Abstract
Leucine-rich repeat kinase 2 (LRRK2), a large and complex protein that possesses two enzymatic properties, kinase and GTPase, is one of the major genetic factors in Parkinson’s disease (PD). Here, we characterize the kinetic and catalytic mechanisms of truncated wild-type LRRK2 (t-wt) and its most common mutant G2019S (t-G2019S) with a structural interpretation of the kinase domain. First, the substitution of threonine by serine in the LRRKtide peptide results in a much less efficient substrate as demonstrated by a 26-fold decrease in kcat and a 6-fold decrease in binding affinity. The significant decrease in kcat is attributed to a slow chemical transfer step as evidenced by the inverse SKIE in the proton inventory and pL- (pH or pD) dependent studies. The shape of the proton inventory and pL profile clearly signals the involvement of a general base (pKa of 7.5) in the catalysis with a low fractionation factor in the ground state. Next, we report for the first time that the increased kinase activity of the mutant G2019S is substrate dependent. Homology modeling of the kinase domain (open and closed forms) and structural analysis of the docked peptide substrates suggest that electrostatic interactions play an important role in substrate recognition, which is affected by G2019S and may directly influence the kinetic properties of the enzyme. Finally, GTPase activity of the mutant t-G2019S was characterized and the mutation modestly decreases GTPase activity without significantly affecting GTP binding affinity.
Parkinson’s disease (PD), characterized by tremor, rigidity, bradykinesia and postural instability, is the second most common neurodegenerative disorder after Alzheimer’s disease (AD). It affects over 1 million Americans and more than 60,000 patients are newly diagnosed each year. PD is caused by a loss of dopaminergic neurons in the substantia nigra. Once damaged, these neurons stop producing dopamine, an essential neurotransmitter, and compromise the brain's ability to control movement. Mutations in several genes have been genetically linked to PD in recent years (1). Among them, the leucine-rich repeat kinase2 (LRRK2) has emerged as the most relevant player in PD pathogenesis. At least 40 mutations in LRRK2 have been identified in the most common familial forms and some sporadic forms of PD, and have been associated with typical idiopathic, late-onset PD (2–6). LRRK2 is a large and complex protein containing several distinct domains, including a leucine-rich repeat (LRR) domain, a Roc domain followed by its associated COR domain, a kinase domain, and a C-terminal WD40 domain (2, 7). LRRK2 is unusual in that it encodes two distinct but functionally linked enzymes: a protein kinase and a GTPase (7–11). Several lines of evidence have suggested that kinase activity of LRRK2 plays a critical role in the pathogenesis of PD: (i) Several mutations demonstrate increased kinase activity that is correlated with increased neurotoxicity in neurons (7, 12, 13); and (ii) Kinase inhibitors protect dopaminergic neuron loss in PD animal models (14). However, most of the LRRK2 mutations do not manifest their effects by simply increasing kinase activity. Even with the considerable debate about the role of LRRK2 kinase activity played in the pathogenesis of PD, identification of LRRK2 inhibitors has still become a priority in drug discovery for the treatment of PD. In addition, the identification of LRRK2 inhibitors can provide valuable information for our understanding of LRRK2’s functions. Determining the kinetic and catalytic mechanisms of LRRK2 will support structure-based inhibitor design. Herein, we integrate steady-state kinetics with both solvent kinetic isotope effects and molecular modeling studies for both truncated LRRK2 and the most common mutant G2019S. Specifically, we report results of studies aimed at (i) understanding the kinetic and catalytic mechanisms of LRRK2 and (ii) revealing critical structural features of LRRK2.
MATERIALS AND METHODS
Materials
ATP, ADP, AMP-PNP, DTT, magnesium chloride, HEPES, and bovine serum albumin were purchased from Sigma (St. Louis, MO). GTP was from Bioline (Taunton, MA). Peptides LRRKtide (RLGRDKYKTLRQIRQ), LRRKtideS (RLGRDKYKSLRQIRQ) and LRRKtideA (RLGRDKYKALRQIRQ) and PO4-LRRKtide (RLGRDKYK(PO4)TLRQIRQ) were purchased from American Peptide (Sunnyvale, CA). PLK-peptide (PLK-derived peptide with a motif of RRRSLLE), Eu-anti-phospho-PLK, [γ-33P]-ATP, and [α-33P]-GTP were from Perkin Elmer (Boston, MA). Truncated wild-type LRRK2 (aa 970–2527) and mutant t-G2019S (aa 970–2527) expressed in baculovirus system were purchased from Invitrogen.
TR-FRET Assay of LRRK2-Catalyzed PLK-peptide Phosphorylation
The kinase assay for phosphorylation of PLK-peptide (PLK-derived peptide with a motif of RRRSLLE) was conducted in buffer containing 20 mM HEPES (pH 7.4), 50 mM NaCl, 10 mM MgCl2, 1 mM DTT, BSA 0.5 mg/ml, 1 mM beta-Gly-PO4, PLK-peptide, and ATP. Beta-Gly-PO4 is a phosphatase inhibitor and was added to block phosphatases. PLK-peptide and ATP were used at various concentrations as indicated in the results section. The reactions were conducted in duplicate, initiated by the addition of 4 nM LRRK2, and incubated at room temperature for 4 h. The reactions were stopped by the addition of 10 mM EDTA and incubated with 2 nM Eu-anti-phospho-PLK for 2 h. The TR-FRET signal was read by an EnVision plate reader (PerkinElmer). In all cases, reaction progress curves for production of phospho-PLK-peptide were linear over at last 6 h and allowed calculation of initial velocities.
Kinetic Analysis of LRRK2-Catalyzed LRRKtide and LRRKtideS Phosphorylation
The kinase assays for phosphorylation of LRRKtide (RLGRDKYKTLRQIRQ) or LRRKtideS (RLGRDKYKSLRQIRQ) were conducted in buffer containing 20 mM HEPES (pH 7.4), 50 mM NaCl, 10 mM MgCl2, 1 mM DTT, BSA 0.5 mg/ml, 1 mM beta-Gly-PO4, LRRKtide, ATP, and [γ-33P]-ATP. Peptidic substrates and ATP were used at various concentrations as indicated in the results section and the ratio of ATP to [γ-33P]-ATP was kept constant at all ATP concentrations (250 µM ATP/5 µCi [γ-33P]-ATP). The reactions were conducted in duplicate, initiated by the addition of 6 nM LRRK2, and incubated at room temperature for 150 min. The reactions were stopped by the addition of 20 mM EDTA and the mixture was transferred to a multiscreen PH filtration plate (Millipore, Billerica, MA) and washed six times with 75 mM H3PO4. The plate was dried, filters were removed, and the samples were counted with a scintillation counter. Background reactions were conducted in the absence of LRRK2. In all cases, reaction progress curves for production of phospho-LRRKtide or phospo-LRRKtideS were linear over at last 60 minutes and allowed calculation of initial velocities.
Isotope Exchange Studies of LRRK2-Catalyzed LRRKtide Phosphorylation
[γ-33P]-ATP was used as the labeled substrate in these studies and the exchange rates of radioactive ATP with PO4-LRRKtide were measured in buffer containing 20 mM HEPES (pH 7.4), 50 mM NaCl, 10 mM MgCl2, 1 mM DTT, BSA 0.5 mg/ml, and 1 mM beta-Gly-PO4 at several different concentrations of reactants while maintaining the concentration of the other reactants constant. The varied reactants were maintained at a constant ratio of 20. The measurements were repeated for different pairs of reactants. In general, the study was carried out by first mixing all the reactants of ADP, ATP, LRRKtide, and PO4-LRRKtide at the desired concentrations. Then a trace amount of [γ-33P]-ATP was added before the reaction was initiated by the addition of the enzyme. The exchange reactions were carried out in duplicate, incubated at room temperature for 45 min and stopped by the addition of 20 mM EDTA. The mixtures were transferred to a multiscreen PH filtration plate (Millipore, Billerica, MA) and washed six times with 75 mM H3PO4. The plates were dried, filters were removed, and the samples were counted with a scintillation counter. Background reactions were conducted in the absence of LRRK2. In all cases, exchange rate was linear over at least 60 minutes and allowed calculation of the initial exchange velocity.
Kinetic Analysis of LRRK2-Catalyzed GTP Hydrolysis
The GTPase assay was conducted in buffer containing 20 mM Tris (pH 7.4), 50 mM NaCl, 10 mM MgCl2, 1 mM DTT, BSA 0.5 mg/ml, GTP, and [α-33P]-GTP. The reactions were conducted in triplicate, initiated by the addition of 30 nM LRRK2, and incubated at room temperature for 20 min. The reactions were stopped by the addition of 20 mM EDTA, and the product [α-33P]-GDP was separated from [α-33P]-GTP by PEI-cellulose thin layer chromatography (TLC) (Sigma, ST. Louis, MO) developed with 0.5 M KH2PO4 (pH3.4) developing buffer and analyzed by scintillation counter. In all cases, reaction progress curves for GTP hydrolysis were linear over at least 30 minutes and allowed calculation of initial velocities.
