Abstract
Endogenous Bone Morphogenetic Protein (BMP) signaling plays a significant role in the kidney’s recovery from acute injury and exogenous administration of BMP7 has therapeutic potential in numerous rodent models of renal injury and disease. However, in the healthy kidney endogenous BMP7 ligand is vigorously counteracted by extracellular antagonists such as USAG1 and CHRDL1. Little is known about the degree of BMP signaling and the ligands driving it in the healthy adult kidney. In this study we characterize basal BMP signaling in the healthy tubular nephron, and show that BMP2 is expressed in proximal nephron epithelial cells. Comparative gene profiling of proximal tubule cell responses to BMP2 and BMP7 does not reveal any qualitative difference, suggesting that identical BMP gene targets may be activated in healthy and injured organs. Interestingly, our gene profiling analysis shows that BMP signaling activates a number of Notch regulated transcription factors, including HEY1. As in other biological systems, HEY1 functions as a negative feedback regulator of BMP2 expression in the proximal tubule. In summary, this work reveals endogenous BMP signaling patterns in the healthy human and mouse kidney, and identifies novel gene targets, some of which are involved in the complex regulation of BMP signaling in the adult kidney.
Keywords: BMP2, BMP7, Acute kidney injury, Smad, Proximal tubule
1. Introduction
Acute kidney injury (“AKI”), defined as a rapid increase in serum creatinine or reduction in urine output, is diagnosed in over 2% of hospital admissions and is associated with a high mortality rate [1]. As the population ages, the prevalence of AKI will increase, providing a strong incentive to find directed therapies [2]. Following AKI, kidneys posses the potential to regenerate and lineage marking experiments show that lost cells are replaced by surviving tubular epithelial cells of the nephron [3–6]. Moreover, it has recently been demonstrated that surviving injured epithelial cells, rather than uninjured bystander cells are responsible for tubule regeneration [7]. In rodent models of kidney injury and disease, treatment with BMP7 both protects the kidney from injury and accelerates regeneration [8–12]. More significantly, endogenous BMP signaling has demonstrable protective and pro-regenerative functions in both acute and chronic kidney injury [13, 14], indicating that this pathway may provide tractable therapeutic targets.
BMP ligands initiate signaling by binding to cell surface serine-threonine kinase receptors that phosphorylate receptor-associated SMAD transcription factors (“R-SMADs”) 1, 5, and 8. RSMADs associate with the co-SMAD, SMAD 4, and the complex moves to the nucleus where it associates with other transcription factors and binds DNA in regulatory regions of target genes [15]. Inhibitor of DNA binding 1 (Id1) is one of several immediate early, transcriptional targets of BMP signaling [16]. The Id1 SMAD-binding element has been used to generate both cell-based and in vivo transcriptional reporters for BMP signaling [16–18].
In this study we further define the extent and localization of endogenous BMP signaling in the kidney, and identify targets of pathway activation in human proximal tubule cells, the cell type in the nephron that is most prone to damage in commonly used AKI disease models [19, 20]. We show that BMP signaling occurs in the largely quiescent cells of the proximal tubule of both the healthy mouse and human kidney. This signaling is likely driven, at least in part, by BMP2 produced at a relatively high level in the proximal tubule. We analyzed immediate early gene targets of BMP in human proximal tubule cells by gene profiling. Comparing stimulation with BMP 2 or 7, we find regulation of a qualitatively indistinguishable set of genes. In addition to well-described transcriptional targets such as IDs, SMAD7, and BAMBI, we observed strong induction of the typical Notch responsive transcription factors HES1, HEY1, SNAI1, and SNAI2. Interestingly, these are expressed in the absence of Notch pathway signaling. In vitro over-expression of one of these factors, HEY1, suggests that this transcription factor may be part of a regulatory feedback mechanism that modulates BMP transcription.
2. Materials and Methods
2.1. Cell Culture
Primary human renal proximal tubule epithelial cells (“RPTECs”) (Lifeline Cell Technology) were cultured in complete RenaLife Cell Culture Medium (Lifeline Cell Technology). HK-2 human proximal tubule epithelial cells (ATCC) were cultured in Keratinocyte Serum Free Media (“KSFM”) supplemented with bovine pituitary extract and Epidermal Growth Factor (“EGF”) (Gibco), Penicillin, Streptomycin and Amphotericin B according to the manufacturer’s instructions. For BMP stimulation assays, cells were serum starved for four hours before BMP addition. Adenoviruses expressing GFP, HEY1 (kindly provided by Dr. Lucy Liaw, Maine Medical Center Research Institute [21]) and SNAI1 (kindly provided by Dr. Paul A. Wade, NIH [22]) were transduced at 250 MOI. Virus was removed after 24 hours.
2.2. Gene profiling
Triplicate wells of RPTECs were treated with 50 ng/ml BMP2, BMP7 (R&D Systems), or vehicle. Total RNA was purified using Trizol Reagent (Invitrogen) followed by RNeasy Mini-kit (Qiagen) with DNase after 2 hours. Microarray analysis using GeneChip Human Gene 1.0 ST Arrays (Affymetrix) was performed by the University of Vermont DNA Microarray and Flow Cytometry Core Facilities. The XRay analysis package (Biotique Inc) was used to make pairwise comparisons between untreated vs. BMP2-treated, untreated vs. BMP7-treated and BMP2-treated vs. BMP7-treated.
2.3. Immunoblot
Analysis was performed as previously described [23], using rabbit antibodies against PhosphoSmads 1, 5, and 8 (Cell Signaling Technology), SNAI2 (ProSci), Beta Tubulin (Santa Cruz Biotechnology) and mouse antibodies against BMP2 (Abcam) and SNAI1 (Origene Technologies) overnight at 4°.
