol) (16), and multiple enzymes (17, 18). Most of these approaches have been applied to analyze plasma membrane and integral membrane proteins (6, 13, 14, 19, 20). However, studies with microsomal membranes using proteomic approach are relatively limited (21, 22).
The microsomal membranes, composed of endoplasmic reticulum (ER)1 and Golgi apparatus, represent the cellular compartments where synthesis, post-translational modification, assembly, and secretion of various proteins and lipoproteins occur. In hepatic cells, the ER and Golgi microsomes play a major role in the assembly and secretion of very low density lipoproteins (VLDL) rich in triglycerides (TAG) and cholesterol (23–27). Multiple protein factors are required for the process of hepatic VLDL assembly and secretion, of which apolipoprotein (apo) B-100, an extremely large (4536 amino acids) and hydrophobic protein serves as a structural backbone in TAG recruitment during the VLDL assembly process. Another protein, termed microsomal triglyceride-transfer protein (MTP) that is encoded by the abetalipoprotein gene MTTP, is a microsome-resident protein that plays an obligatory role in facilitating VLDL assembly and secretion (25). Expression of other proteins, such as the low density lipoprotein (LDL) receptor, also influences the VLDL assembly and secretion process by regulating degradation of apoB-100 (28). The recruitment of lipids in VLDL assembly starts immediately during and after apoB-100 translation and translocation across the ER membrane (29). However, microsomal membrane proteins that may play a role in the initial VLDL assembly process that occurs in association with membranes are not fully characterized.
The purpose of the current work is to determine the proteome of ER and Golgi microsomal membranes in hepatic cells. Recently, we have developed a microfluidic device, termed the proteomic reactor, for rapid preconcentration, derivatization, and enzymatic digestion (30). This proteomic reactor has been successfully applied to the analysis of complex protein mixtures (31–33) and post-translational modifications (34–36). Moreover, a multiplexed proteomic reactor has been developed for parallel processing of minute amounts of protein sample (37). In this study, we developed a simplified reactor compatible with bench-top centrifuges. The centrifugal proteomic reactor allowed improved recovery as well as identification of microsomal membrane proteins from the rat hepatoma cell line McA-RH7777. The centrifugal proteomic reactor identified 945 plasma membrane proteins and 955 microsomal membrane proteins with respectively 63 and 47% predicted as bona fide membrane proteins. Among these proteins, >800 proteins were undetectable by the conventional in-gel digestion approach.
EXPERIMENTAL PROCEDURES
Materials
Ammonium bicarbonate, dithiothreitol, and sodium carbonate were purchased from EMD Chemicals, Inc. (Darmstadt, Germany). Acetonitrile, with 0.1% formic acid, and water, with 0.1% formic acid, were purchased from J.T. Baker (Phillipsburg, NJ). Trypsin was purchased from Promega (Madison, WI). Strong cation exchange beads were obtained from Polymer Laboratories, Varian, Inc. (Palo Alto, CA). CHAPS (3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate, BP 571) and methanol were purchased from Fisher Scientific (Hampton, NH). Iodoacetamide was obtained from Sigma-Aldrich (Saint Louis, MO). Medium and reagents used for cell culture studies were obtained from Invitrogen (Burlington, ON).
Cell Culture
The rat hepatoma McA-RH7777 cells were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum and 10% horse serum as described previously (38).
Plasma and Microsomal Membrane Isolation
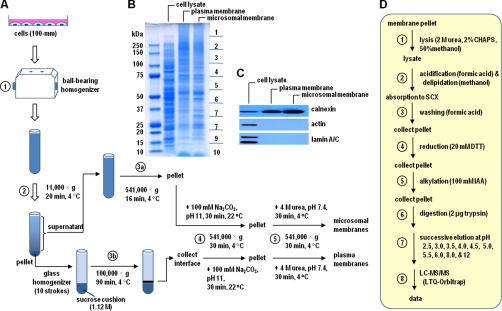
The flowchart of membrane preparation is shown in Fig. 1. Cells at 80% confluence (5 × 106 cells were plated in 10-cm dish) cultured in the Dulbecco's modified Eagle's medium containing 20% fetal bovine serum were washed with ice-cold phosphate-buffered saline three times. Total of four dishes of cells were collected into homogenization buffer (20 mm Tris-HCl, pH 7.4, 250 mm sucrose, 1 mm EDTA, and protease inhibitor mixture), and homogenized by 20 passes through a ball bearing homogenizer (H and Y Enterprise, Redwood City, CA) (Step 1 in Fig. 1A). The homogenates were centrifuged (11,000 × g, 10 min, 4 °C) to pellet nucleus, mitochondria, plasma membrane, and unbroken cells (Step 2). The post-nuclear supernatant was centrifuged (541,000 × g, 16 min, 4 °C) to recover the microsome (Step 3a), whereas the pellet was resuspended in the homogenization buffer (devoid of sucrose) using a glass homogenizer, layered onto a 1.12 m sucrose cushion, and centrifuged (100,000 × g, 90 min, 4 °C) to sediment plasma membrane at the interface (Step 3b). The respective plasma and microsomal membranes (obtained from 3a and 3b) were subjected to two additional washing steps, namely high pH (sodium carbonate 100 mm, pH 11.0; Step 4) and 4 m Urea (20 mm Tris-HCl pH7.4, 1 mm EDTA, protease inhibitor mixture; Step 5), in an attempt to remove loosely associated peripheral membrane proteins.
Fig. 1.
Flow chart of centrifugal proteomic reactor for membrane proteins. A, Plasma and microsomal membrane isolation; B, Colloidal blue-stained gel of total cell lysate, plasma membrane, and microsomal membrane. Twenty micrograms of each sample was loaded on a 7-cm 4∼12% SDS-PAGE, then the gel was stained by Colloidal blue. C, Western blots of calnexin, actin and lamin A/C using total cell lysate, plasma, and microsomal membrane samples. D, Flow chart for steps involved in centrifugal proteomic reactor.
Centrifugal Proteomic Reactor
The flowchart for the centrifugal reactor procedures is shown in Fig. 1D. Briefly, 20 μg of plasma or microsomal membrane protein was solubilized using an extraction buffer containing 50% methanol, 2 m urea and 2% CHAPS, and the samples were transferred into 1.5 ml Eppendorf tubes (Step 1 in Fig. 1D), mixed with 10 μl of strong cation exchange slurry in the presence of 1.2 ml of 50% methanol and 5% formic acid by vigorous vortexing (for 1 min) (Step 2). The samples were centrifuged (16,100 × g, 2 min), and the pellet was again treated with 1.2 ml 0.5% formic acid (Step 3). The samples were reduced by mixing with 20 μl of 150 mm NH4HCO3, 20 mm DTT (shaking at 800 rpm, 56 °C, 15 min), and after reduction the dithiothreitol in the sample was diluted using 1.2 ml 0.5% formic acid and followed by centrifugation (Step 4). The samples were then subjected to alkylation by mixing with 20 μl of 150 mm NH4HCO3, 100 mm iodoacetamide in darkness (15 min, room temperature), and the reaction was stopped by adding 1.2 ml of 0.5% formic acid containing 2 μg trypsin (Step 5). After centrifugation, the resulting pellet was dissolved in 20 μl of 1 m NH4HCO3, and followed by trypsin digestion on a shaker at 800 rpm at 37 °C for 2 h (Step 6). Finally, 1.2 ml of 5% formic acid was added to elute the pH 2.5 fraction by centrifugation, followed by additional nine pH fractions (pH 3.0, 3.5, 4.0, 4.5, 5.0, 5.5, 6.0, 8.0, 12) with 300 μl of respective pH buffers (Step 7) (39). The fractionated samples were subjected to liquid chromatography-tandem MS (LC-MS/MS) analysis (Step 8).
