Abstract
Phospholipase Cβ2 (PLCβ2) is a large, multidomain enzyme that catalyzes the hydrolysis of the signaling lipid phosphoinositol 4,5 bisphosphate (PIP2) to promote mitogenic and proliferative changes in the cell. PLCβ2 is activated by Gα and Gβγ subunits of heterotrimeric G proteins, as well as small G proteins and specific peptides. Activation depends on the nature of the membrane surface. Recent crystal structures suggest one model of activation involving the movement of a small autoinhibitory loop upon membrane binding of the enzyme. Additionally, solution studies indicate multiple levels of activation that involve changes in the membrane orientation as well as interdomain movement. Here, we review the wealth of biochemical studies of PLCβ2– G protein activation and propose a comprehensive model that accounts for both the crystallographic and solution results.
1. Introduction
Mammalian phospholipase C-β2 (PLCβ2) can be thought of as a prototype signaling enzyme. This multidomain enzyme is the main effector of signals emanating from activation of the Gαq family of heterotrimeric G proteins and has several established activators [1, 2]. However, only its basic catalytic core can be found in single cell organisms, leading to the conclusion that as the enzyme evolved in higher organisms, regulatory domains were attached to the catalytic domain to control the interaction with other macromolecules, cellular localization and catalytic regulation. As the number of domains increased, so too did the number of activators of this enzyme. Here we focus on the implications of this increase in the number of activation mechanisms due to the increased number of activators. Structural studies have suggested a single universal activation mechanism as discussed below, whereas solution studies suggest multiple activation pathways. In this review, we highlight the role of regulatory domains in enzyme activity and the attendant activation mechanisms. After a brief overview of the domain organization of the enzyme, we describe the catalytic mechanism and show how regulatory domains may produce different modes of enzyme activation.
2. PLCβ2 is composed of conserved structural domains
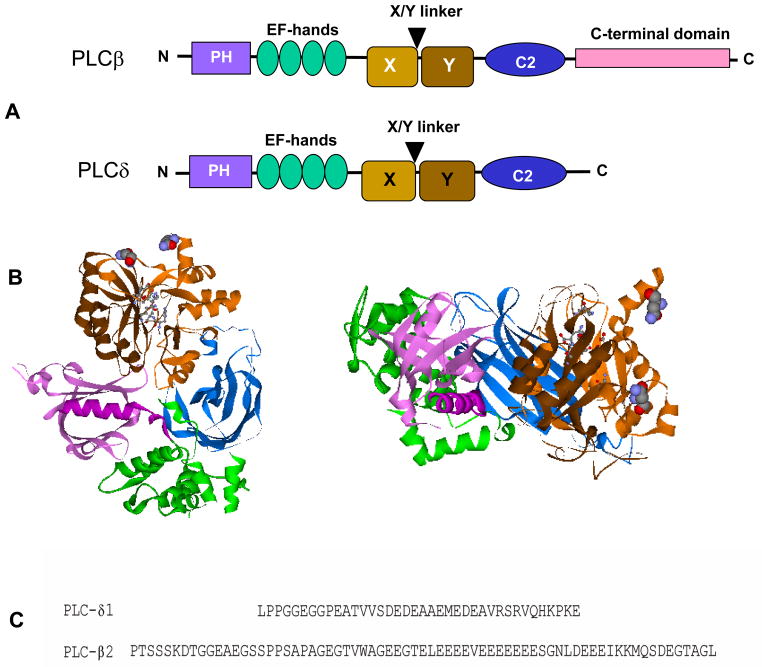
The regulatory domains of PLCβ2 are involved in protein-protein, protein-lipid and protein-substrate interactions including those that control activation and cellular localization (for review see [1–3]). The organization of these domains is shown in Fig.1 in comparison to the cognate mammalian PLC, PLCδ. The catalytic domain is close to the center of the enzyme sequence. The core of this domain consists of two halves, referred to as X and Y and composed of alternating α helices and β strands that form an α/β barrel with the catalytic end on one side of the barrel [4–6]. Between the two halves of that core is an intervening sequence that is not integral to the core s structure. The nature of this insertion loop varies for different mammalian PLCs. In PLCβ2, this region contains a long, almost consecutive stretch of ~18 D/E residues (Fig. 1c). In the crystal structure of the homologous enzyme PLCδ1 [7] and of Rac1- PLCβ2 [4], this region is disordered, but in the crystal structure of isolated PLCβ2 [5], the loop occludes the active site. This positioning of the linker led to the suggestion that activation involves detachment of the loop, presumably due to charge repulsion from the membrane surface [5]. Interestingly, in PLCγ the insertion region contains a split PH domain, two SH2 domains and an SH3 domain, which are integral to enzyme activation [8]. Comparison of the catalytic residues of the bacterial PLC to those of mammalian PLCδ1 and PLCβ2 shows a very similar placement of active site amino acids; the active site utilizes a Ser/His triad with water as an active participant in the reaction [9–11]. Calcium functions as a chelator in the reaction and is absolutely required for activity. PLCβ2 is fully active at basal calcium concentrations whereas PLCδ is active only at elevated Ca2+ generated by the activity of other PLCs.
Figure 1.
A. Comparison of the domain organization of PLCβ and PLCδ enzymes; B. Top and side views of the domain structure of PLCβ2 from [5] where the PH domain is in purple, the EF hands are in green, the catalytic domain is in medium (X) and dark (Y) brown and the C2 domain is in blue. Active site residues are depicted in ball and stick, and the two residues attached to the insertion region are in CPK; C. Sequence comparison of the X/Y linker insertion regions of PLCδ1 and PLCβ2.
