Abstract
This Teaching Resource provides lecture notes, slides, and a problem set for a series of lectures introducing the mathematical concepts behind gene-set enrichment analysis (GSEA) and were part of a course entitled “Systems Biology: Biomedical Modeling.” GSEA is a statistical functional enrichment analysis commonly applied to identify enrichment of biological functional categories in sets of ranked differentially expressed genes from genome-wide mRNA expression data sets.
Keywords: Enrichment analysis, random walks, Kolmogorov-Smirnov, ranked gene list, gene expression microarrays
Introduction
This Teaching Resource is intended for instructors who have some knowledge of statistics. Familiarity with the following programs is useful: R (http://www.rproject.org/) and the GSEA software http://www.broadinstitute.org/gsea/). GSEA is an analysis method that is increasingly gaining acceptance for analyzing genome-wide molecular profiling data.
Lecture Notes
Gene-Set Enrichment Analysis
Transcriptional profiling, by methods such as microarrays or RNA-seq experiments, measures the changes in expression of a large number of genes. These are used to investigate the changes in mRNA abundance that occurs in response to a stimulus or the differences in mRNA status between two different samples. For example, tissue samples from two groups of people—one with a particular disease and the other healthy—may be compared to identify the tissue-specific differences in gene expression.
Traditionally, transcriptional microarray data have been analyzed by a “single-gene approach” to determine the individual genes exhibiting the greatest differences in expression between the two sets of samples. However, this single-gene approach is limited. Gene-set enrichment analysis (GSEA) is a means of identifying, not just individual genes, but groups of genes that are known to be functionally related (Slide 2). Sets of functionally related genes can be obtained from various preestablished libraries, such as libraries of genes encoding proteins involved in metabolic pathways, cell signaling pathways, or kinases; genes that are targeted by particular microRNAs; or genes that produce a common phenotype when knocked down in a model organism, such as mice or yeast. The basic idea of GSEA is that differences in the expression of a set of genes will “stand out” in the data more clearly than differences in the expression of an individual gene.
The lecture follows a step-by-step approach to explain the method of GSEA and the mathematical basis for this type of data analysis (1, 2). The lecture begins with an explanation of “random walks” and then describes a statistical test (the Kolmogorov-Smirnov test) that is based on the idea of a random walk (Slide 3). The two concepts serve as the foundation for understanding GSEA.
One-Dimensional Random Walks
The simplest kind of random walk takes place on a one-dimensional lattice, such as a simple number line with equally spaced points (Slide 4). The walk occurs by moving randomly forward and backward along the line.
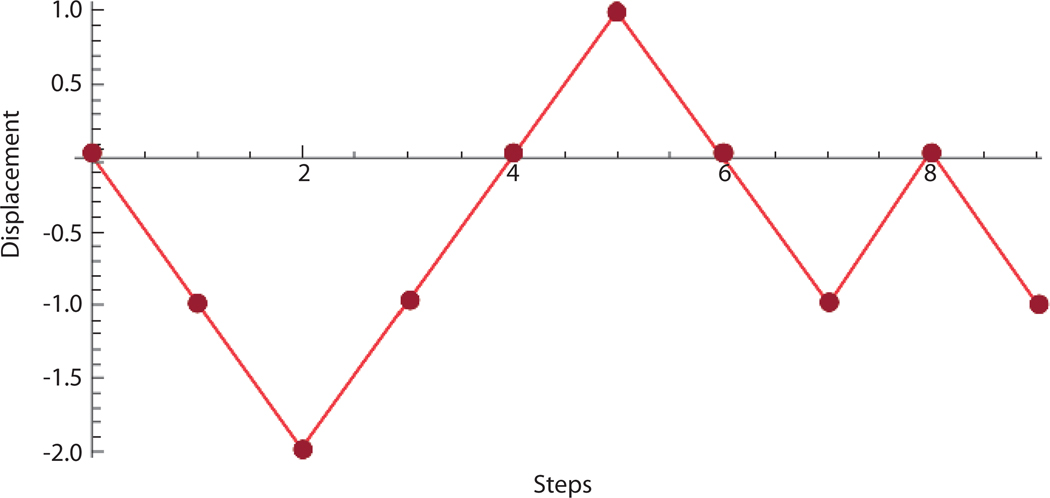
Because it would be difficult to see individual steps that may overlap, the steps of the random walk are plotted on a two-dimensional graph, such that the horizontal (x axis) represents time (or step number) and a vertical (y axis) represents the position along the lattice (Fig. 1; Slide 4). At each step there are two choices, to go left or to go right, which leads to a binomial distribution of the number of ways to reach each possible position at each step (Fig. 2). The mean distance from the starting point, after n steps is proportional to . Long random walks with many steps exhibit fluctuations and these fluctuations can be observed at different resolution scales (Slide 4). If the number of steps goes to infinity and the size of each step goes to zero (the steps become infinitely close together), this becomes a continuous walk in time t, which is called a Weiner process W(t). In stochastic differential equations, the Weiner process is used as an approximation of Brownian motion (Slide 4).