Mass Spectrometry Analysis of Cysteine Residues Modified by N-ethylmaleimide (NEM)
t-Wt LRRK2 was treated with 2 mM cysteine-specific inhibitor N-ethylmaleimide (NEM) at room temperature for 30 min. The sample was then reduced with DTT and alkylated with iodoacetamide and subjected to SDS-PAGE for mass spectrometry analysis.
Modeling of LRRK2 Kinase Domain
LRRK2 kinase domain between residues 1879–2138 was modeled using MODELLER (15). Briefly, the main criteria in homology modeling were template selection and sequence alignment between the target and the template. The top three hits based on sequence identity were yeast snf1 (39% identity), ack1 kinase (33% identity) and B-Raf (33% identity). In this case, the template of B-Raf kinase which had 33% sequence identity was used for homology modeling since this enzyme had higher sequence conservation around the ATP binding site region compared to the other kinases. The Cα RMSD and the backbone RMSD deviations for the model and the template crystal structure were < 1.0 Å and < 1.2 Å respectively. The best model was subjected to geometric evaluations using PROCHECK by calculating the Ramachandran plots (16). Standard bond lengths and bond angles of the model were determined using WHAT IF (17) with an RMS-Z score of 0.889 and 0.91 suggesting that the model is of high quality. ATP was docked into the binding pocket and LRRKtide, LRRKtideS and PLK were modeled based on the x-ray structures of protein kinase A (PKA) complexed with a peptidic inhibitor (1atp.pdb). Individual structues with bound peptides were subjected to 2000 cyles of energy minimization using a steepest descent protocol using DESMOND 3.0 (Schrödinger inc. and D.E Shaw research). The final structures obtained were used for further analysis.
Data Analysis – Basic Equations
Data were analyzed by nonlinear least-squares using either Sigma-Plot or Grafit software packages. Standard kinetic mechanisms for two-substrate reactions and their rate equations are shown below:
- Ping-Pong:
KA and KB are Michaelis constants.(1) - Rapid Equilibrium Ordered:
KA and KB are substrate dissociation constants from EA and EB, respectively.(2) - Rapid Equilibrium Random/Steady-State Ordered:
(3)
For rapid equilibrium systems, KA, KB, αKA, and αKB are substrate dissociation constants from EA, EB, and EAB; for steady-state systems, KA is substrate dissociation constant from EA, αKA and αKB are Michaelis constants. See Segel for definitions of mechanisms, substrate dissociation constants, and α (18).
RESULTS
t-WT LRRK2- and the Mutant t-G2019S-Catalyzed Phosphorylation of LRRKtide– Initial Velocity Studies
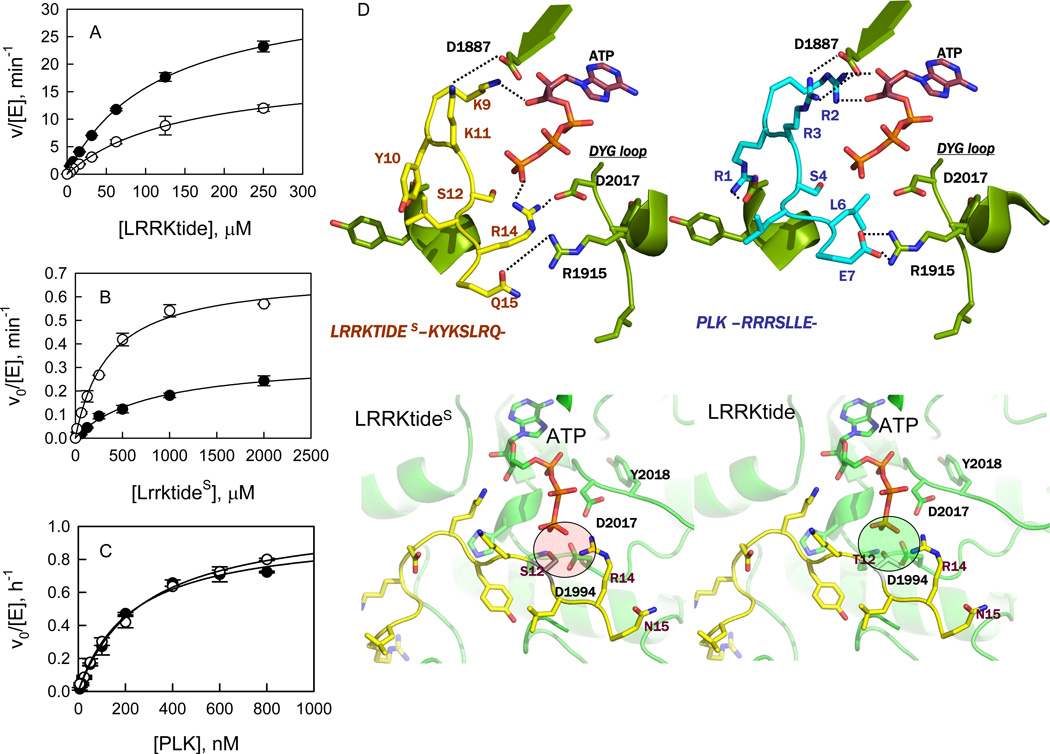
To determine the kinetic mechanism for the phosphorylation of LRRKtide (RLGRDKYKTLRQIRQ), initial velocities were measured as a function of LRRKtide concentration at several fixed concentrations of ATP for t-wt LRRK2 and the t-G2019S mutant (see Supporting Information, S1), respectively. The complete data sets were subjected to global analysis by nonlinear least-squares fits to the three standard mechanisms (ping-pong, rapid equilibrium order, and random/steady-state ordered) using eqs 1–3. Statistically the data fits the random mechanism (or steady-state ordered mechanism) the best for both t-wt LRRK2 and the mutant t-G2019S, yielding the following estimates averaged from two independent experiments: kcat = 8.1 ± 0.7 min−1, KATP = 69 ± 6.6 µM, KLRRKtide = 87 ± 11 µM, and α = 1.8 ± 0.1 for t-wt LRRK2; kcat = 17 ± 1.6 min−1, KATP = 101 ± 16 µM, KLRRKtide = 79 ± 11 µM, and α = 2.0 ± 0.5 for the mutant t-G2019S, as summarized in Table 1. The mutant t-G2019S increases the kcat by 2-fold without significantly affect the binding affinity of ATP and LRRKtide. A direct comparison of the t-G2019S mutant with t-wt LRRK2 is shown in Figure 1A, where the initial velocities were measured as a function of [LRRKtide] at a saturating concentration of ATP.
Table 1.
Initial Velocity Analysis for LRRK2-Catalyzed Phosphorylation
| LRRKtide | LRRKtideS | PLK-peptide | ||||
|---|---|---|---|---|---|---|
| t-WT | t-G2019S | t-WT | t-G2019S | t-WT | t-G2019S | |
| kcat, min−1 | 8.1 ± 0.7 | 17 ± 1.6 | 0.31 ± 0.06 | 0.87 ± 0.04 | 0.018 ± 0.004 | 0.016 ± 0.001 |
| KA, µM | 69 ± 6.6 | 101 ± 16 | 77 ± 19 | 161 ± 38 | 7.1 ± 0.7 | 10 ± 3.9 |
| KB µM | 87 ± 11 | 79 ± 11 | 554 ± 187 | 470 ± 104 | 0.4 ± 0.05 | 0.5 ± 0.2 |
| α | 1.8 ± 0.1 | 2.0 ± 0.5 | 2.6 ± 1.5 | 0.7 ± 0.2 | 0.5 ± 0.1 | 0.6 ± 0.3 |
A represents ATP, B represents phosphoryl acceptors.
Figure 1.