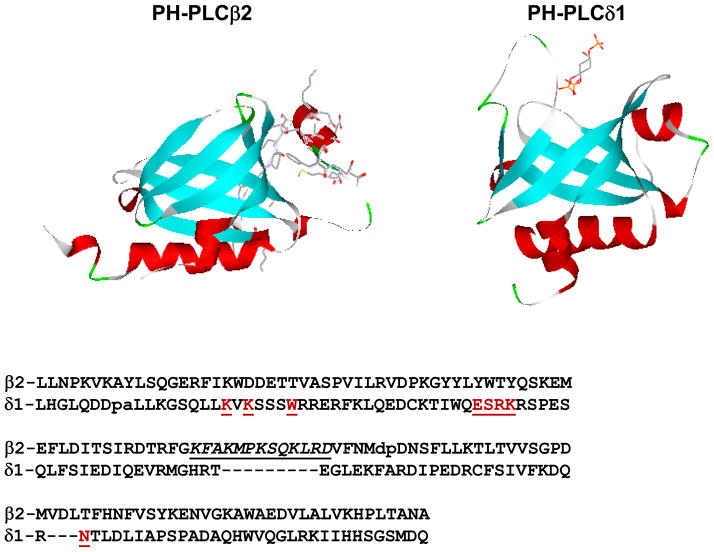
The PLCβ enzymes contain a pleckstrin homology (PH) domain in their N-terminal region. PH domains are highly conserved motifs of ~120 amino acids that, in general, confer specificity for different lipids or proteins [12]. These domains, which were discovered almost two decades ago, are among the most common motifs in mammalian proteins (Fig. 2). It was previously shown that the PH domain of PLCδ1 targeted the enzyme to membranes containing its substrate, PIP2 [13] through a series of specific hydrogen bonds and ionic interactions [14]. These interactions ensure that the enzyme remains membrane-bound and close to its substrate until PIP2 is depleted. In contrast, the PIP2-binding residues are absent in the PH domain of PLCβ2 enzymes (Fig. 2) but the protein binds strongly to membranes with little specificity [15], as discussed further below. In addition to membrane binding, the PH domain of PLCβ2 mediates association to monomeric G proteins and to Gβγ subunits [16–19].
Figure 2.
Comparison of the structures (TOP) of PH-PLCβ2 with residues 71–88 rendered in stick format, and PH-PLCδ1 complexed with Ins(1,3,5)trisphosphate (in stick format); (BOTTOM) sequences of the PH domains of PLCβ2 and PLCδ1 where the residues of PHδ1 important for IP3 binding are in red and underlined, and the residues in the insertion unique to PHβ2 are in underlined italics. Adapted from [9].
Following the PH domain are two EF hands (Fig 1A). The role of these domains in the regulation of PLCβ2 is unclear; while little work has been done to elucidate their function, we note that deletion of this region of PLCδ1 inactivates the enzyme [20]. Additionally, the EF-hands of PLCδ1 do not appear to bind Ca2+ (see discussion in [1]) but could interact with lipids [21]. There is no evidence that this region of PLCβ2 binds Ca2+ as none is observed in the crystal structures [4–6]. A recent structure of a Gαq construct in complex with truncated PLCβ2 shows several contact sites between the EF hands and Gαq suggesting that this region may play a role in Gαq binding [6].
Downstream of the catalytic domain is a C2 domain (Fig 1A). Although C2 domains are usually associated with Ca2+-dependent membrane targeting, this is not the case for PLCβ2 since the C2 domains lack the appropriate residues to chelate Ca2+ (see [22]). Although the C2 domains do not bind strongly to membranes in isolation, they bind strongly and specifically to activated Gαq subunits, as discussed below [22].
PLCβ enzymes have a unique 400 residue extension at their C-terminal end. This region is necessary for activation by Gαq subunits [23–25]. Interestingly, this region contains an RGS domain that promotes the deactivation of Gαq subunits [26], and the kinetic aspects of the activation of these G proteins have been reviewed recently [27]. The C-terminal region also contains other segments that regulate the cellular activity of PLCβ, including two phosphorylation sites and a nuclear localization signal [1]. While the structure of the isolated C-terminal region has been solved [28], this region was absent in the crystal structures of the PLCβ enzymes solved so far. The C-terminal region crystallizes as an intertwined dimer although the purified form of PLCβ appears monomeric up to 200 nM as determined using Forster resonance energy transfer methods (Scarlata, unpublished). Computational studies and mutagenesis of the C-terminal domain correlate with Gαq activation of the dimeric form [29]. This region is not required for enzyme activation by Gβγ subunits [30] and monomeric G proteins [4].
3. PLCβ catalyzes the hydrolysis of PIP2 in a two step reaction pathway
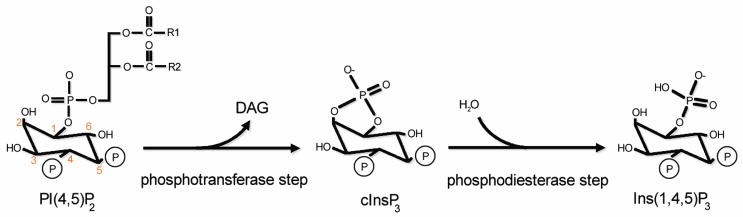
While PLCβ2 can catalyze the hydrolysis PI and PIP, PIP2 is the preferred substrate. Catalysis has been shown to occur via a two step pathway [11] as shown in Fig. 3. The first step yields a cyclic phosphoinositide and this is the terminal product of bacterial enzymes under most conditions. In mammalian PLCs, this cyclic intermediate is held in the catalytic site long enough to be attacked by water to yield a mixture of linear and cyclic products [31–33]. PLCβ, unlike PLCδ and PLCγ enzymes, only produces linear product suggesting stronger binding of cyclic IP in the catalytic site [34]. It has been shown that the presence of Gβγ increases the rate of the second step by increasing the rate of product release [35]. It is not clear whether cells utilize the cyclic and linear IP3 differently, and so the ability of PLCβ2 to produce only linear IP3 has as yet unknown significance.