Fig. 1.
Plot of a random walk.
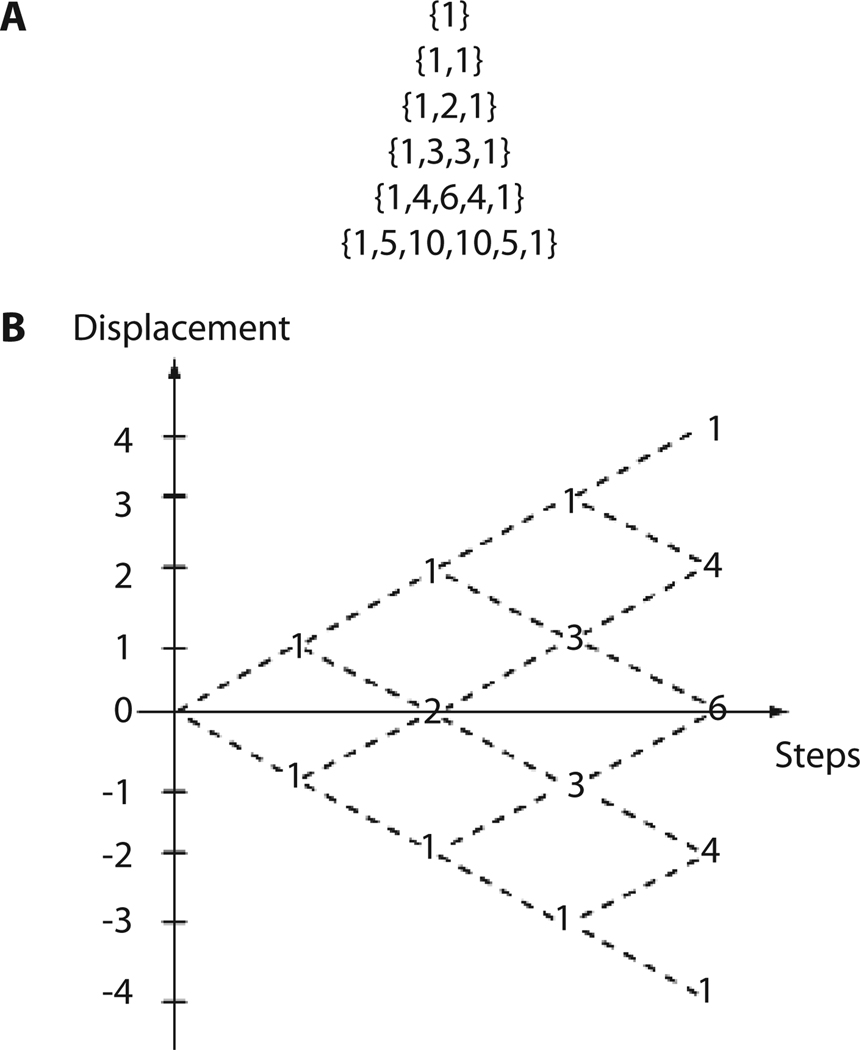
Fig. 2.
Random walks results in a binomial distribution. (A) Example of the binomial distribution produced by five steps. (B) Possible routes for an unbiased random walk starting at the origin (0). Numbers represent the number of possible walks to reach a specific distance from the origin.