Steady-state kinetic studies of LRRK2-catalyzed phosphorylation. (A) Phosphorylation of LRRKtide by t-wt LRRK2 (○) and the mutant t-G2019S (●). (B) Phosphorylation of LRRKtideS by t-wt LRRK2 (●) and the mutant t-G2019S (○). (C) Phosphorylation of PLK-peptide by t-wt LRRK2 (●) and the mutant t-G2019S (○). (D) Structural details of peptide binding (LRRKtide on the top-left and PLK-peptide on the top-right) near the ATP binding site of LRRK2. Both peptides show a series of conserved interactions involving residues such as D1887 and R1915. In case of LRRKtide, R14 makes hydrogen bonds with ATP as well as D2017 (DGY loop) of LRRK2. The structural equivalent position in PLK-peptide is occupied by L6 and does not participate in hydrogen bonding with ATP or D2017. This suggests that LRRKtide is likely to be more sensitive to mutations in the DYG-loop region compared to PLK-peptide. In the bottom-left panel, the interaction of LRRKtideS with the active site of LRRK2 is shown in details. S12 of LRRKtideS is in close proximity of charged residues including D2017 and D1994. In the bottom right-panel, the interaction of LRRKtide (contains Thr instead of Ser) with the active site of LRRK2 is shown in detail. The methyl group of T12 is placed between the oxygen of T12 and D1994 leading to charge shielding in this region.
Initial Velocity Studies of LRRK2-Catalyzed LRRKtideS Phosphorylation
A peptide LRRKtideS (RLGRDKYKSLRQIRQ) was derived from LRRKtide, in which the phosphorylatable threonine was replaced by a serine residue. The initial velocities were measured as a function of LRRKtideS concentration, at several fixed concentrations of ATP for t-wt LRRK2 and the t-G2019S mutant (see Supporting Information, S2), respectively. Statistically the data fits the random mechanism (or steady-state ordered mechanism) the best for both t-wt LRRK2 and the mutant t-G2019S. The kinetic parameter estimates averaged from two independent experiments were generated: kcat = 0.31 ± 0.06 min−1, KATP = 77 ± 19 µM, KLRRKtideS = 554 ± 187 µM, and α = 2.6 ± 0.5 for t-wt LRRK2; kcat = 0.87 ± 0.04 min−1, KATP = 161 ± 38 µM, KLRRKtideS = 470 ± 104 µM, and α = 0.7 ± 0.2 for the mutant t-G2019S, as summarized in Table 1. A direct comparison of the phosphorylation of LRRKtideS by the t-G2019S mutant and t-wt LRRK2 is shown in Figure 1B, where the initial velocities were measured as a function of [LRRKtideS] at a saturating concentration of ATP.
t-WT LRRK2- and the Mutant t-G2019S-Catalyzed Phosphorylation of PLK-peptide–Initial Velocity Studies
A TR-FRET assay has been developed for measuring the phosphorylation of PLK-peptide (a motif of RRRSLLE). Previously, the reaction was evaluated using a radiometric assay and linear correlation between the units of FU and nM allowed the conversion of initial velocities from FU/h to nM/h (19). To determine the kinetic mechanism of t-wt LRRK2- and the mutant t-G2019S-catalyzed phosphorylation of PLK-peptide, initial velocities were measured over a range of ATP concentrations at several fixed concentrations of PLK-peptide (see Supporting Information, S3). The data fits the random (or steady-state ordered) mechanism the best for both t-wt LRRK2 and the t-G2019S mutant, yielding the following estimates averaged from two independent experiments: kcat = 0.018 ± 0.004 min−1, KATP = 7.1 ± 0.7 µM, KPLK-peptide = 0.43 ± 0.05 µM, and α = 0.5 ± 0.1 for t-wt LRRK2; kcat = 0.016 ± 0.001 min−1, KATP = 10 ± 3.9 µM, KPLK-peptide = 0.5 ± 0.2 µM, and α = 0.6 ± 0.3 for the mutant t-G2019S, as summarized in Table 1. Unlike LRRKtide and LRRKtideS results, the mutant t-G2019S shows similar activity compared with t-wt LRRK2. The mutant t-G2019S also had little effect on the binding affinity of ATP and PLK-peptide. A direct comparison of the phosphorylation of PLK-peptide by the t-G2019S mutant and t-wt LRRK2 is shown in Figure 1C, where the initial velocities were measured as a function of [PLK-peptide] at a saturating concentration of ATP.
Isotope Exchange and Product Inhibition Studies for t-WT LRRK2- and the Mutant t-G2019S-Catalzyed LRRKtide Phosphorylation
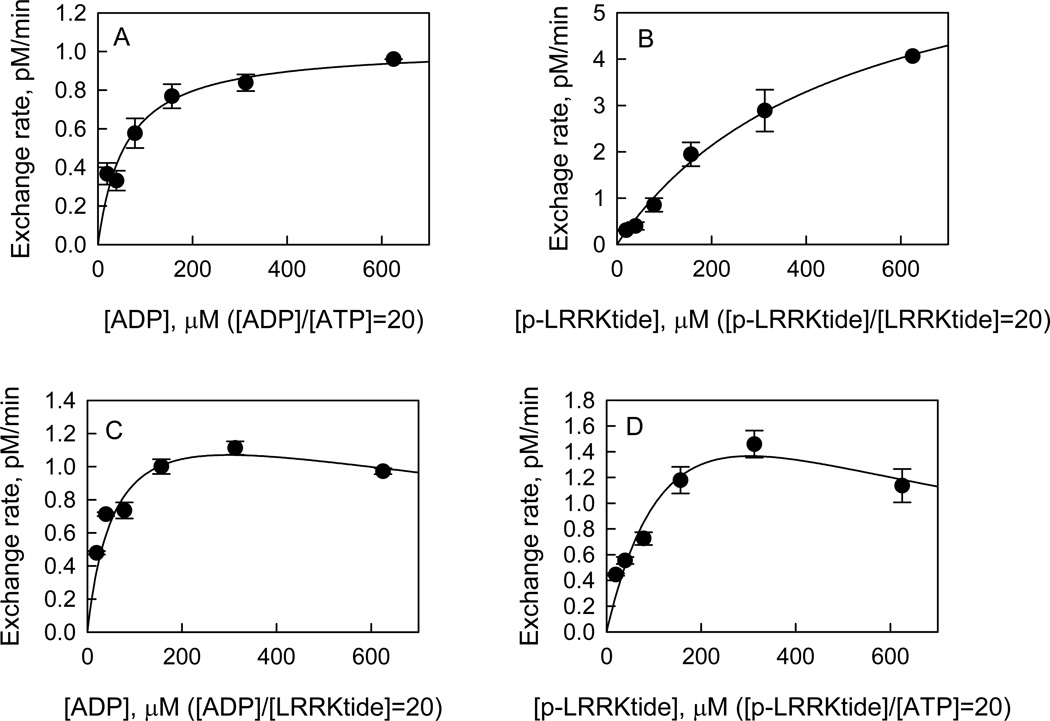
Attempts to measure the exchange rate of radioactive ATP with PO4-LRRKtide at equilibrium were not successful due to the inaccurate determination of the equilibrium constant of the reaction. To keep the system at the same distance from equilibrium under all the conditions, the concentrations of the varied reactants were maintained at a constant ratio while keeping the other reactant concentration constant (18). The concentrations of the varied reactants were kept at a ratio of 20 ([product]/[substrate] = 20), while the other reactants were kept at 1 and 20 µM for the substrate and product, respectively. The exchange was initiated by the addition of the enzyme. The effects of increasing concentrations of the varied reactants on exchange rates were tested for both t-wt LRRK2 and the mutant t-G2019S. First, in all the cases the exchange was linear for at least 60 min (see Supporting Information, S4) and was stopped after 45 min incubation, which allowed the calculation of the initial exchange rate. Next, the initial exchange rate was tested as a function of enzyme concentration and found to be linearly proportional to the enzyme concentration (see Supporting Information, S5). Finally, the effects of increasing concentrations of reactants were examined on the initial exchange rate of radioactive ATP with PO4-LRRKtide. For the mutant t-G2019S, increasing the concentrations of structurally related reactants (i.e., ADP/ATP or PO4-LRRKtide/LRRKtide) resulted in increased ATP with PO4-LRRKtide exchange (Figures 2A and 2B). However, substrate inhibition was observed when structurally unrelated reactants (i.e., PO4-LRRKtide/ATP or ADP/LRRKtide) were varied: the exchange rate initially rose with increasing reactant concentration and gradually and partially decreased (Figures 2C and 2D). Identical patterns were revealed for t-wt LRRK2 as summarized in Table 2. These patterns are consistent with a random mechanism for both t-wt LRRK2 and the mutant t-G2019S with abortive complex formed, i.e. E:ADP:LRRKtide or E:ATP:PO4-LRRKtide.
Figure 2.