Figure 3.
Reaction scheme of PIP2 hydrolysis catalyzed by PLC enzymes.
4. Enhancement of PLCβ activity can occur through several steps in the reaction pathway
Since PLCβ2 is a soluble enzyme that hydrolyzes a lipid substrate, there are many potential steps in the catalytic mechanism that can be influenced by activators. One or all of these steps may involve conformational changes in the enzyme. Importantly, PLCβ2 must bind to the membrane surface to access its substrate, and this access might involve penetration of the enzyme into the lipid surface. Catalytic steps that involve substrate access are expected to depend on the physical properties of the lipid aggregate, and in particular the lipid head groups. The potential role of these steps in the activation process is discussed below.
PLCβ1–3 enzymes have been shown to bind strongly and non-specifically to membranes with varying composition [15]. Unlike PLCδ, which only binds strongly to membranes containing PIP2, PLCβ1–3 enzymes bind with similar affinity to bilayers with electrically neutral and negatively charged head groups, although PIP2–specific binding has been noted in some reports [36]. It is notable that binding is slightly stronger to membranes containing lipids with phosphoethanolamine (PE) head groups which, depending on the mixture composition and temperature, have been observed to form hexagonal phases to various extents [37].
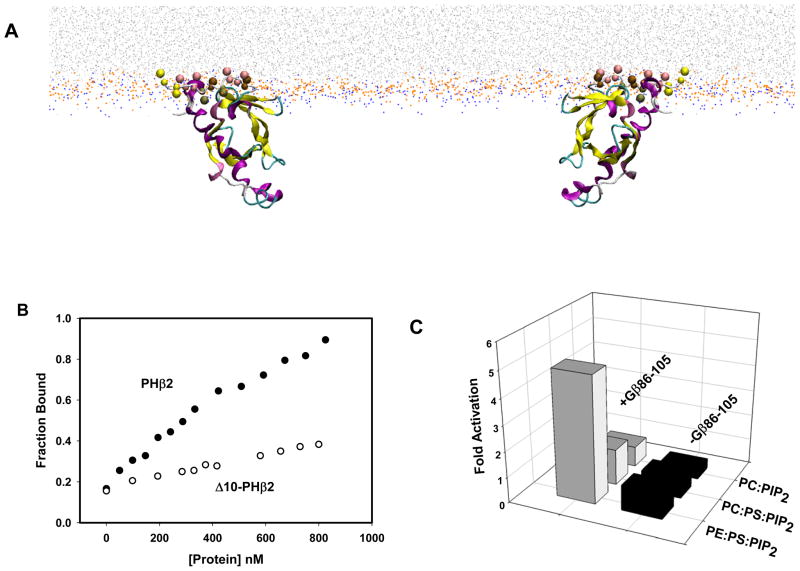
Several regions of PLCβ2 may contribute to membrane binding. The C-terminal region of PLCβ2 has some contribution to binding affinity but its impact does not appear to be substantial [15]. The isolated catalytic domain of the highly homologous PLCδ1 does not interact with membranes and its intrinsic affinity to PIP2 is very low [38]. As mentioned above, PH-PLCβ2 has been shown to play a major role in membrane binding [16, 39]. However, based on the crystal structure of PLCβ2 [4, 5], and the structure of PLCδ-IP3 [40], the manner in which PLCβ2 binds to membranes is unclear. Theoretical studies of membrane binding of the PH-PLCβ2 domain suggest that the binding involves penetration of the first 10 residues of the domain into the lipid matrix (Fig. 4A) [41] . This prediction was substantiated by experimental studies showing that removal of the first 10 residues results in a significant loss in binding affinity (Fig. 4B and Han et al., in press). These residues were absent in the PLCβ constructs that were crystallized.
Figure 4.
A. (Left) Snapshot from the coarse grained simulation of PH-PLCβ2 (residues 1–144) interacting with a membrane model. The beads representing the lipid chains are shown in light gray, the choline group bead is shown in orange and the phosphate group bead is in blue. The larger colored beads are the first 10 N-terminal residues as they insert in the membrane. (Left) In the top left structure the beads of the N-terminal Met are in yellow and the beads of the four leucines oriented into the membrane are rendered in pink. The rest of the protein is shown as a cartoon, colored according to secondary structure. (Right) The same structure rotated 180°. The open space in the center of the structure corresponds roughly to the PIP2 binding site of PLC-delta1 if it were superimposed onto this structure. B. Binding curve to POPC membranes for PHβ2 and Δ10-PHβ2. Binding was measured as a function of concentration using fluorescence methods (adapted from Han et al., in press). C. Activation of the PHβ2-PLCδ1 chimera by Gβ86–105 measured in membranes of different composition composition (adapted from [41])
In searching for regions of PH-PLCβ2 that might confer activation by Gβγ subunits, an insertion (71–88) was identified that is absent in PH-PLCδ (see alignment in Fig. 2). A peptide generated from this sequence (underlined and italicized in Fig.2) bound strongly to lipid membranes containing PE lipids [41]. Surprisingly, when added to PLCβ2, this segment (PH71–88) is able to activate the enzyme to the same level as Gβγ subunits.