In GSEA, which is a modified form of the random walk, the start and end points of the walk are fixed at zero, but the intervening steps are random (Slide 5). In this case, when the number of steps goes to infinity and the step size goes to zero, the random walk becomes another type of continuous process called the Brownian bridge B(t) (Slide 5). In the Weiner process, variation increases throughout the walk, whereas in a Brownian bridge, the variation occurs in the middle of the walk. Although for a random walk without fixed end points, the mean distance from the starting point increases in proportion to ; for a random walk with fixed end points, it is more meaningful to determine the maximum distance any step is from the starting position. In mathematical terminology, this maximum distance traveled is called the “supremum” of the random walk. Kolmogorov (3) reported a recurrence relation that expresses the probability distribution of the supremum of a discrete random walk, and for the continuous walk the probability distribution is given by
| (1) |
where CDF(x) is the probability that the supremum of the Brownian bridge is less than or equal to x. This is called the Kolmogorov distribution (Slide 5).
The Kolmogorov-Smirnov test
GSEA is based on the Kolmogorov-Smirnov test, which is a statistical test of “goodness of fit.” It is an alternative to the commonly applied χ2 test, but it is better than the χ2 test when the sample size is small (Slide 7). In order to understand the Kolmogorov-Smirnov test, it is first important to understand the probability density function and the cumulative distribution function (Slide 6).
For a random variable x, the probability density function, p(x), provides the probability of measuring x in a given range. For example, the probability of measuring x between the values of a and b is calculated from the integral of p(x), as shown in Eq. 2:
| (2) |
For example, for the Gaussian probability distribution function with a mean of 1 and variance of 0.5, the area under the curve between any two points on the x axis provides the probability (P value) of observing measurements in that range (Slide 6).
The cumulative distribution function, CDF(x), which gives the probability of measuring the variable at a value of x or smaller, is related to the probability density function as defined in Eq. 3:
| (3) |
The Kolmogorov-Smirnov test is a mathematical way of answering the question “Given my set of data, how sure can I be that the cumulative distribution function is given by CDF(x)?”
To illustrate the basic idea of this test, we use two simple data sets, one for which the data fit the cumulative distribution function (Slide 8) and one for which the data fail to fit the cumulative distribution function (Slide 9). For the purposes of illustration, we assume that the data should be compared with Gaussian curves with a mean of 1 and a variance of 0.5 because random walks follow this Gaussian distribution. Thus, by comparing the data with this Gaussian distribution, deviations from a random walk can be analyzed. These examples show that at any point x, the curve corresponds to the total fraction of our data that has a value less than or equal to x, and the top graphs in Slides 8 and 9 show the cumulative distribution for the sample data, along with cumulative distribution functions based on a Gaussian distribution with a mean of 1 and a variance of 0.5, like that shown in Slide 6.
Even for the data that appear to match the cumulative distribution function well, there are differences, because the data set only has six points and would be expected to have some random scatter (Slide 8).
The Kolmogorov-Smirnov test is used to determine whether the data’s cumulative distribution function is consistent with the hypothesized distribution by showing that, when the two curves are the same, the difference between these two curves is a random walk (Slide 8, bottom graph). Although the difference plot does appear to be a random walk, this can be quantified using the supremum, which states that if the number of data points were to increase infinitely, then the supremum of this random walk would tend toward zero when the two distributions were identical. However, gene expression data are not infinite. Therefore, in the case of a finite number of data points (in the example, the number is 6, but typically, the number can be as much as 20,000 to 30,000 for gene expression data), as the number of steps (data points) increases, the supremum of this random walk, multiplied by to account for the finite step size of the walk, becomes closer and closer to the supremum of the continuous Brownian bridge for which the probability distribution is given by Eq. 4:
| (4) |
For GSEA, it is not necessary to calculate the distribution from Eq. 4; instead simply calculating the value of and then comparing this value, called the enrichment score, with the critical values would compute whether the data are random and follow a random walk. This critical value is typically computed for random data sets and is provided by the software that computes the test. When the value of is within the confidence interval and is less than the critical value, then the hypothesis that the data match the Gaussian distribution with the defined parameters is considered true.
For the data plotted in Slide 8, the supremum of the random walk is about 0.15, and the data consist of six data points (Eq. 5):
| (5) |
According to a table of precalculated values computed for random data, the critical value at the 1% confidence level is 0.41, so the hypothesis that the data come from the Gaussian distribution with mean 1.0 and variance 0.5, with a 1% error level, is accepted.