LC-MS/MS
The LC-MS/MS was performed as previous described (35). Briefly, samples were acidified with formic acid to a final concentration of 5% (v/v) and loaded on a 200 μm × 100 mm fused silica precolumn packed in-house with 10 cm of 5-μm ReproSil-Pur C18 beads (200 Å; Dr. Maisch GmbH, Ammerbuch, Germany) using an 1100 micro-HPLC system (Agilent Technologies, Santa Clara, CA). After the desalting step, the flow was split, and peptides were eluted through a second 75 μm × 150 mm column packed with 3-μm C18 beads at ∼150 nL/min. The peptides were eluted using a gradient (5–80% acetonitrile with 0.1% formic acid) over a 2 h period into an ESI linear trap quadrupole (LTQ)-Orbitrap mass spectrometer (Thermo Electron, Waltham, MA). MS/MS spectra were acquired in a data-dependent acquisition mode that automatically selected and fragmented the 10 most intense peaks from each MS spectrum generated.
In-gel Digestion
Protein samples (20 μg) were dried, reconstituted in gel loading buffer and separated by polyacrylamide gel electrophoresis in the presence of SDS (SDS-PAGE) (4∼12% NuPAGE gel) at a constant voltage setting of 80 V for 30 min and then 120 V thereafter until the bromphenol blue dye marker reached the bottom of the gel. Proteins were visualized using colloidal blue staining (Invitrogen, Carlsbad, CA), and the gels were sliced into ten pieces of equal length and the in-gel digestion was performed as reported previously (40).
Western Blot
Proteins resolved in 4∼12% NuPAGE gels were transferred onto nitrocellulose membrane, blocked with 5% nonfat milk (in phosphate-buffered saline), and incubated with the respective anti-calnexin, (Stress gen San Diego, CA, SPA-860, rabbit polyclonal), anti-actin (Cedarlane Burlington, ON, Canada, CLT9001, mouse monoclonal), and anti-lamin A/C (Santa Cruz Biotechnology (Santa Cruz, CA), sc-6215, goat polyclonal) antibodies (1:1,000) overnight at 4 °C. After washing with phosphate-buffered saline, the blots were incubated with anti-rabbit, mouse, and goat HRP-conjugated secondary antibodies, respectively.
Data Analysis
Maxquant (41) (http://maxquant.org/, version 1.0.13.13) was used to generate peak lists from raw files, and Mascot (version 2.2.02 Matrix Science, London, UK) was used to search the protein sequence database using the following parameters: carbamidomethyl (C) was set as a fixed modification, whereas oxidation (M, +15.99492 Da) was set as a variable modification. Acquired MS/MS spectra were searched against the decoyed rat International Protein Index protein sequence database (version 3.52, 39,906 protein sequences; European Bioinformatics Institute) augmented with the reversed sequence of each entry in the forward database. The precursor and fragment mass tolerances for the searching were set at 7 ppm and 0.5 Da, respectively. Trypsin was selected as the digestive enzyme with two potential missed cleavages. Only fully tryptic peptides ranked first were accepted in our database searching results. The false positive rate was controlled at <1% using the equation false positive rate = # of false peptides/(# of true peptides + # of false peptides) × 100. The proteins with at least two unique peptides were accepted for following analysis. The mascot cut-off score is 24 for both reactor and in-gel digestion data sets. Membrane protein prediction was achieved using the DAVID Bioinformatics Resources (http://david.abcc.ncifcrf.gov/)(42) and the Gene Ontology (GO) annotation (www.geneontology.org) and the TransMembrane Hidden Markov Model (TMHMM, http://www.cbs.dtu.dk/services/TMHMM/). We used PANTHER (http://www.pantherdb.org/) to do the biological process annotation. The protein sequence comparison between this study and others was performed using Protein Mapping and Comparison Tool (PROMT, http://www.geneinfo.eu/prompt/), and the two protein sequences with > = 80% identity were consider as overlapped proteins. Maxquant (41) (version 1.1.1.36) was used for label-free quantitation of the proteins from in-gel and on-reactor digestion.
RESULTS
Plasma and Microsomal Membrane Preparation
We developed a method that combines membrane fractionation, a centrifugal proteomic reactor, and HPLC-ESI-MS/MS to study membrane proteins with a particular focus on plasma and microsomal membranes. This method consists of first separating plasma and microsomal membranes through multistep centrifugations (Fig. 1A). The proteins from the plasma and microsomal membranes were captured and processed on a centrifugal proteomic reactor prior to analysis by HPLC-ESI-MS/MS. In contrast to our previously reported proteomic reactor for minute amounts of sample, the centrifugal proteomic reactor was constructed in such a way that the amount of samples to be processed can be scaled up to hundreds of micrograms. In addition, all of the processing steps involved centrifugation that effectively minimized sample loss during multiple washing steps (see Experimental Procedures for details). As a proof-of-principle, we compared the results obtained from the membrane enrichment method followed by either the centrifugal proteomic reactor or conventional SDS-PAGE and in-gel digestion method. Plasma and microsomal membranes were isolated from the rat hepatoma McA-RH7777 cells, a cell line that has been used extensively in the studies of hepatic TAG-rich lipoprotein assembly and secretion (43). Approximately 100 μg proteins were obtained from respective plasma and microsomal membrane samples derived from four 10-cm dishes of cells (total of about 20 mg cells proteins) using the protocol depicted in Fig. 1A. Staining of SDS-PAGE resolved protein samples derived from equal amounts of cell lysate, plasma membranes, and microsomal membranes, respectively, showed noticeable difference in protein banding patterns (Fig. 1B). Western blot analysis showed (Fig. 1C) that calnexin (an ER membrane protein) was enriched in both plasma and microsomal membrane fractions as compared with that in cell lysate, whereas actin beta (actb, a cytoplasmic protein) and lamin A/C (a nuclear membrane protein) were not detected in these samples. Thus, contamination of plasma or microsomal membranes by cytosol or nucleus was minimal.