Because PLCβ2 is soluble and G proteins reside on membrane surfaces, activation of PLCβ2 by G proteins was thought to occur through the recruitment of the enzyme to the membrane by activated G proteins. We had explicitly tested this idea using model systems where the concentrations of proteins and lipids can be carefully controlled [42, 43], and found that, under excess lipid conditions, association between G protein subunits and PLCβ2 occurs between the membrane-bound proteins on the surfaces of membranes. Similar findings have been reported from other studies [36]. Association between the pre-bound proteins is promoted by the effective reduction of dimensionality and a more limited number of available protein association sites [43]. Notably, this mode of interaction between membrane bound proteins differs from an alternative mechanism, of simultaneous membrane recruitment of PLCβ2 by G protein binding (see discussion in [43]) .
5. G protein activators bind at different regions of the enzyme
In the early 1990s it was established that Gαq and Gβγ interact with different regions of PLCβ2 to promote activation ([25] and see [44, 45]). Specifically, it was shown that deletion of the C-terminal region ablates activation by Gαq subunits even though the truncated enzyme will still bind Gαq with a reduced affinity [22]. It was further shown that the C2 domain located next to the C-terminal region was responsible for specific binding to activated Gαq [22]. The structure of a complex between an activated Gαq construct and the C-terminal truncated PLCβ3 has recently been solved and shows interaction between the C2 domain and Gαq, although activation of the enzyme by Gαq was not demonstrated [6]. More recent studies suggest that Gαq activation occurs when the G protein displaces an inhibitory interaction between the C-terminal extension and the catalytic domain [46].
Many studies have addressed the interaction between PLCβ2 and Gβγ. Contact sites have been mapped to residues in the catalytic domain, and in the PH domain [47–52]. Additionally, the PH domain has been shown to bind to Gβγ subunits with an affinity only 5 fold weaker than the full length enzyme [16]. Swapping this domain into PLCδ1, which is not activated by Gβγ, results in an enzyme that is fully activated by Gβγ subunits [53]. As mentioned, PLCδ1 specifically binds to PIP2 through residues in its PH domain, and binding to PIP2 results in enzyme activation [54]. Interestingly, swapping the PH domain of PLCδ1 into PLCβ2 results in an enzyme that specifically targets PIP2-containing membranes and allows for PIP2-mediated activation [52].
Some additional activators of PLCβ2 have been identified, including by Rac1 from the Rac family. These small G proteins are thought to activate the enzyme by promoting membrane binding, which is most likely the reason for making this activation difficult to assess in vitro (see [55, 56], and for review [57]). A recent crystal structure of the Rac1- PLCβ2 complex shows that association between the proteins is mediated entirely by the PH- PLCβ2 domain, with no additional contacts in other regions of the enzyme [4]. Since Rac1 does not contact the catalytic residues and does not appear to induce conformational changes, it is unclear how it activates the enzyme. It has been postulated that in vitro activation by Rac1 is only seen under very limited experimental conditions, and that the Rac1- PLCβ2 complex can be activated further by G protein subunits (e.g. [4]).
PLCβ2 can also be activated by several types of peptides. Peptides with sequences corresponding to regions in the catalytic domain have been shown to increase catalytic activity by some unknown mechanism [58, 59]. A dodecapeptide with a sequence corresponding to a blade of the Gβγ propeller, Gβ86–105, not only activates the enzyme but will also increase level of activation of the Gβγ PLCβ2 complex, implying that multiple levels of activation exist [60]. Peptide studies have identified other Gβ regions that are involved in PLCβ2 binding [61]. Probably one of the most interesting activators is the peptide derived from the 71–88 segment of PH-PLCβ2 [41], as mentioned above. The region corresponding to this peptide is thought to induce activation by favorably positioning the enzyme on the membrane surface (see below) although it is possible that this region forms an amphipathic, mostly cationic helix that might compete with the anionic linker for activation.
6. Both the activity, and G protein activation, of PLCβ2 strongly depend on the properties of the membrane surface
The activity of PLCβ2 is almost always measured using a radiometric assay that monitors the release of soluble 3H-IP3 from 3H-PIP2 hydrolysis. PIP2 is usually dispersed in either mixed micelles or lipid bilayers. Little change in PLCβ2 activity is observed as a function of the charge of the membrane surface. Although PE lipids support slightly higher levels of activity (see [41]), this increase is thought to be due to a reduction of membrane surface tension caused by PE (since activity towards substrate immersed in micelles is lower than in bilayers).
The dependence of Gαq activation on membrane composition has not been thoroughly investigated, and activation by monomeric G proteins can be seen only under very limited experimental conditions, but Gβγ activation has been shown to have a significant dependence on membrane composition. Activation of PLCβ2 or the PHβ2-PLCδ1 chimera does not occur when PIP2 is embedded in PC bilayers, and activation is very low in negatively charged PC:PS membranes. However, incorporation of PE lipids allows for enzyme activation [41]. Similarly, activation by Gβ86–105, which does not bind membranes, does not occur on PC membranes [41] showing that the inability of Gβγ to active the enzyme on PC membranes (compared to PE-containing surfaces) is not due to differences in the orientation relative to the membrane, or to binding of the G protein subunit.