Isotope exchange analysis for the mutant t-G2019S-catalyzed LRRKtide phosphorylation. Effect of [ADP]/[ATP] (A), [PO4-LRRKtide]/[LRRKtide] (B), [ADP]/[LRRKtide] (C), and [PO4-LRRKtide]/[ATP] (D) concentrations on the initial rates of the ATP to PO4-LRRKtide isotopic exchanges. The concentrations of the varied reactants were maintained at a constant ratio of 20 while the other reactants were kept as 1 and 20 µM for the substrate and product, respectively. The trace amount of radioactive ATP was added prior to the initiation of the reaction by the addition of enzyme. The data in panel A and B were fit to the simple Michaelis-Menton equation, and data in panel C and D were fit to the equation reflecting the substrate inhibition: v = vmaxS/(Km + [S](1 + [S]/Ki)).
Table 2.
Isotope Exchange Analysis for t-WT LRRK2- and the Mutant t-G2019S-Catalyzed LRRKtide Phosphorylation
| Varied substrate-product pair(s) | |||||
|---|---|---|---|---|---|
| Exchange | AQa | B-Q | B-P | A-P | |
| t-WT | ATP-(PO4)LRRKtide | Hb | HPD | H | HPD |
| T-t-G2019S | ATP-(PO4)LRRKtide | H | HPD | H | HPD |
A represents ATP, B represents LRRKtide, P represents (PO4)LRRKtide, and Q represents ADP.
Hyperbolic (H), hyperbolic with partial depression (HPD)
The product inhibition studies were also conducted with products ADP and PO4-LRRKtide (see Supporting Information, S6). For the mutant t-G2019S-catalyzed LRRKtide phosphorylation, symmetric inhibition patterns were revealed where ADP was competitive with ATP and noncompetitive with LRRKtide; PO4-LRRKtide was competitive with LRRKtide and noncompetitive with ATP as summarized in Table 3. These product inhibition patterns are also consistent with a rapid equilibrium random mechanism with abortive complexes E:PO4-LRRKtide:ATP and E:LRRKtide:ADP formed. Identical inhibition patterns were revealed for t-wt LRRK2.
Table 3.
Inhibition of LRRK2 t-G2019S Mutant by Substrate Analogues and Products
| substrate |
inhibition const (mM) |
||||
|---|---|---|---|---|---|
| inhibitor | variable | mechanism | Ki,e | Ki,ea | Ki,eb |
| ADP | ATP | C | 0.1 ± 0.02 | ||
| LRRKtide | NC | 0.3 ± 0.03 | 0.1 ± 0.01 | ||
| PO4-LRRKtide | ATP | NC | 2.1 ± 0.3 | 1.7 ± 0.1 | |
| LRRKtide | C | 1.8 ± 0.1 | |||
| AMP-PNP | ATP | C | 0.4 ± 0.2 | ||
| LRRKtideS | NC | 1.3 ± 0.3 | 1.3 ± 0.2 | ||
| LRRKtideA | ATP | NC | 0.8 ± 0.3 | 0.5 ± 0.1 | |
| LRRKtideS | C | 0.5 ± 0.1 | |||
| AMP-PNP | ATP | C | 0.4 ± 0.2 | ||
| PLK-peptide | NC | 1.3 ± 0.3 | 1.3 ± 0.2 | ||
| LRRKtideA | ATP | NC | 0.8 ± 0.3 | 0.5 ± 0.1 | |
| PLK-peptide | C | 0.5 ± 0.1 | |||
Inhibition Studies of t-WT LRRK2- and the Mutant t-G2019S-Catalyzed LRRKtideS and PLK-Peptide Phosphorylation
The kinetic mechanism of t-wt LRRK2- and the mutant t-G2019S-catalyzed PLK-peptide and LRRKtideS phosphorylation was determined by conducting the inhibition studies using substrate analogues AMP-PNP and LRRKtideA (RLGRDKYKALRQIRQ), in which the phosphorylatable threonine was replaced by alanine. Symmetric inhibition patterns were revealed for all the reactions as summarized in Table 3 suggesting that both t-wt LRRK2 and the mutant t-G2019S follow a random mechanism for the phosphorylation of PLK-peptide and LRRKtideS.
Proton Inventory Studies
The proton inventory studies of both t-wt LRRK2- and the mutant t-G2019S-catalyzed phosphorylation of LRRKtide were carried out for kcat (initial velocity was measured at saturating concentrations of both ATP and LRRKtide) in a mixture of H2O and D2O at pH 8.2 and pD equivalent. The dependence of the ratio of kcat (kn/k0) in the presence and absence of varying atom fractions of D2O (n) on n is presented in Figure 3A for t-wt LRRK2-catalyzed LRRKtide phosphorylation. The solvent deuterium isotope effect on kcat can be expressed by the Gross-Butler equation (20):
| (4) |
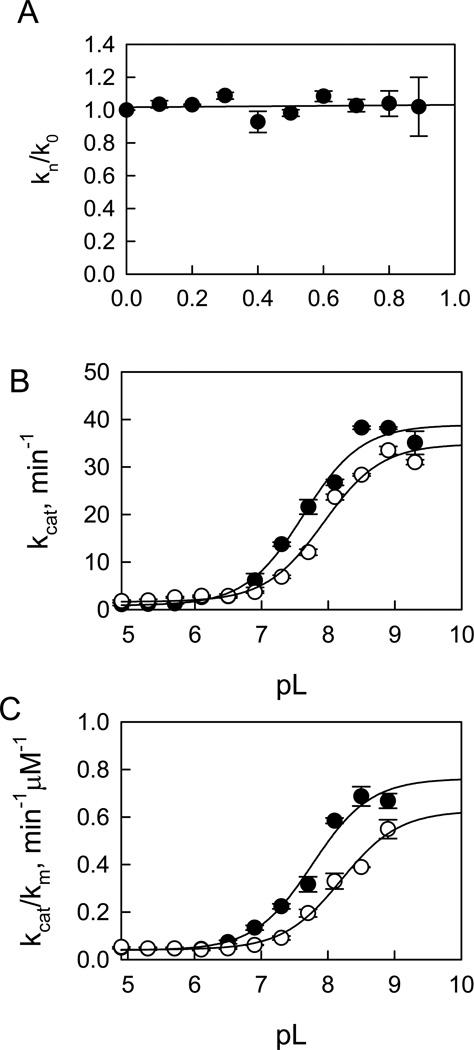
where ϕR and ϕT are fractionation factors for the exchangeable hydrogen in the reactant and transition state, respectively. Fitting the data in Figure 3A to the equation gives the following estimates: ϕR = 1, ϕT = 1.03. Given this, we can assign the estimate to SKIE on kcat, Dkcat = ϕR /ϕT = 1.1 ± 0.1 for t-wt LRRK2 determined from three independent experiments. A similar proton inventory was observed on kcat for the mutant t-G2019S-catalyzed LRRKtide phosphorylation and a SKIE of 1.2 ± 0.2 was determined from three independent experiments.
Figure 3.
Proton inventory and pL-dependent studies for t-wt LRRK2-catalyzed LRRKtide phosphorylaiton. For the proton inventory study, the initial velocities were measured at saturating concentrations of both ATP and peptide substrates for kcat in the mixture of H2O and D2O at pH 8.2 and pD equivalent. The dependence of the ratio of kcat (kn/k0) in the presence and absence of varying atom fractions of D2O (n) on n revealed a normal SKIE of 1.1 (A). pL-dependent studies were carried out using a triple buffer consisting of 50 mM MES, 100 mM Tris, and 50 mM acetic acid. Panel B revealed pH (●) and pD (○) dependencies of kcat with a SKIE of 1.1 and panel C revealed pH (●) and pD (○) dependencies of kcat/Km with a SKIE of 1.2.