2.4. Immunofluorescence and immunohistochemistry
Immunofluorescent staining was as previously described [23]. Markers used were: Proximal tubules, Lotus Tetragonolobus lectin; Collecting ducts, Dolichos Biflorus Agglutinin (Vector Laboratories); β-galactosidase, rabbit anti-β-gal antibody (Cappel); pSmad1/5/8, clone Vli-49 rabbit monoclonal antibody raised against the NPIS(pS)V(pS) phosphopeptide from human SMAD1. Animal care in accordance with the National Research Council Guide for the Care and Use of Laboratory Animals was approved by the Institutional Animal Care and Use Committee of Maine Medical Center. Distal tubule,mouse anti-E. cadherin (BD Transduction Laboratories); thick ascending limb, sheep anti-Tamm Horsfall protein (Biodesign International). BMP2 was marked with a rabbit antibody (Abcam). Formalin fixed paraffin embedded human kidney sections were purchased from Zyagen. Our use of human tissue was reviewed and approved by the Institutional Review Board at Maine Medical Center. Nuclei were visualized with Dapi (Molecular Probes). Immunohistochemistry was performed as previously described [23] using a polyclonal rabbit anti-pSmad1/5/8 (Cell Signaling Technology).
2.5. Luciferase Assay
Renilla control plasmid pRLCMV (Promega) and the pBRE-Luc BMP transcriptional reporter [16] (provided by Peter ten Dijke, Leiden University, Netherlands) were transfected into HK-2 cells and assayed as previously described [23].
2.6. Miscellaneous Reagents
DAPT (Gamma Secretase Inhibitor IX) was obtained from Calbiochem. Cycloheximide was obtained from Sigma. The LDN-193189 BMP Inhibitor was obtained from Stemolecule. BMP2, BMP4, BMP7 and TGFβ1 were obtained from R&D Systems.
2.7. Quantitative PCR
RNA was prepared as for microarray. cDNA was synthesized using qScript cDNA SuperMix (Quanta Biosciences). QPCR was run in replicate wells on an iCycler IQ (BioRad) using iQ SYBR Green Supermix (Bio-Rad). Primers used are listed in Supplemental Table T3. Fold change analysis was computed using the ΔΔCT method normalizing to the expression of β-Actin. To determine relative basal levels of BMP expression in RPTECs, primer sets were designed to amplify targets within a single exon. Reaction efficiency for each primer set was determined using a standard curve generated from a dilution series of human genomic DNA. Cycle threshold values obtained from cDNA amplified from RPTECs were adjusted based on the reaction efficiency determined from genomic amplification of Beta actin, BMP2, 4 and 6
2.8. Statistical analysis
Statistical analysis was performed using Student’s t test, with a significant difference determined as P-value < 0.05. Data are presented as means +/− 1 standard deviation.
3. Results and discussion
3.1. Proximal tubule cells of the healthy kidney maintain basal BMP signaling in vivo
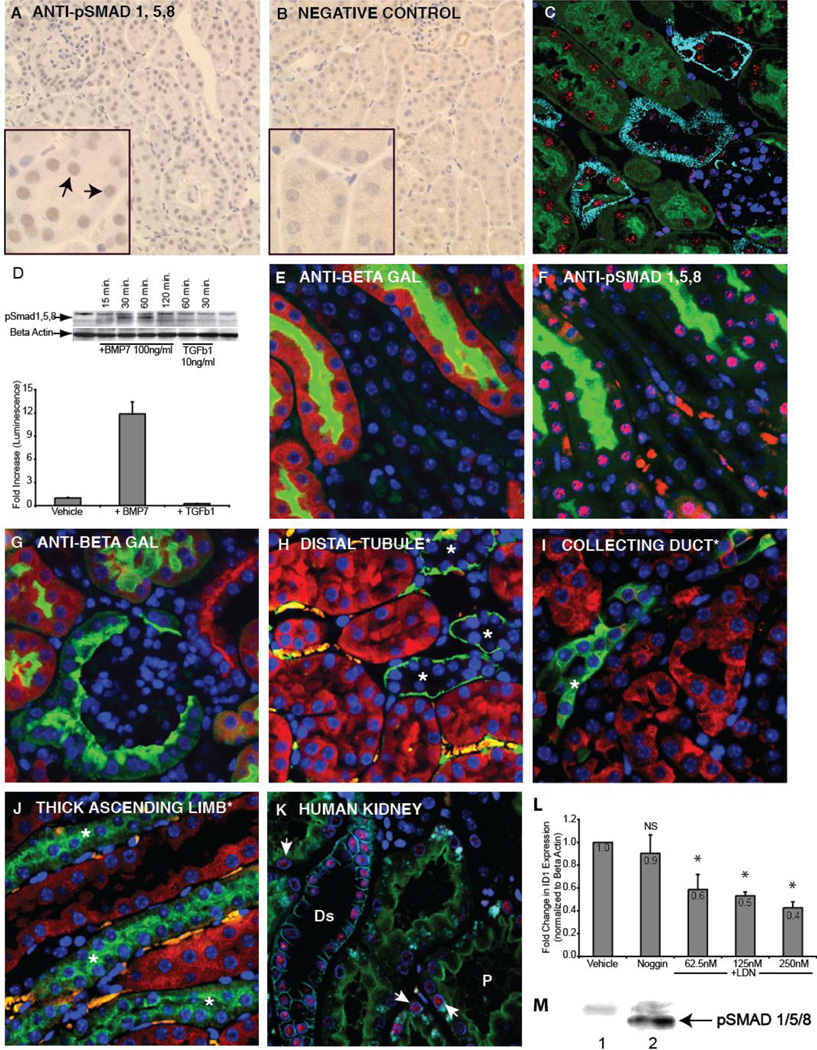
To define the extent and localization of BMP signaling in the healthy adult kidney, we analyzed both protein phosphorylation and pathway transcriptional activation. BMP ligands initiate signaling by binding to cell surface receptors that phosphorylate SMAD transcription factors 1, 5, and 8. These SMADs associate with the co-SMAD, SMAD 4, and the complex translocates to the nucleus where it associates with regulatory regions of target genes [15]. We monitored pathway activation using antibodies directed against phosphorylated SMAD1/5/8 (pSMAD1/5/8). Consistent with previous observations using commercial polyclonal antiserum [13, 24], active signaling was seen in mouse renal epithelium, but little or no signaling was detected in glomeruli and interstitium (Figure 1A,B). We confirmed this analysis using a newly developed monoclonal pSMAD1/5/8 antibody, and were able to immunofluorescently localize activation primarily to mouse proximal and distal tubules by marker costaining (Figure 1C). While some staining was observed in glomeruli, it was at a significantly lower level. Because recent reports show that SMAD1/5/8 phosphorylation may be driven by TGFβ1 [25–27], we evaluated pathway and transcriptional activation in TGFβ1 treated proximal tubule cells. No pSMAD1/5/8 or transcriptional activation can be detected in response to TGFβ1, indicating that pathway activation in the proximal tubule is a result of BMP signaling (Figure 1D). To evaluate BMP pathway specific transcriptional activation, we utilized a reporter mouse that drives expression of β-galactosidase under the control of a minimal promoter with a pSMAD1/5/8 binding element [16, 17]. Transcriptional activation shown by the reporter colocalized with pathway activation shown by nuclear pSMAD1/5/8 accumulation, with robust signaling activity in tubule epithelia and little in glomeruli (Figures 