Even though negatively charged lipids do not affect Gβγ activation, it has been found that large quantities of anionic PA lipids, but not anionic PG lipids can activate PLCβ1 [62]. This apparent electrostatic mode of activation has been thought to be mediated through a region located in the C-terminal region of PLCβ1 that targets PA lipids.
7. Detachment of the insertion loop as a model for activation
For almost two decades biochemical studies have sought to understand the pathway through which G protein subunits activate PLCβ enzymes, sustained by the expectation that structural studies would ultimately reveal the various conformational changes that seemed to be associated with activation. However, crystal structures of PLCβ enzymes alone [5], complexed with Rac1 [4] or with a Gαq construct [6], showed little differences in backbone conformations. As mentioned, the only significant difference in these structures was the position of an anionic linker loop between the two major folds of the catalytic domain (Fig. 1), which contacts the catalytic domain in the absence of G proteins and is unresolved in the presence of activator. While movement of this linker will certainly allow for increased activity [39], it is not clear how G proteins would promote this movement since it appears that they do not directly contact this region of the catalytic domain, and since no other movement is observed to occur in the backbone residues. Because the linker contains so many negatively charged residues (Fig. 1C), the detachment of this anionic loop from the active site was proposed to occur when the enzyme binds to membrane surfaces and to be due to charge repulsion from the phosphates of the lipid head groups [5]. Thus, activation by G proteins could occur if the G proteins recruited the enzyme to the membrane surface. However, with this explanation it remains unclear how the detachment of the loop observed in the crystals of the complex would occur in the absence of the negatively charged membrane headgroups, and leaves open the questions of how G proteins alone can affect the interaction between the loop and the catalytic site, or how Gβγ activation occurs in the PHβ2-PLCδ1 chimera which has a very different insertion sequence (Fig. 1C). Activation through changes in the insertion region has been proposed for PLCγ which has a much larger and more complex insertion [8].
While the anionic insertion is autoinhibitory and its removal plays a role in the activation of PLCβ2 and other PLC families [5], it is clearly not the only pathway of PLCβ2 activation and there are several reasons to suggest that activation is more complex. First, although deletion of this anionic linker in PLCβ2 results in a more active enzyme, it does not affect the enzyme s ability to be activated by G protein subunits [5, 39]. Second, the observation that Gβγ can further activate Rac1- PLCβ2, whose crystal structure is thought to correspond to this insertion-activated form, does not fit with this model as the sole activation mode [4]. Third, the linker-based activation model predicts that the catalytic domain, but not the PH domain, interacts with the membrane surface and this contradicts studies showing that the PH domain is required for membrane binding of the host enzyme [38, 39]. Fourth, the simple structure-based model cannot account for the many solution studies of PLCβ2 activation by Gβγ subunits, and the analogous activation of PLCδ1 by PIP2, that show that Gβγ activation occurs through PH domain membrane binding and changes in membrane and domain orientation (e.g. [41, 54]); nor can this model account for activation by the Gβ86–105. Fifth, this model cannot account for the observation that Gβγ activates PLCβ2 catalysis of soluble cIPs as well as membrane-bound PIP2 [35]. Finally, there appear to be multiple levels of PLCβ2 activation, which is inconsistent with a simple model based on the loop detachment [60].
8. Comprehensive model of PLCβ activation states
In searching for a more comprehensive understanding of the multiple potential levels of PLCβ2 activation, we reasoned that a model that would agree with both solution and crystallographic data would have both the isolated PLCβ2 structure, and the bound G protein, correspond to an activated form of the enzyme. One of the inactive forms would maintain the contacts between the loop region and the catalytic site, and the other would not. This idea is consonant with the observed multiple levels of PLCβ2 activation and implies that other deactivated forms of the enzyme exist. Others have also made this suggestion [27].
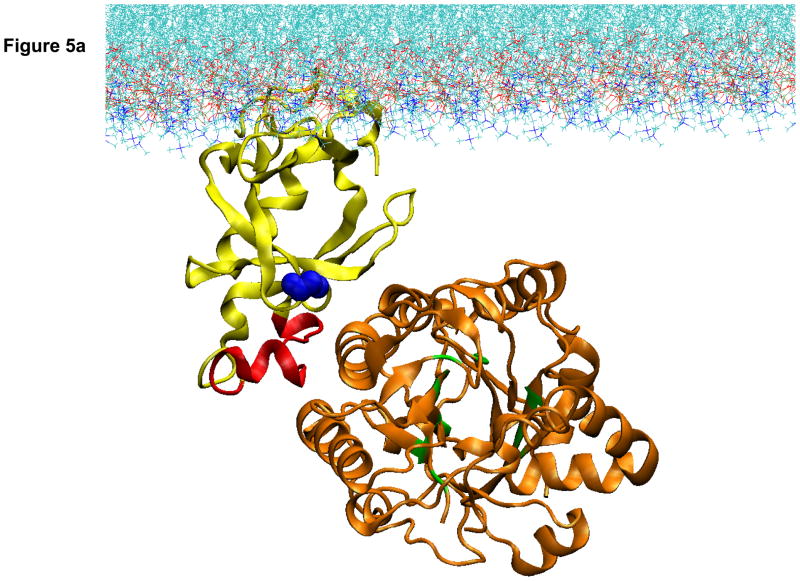
In searching for deactivated structures we began by considering previous and ongoing work showing that the PH domain targets the enzyme to membrane surfaces through penetration of the first 10 amino acids ([41] and Han et al. in press). However, it became clear from the structural constraints that directing the enzyme to the membrane by interaction with the PH domain anchored through its N-terminal region positions the catalytic domain too far from the membrane surface to enable substrate access (Fig. 5A).