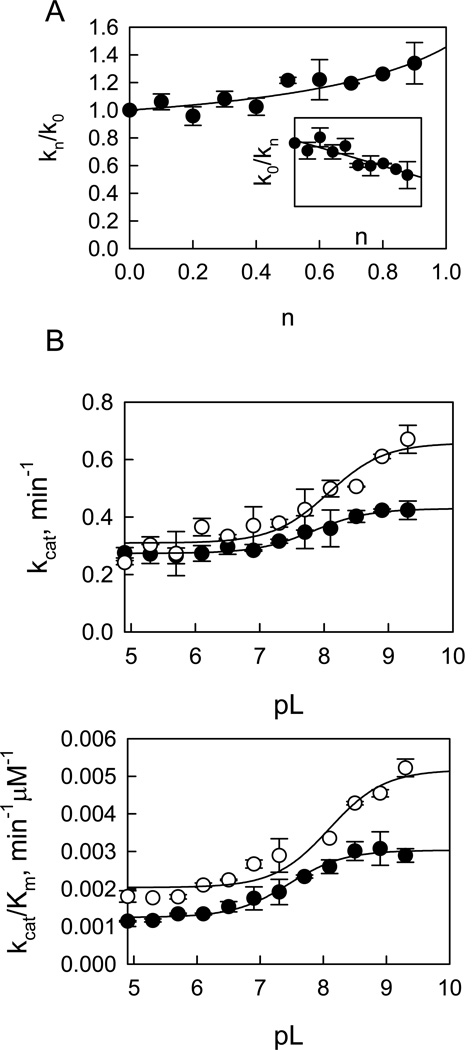
Proton inventory studies for LRRKtideS phosphorylation were also carried out for kcat (initial velocities were measured for both ATP and LRRKtideS at saturating concentrations) for both t-wt and the mutant t-G2019S. Figure 4A shows the dependence of the ratio of kcat (kn/k0) on atom fractions of D2O (n) in the presence and absence of varying D2O for t-wt LRRK2. The plot revealed a nonlinear dependence of kn/k0 on n and was fit to equation 4 well, yielding the following estimates: ϕT =1 and ϕR = 0.7. Given this, we can assign the estimate to SKIE for kcat: Dkcat = ϕR/ϕT = 0.7 and also expect that the reciprocal of the equation should be linear in n. The inset in Figure 4A shows that this expectation is met (the reciprocal of the ratio of kcat (k0/kn) linearly depends on n). Similar proton inventory was observed on kcat for the mutant t-G2019S-catalyzed LRRKtideS phosphorylation and an inverse SKIE of 0.7 ± 0.1 was determined from three independent experiments.
Figure 4.
Proton inventory and pL-dependent studies for t-wt LRRK2-catalyzed LRRKtideS phosphorylation. Panel A shows proton inventory studies of LRRKtideS phosphorylation. The inset shows the reciprocal of the ratio of kcat (k0/kn) dependence on n. Panel B reveals pH (●) and pD (○) dependencies of kcat with a SKIE of 0.65 and panel C reveals pH (●) and pD (○) dependencies of kcat/Km with a SKIE of 0.57.
pL-Dependence of Steady-State Kinetic Parameters and SKIE
The steady-state kinetic parameter-pL (pH or pD) profile of LRRKtideS phosphorylation was carried out in a triple buffer consisting of 50 mM MES, 100 mM Tris, and 50 mM acetic acid along with other components as described in Material and Methods section. The combination of the three buffers has the advantage of providing a constant ionic strength for the entire pL range studied. The pH stability of LRRK2 was determined by incubating the enzyme at the desired pH and assaying aliquots at pH 7.5. The enzyme was determined to be stable from pH 6 to 9.3. Small activity loss occurs at pH < 5.5. However, the small activity loss does not affect the measurement of initial velocities, since enzyme was added from a stock solution at pH 7.5 to the assay mixture and assayed for 30 min in the presence of substrate which is likely to provide some protection against denaturation. The LRRK2-catalyzed phosphorylation of LRRKtideS was monitored in a pL range of 4.9–9.3 for kcat (initial velocities were measured at saturating ATP and LRRKtideS concentrations of 1 mM and 2 mM, respectively) and for kcat/Km (initial velocities were measured at ATP and LRRKtideS concentrations 10-fold less than the Km’s, 10 µM and 50 µM, respectively). The pL profile of kcat for t-wt LRRK2 revealed an acidic limb (Figure 4B) and was fit to equation 5, yielding one apparent pKa of 7.7 and 8.2 from pH and pD profile, respectively.
| (5) |
kcat/Km exhibited a similar pL dependence as kcat, revealing one apparent pKa of 7.5 and 8.0 from pH and pD profile, respectively (Figure 4C). The pL-independent steady-state parameters revealed SKIE values of 0.65 and 0.57 for kcat and kcat/Km, respectively. Similar pL profile and SKIE values were observed for the mutant t-G2019S (see Supporting Information, S7).
The same pL-dependent studies were conducted for LRRKtide as well. The pL profiles revealed similar shape with one apparent pKa of 7.6 and 7.9 from pH and pD profile, respectively for kcat; and one apparent pKa’s of 7.7 and 8.2 from pH and pD profile, respectively for kcat/Km (Figure 3B and 3C). The pL-independent steady-state parameters revealed SKIE values of 1.1 and 1.2 for kcat and kcat/Km, respectively. Similar pL profile and SKIE values were observed for the mutant t-G2019S (see Supporting Information, S8).
Mass Spectrometry Analysis of Cysteine Residues Modified by N-ethylmaleimide (NEM)
The mass spectrometry analysis revealed twenty-eight cysteine residues modified by NEM. Twenty-three of them locate in the LRR, COR, and WD40 domains. Only one cysteine C1465 is in the GTPase domain and four (C2024, C2025, C2101, and C2114) are in the kinase domain.
LRRK2-Catalyzed Hydrolysis of GTP – Steady-State Kinetic Study
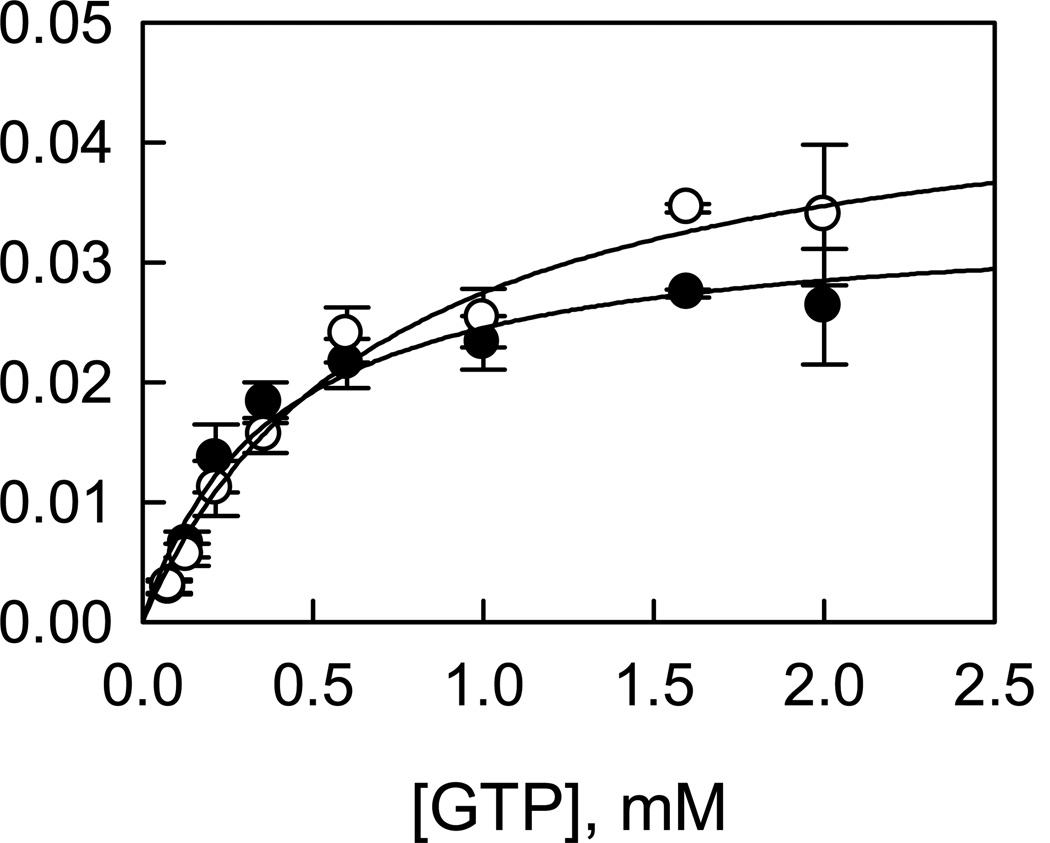
To characterize LRRK2-catalyzed hydrolysis of GTP, a radiometric assay in which [α-33P]-GDP is separated from [α-33P]-GTP by TLC and counted by a scintillation counter was developed. The production of GDP is linearly dependent on both time and enzyme concentration (data not shown). Initial velocities were measured as a function of GTP concentration for t-wt and the mutant t-G2019S (Figure 5). The dependence of initial velocities on GTP concentration adheres to the simple Michaelis-Menten equation, which allowed us to calculate the steady-state kinetic parameters: kcat = 0.04 ± 0.004 s−1, KGTP = 0.7 ± 0.1 mM for t-wt LRRK2; and kcat = 0.03 ± 0.002 s−1, KGTP = 0.4 ± 0.1 mM for the mutant t-G2019S. These results suggest that the mutation decreases the GTPase activity by 20% without significantly affects the binding of GTP.
Figure 5.
LRRK2-catalyzed GTP hydrolysis. Initial velocities were measured as a function of [GTP] for t-wt LRRK2 (○) and the mutant t-G2019S (●).