1E,F,G). To localize transcriptional activation using molecular markers, we stained for β-galactosidase and segment markers immunofluorescently. While high levels of signaling were present in the proximal tubule, there was no visible signaling at the same exposure in the distal tubule, collecting duct or thick ascending limb (Figures 1H – J). Having confirmed the correlation between pSMAD1/5/8 immunodetection and transcriptional activation, we evaluated pathway activation in human kidney tissue using the monoclonal pSMAD1/5/8 antibody. As in the mouse, pathway activation can be detected in the proximal tubule, but in contrast to the mouse signaling appears to be more robust in the distal tubule (Figure 1K). Some signaling was also observed in human glomeruli (data not shown). To understand if basal BMP pathway activation is maintained in cultured primary human renal proximal tubule epithelial cells (RPTECs), we measured transcription of the BMP target gene ID1 in cells treated with the LDN-193189 small molecule inhibitor of the ALK2 and ALK3 BMP receptors [28, 29]. Inhibitor treatment significantly reduced ID1 transcription, indicating that basal BMP signaling indeed does occur in isolated RPTECs (Figure 1L). To test the possibility that the culture medium was a source of BMP ligand, we incubated the cultures with Noggin protein, a potent extracellular BMP antagonist [23]. Noggin did not significantly reduce ID1 expression, indicating that endogenous pathway activation in RPTECs may be due to autocrine/intracrine signaling. The specificity of the monoclonal antibody against pSMAD 1/5/8 is demonstrated in Figure 1M.
Figure 1. BMP signaling in the adult mouse and human kidney.
A: Immunohistochemical staining for SMAD 1/5/8 (Cell Signaling Technology) reveals nuclear accumulation in mouse nephron epithelia. B: In the negative control, the primary antibody was omitted. C: Immunofluorescent staining of mouse kidney using monoclonal antibody against pSMAD1/5/8 (clone Vli-49), red), antibody against E-cadherin marking distal tubule (light blue), Lotus lectin marking proximal tubule (green) and DAPI (dark blue). D: Immunoblot comparing SMAD1/5/8 phosphorylation following BMP7 or TGFβ1 treatment of human primary renal proximal tubule epithelial cells (top panel). pBRE-Luc BMP transcriptional reporter assay in HK-2 human proximal tubule cells stimulated either with BMP7 (50 ng/ml) or TGFβ1 (5ng/ml) for 18 hours. E: Immunofluorescent staining of the reporter mouse kidney using an antibody against β-galactosidase (red) indicates that the highest level of BMP signaling is within the proximal tubule (green, lotus lectin). F: An adjacent section stained with a monoclonal anti pSMAD1/5/8 (Vli-49, red) shows pathway activation in a pattern consistent with reporter activation. Lotus lectin (green) labels proximal tubules. G: Staining for β-galactosidase (red) shows absence of staining in the glomerulus. Panels H, I, and J show BMP signaling (red) relative to molecular markers. H: Distal tubule (green, anti-E-cadherin). I: Collecting duct (green, DBA lectin). J: Thick ascending limb of the distal tubule (green, anti-Tamm Horsfall protein). K: In the human kidney, BMP signaling (red, monoclonal anti-pSMAD1/5/8 Vli-49) is most intense in the distal tubule (light blue, anti-E-cadherin), and weaker signal is seen in the proximal tubule (green, Lotus Lectin, arrows). L: Expression of ID1 was measured in RPTECs incubated for 14 hours with the BMP signaling inhibitor, LDN-193189. To control for BMP in medium, Noggin was added. Asterisk indicates a p value (relative to vehicle) of less than .05. M: Western blot analysis on MLFM4 cell lysate using rabbit monoclonal (Vli-49) anti-pSmad 1/5/8: Lane 1, untreated, lane 2 treated with BMP7 (50ng/ml). Abbreviations: Ds, distal tubule; P, proximal tubule, NS, not significant (p value greater than .05).
3.2 BMPs 2, 4, and 6 are expressed in human proximal tubule cells, with BMP2 being the most abundant
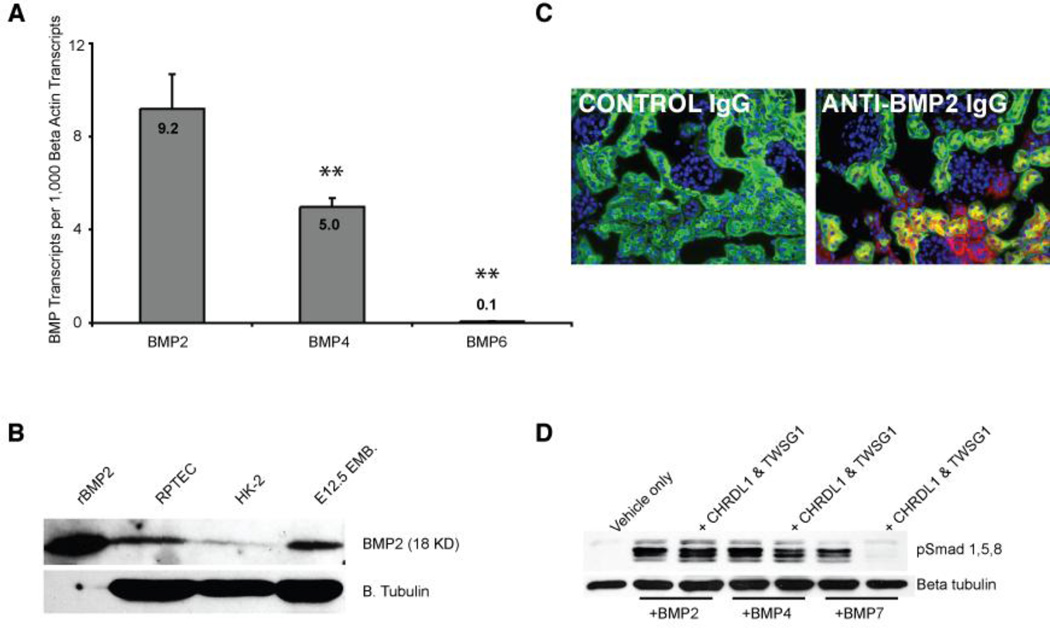
While the full complement of receptors required for BMP signaling is present on proximal tubule epithelia [30–32], it is not known which BMPs are responsible for stimulating basal signaling in these cells. We assayed expression levels of BMP 2, 3, 4, 5, 6, 7 and 9 in RPTECs, and observed detectable amplification of only BMP 2, 4, and 6. Relative to expression of β-actin, BMP2 was found to be most abundant (Figure 2A). Immunoblotting confirmed the expression of BMP2 in these cells (Figure 2B). In kidney tissue, immunostaining clearly demonstrates BMP2 expression in the proximal tubule (Figure 2C), strongly suggesting that autocrine BMP2 signal contributes to basal pathway activation in the proximal tubule.