For the second sample data set (Slide 9), performing the same analysis and assuming that the data fit a Gaussian distribution with a mean of 1 and a variance of 0.5 shows that the supremum looks large for a random walk. Most random walks would have a supremum less than 0.41. Comparing the supremum to that expected for a Brownian bridge yields Eq. 6:
| (6) |
According to the table of precalculated values we created for random data, 0.98 is larger than the critical value, which indicates that these data do not fit a Gaussian distribution with mean of 1 and variance of 0.5. Hence, the hypothesis that the data are coming from a random walk is rejected, and it is likely that there is a biologically meaningful pattern in the data.
The GSEA Test
Typically gene expression profiles are measured for a few samples from two different conditions, such as diseased and healthy cells or treated and control cells. The expression profiles are then examined for differences that correspond to the specific condition, with the goal that understanding these gene expression differences may help explain the molecular basis between the two conditions (Slide 10).
One method for analyzing these kinds of data are to rank the genes according to their differential expression across the different conditions and, then, to focus attention on those genes that are found at the top and bottom of the list. The genes at the top are those that are differentially expressed less in the 1st condition than the 2nd (up-regulated in condition 2), and genes at the bottom are those that are expressed more in the 1st condition than the 2nd (down-regulated in condition 2). Those genes exhibiting a fold change in expression greater than a set amount (for example, greater than twofold change) are considered to contribute to the differences in the two conditions. However, there are some problems with this approach. First, no individual genes may exhibit a change in expression that is above the noise in a statistically significant way. Second, there may be a large number of statistically significant genes, but no apparent unifying biological theme connecting them, which makes it difficult to understand the meaning of the differentially expressed genes. Third, functionally important genes may be missed in this approach. For example, a small change in the expression of genes encoding multiple members of a biological pathway can sometimes have a larger biological effect than a large change in a gene encoding a single member of the pathway. Therefore, it is important to look not only for genes that exhibit large changes in expression but also for those that may only exhibit a relatively small change; otherwise, important aspects of the biology may be missed. A final issue is that there can be large variations in the expression of individual genes across relevant related experiments.
GSEA was developed to overcome these problems by looking for statistically significant changes in the expression of sets of genes as opposed to individual genes. These gene sets are defined on the basis of existing knowledge of related genes. GSEA quantifies the degree to which these gene sets occur toward the top of a ranked list of genes (up-regulated) or toward the bottom (down-regulated) and then estimates the significance of the finding. It is arguable that GSEA is statistically more sensitive than analysis of the fold change in expression of individual genes, because the signal-to-noise ratio is larger for a set of genes than it is for a single gene. Thus, GSEA should be sensitive to small but consistent changes within a gene set, and these changes may be biologically important.
The first time GSEA was applied to analyze gene expression changes, it was used to identify a set of genes relevant to oxidative phosphorylation, which are a factor in type II diabetes (1). We describe the method of GSEA by applying it to a simple example (Slides 11 to 14). The example includes samples from two groups, called Groups A and B. Group A samples represent the relative expression of five genes (Genes A to E) in muscle biopsies from two normal patients (Sample 1 and Sample 2) and Group B samples (Sample 3 and Sample 4) represent the relative expression of the same five genes in samples from two patients with diabetes (Slide 11). The first step is to rank the genes on the basis of their level of differential expression. Here, we simply averaged the expression of the two samples and then ranked them on the basis of the difference in expression across samples. The next step is to identify, from the genes analyzed, the ones that belong to a particular biological category. Then the significance in the difference between the two groups is tested. In the example, Genes B and C belong to a gene set of interest. To determine whether these genes appear significantly toward the top or bottom of the ranked list (that is if the expression of this set of genes is up-regulated or down-regulated between the two conditions), we calculate a running sum in the relative expression of the genes by moving down the list. The sum is increased (up-step) when a gene that is part of the set (Gene B or C) is encountered, and the sum is decreased (down-step) when a gene that is not part of the set (Gene A, D, or E) is encountered. The size of the up-step is given by
where N is the number of genes whose expression is measured (in the example, five) and G is the number of genes in the set (in the example, two) (Slide 12). N is always greater than G. The size of the down-step is given by
There are other formulations for performing a similar analysis in a more complicated way where the weight of each gene in the set is accounted for by its correlation with the group label, but we use the simpler approach as a demonstration with the example having five genes of which two are in the set (Slides 11 to 13). The more complicated versions of GSEA are described in (2).