Process of Membrane Proteins with the Centrifugal Reactor
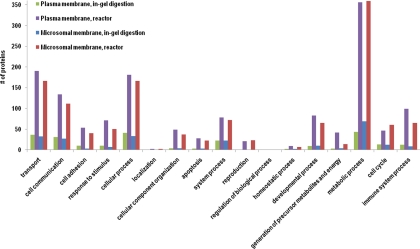
Plasma and microsomal membrane proteins obtained above were processed by the centrifugal proteomic reactor prior to LC-MS/MS analysis (Fig. 1D), and the results were contrasted with those derived from conventional in-gel digestion methodology (Fig. 2). There are 62,958 peptides (4651 unique peptides) and 101,794 peptides (5577 unique peptides), resulting in 945 and 955 proteins identified by centrifugal proteomic reactor from plasma and microsomal membranes, respectively; whereas there are only 9843 peptides (297 unique peptides) and 9947 peptides (397 unique peptides), resulting in 110 and 128 proteins identified by conventional in-gel digestion from plasma and microsomal membranes, respectively (Table I, supplemental Table S4). Notably, greater than 95% of membrane proteins detected by the in-gel method were also identified by the reactor approach, authenticating the efficacy of the centrifugal reactor (supplemental Table S1). Moreover, the centrifugal proteomic reactor method exhibited less contamination by cytosolic proteins. For instance, actin (actb), identified in plasma and microsomal membranes by in-gel digestion, was not seen either by Western blot or by the centrifugal proteomic reactor. The heatmaps were generated to compare the identified proteins between on-reactor digestion and in-gel digestion using peptide hits and peak intensities from label-free quantitation (supplemental Fig. S6). The patterns of the heatmap from peptide hits and peak intensities were similar to each other. Furthermore, some proteins not identified by MS/MS (no peptide hits because of low quantity) from in-gel digestion were still present in the MS scans and quantified by peak intensity from label-free quantitation.
Fig. 2.
Biological process classifications of the proteins identified by reactor and in-gel digestion from plasma and microsomal membrane.
Table I. Peptides, unique peptides, protein groups, predicted membrane proteins obtained from plasma and microsomal membrane using centrifugal reactor and in-gel digestion.
| Method samples | Centrifugal proteomic reactor |
In-gel digestion |
||
|---|---|---|---|---|
| Plasma membrane | Microsomal membrane | Plasma membrane | Microsomal membrane | |
| Peptides | 62958 | 101794 | 9843 | 9947 |
| Unique peptides | 4651 | 5577 | 297 | 397 |
| Protein groups | 945 | 955 | 110 | 128 |
| Membrane proteinsa | 591 (63%) | 447 (47%) | 72 (65%) | 70 (55%) |
| Integral to membranea | 391 (41%) | 300 (31%) | 56 (51%) | 50 (39%) |
| Proteins with TMsb | 500 (53%) | 396 (42%) | 64 (58%) | 66 (52%) |
a Predicted by the DAVID bioinformatics resources.
b Predicted by the TMHMM software.
The plasma and microsomal membrane fractions obtained were enriched for authentic membrane proteins. The DAVID Bioinformatics Resources suggested that of all proteins identified in the plasma and microsomal membranes, ∼50–60% were bona fide membrane proteins and ∼30–50% were integral membrane proteins, regardless whether the processing was done by in-gel digestion or proteomic reactor method (Table I). Additional analysis using the TMHMM software suggested that ∼40–50% of the identified membrane proteins contained transmembrane domain(s) (Table I). There was considerable overlap (59% of total proteins identified) between plasma and microsomal membrane fractions (supplemental Fig. S1). However, clear distinctions in protein composition did occur between the two fractions. Thus, of total proteins identified, 20% were found only in the plasma membranes, whereas 21% were found only in the microsomal membranes. The percentage of bona fide membrane proteins predicted by the DAVID bioinformatics resources also showed a distinction between the two fractions; whereas 77% in plasma membrane fraction were membrane proteins, the number for microsomal membrane fraction was only 16%. The nonintegral membrane proteins associated with the microsomal membrane fraction were mostly ribosome- and proteasome-related proteins that are involved in protein synthesis and degradation (supplemental Table S1B). Apparently, these proteins are abundant and bind tightly to the microsomal membrane even after high pH and 4 m urea wash. The membrane proteins involved in lipid metabolism and in ER and Golgi trafficking were identified exclusively in the microsomal membrane fraction and not in the plasma membrane fraction. The membrane proteins related to lipid metabolism included elongation of very long chain fatty acids protein 5 (Elovl5, lipid synthesis), lysophosphatidylglycerol acyltransferase 1 (LOC683760, lipid synthesis), solute carrier family 27 (fatty acid transporter), member 6 (Slc27a6, lipid transporting), arylacetamide deacetylase-like 1 (Aadacl1, lipid hydrolysis), and cytochrome P450 2D4 (Cyp2d4v1, lipid oxidation). The membrane proteins involved in ER and Golgi trafficking included ERGIC and golgi 3 (Ergic3), sec1 family domain-containing protein 1 (Scfd1), isoform 1 of protein YIF1B (Yif1b), coatomer subunit beta (Copb1), AP-2 complex subunit mu-1 (Ap2m1). On the other hand, membrane proteins detected exclusively in the plasma membrane fraction (and not in the microsomal membrane fraction) included ATPase and transporters, such as copper-transporting ATPase 1 (Atp7a), isoform D of plasma membrane calcium-transporting ATPase 1 (Atp2b1), sodium and hydrogen exchanger 1 (Slc9a1), sulfate anion transporter 1 (Slc26a1), similar to solute carrier family 37 (glycerol-3-phosphate transporter), member 2 (Slc37a2), sodium- and chloride-dependent glycine transporter 1 (Slc6a9), and chloride channel protein 5 and 7 (Clcn5, Clcn7), to name a few.
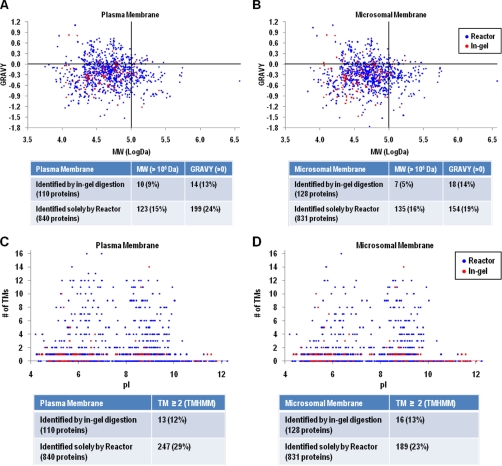
Membrane proteins that were only identified by the centrifugal proteomic reactor covered a broad spectrum of functions, ranging from cell communication, adhesion, apoptosis, energy, and intermediate metabolism (Fig. 2). Detailed analysis was performed to determine if the membrane proteins that were only identified by the centrifugal proteomic reactor method had particular characteristics such as size (MW), hydrophobicity (GRAVY), isoelectric point (pI), or the presence of transmembrane (TM) segments. As shown in Fig. 3A, among 110 proteins identified by in-gel digestion in plasma membranes, only 10 (9%) had MW >100 kDa and 14 (13%) with GRAVY > 0. The largest MW and GRAVY values identified by in-gel method were 251 kDa and 0.82, respectively. In comparison, the reactor approach identified 123 (15%) proteins with MW >100 kDa and 199 (24%) proteins with GRAVY > 0. The largest MW and GRAVY values identified by reactor approach were 3704 kDa and 1.10, respectively (supplemental Fig. S2A and S2B). Likewise, proteins with high MW and GRAVY values were detected by the reactor approach for microsomal membrane samples (Fig. 3B). These data indicate that the centrifugal proteomic reactor can identify the vast majority of the proteins observed by in gel digestion, but moreover can detect membrane proteins with high MW and hydrophobicity.