Figure 2. BMP is expressed in the proximal tubule.
A: QPCR showing the number of BMP transcripts per 1,000 copies of β-actin transcripts. Double asterisks indicate a p value of less than .01 (when compared to BMP2 expression). B: Immunoblot showing BMP2 expression in RPTECs. Recombinant BMP2 protein and lysate from an E12.5 embryo were used as positive controls. C: Immunofluorescent micrographs of the kidney showing BMP2 (red) expression in renal epithelia, including proximal tubules (green, lotus lectin). In the left panel, nonspecific IgG was used as a negative control. D: The proximal tubule expressed BMP antagonist CHRDL1 antagonizes BMP7 but not BMP2 or 4 in conjunction with Twisted gastrulation (TWSG1). RPTECs were serum starved for 4 hours and incubated with BMPs (50ng/ml) and antagonists (400 ng/ml) for 30 minutes prior to lysis and western blotting for pSMAD1/5/8.
3.3 BMP7, but not BMP2 or 4 is modulated by CHRDL1, a BMP antagonist specific to the proximal tubule
It has previously been shown that BMP7 mediated signaling is strongly antagonized in both distal [13] and proximal tubules. We have previously shown that the antagonist CHRDL1 is specifically expressed in the proximal tubule and antagonizes BMP7 in conjunction with the ubiquitous TWSG1 protein [23]. To understand how BMP2 signaling in the proximal tubule may be affected by this antagonist, we conducted a series of interaction studies using RPTECs. We show that CHRDL1 and TWSG1 together do not antagonize SMAD dependent BMP signaling driven by BMP2 or 4 (Figure 2D), suggesting that paracrine BMP7 signaling is blocked, but autocrine/intracrine BMP2/4 signaling is permitted in the healthy proximal tubule. In the injured kidney however, CHRDL1 is lost from the proximal tubule allowing BMP7 signaling [23]. Biochemical studies have shown differential receptor binding by BMP2 and BMP7 [33, 34], suggesting that these closely related ligands may elicit distinct transcriptional responses. To evaluate this possibility we performed a comparative global transcriptome analysis.
3.4 BMP2 and BMP7 elicit indistinguishable early transcriptional responses
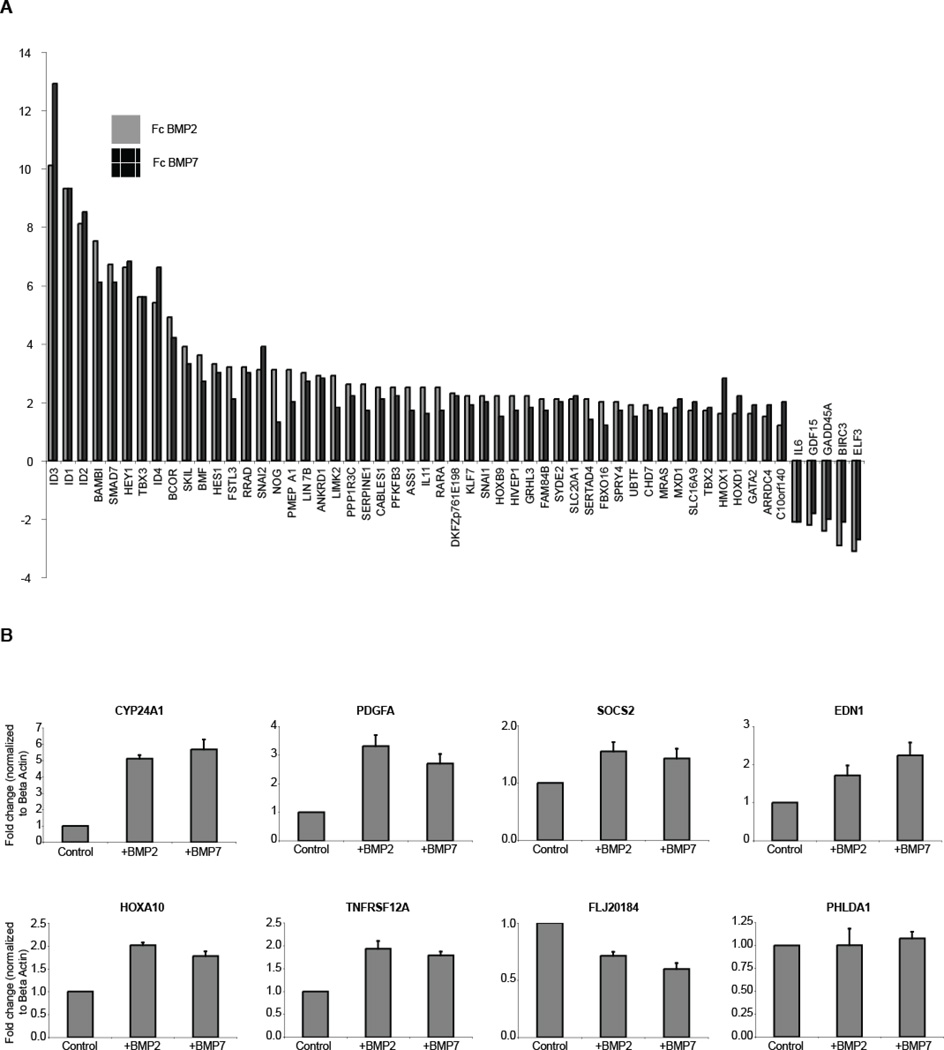
To globally identify transcriptional targets of BMP2 and BMP7 signaling in the proximal tubule, we stimulated primary proximal tubule cells with ligand, and measured transcriptional activation after 2 hours. This time point was chosen because the peak of SMAD1/5/8 activation is seen at 1 hour (Figure 1D), allowing approximately 1 hour for accumulation of transcript. Because we wanted to profile the effects of BMP2 vs. BMP7 unbiased by the BMP7-specific antagonism of CHRDL1, we used experimental conditions that did not favor the accumulation of secreted CHRDL1 in the medium. Comparing untreated versus treated cells with a threshold of p < 0.01, the microarray revealed 55 transcripts that responded to either BMP2 or BMP7 ligand with a minimum fold change of +/− 1.75 and were significantly regulated in the same direction by both ligands (Figure 3A and Supplemental Table T1). Using the same thresholds, 27 genes were putatively regulated solely by BMP2, and 4 genes were putatively regulated by BMP7 only (Supplemental Table T2). To verify these candidate BMP2- and BMP7-specific targets, we performed a direct comparison between BMP2 and BMP7 treated cells, thresholding with p < 0.01. Using this approach the candidate list was reduced to 8 putatively uniquely regulated genes, which were assayed by qPCR (Figure 3B). Although qPCR analysis verified that 7 out of 8 of these genes were indeed BMP-regulated, no material difference in transcriptional activation was seen following BMP2 or BMP7 treatment (Figure 3B), and we conclude that no qualitative differences in global transcriptional activation can be seen between BMP2 and BMP7 in primary proximal tubule cells using our stringency thresholds. Previous genetic studies from our laboratory have shown that BMPs 4, 6, and 7 are functionally interchangeable in kidney development [35], and the finding that BMP2 