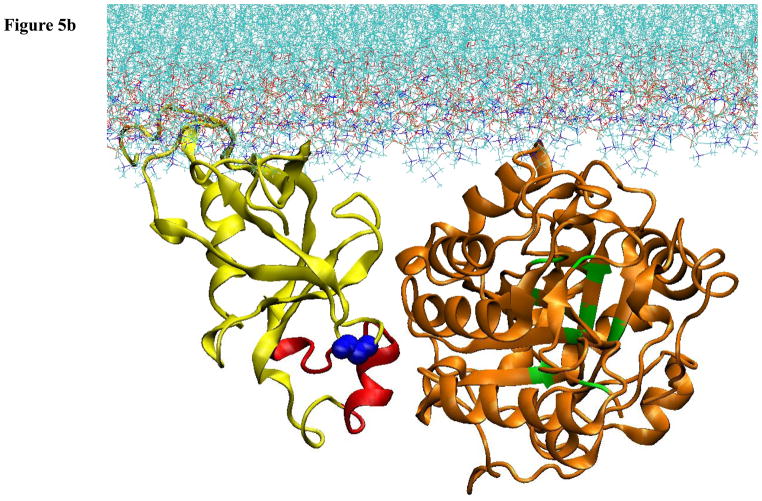
Figure 5.
Computational model of the docking of the PH and catalytic domains of PLCβ2 to a lipid membrane. A. Result of computational docking with the first 10 residues of the PH domain penetrating into the lipid bilayer. B. Model of the pre-active form of PLCβ2 in which the PH domain has reoriented to allow for active site contact with the membrane surface.
While the structure in Fig. 5A could well correspond to an inactive form of the enzyme, it does not make it immediately clear how the enzyme would be activated. We approached a solution to this problem by keeping in mind two key experimental findings – that Gβγ activation is greatly promoted by PE-containing membrane surfaces, but not by PC surfaces, and that the orientation of the PH domain relative to the catalytic domain changes upon binding to PE surfaces and upon Gβγ binding ([41] and Han et al, in press.). Insight into the molecular details that would support an activation model that agrees with these key experimental findings came from comparative coarse-grained molecular dynamics simulations of the interaction of the isolated PH-PLCβ2 with PC membranes containing increasing amounts of PE lipids (Han et al., in press). Binding to pure PC membranes occurred with the predicted penetration of the N-terminus of the PH domain leading to a stable membrane-bound structure. However, the presence of PE membranes was shown to change the mode of interaction, leading to the appearance of a second orientation of the PH domain relative to the membrane surface (Fig. 5B). In this orientation, PH71–88, the region of the PH domain that is found in PLCβ2, but not PLCδ[41], could interact with the catalytic domain. Direct interaction of PH71–88 with the catalytic domain is consistent with the ability of this peptide to interfere with membrane interactions of the catalytic domain but not the PH domain [41]. Complementing the complex by attaching the catalytic domain to such a “pre-active state” orientation of the PH domain, and computationally optimizing the domain contacts, results in the configuration presented in Fig. 5B. This orientation of the PLCβ2 domains places the catalytic domain close to the membrane surface and allows the enzyme to access substrate
Bringing together the results of the computational modeling and experimental studies, we propose the following model of activation. The initial interaction of the enzyme to the membrane surface is through the N-terminal region that comprises the PH domain (Fig 5A). If the membrane is composed of PC lipids, then the penetrating N-terminus remains trapped and the catalytic domain does not achieve proximity to the membrane substrate. Because the smaller and more hydrophobic PE head groups do not anchor the N-terminus as strongly, the PH domain can adopt different membrane orientations. The change in membrane orientation of the PH domain to such a pre-active orientation triggers a change in the relative orientation of the catalytic domain that brings the active site close to the membrane surface (Fig 5B). This change of the interdomain orientation is seen experimentally as a decrease in the distance between the PH and catalytic domains upon binding to PE membranes and Gβγ subunits, and is evidenced by changes in the accessibility of Cys and Trp residues ([41] and Han et al., in press). In this later conformation (Fig 5B), the PLCβ2 can interact productively with G proteins and this can further modulate the PH-catalytic domain interactions to enhance activity.
We note that with enzyme positioned with the PH domain in the pre-active orientation (Fig 5B), the complex is poised to bind to G proteins and move into the fully activated form, which is expected to be stable only when the G protein is productively bound. This view of the membrane binding sequence of steps that enables activation agrees with the experimental findings suggesting that activation occurs upon association of the membrane-bound species, as discussed above. As the result of one of several activation mechanisms, the nature of this active form is still unclear but it is likely to be distinct from Rac1-PLCβ because the latter complex can be further activated by Gβγ subunits; it is also distinct from the very similar structure of the Gαq-PLCβ2 complex since solution studies argue against activation of the truncated enzyme [63, 64]. We propose that the binding of Gβγ and Gαq subunits induce further movement between the domains as indicated by FRET measurements ([41] and Han et al.,in press). Based on the enzymatic reaction (Fig. 3), it is tempting to speculate that the insertion region on the PH domain regulates substrate entry into the active site, whereas Gβγ catalyzes product removal from the active site. This model is presently being tested using single molecule fluorescence methods.