Fig. 3.
The grand average hydrophobicity (GRAVY) index and molecular weight (MW) (A, B), transmembrane domains (TMs) and isoelectric point (pI) (C, D) distributions of proteins identified by reactor and in-gel digestion from plasma and microsomal membrane. A, B, y-axis, GRAVY index value; x-axis, log MW (Da); C, D, y-axis, Number of transmembrane domains; x-axis, theoretical pI.
With respect to the number of TM segments, only 13 out of total 110 plasma membrane proteins (12%) identified by in-gel digestion had TMs ≥ 2. Whereas of total 840 proteins from plasma membrane identified only by the centrifugal proteomic reactor, 247 (29%) had TMs ≥ 2 (Fig. 3C). Similarly, detection of the proportion of microsomal membrane proteins having TMs ≥ 2 were increased from 13% by in-gel digestion method to 23% by the reactor approach (Fig. 3D). Thus, the majority of membrane proteins that failed to be detected by the in-gel digestion method were those proteins that contain two or more TM segments. Notably, the proteins with highest transmembrane domains (16 TMs), isoform 1 of multidrug resistance-associated protein 1 (Abcc1) and ATP-binding cassette, subfamily C (CFTR/MRP), member 3 (Abcc3), were only identified by the centrifugal reactor approach (supplemental Fig. S2C and S2D), which are involved in transporting various molecules across extra- and intra-cellular membranes. Another multiple transmembrane spanner protein involved in lipoprotein metabolism, the ATB-binding cassette A1 (Abca1, with 13 TMs predicted by TMHMM software), was also only detected by the reactor approach (supplemental Table S2). On the contrary, pI values (ranging from 4 to 12) of membrane proteins did not appear to have an impact on the recovery between the in-gel digestion and the reactor approach (Figs. 3C and 3D).
Microsomal Membrane Protein Analysis
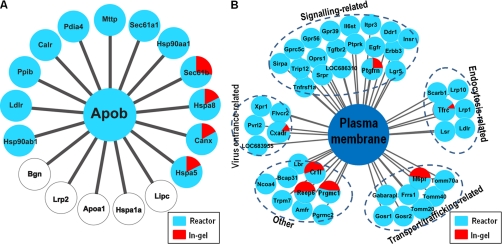
The ER and Golgi microsomes in hepatic cells act as a cradle for the assembly and secretion of VLDL. Each VLDL particle is composed of a single copy of apoB-100 and various amounts of lipids (such as phospholipids, TAG and cholesterol). Although apoB-100 does not contain any TM segments, the protein has been found in close association with ER microsomal membranes. It has been postulated that the association of apoB-100 with ER membrane represents the initial stage of lipid recruitment during VLDL assembly. Hence, we examined closely the microsomal membrane proteins identified between the in-gel digestion and the reactor methods. It was reported previously that there are 17 proteins interacting with apoB-100 as determined by in vivo and in vitro screening (www.hprd.org). The reactor approach identified 12 of the 17 proteins whereas the in-gel digestion method only detected four (Fig. 4A). The identified apoB-100 interactors included MTP (Mttp) and LDL receptor (Ldlr), which play key roles in apoB-100 lipidation and degradation processes (28, 44). Both of the proteins were detected only by the reactor approach and were missed by the in-gel digestion method. Other proteins that interacted with apoB-100 within the microsomes include molecular chaperones that play a role in apoB-100 folding (e.g. calnexin, calreticulin, GRP78/BiP, and protein disulfide isomerise) (Table II). Association of these chaperone proteins with apoB-100 has been observed previously by cross-linking experiments in hepatic cells (45). Hepatic lipase (Lipc), megalin (Lrp2), apoA-I (Apoa1), and two other proteins that were reported to interact with apoB-100 were not detected in the current study, suggesting that these proteins may not interact with membrane-associated apoB-100 during VLDL assembly, or are present in McA-RH7777 cells at low abundance.
Fig. 4.
Selected proteins identified from microsomal and plasma membranes. A, Proteins identified from microsomal membrane are mapped to the apob interactome in the human protein reference database (www.hprd.org). Twelve of 17 proteins apob-interacting proteins in human protein reference database (71%) were identified from the microsomal membrane fraction. The proteins identified by membrane proteomic reactor are shown in blue color; and the proteins identified by in-gel digestion are shown in red color. B, Receptors identified from plasma membrane.
Table II. List of proteins identified from microsomal membrane overlapped with apob-interacting proteins.