and 7 elicit identical responses in the adult proximal tubule cell supports the model that transcriptional responses to distinct BMPs in any particular cell type may be equivalent. Our results here are also consistent with earlier findings by Korchynskyi, et al. showing a virtually identical transcriptional response driven by the three different constitutively active BMP Type I receptors in C2C12 cells [36]. Rather than initiating distinct cellular responses, BMPs may instead distinguish themselves functionally by associating with different combinations of extracellular modulators such as CHRDL1, which we [23] and others [37] have shown can be highly specific for distinct BMP ligands. In a similar vein, Sclerostin, encoded by the SOST gene, has been found to antagonize BMP6 and 7, but not BMP 2 or 4 [38].
Figure 3. BMP2 and BMP7 stimulation of RPTECs produce closely related expression profiles.
A: Gene profiling of RPTECs stimulated for two hours with 50ng/ml of BMP2 or BMP7 shows extensive overlap. All candidates with a p-value < 0.01 are shown where stimulation with either ligand resulted in a > +/− 1.75 fold change and both ligands drove expression in the same direction. B: A subset of genes suggested by the profiling to be differentially regulated by BMP2 and BMP7 were compared by qPCR and were regulated in a similar fashion.
3.5 BMP activates a cassette of genes typically regulated by the Notch pathway
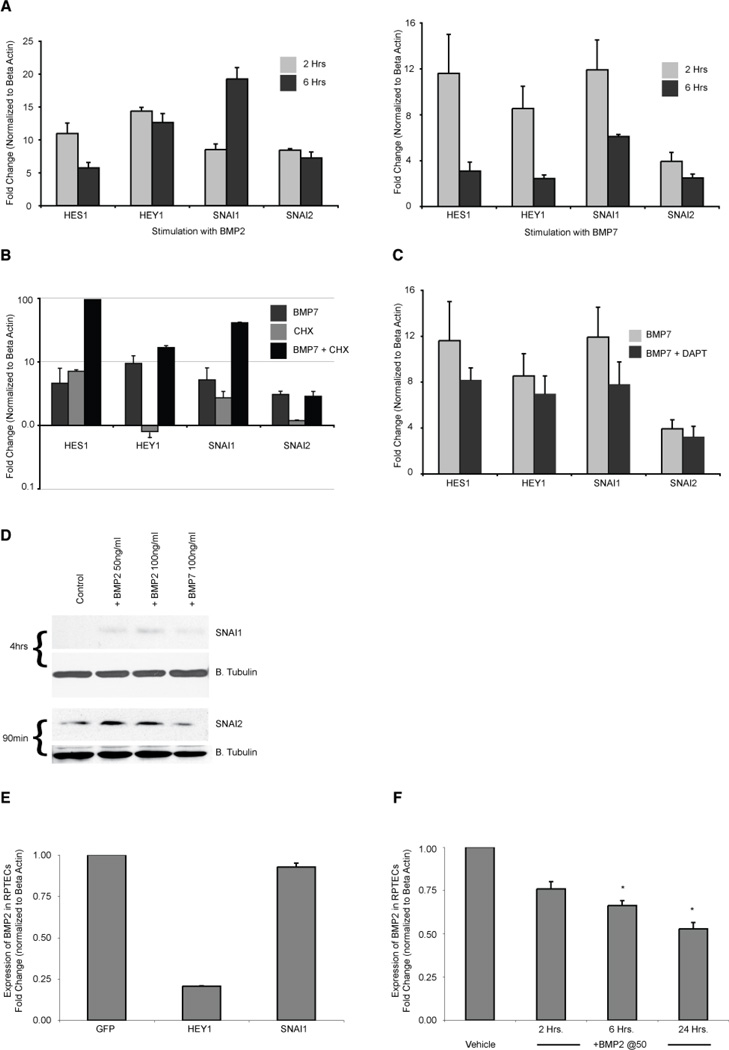
The BMP transcriptional profile included several prototypical BMP response genes such as ID1 – 4, SMAD7, NOG and BAMBI. Interestingly, a cassette of genes typically associated with Notch signaling was found among the more highly regulated genes: HEY1, HES1, SNAI1 (SNAIL), and SNAI2 (SLUG) [39–41] (Figures 3A, 4A). While some reports suggest that Notch signaling plays a beneficial role in recovery from AKI [42, 43], others identify a pathological role for this pathway in kidney injury and disease [44, 45]. The finding that Notch targets are activated by BMP signaling suggests an intriguing and potentially important relationship between the BMP and Notch pathways in the proximal tubule. By repeating our BMP stimulation experiment in the presence of the translation inhibitor cycloheximide, we verified that Notch targets were primary transcriptional targets of BMP (Figure 4B). Furthermore, stimulation in the presence of a Notch pathway inhibitor verified that simultaneous Notch signaling is not required for BMP activation of these Notch target genes (Figure 4C). Thus, BMP signaling in proximal tubule epithelial cells directly activates not only conventional BMP targets, but also Notch targets. Immunoblotting demonstrated that SNAI1 and SNAI2 proteins are induced by BMP stimulation, indicating that the transcriptional responses that we have identified are mirrored by increases in protein abundance (Figure 4D). Interestingly, the response to BMP2 appears stronger than that to BMP7, a feature that we have noted in previous experiments (unpublished observations). One key difference between the response to BMP2 and BMP7 seen at the transcriptional level is the duration of transcriptional activation: between 2 and 6 hours of stimulation there is maintenance or even accumulation of BMP2 driven transcript, whereas the transcript level declines in BMP7 treated cells (Figure 4A). This difference may be due to accumulation of CHRDL1 in cell culture medium, leading to specific antagonism of BMP7. However, we cannot exclude the possibility that distinct stabilities of the recombinant protein preparations contribute to this difference. Unfortunately, we have been unable to visualize endogenous levels of HEY1 and HES1 proteins using several commercially available antibodies (data not shown).
Figure 4. BMP signaling directly activates a set of typical Notch response genes.