For the sample data, the running sum is plotted as a random walk with the x axis as the genes (1 representing the first gene in the data set, 2 the second, and so on) and the y axis as the running sum (Slide 13). If the genes in the set (Genes B and C) are negatively differentially expressed, then those two genes will tend to be close to the bottom of the ranked list (Slide 11), and the running sum should become significantly negative before the values for the members of the gene set (B and C) are included, and the running sum should begin to increase as those are included (Slide 13). Conversely, if the test genes (B and C) are positively differentially expressed, then the running sum should become positive before returning to zero, because there should be more positive steps earlier in the running sum.
The supremum of this curve is called the “enrichment score,” and it is a measure of the position of the genes that are part of the set in the random walk. For the sample gene set (B and C), the enrichment score is ~−2.5 (Slide 13), which suggests that the genes in the test set are down-regulated.
From this example and description, the relation to the Kolmogorov-Smirnov test becomes evident. If the genes in the set that are tested for significance are distributed uniformly through the ranked list (there is no general trend for the members of the set to be high or low in the ranked list), then the running sum will look like a random walk and will have a supremum of a random walk. However, if the genes in the test set tend to be high or low in the ranked list, then the supremum will be larger in magnitude that what is expected for a random walk.
If a set of genes has a large enrichment score and large supremum, then the significance of the result can be determined by randomly ordering the genes and then repeating the ranking and plotting of the random walks again and again (Slide 14). Note that the samples are shuffled among the groups; the genes are not shuffled with each other because the correlations between the genes need to be preserved. This process is repeated many times to produce many running sums and enrichment scores for all these random permutations of the samples. In the example, Samples 1 and 3 are randomly assigned to Group A and Samples 4 and 2 to Group B (Slide 14).
Statistical significance is determined from how many times the enrichment score for genes in the set from the first nonrandomized analysis is greater than the enrichment scores determined for the randomly shuffled data. For larger data sets containing more genes, there are more possible permutations, and the walks would have more steps.
For the sample set of genes B and C, the enrichment score of −2.5 is larger in magnitude than all the scores for randomly permuted data; thus, the test gene set (B and C) is expressed significantly lower in condition B than in condition A. On the basis of this information, biological implications can be inferred.
If the enrichment scores for the randomly shuffled data rarely exceed (less than 1% of the time) the score for the original, unshuffled data, then the set of genes can be confidently considered to play a role in the difference between the two conditions.
In order to be confident that the differences are statistically significant and thus likely to be biologically meaningful, the significance of the result has to be corrected for multiple hypotheses testing to account for the frequency that false-positive differences occur in the data. There are various ways to control for the rate of false-positives, and one of the simplest is the Bonferroni procedure, which is not described in detail as part of the lecture [see (2) for details].
An Example of the Use of GSEA in the Literature
Mootha et al. took muscle samples from 43 age-matched males: 17 had normal glucose tolerance (Group 1), 8 had impaired glucose tolerance but were not considered diabetic yet (Group 2), and 18 had type II diabetes (Group 3) (1). No single gene could be identified as statistically significant between any pair of these groups (Slide 15).
The authors performed a GSEA analysis, looking for the significance of 149 gene sets. Of these sets, 113 were grouped according to metabolic pathways, and 36 were coregulated as described in a mouse expression atlas of 46 tissues. The sets were selected without regard to the data. The gene set that had the highest enrichment score represented a set of 106 genes involved in oxidative phosphorylation. Although each gene in the set was only down-regulated by a small amount (~20%), this reduction in expression was consistent across 80% of the genes in the set. When they randomly permuted the data and recalculated the enrichment score 1000 times, they found that only 29 of these 1000 scores were higher than the observed enrichment score before shuffling, giving a significance of P = 0.029, which is less than the commonly used cutoff of P < 0.05, which is used as a threshold for statistical significance.
Having identified this set of genes as significant, Mootha et al. used the analysis to look more closely at the set and found that there was a subset, consisting of about two-thirds of the original 106 genes in the set, which accounted for most of the significance. The identification of this set of genes led to their investigation of the biological mechanisms that might be involved in the down-regulation of this biological function and, therefore, its molecular contribution to the phenotype of type II diabetes. The method has subsequently been applied many times to address many different problems (4).
From this lecture, students should be able to understand GSEA and its application, realize the basis for the sensitivity of the method, and understand how this method can lead researchers toward biological mechanisms for differences in gene expression among different groups (Slide 16).