| IPI number | Gene | Protein name | Centrifugal reactor |
In-gel digestion |
||||
|---|---|---|---|---|---|---|---|---|
| Peptide | Unique peptide | Coverage | Peptide | Unique peptide | Coverage | |||
| IPI00555161 | Apob | Apolipoprotein B-100 | 135 | 18 | 5% | × | × | × |
| IPI00206624 | Hspa5 | 78 kDa glucose-regulated protein/Bip | 1803 | 43 | 56% | 385 | 14 | 29% |
| IPI00199636 | Canx | Calnexin | 1295 | 30 | 52% | 267 | 6 | 14% |
| IPI00208205 | Hspa8 | Heat shock cognate 71 kDa protein | 442 | 25 | 46% | 95 | 6 | 13% |
| IPI00364983 | Sec61b | Sec61 beta subunit | 57 | 3 | 38% | 22 | 2 | 26% |
| IPI00213644 | Ppib | Peptidyl-prolyl cis-trans isomerase B | 147 | 11 | 51% | × | × | × |
| IPI00191728 | Calr | Calreticulin | 433 | 15 | 45% | × | × | × |
| IPI00212220 | Pdia4 | Protein disulfide-isomerase A4/ERP72 | 244 | 23 | 42% | × | × | × |
| IPI00212316 | Mttp | Microsomal triglyceride transfer protein | 148 | 17 | 23% | × | × | × |
| IPI00231341 | Sec61a1 | Protein transport protein Sec61 subunit alpha isoform 1 | 182 | 7 | 15% | × | × | × |
| IPI00210566 | Hsp90aa1 | Heat shock protein HSP 90-alpha | 13 | 4 | 6% | × | × | × |
| IPI00203747 | Ldlr | Low-density lipoprotein receptor | 21 | 4 | 5% | × | × | × |
| IPI00471584 | Hsp90ab1 | Heat shock protein HSP 90-beta/Tumor rejection antigen 1 | 81 | 10 | 13% | × | × | × |
| Lipc | Hepatic lipase | × | × | × | × | × | × | |
| Bgn | Biglycan | × | × | × | × | × | × | |
| Lrp2 | Megalin | × | × | × | × | × | × | |
| Apoa1 | Apolipoprotein A I | × | × | × | × | × | × | |
| Hspa1a | Heat shock 70 kDa protein 1A | × | × | × | × | × | × | |
Microsomal membrane proteins that were detected only by the proteomic reactor also included those involved in glycerolipid biosynthesis, such as diacylglycerol acyltransferase (Dgat1), 1-acylglycerol-3-phosphate acyltransferases (Agpat1, Agpat2, and Agpat9), lysophosphatidylcholine acyltransferase (Lpcat3), lysophosphatidylglycerol acyltransferase (LOC683760), phosphatidylserine synthase (Ptdss1), phosphatidate cytidylyltransferase (Cds2), lysocardiolipin acyltransferase isoform (Lycat), and CDP-diacylglycerol-inositol 3-phosphatidyltransferase (Cdipt) (supplemental Table S1B). Those involved in sphingolipid synthesis included serine palmitoyltransferase subunit 1 (Sptlc1) and sphingolipid desaturase (Degs1). Proteins involved in fatty acid synthesis included fatty acid synthase (Fasn), very long-chain acyl-CoA synthetase (Slc27a2), acyl-CoA desaturase (Scd1), fatty acid desaturase (Fads2), elongation of very long chain fatty acids protein (Elovl5), long-chain-fatty-acid-CoA ligases (Acsl1 and Acsl5). Also, enzymes involved in cholesterol metabolism included HMG-CoA reductase (Hmgcr), 24-dehydrocholesterol reductase (Dhcr24), and 17-beta hydroxysteroid dehydrogenase (Hsd17b13). The products of these genes (such as Dgat1, Agpat1, and Scd1) have been shown to play an important role hepatic TAG production and VLDL metabolism (46).
Plasma Membrane Protein Analysis
Finally, we analyzed proteins associated with plasma membranes, particularly proteins with receptor function ranging from signaling, endocytosis, virus entrance, transport, and trafficking, and others (Fig. 4B). Of note, most of these receptor proteins failed to be detected by the in-gel method, among them several play important roles in lipoprotein metabolism, such as LDL receptor (Ldlr), lipolysis stimulated lipoprotein receptor (Lsr), scavenger receptor B1 (Scarb1), and LDL receptor-related protein 1 (Lrp1) (supplemental Table S3). Additionally, the important receptors for signaling transduction, such as tumor necrosis factor (TGF) receptor type 2, epidermal growth factor receptor and insulin receptor, were exclusively detected by centrifugal reactor (supplemental Table S3).
DISCUSSION
Development of Centrifugal Proteomic Reactor
The centrifugal proteomic reactor described in the current study has several operational advantages over the previously described column-based proteomic reactor (30, 32–36, 39). First, because all the processing steps, including delipidation, acidification, reduction, alkylation, digestion, and pH gradient elution, were conducted with proteins absorbed onto the SCX beads, the quantity of protein sample loading could be scaled up to hundreds of micrograms. Second, because the transition between each step was achieved by simple centrifugations, the entire process prior to LC-MS/MS could be accomplished in 2½ hours. Third, the bench-top microfuge-based proteomic reactor eliminated the need for specialized equipment allowing the procedure to be performed in any proteomic laboratory. Using this centrifugal proteomic reactor, we were able to identify over 900 proteins associated with plasma or microsomal membranes from 20 μg starting materials, which was eight times more than the number of proteins identified by our conventional in-gel digestion method. The in-gel digestion method conducted in the current study and reported previously yielded as low as 2.22 (47) or 0.65 (19) transmembrane proteins per microgram membrane sample. In contrast, our centrifugal proteomic reactor identified 25 and 19.8 transmembrane proteins per μg of plasma and microsomal membrane samples, respectively. Moreover, our centrifugal proteomic reactor identified 50–60% authentic membrane proteins from the samples, which is far greater than the 20–30% identification reported previously using combined SDS-PAGE, 2-D LC, and 3-D LC methods for mouse liver microsomes (22). The low recovery of membrane proteins with the in-gel digestion method is most likely attributable to (1) inefficient delipidation or solubilization of proteins during gel electrophoresis, (2) inadequate resolution of proteins with high MW and/or hydrophobicity by the SDS-PAGE, and (3) sample loss as a result of incomplete transfer and extraction of proteins from the polyacrylamide gel. All of these shortcomings associated with in-gel digestion were circumvented by the proteomic reactor method. Moreover, this approach is applicable to other samples, for example we are currently using this reactor for total cell and tissue lysate and secreted protein analysis (unpublished results).
Improved Identification of Microsomal Membrane Proteins
The ER and Golgi microsomes are the major subcellular organelle for the biosynthesis of various lipids, including glycerolipids, TAG, cholesterol, and sphingolipids. Regulation of the biosynthesis of these lipids has a profound impact on hepatic lipid homeostasis; one of the homeostatic aspects of hepatic lipid metabolism is VLDL assembly and secretion (48). A unique feature associated with VLDL assembly process is that although the structural protein apoB-100 contains no transmembrane segments the protein behaves like a membrane protein, thus tightly binds to membranes during the VLDL assembly process (23). For this reason, apoB-100 has been dubbed as a “secretory membrane protein.” The centrifugal proteomic reactor method developed in the current study has identified multiple enzymes that are involved in glycerolipid biosynthesis (mainly acyltransferases) as well as cholesterol and sphingolipid biosynthesis. Some of these lipid synthesis enzymes are known to play an important role in VLDL assembly and secretion. The centrifugal proteomic reactor method has also identified several protein factors that play key roles in VLDL assembly and secretion, including MTP, LDL receptor, and apoE in addition to apoB-100. None of these proteins were detected by the conventional in-gel digestion method. Altogether, studies with proteins associated with the McA-RH7777 microsomal membranes provide strong evidence for the analytical power of the centrifugal proteomic reactor.