A: QPCR confirming that HES1, HEY1, SNAI1 and SNAI2 are strongly up-regulated in RPTECs after treatment for 2 or 6 hours with 50ng/ml of BMP2 (left panel) and BMP7 (right panel). The less durable response of BMP7 at 6 hours may result from increased amounts of secreted CHRDL1 accumulating in the medium and resulting in greater antagonism of BMP7, but not BMP2. B: RPTECs were incubated for 30 minutes with cycloheximide (5µM) or vehicle prior to the addition of BMP7 (50ng/ml) for 2 hours and analyzed by RT-qPCR. Cycloheximide did not reduce target gene activation showing that protein synthesis is not required for BMP7 induction of expression. C: Cells were incubated with vehicle or 5µM DAPT for 30 minutes prior to adding BMP7 at 50ng/ml. qPCR shows that the inhibition of Notch signaling did not significantly reduce gene induction at two hours. D: REPTCs were serum starved for 2 hours, then treated for the time specified with BMP2 at 50 and 100ng/ml or with BMP7 at 100ng/ml. Immunoblots were probed for SNAI1 (Snail) and SNAI2 (Slug). Blots for Beta tubulin below each protein of interest show relative loading of sample. E: qPCR showing BMP2 expression in RPTECs transduced for 32 hours with adenoviral vectors expressing GFP (control), HEY1 or SNAI1. F: qPCR showing reduction in BMP2 expression in RPTECs incubated with 50ng/ml of BMP2 for 2, 6, and 24 hours. Asterisk indicates p value < 0.05.
3.6 HEY1 functions as a negative feedback regulator of BMP2 expression in the proximal tubule
Studies have shown that Notch signaling reduces BMP2 expression in other cell types through the expression of HEY1 [46–48]. To assess whether this also is a feature of the proximal tubule, we measured BMP2 expression in HEY1 transduced cells (Figure 4E). We find that HEY1, but not SNAI1, strongly and specifically attenuates BMP2 expression, indicating that this BMP induced transcription factor limits BMP production by the proximal tubule. As predicted, this downregulation of BMP2 can be recapitulated by BMP treatment (Figure 4F).
4. Conclusion
Our findings highlight the complexity of BMP signaling in the adult nephron. In the healthy kidney, BMP7 is produced in the distal tubule but it is vigorously counteracted by the extracellular antagonists USAG1 [13] and CHRDL1 [23], suggesting that BMP7-elicited signaling is highly restricted in the healthy kidney. Our present study indicates that endogenously produced BMP2 contributes to signaling in the healthy proximal tubule, with HEY1 acting as a feedback regulator to limit BMP2 production and pathway activation. Following kidney injury, extracellular antagonism of BMP7 is lost in both proximal and distal tubules [13, 23] concomitant with upregulation of the signaling amplifier KCP [14]. Paracrine BMP7 signaling from the distal tubule is thus permitted, eliciting BMP target gene activation in the proximal tubule. Considering the similarity of BMP2 and BMP7 transcriptional targets, we propose that BMP7 from the distal tubule may compensate for the loss of BMP2 following proximal tubule injury. Further functional analysis of these target genes will identify their roles in homeostasis, and in the regenerative process.
Highlights.
Extensive BMP signaling in healthy adult kidney, peak levels in proximal tubule in mouse, and distal tubule in human
BMP 2 is expressed in the proximal tubule
Transcriptional profiling shows BMP2 and BMP7 elicit indistinguishable responses
Transcriptional response of proximal tubule cells to BMP includes a cassette of genes typically downstream of Notch
Notch response gene HEY1 suppresses BMP2 expression in proximal tubule cells
Supplementary Material
Acknowledgments
This work was supported by NIH/NCRR 2P20RR18798 (project 11, PI: LO). Additional technical core support was provided by MMCRI's Center of Excellence in the Stem and Progenitor Cell Biology core facility in bioinformatics (2P20RR18798) and the Center of Excellence in Vascular Biology cell culture and viral vector core facility (2P20RR15555). We are grateful to Dr. Aaron C. Brown, Ph.D for his thoughtful comments on this manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Statement of competing financial interests
The authors have no competing financial interests.