Problem Set
The expression of eight genes, labeled G1 to G8, is measured (Table 1). The measurements are taken from four samples from a nondiseased, healthy group, and four samples from a diseased group. You would like to test the hypothesis that the gene set {G2, G5} plays a significant role in the disease.
Table 1.
Gene expression levels for the problem set.
| Healthy | Diseased | |||||||
|---|---|---|---|---|---|---|---|---|
| A | B | C | D | E | F | G | H | |
| G1 | 1.0 | 1.0 | 0.8 | 0.7 | 2.0 | 1.9 | 1.5 | 1.6 |
| G2 | 1.2 | 1.1 | 1.0 | 1.1 | 0.6 | 0.7 | 0.5 | 0.7 |
| G3 | 0.5 | 0.6 | 0.4 | 0.7 | 0.1 | 0.2 | 0.2 | 0.3 |
| G4 | 0.7 | 0.4 | 0.7 | 0.9 | 2.2 | 2.0 | 2.0 | 1.3 |
| G5 | 0.2 | 0.4 | 0.3 | 0.2 | 0.3 | 0.2 | 0.5 | 0.4 |
| G6 | 0.9 | 0.7 | 0.9 | 0.5 | 0.1 | 0.5 | 0.2 | 0.2 |
| G7 | 1.3 | 1.1 | 1.0 | 1.3 | 0.9 | 1.5 | 0.7 | 1.0 |
| G8 | 0.1 | 0.3 | 0.5 | 0.1 | 1.4 | 1.9 | 1.6 | 1.7 |
The following problems are from the steps of performing GSEA on the data. The software R may be used to solve the problems, and the answer key is available in that form. However, any similar tool, such as MATLAB, or any computer programming language development environment may also be used.
Calculate the mean differential expression for each gene between the two groups.
Generate an ordered list by ranking the genes in order of decreasing differential expression and then determine whether the gene set {G2, G5} is at the top or bottom of the ranked list.
Calculate the appropriate sizes of the up-and down-steps for the GSEA running sum.
Using the ordered list generated in Problem 2, calculate the running sum, adding the appropriate up- and down-steps as you move down the ranked list of genes. What is the supremum (enrichment score) of this running sum?
-
Permute the samples by randomly assigning them to the Healthy or Diseased group and repeat problems 1 through 4 to calculate enrichment scores for the randomly shuffled data.
Note: If the students find this too difficult, the instructor can provide a set of Enrichment Scores from randomly permuted data.
Using the enrichment score for the original data and the enrichment scores for the randomly shuffled data, determine whether the set of genes {G2, G5} are significantly differentially expressed between the two sets.
Supplementary Material
Acknowledgments
We thank Sherry L. Jenkins for comments and suggestions and Y.-S. Lee for validating the problem sets. Funding: This work was supported by NIH grants 5P50GM071558-03, 1R01DK088541-01A1, KL2RR029885-0109, and RC2OD006536-01.
Footnotes
Educational Details
Learning Resource Type: Lecture, assignment, digital presentation
Context: Graduate
Intended Users: Teacher, learner
Intended Educational Use: Learn, plan, teach
Discipline: biocomplexity; bioinformatics, genomics and proteomics; biostatistics; biotechnology; molecular biology; systems biology;
Technical Details
Software: R
Requirements: Platform-independent, open-source
Download: http://www.r-project.org/
Software: GSEA software
Requirements: Platform-independent, open-source
Supplementary Materials
(http://stke.sciencemag.org/cgi/content/full/sigtrans;4/190/tr4/DC1)
Slides: Introduction to Statistical Methods for Analyzing Large Data Sets: Gene-Set Enrichment Analysis (GSEA)
Problem set key is available upon request.
References and Notes
- 1.Mootha VK, Lindgren CM, Eriksson KF, Subramanian A, Sihag S, Lehar J, Puigserver P, Carlsson E, Ridderstråle M, Laurila E, Houstis N, Daly MJ, Patterson N, Mesirov JP, Golub TR, Tamayo P, Spiegelman B, Lander ES, Hirschhorn JN, Altshuler D, Groop LC. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet. 2003;34:267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 2.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: A knowledge-based approach for interpreting genomewide expression profiles. Proc. Natl. Acad. Sci. U.S.A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kolmogorov AN. Sulla determinazione empirica di una legge di distribuzione (On the empirical definition of a distribution function) G. Istit. Ital. Attuari. 1933;4:83–91. [Google Scholar]
- 4.Huang W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.