Several previously published reports have described the analysis of plasma membrane (47, 49, 50) and microsomal membrane proteins (49, 51, 52) of mouse or rat liver origin. Two observations are noteworthy when the current data are compared with those reported previously. First, the number (257) and percentage (56%) of transmembrane proteins identified in the plasma membrane fraction using the current reactor method are much greater than previously reported (supplemental Fig. S4A). Close inspection of our dataset in comparison with the reported datasets revealed marked improvement in identifying proteins involved in lipid and lipoprotein metabolism, such as the extremely large and hydrophobic apolipoprotein B-100 (Apob), lysocardiolipin acyltransferase isoform 1 (Lycat), lysophosphatidylglycerol acyltransferase 1(LOC683760), 1-acylglycerol-3-phosphate O-acyltransferase 1, 2, and θ(Agpat1, Agpat2, Agpat9). Elongation of very long chain fatty acids protein 5 (Elovl5), long chain fatty acid CoA ligase 3 and 4 (Acsl3, Acsl4) and sphingolipid Δ(4)-desaturase DES1 (Degs1). These proteins were identified exclusively in our microsomal membrane data. Many membrane proteins involved in transportation and signaling transduction, for example, proteins from ATP-binding cassette transporter family (Abca1, Abcb1, Abcb1b, Abcc1, Abcc4, Abcc5 and Abcf2), proteins from solute carrier (SLC) family (Slc1a4, Slc1a5, Slc2a1, Slc3a2, Slc4a2, Slc4a4, Slc6a6, Slc6a8, Slc6a9, Slc7a1, Slc7a5, Slc7a11, Slc9a1, Slc9a3r1, Slc12a7, Slc16a10, Slc20a1, Slc23a1, Slc25a24, Slc27a4, Slc29a2, Slc30a7, Slc35e1, Slc37a2, Slc38a2, Slc38a10, Slc39a1, Slc39a7, and Slc39a10) and proteins from G protein-coupled receptor family (Gprc5c, Gpr39, and Gpr56) were identified only in our plasma membrane data (see supplemental Fig. S3, in which “unique” means proteins not overlapping between different datasets). Second, the percentage of microsomal membrane proteins with TMs ≥ 1 identified by the current reactor method was also much greater than the published value (supplemental Fig. S4B), with one exception of rat microsomal membrane proteins (52). In this report (“supplemental Fig. S4B: Total 321 proteins (Ref. 52)”), only protein with TMs ≥ 2 in the range of pH 4.0–8.5 fractions were available (supplemental Table in (52)). The lack of complete information of all proteins with TMs in this report (52) precludes an adequate comparison of analytical efficacy between the two studies. We compared our 955 proteins from the microsomal membrane (from one single microsomal membrane reactor experiment) to the 1269 total proteins by Mathias et al. (12) (which were identified by a combination of four distinct triton X-114 phase separation fractions: microsome fraction <MF>, pellet fraction <PF>, detergent phase <DP>, and aqueous phase <AP>), using PROMT. As shown in supplemental Fig. S5, 538 proteins overlapped between the two datasets, and the proteins predicted with TMs were about 40% in the total proteins.
The grand average of hydropathicity index (GRAVY) and number of transmembrane domains (TMs) are the two major parameters for assessing protein hydrophobicity. The GRAVY indicates the hydrophobicity of the proteins, calculated by adding the hydropathy value for each residue and dividing by the length of the sequence: positive GRAVY (hydrophobic), negative GRAVY (hydrophilic). TMs are the number of transmembrane domains predicted by the TMHMM software. We calculated the correlation coefficients of GRAVY and TMs for our data and others. The correlation coefficients between GRAVY and TMs of plasma membrane from our data, Cao et al. (47), MPIR (49) and Lee et al. (50) are 0.58, 0.66, 0.50, and 0.52, respectively, and the correlation coefficients between GRAVY and TMs of microsomal membrane from our data, Mathias et al. (12), MICR (49), Ma et al. (51) and Lee et al. (52) are 0.56, 0.54 0.33, 0.49, and 0.55, respectively. Taken together, there is a relatively medium correlation between GRAVY and TMs.
In summary, the present centrifugation-based proteomic reactor provides an improved method in identifying membrane proteins with high degree of hydrophobicity and number of transmembrane domains. The availability of this novel technology will facilitate qualitative and quantitative analysis of hydrophobic and membrane proteins involved in lipid and lipoprotein metabolism.
Acknowledgments
We thank Dr. Janice Mayne for editorial assistance.
Footnotes
* This work was supported by grants from NSERC and the J.-Louis Lévesque Foundation. DF would like to acknowledge a Canada Research Chair in Proteomics and Systems Biology. ZY would like to acknowledge a China-Canada Health Research Initiative Grant (CCI 92211) and an Operating Grant (NMD 15486) from Canadian Institute for Health Research. HZ and ZN would like to acknowledge the post-doctorate fellow award from the CIHR training program in neurodegenerative lipidomics.
 This article contains supplemental Figs. S1 to S6 and Tables S1 to S4.
This article contains supplemental Figs. S1 to S6 and Tables S1 to S4.
1 The abbreviations used are:
- ER
- endoplasmic reticulum
- HPLC-ESI-MS/MS
- high performance liquid chromatography coupled electrospray tandem mass spectrometry
- VLDL
- very low density lipoproteins
- TAG
- triglycerides
- CHAPS
- 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonate
- GRAVY
- the grand average hydrophobicity.