Modeling of LRRK2 Kinase Domain
The LRRK2 kinase domain between residues 1879–2138 was modeled using MODELLER (15) (details in materials and method section). ATP was docked into the ATP-binding pocket using GLIDE (21, 22) and the final model in complex with peptide substrates is shown in Figure 1D and also Supporting Information S9. The LRRK2 kinase model shows all of the expected subdomains of a Ser/Thr protein kinase (see Supporting Information, S9c) (23–26). The ATP binding cleft shows a glycine-rich loop (residues 1885–1982) facilitating backbone interactions with the γ-phosphate of ATP (see Supporting Information, S9c) (27). D2017 is part of the regulatory DYG loop and makes a stabilizing interaction with ATP via Mg2+ ion (25, 26). The DYG loop in this case was modeled on the “DYG-in” active form of the kinase. In addition, the catalytic loop shows H1998, K1996 and D1995 correctly positioned for catalysis in close proximity to the γ-phosphate of ATP. The LRRK2 kinase domain shows spatial conservation of residues in the regulatory spine like other kinases in this subfamily (25) and comprises residues L1924, L1935, Y1992, and Y2018. The catalytic spine comprises V1893, L1955, L2001, L2062, and I2066. The adenine group of the ATP molecules completes this catalytic spine by positioning itself between V1893 and L2001. In addition, the model shows C2024 and C2025, which are both on the regulatory loop, exposed to solvent.
DISCUSSION
Source of LRRK2 Enzymes
We have reported the characterization of the kinase and GTPase activities of full-length mouse wt LRRK2 purified from mouse brain (19). Due to the difficulty of generating the full-length G2019S mutant, for this study, we used the truncated version of human LRRK2 and the mutant G2019S containing residues 970–2527 which includes the Roc, COR, kinase, WD40, and entire C-terminal domains expressed in baculovirus system. The truncated protein demonstrated both kinase and GTPase activities.
The Mutant t-G2019S Increases the Kinase Activity in a Substrate-Dependent Manner—Initial Velocity Studies
The natural substrate(s) of LRRK2 are not known. Several potential substrates of LRRK2 have been described, including ezrine, radixin, and moesin (ERM), eukaryotic initiation factor 4E (eIF4E)-binding protein (4E-BP), MAPK kinases, tubulin beta, and α-synuclein (28–32). However, none of the proteins have proven to be efficiently phosphorylated by LRRK2 in cells or animals. Therefore, in this study LRRKtide and PLK-peptide were used as phosphoryl acceptors.
The mutant G2019S has been consistently reported to increase the kinase activity of LRRK2 by 2- to 3-fold (7, 8, 11, 33). In this study, we were able to replicate this finding for LRRKtide phosphorylation. However, when PLK-peptide was used as the phosphoryl acceptor, surprisingly the mutant t-G2019S showed no difference in specific activity to that of t-wt LRRK2. It has been reported that LRRK2 favors threonine over serine for phosphorylation. Replacement of threonine with serine in the Nictide peptide abolished phosphorylation by LRRK2 (34). The lack of increased activity of the mutant t-G2019S might be attributed to the unfavorable serine residue. Therefore, LRRKtideS peptide was prepared, in which phosphorylatable threonine was replaced by serine. The replacement resulted in a much less efficient substrate. This is most likely due to a possible charge repulsion introduced by serine which weakens the electrostatic network between LRRK2 and the peptide substrate (Figure 1D, also see supporting information for detail). However, the mutant t-G2019S still retains a 3-fold increase in kcat compared to t-wt LRRK2.
Modeling the LRRK2 kinase domain in complex with ATP, LRRKtide(T/S) and/or PLK-peptide provides a model for understanding the substrate-dependent effect of the mutant t-G2019S (Figure 1D). PLK-peptide has a higher charge density with excellent charge complementarity with the nearby LRRK2 substrate binding pocket (see supporting information for detail) and is consistent with > 200-fold tighter binding of the PLK-peptide relative to LRRKtide, as demonstrated by differences in Km values (Km of 0.4 µM for PLK-peptide versus 87 µM for LRRKtide). The tight binding of the PLK-peptide minimizes any effect that the t-G2019S mutant may have therefore eliminating any difference between the t-wt and the mutant t-G2019S. The individual interactions between peptide substrates and LRRK2 will be tested by making a series of single amino acid changes in a future study.
The observation that the mutant t-G2019S shows substrate-dependent effect on kinase activity highlights the uncertainty that the mutant G2019S will necessarily have increased kinase activity toward physiological substrates compared to wt LRRK2. The fact that most of the LRRK2 mutations do not simply increase kinase activity, along with our observation on the mutant t-G2019S raises the possibility that there might be complex mechanisms that regulate the functions and enzymatic activities of the mutant G2019S and other mutations. A more comprehensive understanding of the effects of LRRK2 mutations must take into account the complex mechanisms that regulate LRRK2’s functions. The recent discovery of 14-3-3 (33) has begun to reveal the mutant-specific regulating mechanisms of LRRK2.
Formation of a Nonproductive Complex and Its Impact on Inhibitor Efficacy In Vivo
Values of KATP for the reaction of LRRK2 with PLK-peptide and LRRKtide/LRRKtideS dramatically depend on the identity of the phosphoryl acceptor with a notable difference of 10-fold. Given a α value of 2 versus 0.5, this difference in KATP is large. As discussed in more detail previously (35), a mechanism that involves the formation of nonproductive binary complex upon the binding of the phosphoryl acceptor to free enzyme, to which nucleotides bind nonproductively, could account for these results. The measured values of KATP are complex terms that include the contribution from the nonproductive binding of the phosphoryl acceptor and would be dependent on the identity of the phosphoryl acceptor. It would be expected that dissociation constants of the ATP competitive inhibitors could also show phosphoryl acceptor-dependence resulting in a similar potency difference between in vitro and in vivo situations. In other words, inhibitors of LRRK2 currently assessed using peptide substrates may not truly represent efficacy with its physiological substrates. Given the amount of efforts currently dedicated to drug discovery targeting LRRK2 kinase, identifying LRRK2 physiological substrates has become even more urgent for potential therapeutic applications in PD, not to mention the importance for elucidating LRRK2’s functions.
The Mutant t-G2019S Follows the Same Kinetic Mechanism as t-WT— Inhibition and Isotope Exchange Studies
The inhibition and isotope exchange patterns rule out a steady-state ordered mechanism and indicate a rapid equilibrium random mechanism for all three reactions catalyzed by t-wt and the mutant t-G2019S. In our previous study, we had determined that full-length mouse wt LRRK2 follows the same rapid equilibrium random mechanism for LRRKtide and PLK-peptide phosphorylation (19). The results indicate that the truncation of the N-terminus or the source of LRRK2 enzyme does not change the substrate binding order.
There is General Base Involved in the Reaction
Linear proton inventories with small SKIE on kcat of LRRKtide phosphorylation for both t-wt LRRK2 and the mutant t-G2019S suggest that the rate-limiting step of the process governed by kcat is the product release or any conformational change associated with this process. The use of the proton inventory shape and the size of the solvent isotope effect to determine the number of hydrogenic sites and to diagnose the mechanism have previously discussed by Schowen and Venkatasubban (20, 36). In order to understand the catalytic mechanism and to identify the critical residues involved in catalysis, the experiments must be carried out using a rapid-quench instrument to measure the phosphoryl transfer, which is a rapid step. However, here we used the less efficient substrate, LRRKtideS, with the expectation that the 26-fold decrease in kcat would lead to a slow phosphoryl transfer. Proton inventory studies for LRRKtideS phosphorylation revealed a nonlinear dependence of kn/k0 on n with an inverse SKIE of 0.7 for both t-wt and the mutant t-G2019S. The reciprocal of the ratio of kcat (k0/kn) linearly depends on n. These results suggest that (i) the phosphoryl transfer step is rate-limiting for LRRKtideS phosphorylation and is associated with one proton transfer and (ii) the fractionation factor in the ground state is less than unity.