References
- 1.Palevsky PM, Zhang JH, O'Connor TZ, Chertow GM, Crowley ST, Choudhury D, Finkel K, Kellum JA, Paganini E, Schein RM, Smith MW, Swanson KM, Thompson BT, Vijayan A, Watnick S, Star RA, Peduzzi P. N Engl J Med. 2008;359(1):7–20. doi: 10.1056/NEJMoa0802639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderson S, Eldadah B, Halter JB, Hazzard WR, Himmelfarb J, Horne FM, Kimmel PL, Molitoris BA, Murthy M, O'Hare AM, Schmader KE, High KP. J Am Soc Nephrol. 22(1):28–38. doi: 10.1681/ASN.2010090934. [DOI] [PubMed] [Google Scholar]
- 3.Witzgall R, Brown D, Schwarz C, Bonventre JV. J Clin Invest. 1994;93(5):2175–2188. doi: 10.1172/JCI117214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vogetseder A, Picard N, Gaspert A, Walch M, Kaissling B, Le Hir M. Am J Physiol Cell Physiol. 2008;294(1):C22–C28. doi: 10.1152/ajpcell.00227.2007. [DOI] [PubMed] [Google Scholar]
- 5.Fujigaki Y, Goto T, Sakakima M, Fukasawa H, Miyaji T, Yamamoto T, Hishida A. Nephrol Dial Transplant. 2006;21(1):41–50. doi: 10.1093/ndt/gfi035. [DOI] [PubMed] [Google Scholar]
- 6.Humphreys BD, Valerius MT, Kobayashi A, Mugford JW, Soeung S, Duffield JS, McMahon AP, Bonventre JV. Cell Stem Cell. 2008;2(3):284–291. doi: 10.1016/j.stem.2008.01.014. [DOI] [PubMed] [Google Scholar]
- 7.Humphreys BD, Czerniak S, Dirocco DP, Hasnain W, Cheema R, Bonventre JV. Proc Natl Acad Sci U S A. 2011 doi: 10.1073/pnas.1100629108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang S, Chen Q, Simon TC, Strebeck F, Chaudhary L, Morrissey J, Liapis H, Klahr S, Hruska KA. Kidney Int. 2003;63(6):2037–2049. doi: 10.1046/j.1523-1755.2003.00035.x. [DOI] [PubMed] [Google Scholar]
- 9.Wang S, de Caestecker M, Kopp J, Mitu G, Lapage J, Hirschberg R. J Am Soc Nephrol. 2006 doi: 10.1681/ASN.2006030278. [DOI] [PubMed] [Google Scholar]
- 10.Zeisberg M, Bottiglio C, Kumar N, Maeshima Y, Strutz F, Muller GA, Kalluri R. Am J Physiol Renal Physiol. 2003;285(6):F1060–F1067. doi: 10.1152/ajprenal.00191.2002. [DOI] [PubMed] [Google Scholar]
- 11.Hruska KA, Guo G, Wozniak M, Martin D, Miller S, Liapis H, Loveday K, Klahr S, Sampath TK, Morrissey J. Am J Physiol Renal Physiol. 2000;279(1):F130–F143. doi: 10.1152/ajprenal.2000.279.1.F130. [DOI] [PubMed] [Google Scholar]
- 12.Vukicevic S, Basic V, Rogic D, Basic N, Shih MS, Shepard A, Jin D, Dattatreyamurty B, Jones W, Dorai H, Ryan S, Griffiths D, Maliakal J, Jelic M, Pastorcic M, Stavljenic A, Sampath TK. J Clin Invest. 1998;102(1):202–214. doi: 10.1172/JCI2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yanagita M, Okuda T, Endo S, Tanaka M, Takahashi K, Sugiyama F, Kunita S, Takahashi S, Fukatsu A, Yanagisawa M, Kita T, Sakurai T. J Clin Invest. 2006;116(1):70–79. doi: 10.1172/JCI25445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lin J, Patel SR, Cheng X, Cho EA, Levitan I, Ullenbruch M, Phan SH, Park JM, Dressler GR. Nat Med. 2005;11(4):387–393. doi: 10.1038/nm1217. [DOI] [PubMed] [Google Scholar]
- 15.Massagué J. Nat Rev Mol Cell Biol. 2000;1(3):169–178. doi: 10.1038/35043051. [DOI] [PubMed] [Google Scholar]
- 16.Korchynskyi O, ten Dijke P. J Biol Chem. 2002;277(7):4883–4891. doi: 10.1074/jbc.M111023200. [DOI] [PubMed] [Google Scholar]
- 17.Blank U, Seto ML, Adams DC, Wojchowski DM, Karolak MJ, Oxburgh L. BMC Dev Biol. 2008;8:86. doi: 10.1186/1471-213X-8-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Monteiro RM, De Sousa Lopes SM, Korchynskyi O, Dijke Pt P, Mummery CL. J Cell Sci. 2004;117(Pt 20):4653–4663. doi: 10.1242/jcs.01337. [DOI] [PubMed] [Google Scholar]
- 19.Venkatachalam MA, Bernard DB, Donohoe JF, Levinsky NG. Kidney Int. 1978;14(1):31–49. doi: 10.1038/ki.1978.87. [DOI] [PubMed] [Google Scholar]
- 20.Sheridan AM, Bonventre JV. Curr Opin Nephrol Hypertens. 2000;9(4):427–434. doi: 10.1097/00041552-200007000-00015. [DOI] [PubMed] [Google Scholar]
- 21.Tang Y, Urs S, Liaw L. Circ Res. 2008;102(6):661–668. doi: 10.1161/CIRCRESAHA.107.165134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kajita M, McClinic KN, Wade PA. Mol Cell Biol. 2004;24(17):7559–7566. doi: 10.1128/MCB.24.17.7559-7566.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Larman BW, Karolak MJ, Adams DC, Oxburgh L. J Am Soc Nephrol. 2009;20(5):1020–1031. doi: 10.1681/ASN.2008070768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dendooven A, van Oostrom O, van der Giezen DM, Willem Leeuwis J, Snijckers C, Joles JA, Robertson EJ, Verhaar MC, Nguyen TQ, Goldschmeding R. Am J Pathol. 178(3):1069–1079. doi: 10.1016/j.ajpath.2010.