9. Conclusions and Future Challenges
PLCβ is an interesting example of how an increasing the number of regulatory domains increases the number of ways the enzyme activity can be modulated. . While we focused here on activation mechanisms that involve movement between the PH and catalytic domains, the mechanism through which Gαq binds to the C2 domain and induces activation through the C-terminus is unknown. It is clear that each regulatory domain introduces different modes of regulation that can be autoinhibitory, as in the case of the insertion region, or target the protein to membrane surfaces containing substrate and other activation proteins, as is the case for the PH domain. Our model of G protein activation presented above accounts for many experimental and structural facets of PLCβ activation, although an understanding of how interdomain movement produces activation on the molecular scale is not yet available. Nor can our model explain the large activation of PLCβ by Gβ86–105 that is additive to Gβγ stimulation. Studies that combine mutagenesis with advanced spectroscopic and computational methods should provide further insight into the manner in which the various modes of regulation of PLCβ enzymes work together to produce the cellular phenotypes of action of these important proteins.
Highlights.
A model of activation of phospholipase C-beta is proposed; the model shows how activation involves G proteins, membrane surfaces and domain movement; the model fits well with experimental results
Acknowledgments
We thank Sebastian Stolzenberg for expert help with the representation of the molecular models and an anonymous reviewer for very helpful suggestions . This work was supported by NIH GM053132 to SS and U54GM087519 to HW.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Rebecchi M, Pentylana S. Structure, function and control of phosphoinositide-specific phospholipase C. Physiological Reviews. 2000;80:1291–1335. doi: 10.1152/physrev.2000.80.4.1291. [DOI] [PubMed] [Google Scholar]
- 2.Suh P, Park J, Manzoli L, Cocco L, Peak J, Katan M, Fukami K, Kataoka T, Yun S, Ryu S. Multiple roles of phosphoinositide-specific phospholipase C isozymes. BMB reports. 2008;41:415–434. doi: 10.5483/bmbrep.2008.41.6.415. [DOI] [PubMed] [Google Scholar]
- 3.Rhee SG. Regulation of phosphoinositide-specific phospholipase C. Ann Rev Biochem. 2001;70:281–312. doi: 10.1146/annurev.biochem.70.1.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jezyk MR, Snyder JT, Gershberg S, Worthylake DK, Harden TK, Sondek J. Crystal structure of Rac1 bound to its effector phospholipase C-beta2. Nat Struc & Mol Biol. 2006;13:1135–1139. doi: 10.1038/nsmb1175. [DOI] [PubMed] [Google Scholar]
- 5.Hicks SN, Jezyk MR, Gershburg S, Seifert JP, Harden TK, Sondek J. General and Versatile Autoinhibition of PLC Isozymes. 2008;31:383–394. doi: 10.1016/j.molcel.2008.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Waldo GL, Ricks TK, Hicks SN, Cheever ML, Kawano T, Tsuboi K, Wang X, Montell C, Kozasa T, Sondek J, Harden TK. Kinetic Scaffolding Mediated by a Phospholipase C–β and Gq Signaling Complex. Science. 2010;330:974–980. doi: 10.1126/science.1193438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Essen LO, Perisic O, Cheung R, Katan M, Williams RL. Crystal structure of a mammalian phosphoinositide-specific phospholipase C-d. Nature. 1996;380:595–602. doi: 10.1038/380595a0. [DOI] [PubMed] [Google Scholar]
- 8.Bunney TD, Opaleye O, Roe SM, Vatter P, Baxendale RW, Walliser C, Everett KL, Josephs MB, Christow C, Rodrigues-Lima F, Gierschik P, Pearl LH, Katan M. Structural Insights into Formation of an Active Signaling Complex between Rac and Phospholipase C Gamma 2. Molecular cell. 2009;34:223–233. doi: 10.1016/j.molcel.2009.02.023. [DOI] [PubMed] [Google Scholar]
- 9.Drin G, Scarlata S. Stimulation of phospholipase C[beta] by membrane interactions, interdomain movement, and G protein binding -- How many ways can you activate an enzyme? Cellular Signalling. 2007;19:1383–1392. doi: 10.1016/j.cellsig.2007.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Heinz DW, Ryan M, Bullock TL, OHG Crystal structure of the phosphatidylinositol-specific phospholipase C from Bacillus cereus in complex with myo-inositol. EMBO J. 1995;14:3855–3863. doi: 10.1002/j.1460-2075.1995.tb00057.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Essen LO, Perisic O, Katan M, Wu Y, Roberts MF, Williams RL. Structural mapping of the catalytic mechanism for a mammalian phosphoinositide-specific phospholipase C. Biochemistry. 1997;36:1704–1718. doi: 10.1021/bi962512p. [DOI] [PubMed] [Google Scholar]
- 12.Rebecchi MJ, Scarlata S. Pleckstrin homology domains: A common fold with diverse functions. Annu Rev Biophys Biomolec Struc. 1998;27:503–528. doi: 10.1146/annurev.biophys.27.1.503. [DOI] [PubMed] [Google Scholar]