REFERENCES
- 1. Yildirim M. A., Goh K. I., Cusick M. E., Barabási A. L., Vidal M. (2007) Drug-target network. Nat. Biotechnol. 25, 1119–1126 [DOI] [PubMed] [Google Scholar]
- 2. Fagerberg L., Jonasson K., von Heijne G., Uhlén M., Berglund L. (2010) Prediction of the human membrane proteome. Proteomics 10, 1141–1149 [DOI] [PubMed] [Google Scholar]
- 3. Bakheet T. M., Doig A. J. (2009) Properties and identification of human protein drug targets. Bioinformatics 25, 451–457 [DOI] [PubMed] [Google Scholar]
- 4. Lu B., McClatchy D. B., Kim J. Y., Yates J. R., 3rd (2008) Strategies for shotgun identification of integral membrane proteins by tandem mass spectrometry. Proteomics 8, 3947–3955 [DOI] [PubMed] [Google Scholar]
- 5. Helbig A. O., Heck A. J., Slijper M. (2010) Exploring the membrane proteome–challenges and analytical strategies. J. Proteomics 73, 868–878 [DOI] [PubMed] [Google Scholar]
- 6. Zhang L., Xie J., Wang X., Liu X., Tang X., Cao R., Hu W., Nie S., Fan C., Liang S. (2005) Proteomic analysis of mouse liver plasma membrane: use of differential extraction to enrich hydrophobic membrane proteins. Proteomics 5, 4510–4524 [DOI] [PubMed] [Google Scholar]
- 7. Zhang L. J., Wang X. E., Peng X., Wei Y. J., Cao R., Liu Z., Xiong J. X., Yin X. F., Ping C., Liang S. (2006) Proteomic analysis of low-abundant integral plasma membrane proteins based on gels. Cell. Mol. Life Sci. 63, 1790–1804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Avramoglu R. K., Nimpf J., McLeod R. S., Ko K. W., Wang Y., FitzGerald D., Yao Z. (1998) Functional expression of the chicken low density lipoprotein receptor-related protein in a mutant chinese hamster ovary cell line restores toxicity of Pseudomonas exotoxin A and degradation of alpha2-macroglobulin. J. Biol. Chem. 273, 6057–6065 [DOI] [PubMed] [Google Scholar]
- 9. Yao Y., Hong S., Zhou H., Yuan T., Zeng R., Liao K. (2009) The differential protein and lipid compositions of noncaveolar lipid microdomains and caveolae. Cell Res. 19, 497–506 [DOI] [PubMed] [Google Scholar]
- 10. Stolz D. B., Jacobson B. S. (1992) Examination of transcellular membrane protein polarity of bovine aortic endothelial cells in vitro using the cationic colloidal silica microbead membrane-isolation procedure. J. Cell Sci. 103, (Pt 1), 39–51 [DOI] [PubMed] [Google Scholar]
- 11. Schindler J., Lewandrowski U., Sickmann A., Friauf E., Nothwang H. G. (2006) Proteomic analysis of brain plasma membranes isolated by affinity two-phase partitioning. Mol. Cell. Proteomics 5, 390–400 [DOI] [PubMed] [Google Scholar]
- 12. Mathias R. A., Chen Y. S., Kapp E. A., Greening D. W., Mathivanan S., Simpson R. J. (2011) Triton X-114 phase separation in the isolation and purification of mouse liver microsomal membrane proteins. Methods 54, 396–406 [DOI] [PubMed] [Google Scholar]
- 13. Nagaraj N., Lu A., Mann M., Wiśniewski J. R. (2008) Detergent-based but gel-free method allows identification of several hundred membrane proteins in single LC-MS runs. J. Proteome Res. 7, 5028–5032 [DOI] [PubMed] [Google Scholar]
- 14. Lu A., Wiśniewski J. R., Mann M. (2009) Comparative proteomic profiling of membrane proteins in rat cerebellum, spinal cord, and sciatic nerve. J. Proteome Res. 8, 2418–2425 [DOI] [PubMed] [Google Scholar]
- 15. Chen E. I., McClatchy D., Park S. K., Yates J. R., Iii (2008) Comparisons of Mass Spectrometry Compatible Surfactants for Global Analysis of the Mammalian Brain Proteome. Anal. Chem. 80, 8694–8701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zhang N., Chen R., Young N., Wishart D., Winter P., Weiner J. H., Li L. (2007) Comparison of SDS- and methanol-assisted protein solubilization and digestion methods for Escherichia coli membrane proteome analysis by 2-D LC-MS/MS. Proteomics 7, 484–493 [DOI] [PubMed] [Google Scholar]
- 17. Wu C. C., MacCoss M. J., Howell K. E., Yates J. R., 3rd (2003) A method for the comprehensive proteomic analysis of membrane proteins. Nat. Biotechnol. 21, 532–538 [DOI] [PubMed] [Google Scholar]
- 18. Dormeyer W., van Hoof D., Mummery C. L., Krijgsveld J., Heck A. J. (2008) A practical guide for the identification of membrane and plasma membrane proteins in human embryonic stem cells and human embryonal carcinoma cells. Proteomics 8, 4036–4053 [DOI] [PubMed] [Google Scholar]
- 19. Chick J. M., Haynes P. A., Bjellqvist B., Baker M. S. (2008) A combination of immobilised pH gradients improves membrane proteomics. J. Proteome Res. 7, 4974–4981 [DOI] [PubMed] [Google Scholar]
- 20. Leth-Larsen R., Lund R. R., Ditzel H. J. (2010) Plasma membrane proteomics and its application in clinical cancer biomarker discovery. Mol. Cell. Proteomics 9, 1369–1382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Wong D. M., Adeli K. (2009) Microsomal proteomics. Methods Mol. Biol. 519, 273–289 [DOI] [PubMed] [Google Scholar]
- 22. Zgoda V. G., Moshkovskii S. A., Ponomarenko E. A., Andreewski T. V., Kopylov A. T., Tikhonova O. V., Melnik S. A., Lisitsa A. V., Archakov A. I. (2009) Proteomics of mouse liver microsomes: performance of different protein separation workflows for LC-MS/MS. Proteomics 9, 4102–4105 [DOI] [PubMed] [Google Scholar]
- 23. Tran K., Thorne-Tjomsland G., DeLong C. J., Cui Z., Shan J., Burton L., Jamieson J. C., Yao Z. (2002) Intracellular assembly of very low density lipoproteins containing apolipoprotein B100 in rat hepatoma McA-RH7777 cells. J. Biol. Chem. 277, 31187–31200 [DOI] [PubMed] [Google Scholar]
- 24. Tran K., Wang Y., DeLong C. J., Cui Z., Yao Z. (2000) The assembly of very low density lipoproteins in rat hepatoma McA-RH7777 cells is inhibited by phospholipase A2 antagonists. J. Biol. Chem. 275, 25023–25030 [DOI] [PubMed] [Google Scholar]
- 25. Wang Y., Tran K., Yao Z. (1999) The activity of microsomal triglyceride transfer protein is essential for accumulation of triglyceride within microsomes in McA-RH7777 cells. A unified model for the assembly of very low density lipoproteins. J. Biol. Chem. 274, 27793–27800 [DOI] [PubMed] [Google Scholar]
- 26. Bou, Khalil M., Blais A., Figeys D., Yao Z. (2010) Lipin-The bridge between hepatic glycerolipid biosynthesis and lipoprotein metabolism. Biochim. Biophys. Acta 1801, 1249–1259 [DOI] [PubMed] [Google Scholar]
- 27. Sundaram M., Yao Z. (2010) Recent progress in understanding protein and lipid factors affecting hepatic VLDL assembly and secretion. Nutr. Metab. 7, 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Gillian-Daniel D. L., Bates P. W., Tebon A., Attie A. D. (2002) Endoplasmic reticulum localization of the low density lipoprotein receptor mediates presecretory degradation of apolipoprotein B. Proc. Natl. Acad. Sci. U.S.A. 99, 4337–4342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Borén J., Graham L., Wettesten M., Scott J., White A., Olofsson S. O. (1992) The assembly and secretion of ApoB 100-containing lipoproteins in Hep G2 cells. ApoB 100 is cotranslationally integrated into lipoproteins. J. Biol. Chem. 267, 9858–9867 [PubMed] [Google Scholar]