To make certain that changes in side chain pKa values in light and heavy water were not causing a dramatic difference in SKIE, studies of the pL-dependence of kcat and kcat/Km were carried out. For the LRRKtideS phosphorylation, the pL-profile revealed inverse SKIE on kcat at the optimal pL for both the t-wt and mutant t-G2019S and identified a residue with pKa of 7.5. The pK value of the catalytic residue was shifted by + 0.5 unit in D2O and may result from the solvent equilibrium isotope effect on the residue. Similar results were obtained for kcat/Km. First we postulated that SH ionization of a cysteine residue in the active site was the possible source of the low fractionation factor, although a solvent equilibrium isotope effect on pK value of sulfhydryl group is expected to be 0.15–0.18 (20). To test this hypothesis, t-wt LRRK2 was treated with the cysteine-specific irreversible inhibitor N-ethylmaleimide (NEM). A dose-response curve was obtained with signs of complete inhibition at high NEM concentration, suggesting that cysteine residues play an important role in LRRK2 catalysis. A mass spectrometry analysis revealed that four cysteines (C2010, C2014, C2025, and C2114) in the kinase domain of LRRK2 were modified by NEM to different extent. However, modeling of the kinase domain of LRRK2/LRRKtide complex revealed no cysteine in the active site— C2101 and C2114 are away from the active site while C2024 and C2025 are on the catalytic loop of the enzyme. Instead, H1998 was identified in close proximity to the hydroxyl proton of threonine and could function as the possible general base for catalysis. The shift of pK of the catalytic residue by 0.5 unit is consistent with the expected solvent equilibrium isotope effect on pK of histidinyl group of 0.5–0.7 (20). However, it raised a question: what is the source of the less-than-unity fractionation factor in the ground state? Certainly the thiol group of cysteine is not the source. In general, a low-barrier hydrogen bond would give rise to such a fractionation factor and that would be formed only when the pK values of the two groups in the hydrogen bond are similar. There are some examples of low-barrier hydrogen bond for enzyme groups involved in acid-base catalysis (37). In the case of enolase, the base appears to be a water molecule held between two glutamates (38). Identification of the source of the low fractionation factor and the general base involved in catalysis will be greatly facilitated by the future elucidation by x-ray crystal structure of LRRK2 kinase domain. In addition, site-directed mutagenesis may also help elucidate the role and function of H1988 in the future.
Effects of the Mutant t-G2019S on the GTPase Activity
The kinetic study of GTP hydrolysis revealed similar kinetic parameters for the mutant t-G2019S and t-wt LRRK2. The mutant t-G2019S does not significantly affect GTP binding and slightly decreases the turnover of GTP to GDP. We have previously reported that GTP or GDP binding to the GTPase domain does not affect the downstream kinase activity in vitro when full-length protein purified from mouse brain was used (35). Those results raised the question of whether LRRK2 undergoes intramolecular regulation in a GTP-dependent manner. In the current study, we were still unable to detect any effect of GTP or GDP on kinase activity using truncated proteins. Interestingly, we recently identified a GTPase activating protein that downregulates the kinase activity of LRRK2 by increasing GTPase activity (manuscript in preparation). Those findings constitute direct evidence that LRRK2 requires the GTPase activating protein in order to undergo intramolecular regulation.
In summary, the studies reported herein provide important information for understanding the critical interactions in the active site of LRRK2, which provides an enzymology foundation to assist drug discovery efforts for PD. Designing selective kinase inhibitors is a challenge given the fact that all kinases in the TKL sub-family have very similar active sites. The final active site of the enzyme is comprised of residues not only from the kinase but from the substrate as well. The identification of critical residues in LRRK2’s substrate will provides important information for identifying physiologically relevant substrates of LRRK2 via a consensus motif. The identification of LRRK2’s substrates will be critical not only for accurately estimating the efficacy of inhibitors but also for selective inhibitor design.
Supplementary Material
Acknowledgments
Funding: This work was supported by National Institutes of Health (grant number 1U24NS049339).
ABBREVIATIONS
- PD
Parkinson’s disease
- t-wt LRRK2
truncated wild type leucine-rich repeat kinase 2
- t-G2019S
truncated G2019S
- PLK-peptide
PLK-derived peptide with a motif of RRRSLLE
- LRRKtide
RLGRDKYKTLRQIRQ
- LRRKtideS
RLGRDKYKSLRQIRQ
Footnotes
Supporting Information Available. Additional data of initial velocity, inhibition, and pL-dependent studies are provided as supplemental information. This material is available free of charge via the internet at http://pubs.acs.org.
REFERENCES
- 1.Moore DJ, West AB, Dawson VL, Dawson TM. Molecular pathophysiology of Parkinson's disease. Annu Rev Neurosci. 2005;28:57–87. doi: 10.1146/annurev.neuro.28.061604.135718. [DOI] [PubMed] [Google Scholar]
- 2.Paisan-Ruiz C, Jain S, Evans EW, Gilks WP, Simon J, van der Brug M, Lopez de Munain A, Aparicio S, Gil AM, Khan N, Johnson J, Martinez JR, Nicholl D, Carrera IM, Pena AS, de Silva R, Lees A, Marti-Masso JF, Perez-Tur J, Wood NW, Singleton AB. Cloning of the gene containing mutations that cause PARK8-linked Parkinson's disease. Neuron. 2004;44:595–600. doi: 10.1016/j.neuron.2004.10.023. [DOI] [PubMed] [Google Scholar]
- 3.Zimprich A, Biskup S, Leitner P, Lichtner P, Farrer M, Lincoln S, Kachergus J, Hulihan M, Uitti RJ, Calne DB, Stoessl AJ, Pfeiffer RF, Patenge N, Carbajal IC, Vieregge P, Asmus F, Muller-Myhsok B, Dickson DW, Meitinger T, Strom TM, Wszolek ZK, Gasser T. Mutations in LRRK2 cause autosomal-dominant parkinsonism with pleomorphic pathology. Neuron. 2004;44:601–607. doi: 10.1016/j.neuron.2004.11.005. [DOI] [PubMed] [Google Scholar]
- 4.Berg D, Schweitzer KJ, Leitner P, Zimprich A, Lichtner P, Belcredi P, Brussel T, Schulte C, Maass S, Nagele T, Wszolek ZK, Gasser T. Type and frequency of mutations in the LRRK2 gene in familial and sporadic Parkinson's disease*. Brain. 2005;128:3000–3011. doi: 10.1093/brain/awh666. [DOI] [PubMed] [Google Scholar]
- 5.Khan NL, Jain S, Lynch JM, Pavese N, Abou-Sleiman P, Holton JL, Healy DG, Gilks WP, Sweeney MG, Ganguly M, Gibbons V, Gandhi S, Vaughan J, Eunson LH, Katzenschlager R, Gayton J, Lennox G, Revesz T, Nicholl D, Bhatia KP, Quinn N, Brooks D, Lees AJ, Davis MB, Piccini P, Singleton AB, Wood NW. Mutations in the gene LRRK2 encoding dardarin (PARK8) cause familial Parkinson's disease: clinical, pathological, olfactory and functional imaging and genetic data. Brain. 2005;128:2786–2796. doi: 10.1093/brain/awh667. [DOI] [PubMed] [Google Scholar]
- 6.Farrer M, Stone J, Mata IF, Lincoln S, Kachergus J, Hulihan M, Strain KJ, Maraganore DM. LRRK2 mutations in Parkinson disease. Neurology. 2005;65:738–740. doi: 10.1212/01.wnl.0000169023.51764.b0. [DOI] [PubMed] [Google Scholar]
- 7.Smith WW, Pei Z, Jiang H, Dawson VL, Dawson TM, Ross CA. Kinase activity of mutant LRRK2 mediates neuronal toxicity. Nat Neurosci. 2006;9:1231–1233. doi: 10.1038/nn1776. [DOI] [PubMed] [Google Scholar]
- 8.Greggio E, Jain S, Kingsbury A, Bandopadhyay R, Lewis P, Kaganovich A, van der Brug MP, Beilina A, Blackinton J, Thomas KJ, Ahmad R, Miller DW, Kesavapany S, Singleton A, Lees A, Harvey RJ, Harvey K, Cookson MR. Kinase activity is required for the toxic effects of mutant LRRK2/dardarin. Neurobiol Dis. 2006;23:329–341. doi: 10.1016/j.nbd.2006.04.001. [DOI] [PubMed] [Google Scholar]