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tanaka M, Asada M, Higashi AY, Nakamura J, Oguchi A, Tomita M, Yamada S, Asada N, Takase M, Okuda T, Kawachi H, Economides AN, Robertson E, Takahashi S, Sakurai T, Goldschmeding R, Muso E, Fukatsu A, Kita T, Yanagita M. J Clin Invest. 120(3):768–777. doi: 10.1172/JCI39569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Daly AC, Randall RA, Hill CS. Mol Cell Biol. 2008;28(22):6889–6902. doi: 10.1128/MCB.01192-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Luo DD, Phillips A, Fraser D. Am J Pathol. 176(3):1139–1147. doi: 10.2353/ajpath.2010.090459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cuny GD, Yu PB, Laha JK, Xing X, Liu JF, Lai CS, Deng DY, Sachidanandan C, Bloch KD, Peterson RT. Bioorg Med Chem Lett. 2008;18(15):4388–4392. doi: 10.1016/j.bmcl.2008.06.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yu PB, Deng DY, Lai CS, Hong CC, Cuny GD, Bouxsein ML, Hong DW, McManus PM, Katagiri T, Sachidanandan C, Kamiya N, Fukuda T, Mishina Y, Peterson RT, Bloch KD. Nat Med. 2008;14(12):1363–1369. doi: 10.1038/nm.1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gould SE, Day M, Jones SS, Dorai H. Kidney Int. 2002;61(1):51–60. doi: 10.1046/j.1523-1755.2002.00103.x. [DOI] [PubMed] [Google Scholar]
- 31.Kitten AM, Kreisberg JI, Olson MS. J Cell Physiol. 1999;181(3):410–415. doi: 10.1002/(SICI)1097-4652(199912)181:3<410::AID-JCP4>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 32.Wetzel P, Haag J, Campean V, Goldschmeding R, Atalla A, Amann K, Aigner T. Kidney Int. 2006 doi: 10.1038/sj.ki.5001653. [DOI] [PubMed] [Google Scholar]
- 33.ten Dijke P, Yamashita H, Sampath TK, Reddi AH, Estevez M, Riddle DL, Ichijo H, Heldin CH, Miyazono K. J Biol Chem. 1994;269(25):16985–16988. [PubMed] [Google Scholar]
- 34.Macias-Silva M, Hoodless PA, Tang SJ, Buchwald M, Wrana JL. J Biol Chem. 1998;273(40):25628–25636. doi: 10.1074/jbc.273.40.25628. [DOI] [PubMed] [Google Scholar]
- 35.Oxburgh L, Dudley AT, Godin RE, Koonce CH, Islam A, Anderson DC, Bikoff EK, Robertson EJ. Dev Biol. 2005;286(2):637–646. doi: 10.1016/j.ydbio.2005.08.024. [DOI] [PubMed] [Google Scholar]
- 36.Korchynskyi O, Dechering KJ, Sijbers AM, Olijve W, ten Dijke P. J Bone Miner Res. 2003;18(7):1177–1185. doi: 10.1359/jbmr.2003.18.7.1177. [DOI] [PubMed] [Google Scholar]
- 37.Chandra A, Itakura T, Yang Z, Tamakoshi T, Xue X, Wang B, Ueki T, Sato K, Uezato T, Miura N. Biochem Biophys Res Commun. 2006;344(3):786–791. doi: 10.1016/j.bbrc.2006.03.195. [DOI] [PubMed] [Google Scholar]
- 38.Kusu N, Laurikkala J, Imanishi M, Usui H, Konishi M, Miyake A, Thesleff I, Itoh N. J Biol Chem. 2003;278(26):24113–24117. doi: 10.1074/jbc.M301716200. [DOI] [PubMed] [Google Scholar]
- 39.Iso T, Kedes L, Hamamori Y. J Cell Physiol. 2003;194(3):237–255. doi: 10.1002/jcp.10208. [DOI] [PubMed] [Google Scholar]
- 40.Timmerman LA, Grego-Bessa J, Raya A, Bertran E, Perez-Pomares JM, Diez J, Aranda S, Palomo S, McCormick F, Izpisua-Belmonte JC, de la Pompa JL. Genes Dev. 2004;18(1):99–115. doi: 10.1101/gad.276304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Leong KG, Niessen K, Kulic I, Raouf A, Eaves C, Pollet I, Karsan A. J Exp Med. 2007;204(12):2935–2948. doi: 10.1084/jem.20071082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kobayashi T, Terada Y, Kuwana H, Tanaka H, Okado T, Kuwahara M, Tohda S, Sakano S, Sasaki S. Kidney Int. 2008;73(11):1240–1250. doi: 10.1038/ki.2008.74. [DOI] [PubMed] [Google Scholar]
- 43.Gupta S, Li S, Abedin MJ, Wang L, Schneider E, Najafian B, Rosenberg ME. Am J Physiol Renal Physiol. 2009 doi: 10.1152/ajprenal.00451.2009. [DOI] [PubMed] [Google Scholar]
- 44.Huang R, Zhou Q, Veeraragoo P, Yu H, Xiao Z. Ren Fail. 33(2):207–216. doi: 10.3109/0886022X.2011.553979. [DOI] [PubMed] [Google Scholar]
- 45.Murea M, Park JK, Sharma S, Kato H, Gruenwald A, Niranjan T, Si H, Thomas DB, Pullman JM, Melamed ML, Susztak K. Kidney Int. 78(5):514–522. doi: 10.1038/ki.2010.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Minamizato T, Sakamoto K, Liu T, Kokubo H, Katsube K, Perbal B, Nakamura S, Yamaguchi A. Biochem Biophys Res Commun. 2007;354(2):567–573. doi: 10.1016/j.bbrc.2007.01.029. [DOI] [PubMed] [Google Scholar]
- 47.Rutenberg JB, Fischer A, Jia H, Gessler M, Zhong TP, Mercola M. Development. 2006;133(21):4381–4390. doi: 10.1242/dev.02607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Luna-Zurita L, Prados B, Grego-Bessa J, Luxan G, del Monte G, Benguria A, Adams RH, Perez-Pomares JM, de la Pompa JL. J Clin Invest. 120(10):3493–3507. doi: 10.1172/JCI42666. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.