- 30. Ethier M., Hou W., Duewel H. S., Figeys D. (2006) The proteomic reactor: a microfluidic device for processing minute amounts of protein prior to mass spectrometry analysis. J. Proteome Res. 5, 2754–2759 [DOI] [PubMed] [Google Scholar]
- 31. Zhou H., Hou W., Lambert J. P., Tian R., Figeys D. Analysis of low-abundance proteins using the proteomic reactor with pH fractionation. Talanta 80, 1526–1531 [DOI] [PubMed] [Google Scholar]
- 32. Zhou H., Hou W., Lambert J. P., Figeys D. (2010) New ammunition for the proteomic reactor: strong anion exchange beads and multiple enzymes enhance protein identification and sequence coverage. Anal. Bioanal. Chem. 397, 3421–3430 [DOI] [PubMed] [Google Scholar]
- 33. Tian R., Wang S., Elisma F., Li L., Zhou H., Wang L., Figeys D. (2010) Rare cell proteomic reactor applied to SILAC based quantitative proteomic study of human embryonic stem cell differentiation. Mol. Cell. Proteomics 10, [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Vasilescu J., Zweitzig D. R., Denis N. J., Smith J. C., Ethier M., Haines D. S., Figeys D. (2007) The proteomic reactor facilitates the analysis of affinity-purified proteins by mass spectrometry: application for identifying ubiquitinated proteins in human cells. J. Proteome Res. 6, 298–305 [DOI] [PubMed] [Google Scholar]
- 35. Zhou H., Hou W., Denis N. J., Zhou H., Vasilescu J., Zou H., Figeys D. (2009) Glycoproteomic Reactor for Human Plasma. J. Proteome Res. 8, 556–566 [DOI] [PubMed] [Google Scholar]
- 36. Zhou H., Elisma F., Denis N. J., Wright T. G., Tian R., Zhou H., Hou W., Zou H., Figeys D. (2010) Analysis of the subcellular phosphoproteome using a novel phosphoproteomic reactor. J. Proteome Res. 9, 1279–1288 [DOI] [PubMed] [Google Scholar]
- 37. Hou W., Ethier M., Smith J. C., Sheng Y., Figeys D. (2007) Multiplexed proteomic reactor for the processing of proteomic samples. Anal. Chem. 79, 39–44 [DOI] [PubMed] [Google Scholar]
- 38. Zhong S., Magnolo A. L., Sundaram M., Zhou H., Yao E. F., Di Leo E., Loria P., Wang S., Bamji-Mirza M., Wang L., McKnight C. J., Figeys D., Wang Y., Tarugi P., Yao Z. (2010) Nonsynonymous mutations within APOB in human familial hypobetalipoproteinemia: evidence for feedback inhibition of lipogenesis and postendoplasmic reticulum degradation of apolipoprotein B. J. Biol. Chem. 285, 6453–6464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Zhou H., Hou W., Lambert J. P., Tian R., Figeys D. (2010) Analysis of low-abundance proteins using the proteomic reactor with pH fractionation. Talanta 80, 1526–1531 [DOI] [PubMed] [Google Scholar]
- 40. Zhou H., Chen B., Li R. X., Sheng Q. H., Li S. J., Zhang L., Li L., Xia Q. C., Wang H. Y., Zeng R. (2005) Large-scale identification of human biliary proteins from a cholesterol stone patient using a proteomic approach. Rapid Commun. Mass Spectrom. 19, 3569–3578 [DOI] [PubMed] [Google Scholar]
- 41. Cox J., Mann M. (2008) MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26, 1367–1372 [DOI] [PubMed] [Google Scholar]
- 42. Huang da W., Sherman B. T., Lempicki R. A. (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57 [DOI] [PubMed] [Google Scholar]
- 43. McLeod R. S., Wang Y., Wang S., Rusiñol A., Links P., Yao Z. (1996) Apolipoprotein B sequence requirements for hepatic very low density lipoprotein assembly. Evidence that hydrophobic sequences within apolipoprotein B48 mediate lipid recruitment. J. Biol. Chem. 271, 18445–18455 [DOI] [PubMed] [Google Scholar]
- 44. Wang S., McLeod R. S., Gordon D. A., Yao Z. (1996) The microsomal triglyceride transfer protein facilitates assembly and secretion of apolipoprotein B-containing lipoproteins and decreases cotranslational degradation of apolipoprotein B in transfected COS-7 cells. J. Biol. Chem. 271, 14124–14133 [PubMed] [Google Scholar]
- 45. Linnik K. M., Herscovitz H. (1998) Multiple molecular chaperones interact with apolipoprotein B during its maturation. The network of endoplasmic reticulum-resident chaperones (ERp72, GRP94, calreticulin, and BiP) interacts with apolipoprotein b regardless of its lipidation state. J. Biol. Chem. 273, 21368–21373 [DOI] [PubMed] [Google Scholar]
- 46. Coleman R. A., Lee D. P. (2004) Enzymes of triacylglycerol synthesis and their regulation. Prog. Lipid Res. 43, 134–176 [DOI] [PubMed] [Google Scholar]
- 47. Cao R., He Q., Zhou J., He Q., Liu Z., Wang X., Chen P., Xie J., Liang S. (2008) High-throughput analysis of rat liver plasma membrane proteome by a nonelectrophoretic in-gel tryptic digestion coupled with mass spectrometry identification. J. Proteome Res. 7, 535–545 [DOI] [PubMed] [Google Scholar]
- 48. Yao Z., McLeod R. S. (1994) Synthesis and secretion of hepatic apolipoprotein B-containing lipoproteins. Biochim. Biophys. Acta 1212, 152–166 [DOI] [PubMed] [Google Scholar]
- 49. Peng L., Kapp E. A., Fenyö D., Kwon M. S., Jiang P., Wu S., Jiang Y., Aguilar M. I., Ahmed N., Baker M. S., Cai Z., Chen Y. J., Van Chi P., Chung M. C., He F., Len A. C., Liao P. C., Nakamura K., Ngai S. M., Paik Y. K., Pan T. L., Poon T. C., Salekdeh G. H., Simpson R. J., Sirdeshmukh R., Srisomsap C., Svasti J., Tyan Y. C., Dreyer F. S., McLauchlan D., Rawson P., Jordan T. W. (2010) The Asia Oceania Human Proteome Organisation Membrane Proteomics Initiative. Preparation and characterisation of the carbonate-washed membrane standard. Proteomics 10, 4142–4148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Lee A., Kolarich D., Haynes P. A., Jensen P. H., Baker M. S., Packer N. H. (2009) Rat liver membrane glycoproteome: enrichment by phase partitioning and glycoprotein capture. J. Proteome Res. 8, 770–781 [DOI] [PubMed] [Google Scholar]
- 51. Ma J., Hou C., Sun L., Tao D., Zhang Y., Shan Y., Liang Z., Zhang L., Yang L., Zhang Y. (2010) Coupling formic acid assisted solubilization and online immobilized pepsin digestion with strong cation exchange and microflow reversed-phase liquid chromatography with electrospray ionization tandem mass spectrometry for integral membrane proteome analysis. Anal. Chem. 82, 9622–9625 [DOI] [PubMed] [Google Scholar]
- 52. Lee H. J., Kwon M. S., Lee E. Y., Cho S. Y., Paik Y. K. (2008) Establishment of a PF2D-MS/MS platform for rapid profiling and semiquantitative analysis of membrane protein biomarkers. Proteomics 8, 2168–2177 [DOI] [PubMed] [Google Scholar]