- 9.Guo L, Gandhi PN, Wang W, Petersen RB, Wilson-Delfosse AL, Chen SG. The Parkinson's disease-associated protein, leucine-rich repeat kinase 2 (LRRK2), is an authentic GTPase that stimulates kinase activity. Exp Cell Res. 2007;313:3658–3670. doi: 10.1016/j.yexcr.2007.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ito G, Okai T, Fujino G, Takeda K, Ichijo H, Katada T, Iwatsubo T. GTP binding is essential to the protein kinase activity of LRRK2, a causative gene product for familial Parkinson's disease. Biochemistry. 2007;46:1380–1388. doi: 10.1021/bi061960m. [DOI] [PubMed] [Google Scholar]
- 11.West AB, Moore DJ, Biskup S, Bugayenko A, Smith WW, Ross CA, Dawson VL, Dawson TM. Parkinson's disease-associated mutations in leucine-rich repeat kinase 2 augment kinase activity. Proc Natl Acad Sci U S A. 2005;102:16842–16847. doi: 10.1073/pnas.0507360102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.West AB, Moore DJ, Choi C, Andrabi SA, Li X, Dikeman D, Biskup S, Zhang Z, Lim KL, Dawson VL, Dawson TM. Parkinson's disease-associated mutations in LRRK2 link enhanced GTP-binding and kinase activities to neuronal toxicity. Hum Mol Genet. 2007;16:223–232. doi: 10.1093/hmg/ddl471. [DOI] [PubMed] [Google Scholar]
- 13.Xiong Y, Coombes CE, Kilaru A, Li X, Gitler AD, Bowers WJ, Dawson VL, Dawson TM, Moore DJ. GTPase activity plays a key role in the pathobiology of LRRK2. PLoS Genet. 2010;6:e1000902. doi: 10.1371/journal.pgen.1000902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee BD, Shin JH, VanKampen J, Petrucelli L, West AB, Ko HS, Lee YI, Maguire-Zeiss KA, Bowers WJ, Federoff HJ, Dawson VL, Dawson TM. Inhibitors of leucine-rich repeat kinase-2 protect against models of Parkinson's disease. Nat Med. 2010;16:998–1000. doi: 10.1038/nm.2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eswar N, Eramian D, Webb B, Shen MY, Sali A. Protein structure modeling with MODELLER. Methods Mol Biol. 2008;426:145–159. doi: 10.1007/978-1-60327-058-8_8. [DOI] [PubMed] [Google Scholar]
- 16.Vaguine AA, Richelle J, Wodak SJ. SFCHECK: a unified set of procedures for evaluating the quality of macromolecular structure-factor data and their agreement with the atomic model. Acta Crystallogr D Biol Crystallogr. 1999;55:191–205. doi: 10.1107/S0907444998006684. [DOI] [PubMed] [Google Scholar]
- 17.Verind G. WHAT IF: A molecular modeling and drug design program. J Mol Graph. 1996;8:52–56. doi: 10.1016/0263-7855(90)80070-v. [DOI] [PubMed] [Google Scholar]
- 18.Segel IHN. Enzyme Kinetics. New York: John Wiley& Sons; 1975. [Google Scholar]
- 19.Liu M, Poulose S, Schuman E, Zaitsev AD, Dobson B, Auerbach K, Seyb K, Cuny GD, Glicksman MA, Stein RL, Yue Z. Development of a mechanism-based high-throughput screen assay for leucine-rich repeat kinase 2--discovery of LRRK2 inhibitors. Anal Biochem. 2010;404:186–192. doi: 10.1016/j.ab.2010.05.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schowen KB, Schowen RL. Solvent isotope effects of enzyme systems. Methods Enzymol. 1982;87:551–606. [PubMed] [Google Scholar]
- 21.Friesner RA, Banks JL, Murphy RB, Halgren TA, Klicic JJ, Mainz DT, Repasky MP, Knoll EH, Shelley M, Perry JK, Shaw DE, Francis P, Shenkin PS. Glide: a new approach for rapid, accurate docking and scoring. 1. Method and assessment of docking accuracy. J Med Chem. 2004;47:1739–1749. doi: 10.1021/jm0306430. [DOI] [PubMed] [Google Scholar]
- 22.Halgren TA, Murphy RB, Friesner RA, Beard HS, Frye LL, Pollard WT, Banks JL. Glide: a new approach for rapid, accurate docking and scoring. 2. Enrichment factors in database screening. J Med Chem. 2004;47:1750–1759. doi: 10.1021/jm030644s. [DOI] [PubMed] [Google Scholar]
- 23.Gosal D, Ross OA, Wiley J, Irvine GB, Johnston JA, Toft M, Mata IF, Kachergus J, Hulihan M, Taylor JP, Lincoln SJ, Farrer MJ, Lynch T, Mark Gibson J. Clinical traits of LRRK2-associated Parkinson's disease in Ireland: a link between familial and idiopathic PD. Parkinsonism Relat Disord. 2005;11:349–352. doi: 10.1016/j.parkreldis.2005.05.004. [DOI] [PubMed] [Google Scholar]
- 24.Kannan N, Neuwald AF. Did protein kinase regulatory mechanisms evolve through elaboration of a simple structural component? J Mol Biol. 2005;351:956–972. doi: 10.1016/j.jmb.2005.06.057. [DOI] [PubMed] [Google Scholar]
- 25.Kornev AP, Haste NM, Taylor SS, Eyck LF. Surface comparison of active and inactive protein kinases identifies a conserved activation mechanism. Proc Natl Acad Sci U S A. 2006;103:17783–17788. doi: 10.1073/pnas.0607656103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kornev AP, Taylor SS, Ten Eyck LF. A helix scaffold for the assembly of active protein kinases. Proc Natl Acad Sci U S A. 2008;105:14377–14382. doi: 10.1073/pnas.0807988105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hemmer W, McGlone M, Tsigelny I, Taylor SS. Role of the glycine triad in the ATP-binding site of cAMP-dependent protein kinase. J Biol Chem. 1997;272:16946–16954. doi: 10.1074/jbc.272.27.16946. [DOI] [PubMed] [Google Scholar]
- 28.Parisiadou L, Xie C, Cho HJ, Lin X, Gu XL, Long CX, Lobbestael E, Baekelandt V, Taymans JM, Sun L, Cai H. Phosphorylation of ezrin/radixin/moesin proteins by LRRK2 promotes the rearrangement of actin cytoskeleton in neuronal morphogenesis. J Neurosci. 2009;29:13971–13980. doi: 10.1523/JNEUROSCI.3799-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gillardon F. Leucine-rich repeat kinase 2 phosphorylates brain tubulin-beta isoforms and modulates microtubule stability--a point of convergence in parkinsonian neurodegeneration? J Neurochem. 2009;110:1514–1522. doi: 10.1111/j.1471-4159.2009.06235.x. [DOI] [PubMed] [Google Scholar]
- 30.Imai Y, Gehrke S, Wang HQ, Takahashi R, Hasegawa K, Oota E, Lu B. Phosphorylation of 4E-BP by LRRK2 affects the maintenance of dopaminergic neurons in Drosophila. EMBO J. 2008;27:2432–2443. doi: 10.1038/emboj.2008.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gloeckner CJ, Schumacher A, Boldt K, Ueffing M. The Parkinson disease-associated protein kinase LRRK2 exhibits MAPKKK activity and phosphorylates MKK3/6 and MKK4/7, in vitro. J Neurochem. 2009;109:959–968. doi: 10.1111/j.1471-4159.2009.06024.x. [DOI] [PubMed] [Google Scholar]
- 32.Hsu CH, Chan D, Greggio E, Saha S, Guillily MD, Ferree A, Raghavan K, Shen GC, Segal L, Ryu H, Cookson MR, Wolozin B. MKK6 binds and regulates expression of Parkinson's disease-related protein LRRK2. J Neurochem. 2010;112:1593–1604. doi: 10.1111/j.1471-4159.2010.06568.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nichols RJ, Dzamko N, Morrice NA, Campbell DG, Deak M, Ordureau A, Macartney T, Tong Y, Shen J, Prescott AR, Alessi DR. 14-3-3 binding to LRRK2 is disrupted by multiple Parkinson's disease-associated mutations and regulates cytoplasmic localization. Biochem J. 2010;430:393–404. doi: 10.1042/BJ20100483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nichols RJ, Dzamko N, Hutti JE, Cantley LC, Deak M, Moran J, Bamborough P, Reith AD, Alessi DR. Substrate specificity and inhibitors of LRRK2, a protein kinase mutated in Parkinson's disease. Biochem J. 2009;424:47–60. doi: 10.1042/BJ20091035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu M, Dobson B, Glicksman MA, Yue Z, Stein RL. Kinetic mechanistic studies of wild-type leucine-rich repeat kinase 2: characterization of the kinase and GTPase activities. Biochemistry. 2010;49:2008–2017. doi: 10.1021/bi901851y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Venkatasubban KS, Schowen RL. The proton inventory technique. CRC Crit Rev Biochem. 1984;17:1–44. doi: 10.3109/10409238409110268. [DOI] [PubMed] [Google Scholar]
- 37.Cleland WW. Low-barrier hydrogen bonds and low fractionation factor bases in enzymatic reactions. Biochemistry. 1992;31:317–319. doi: 10.1021/bi00117a001. [DOI] [PubMed] [Google Scholar]
- 38.Lebioda L, Stec B. Mechanism of enolase: the crystal structure of enolase-Mg2(+)-2-phosphoglycerate/phosphoenolpyruvate complex at 2.2-A resolution. Biochemistry. 1991;30:2817–2822. doi: 10.1021/bi00225a012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.