- 13.Garcia P, Gupta R, Shah S, Morris AJ, Rudge SA, Scarlata S, Petrova V, McLaughlin S, Rebecchi MJ. The pleckstrin homology domain of phospholipase C-delta 1 binds with high affinity to phosphatidylinositol 4,5-bisphosphate in bilayer membranes. Biochemistry. 1995;34:16228–16234. doi: 10.1021/bi00049a039. [DOI] [PubMed] [Google Scholar]
- 14.Ferguson KM, Lemmon MA, Schlessinger J, Sigler PB. Structure of the high affinity complex of inositol trisphosphate with a phospholipase C pleckstrin homology domain. Cell. 1995;83:1037–1046. doi: 10.1016/0092-8674(95)90219-8. [DOI] [PubMed] [Google Scholar]
- 15.Runnels LW, Jenco J, Morris A, Scarlata S. Membrane binding of phospholipases C-b1 and C-b2 is independent of phosphatidylinositol 4,5-bisphosphate and the a and bg subunits of G proteins. Biochemistry. 1996;35:16824–16832. doi: 10.1021/bi961606w. [DOI] [PubMed] [Google Scholar]
- 16.Wang T, Pentyala S, Rebecchi MJ, Scarlata S. Differential association of the pleckstrin homology domains of phospholipases C-beta 1, C-beta 2, and C-delta 1 with lipid bilayers and the beta gamma subunits of heterotrimeric G proteins. Biochemistry. 1999;38:1517–1524. doi: 10.1021/bi982008f. [DOI] [PubMed] [Google Scholar]
- 17.Illenberger D, Walliser C, Strobel J, Gutman O, Niv H, Gaidzik V, Kloog Y, Gierschik P, Henis Y. Rac2 regulation of phospholipase Cb2 activity and mode of membrane interactions in intact cells. J Biol Chem. 2003;278:8645–8652. doi: 10.1074/jbc.M211971200. [DOI] [PubMed] [Google Scholar]
- 18.Rebecchi MJ, Gershengorn MC. Thyroliberin stimulates rapid hydrolysis of phosphatidylinositol 4,5-bisphosphate by a phosphodiesterase in rat mammotropic pituitary cells. Evidence for an early Ca2+-independent action. Biochem J. 1983;216:287–294. doi: 10.1042/bj2160287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Snyder JT, Singer AU, Wing MR, Harden TK, Sondek J. The pleckstrin homology domain of phospholipase C-beta2 as an effector site for Rac. J Biol Chem. 2003;278:21099–21104. doi: 10.1074/jbc.M301418200. [DOI] [PubMed] [Google Scholar]
- 20.Nakashima S, Banno Y, Watanabe T, Nakamura Y, Mizutani T, Sakai H, Zhao Y, Sugimoto Y, Nozawa Y. Deletion and site-directed mutagenesis of EF-hand domain of phospholipase C-delta 1: effects on its activity. Biochem Biophys Res Commun. 1995;211:365–369. doi: 10.1006/bbrc.1995.1822. [DOI] [PubMed] [Google Scholar]
- 21.Kobayashi M, Gryczynski Z, Lukomska J, Feng J, Roberts MF, Lakowicz JR, Lomasney JW. Spectroscopic characterization of the EF-hand domain of phospholipase C delta1: identification of a lipid interacting domain. Arch Biochem Biophys. 2005;440:191–203. doi: 10.1016/j.abb.2005.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang T, Pentyala S, Elliott JT, Dowal L, Gupta E, Rebecchi MJ, Scarlata S. Selective interaction of the C2 domains of phospholipase C-beta1 and - beta2 with activated Galphaq subunits: an alternative function for C2- signaling modules. Proc Natl Acad Sci U S A. 1999;96:7843–7846. doi: 10.1073/pnas.96.14.7843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Taylor SJ, Chae HZ, Rhee SG, Exton JH. Activation of the beta 1 isozyme of phospholipase C by alpha subunits of the Gq class of G proteins. Nature. 1991;350:516–518. doi: 10.1038/350516a0. [DOI] [PubMed] [Google Scholar]
- 24.Lee CH, Park D, Wu D, Rhee SG, Simon MI. Members of the Gq alpha subunit gene family activate phospholipase C beta isozymes. J Biol Chem. 1992;267:16044–16047. [PubMed] [Google Scholar]
- 25.Lee SB, Shin SH, Hepler JR, Gilman AG, Rhee SG. Activation of phospholipase C-b2 mutants by G protein aq and bg subunits. Journal of Biological Chemistry. 1993;268:25952–25957. [PubMed] [Google Scholar]
- 26.Berstein G, Blank JL, Jhon DY, Exton JH, Rhee SG, Ross EM. Phospholipase C-b1 is a GTPase-Activating Protein for Gq/11, Its Physiologic Regulator. Cell. 1992;70:411–418. doi: 10.1016/0092-8674(92)90165-9. [DOI] [PubMed] [Google Scholar]
- 27.Ross EM. G{alpha}q and Phospholipase C-{beta}: Turn On, Turn Off, and Do It Fast. Sci Signal. 2011;4:pe5. doi: 10.1126/scisignal.2001798. [DOI] [PubMed] [Google Scholar]
- 28.Singer AU, Waldo GL, Harden TK, Sondek J. A unique fold of phospholipase C-beta mediates dimerization and interaction with G alpha q. Nat Struct Biol. 2002;9:32–36. doi: 10.1038/nsb731. [DOI] [PubMed] [Google Scholar]
- 29.Ilkaeva O, Kinch L, Paulssen R, Ross EM. Mutations in the caboxyl-terminal domain of phospholipase C-beta1 delineate the dimer interface and a potential G(alpha)q interaction site. J Biol Chem. 2002;277:4294–4230. doi: 10.1074/jbc.M109612200. [DOI] [PubMed] [Google Scholar]
- 30.Park D, Jhon D, Lee LCK, Rhee SG. Activation of Phospholipase C Isozymes by G Protein bg Subunits. Journal of Biological Chemistry. 1993;268:4573–4576. [PubMed] [Google Scholar]
- 31.Zhou C, Wu Y, Roberts MF. Activation of Phosphatidylinositol-Specific Phospholipase C toward Inositol 1,2-(Cyclic)-Phosphate. Biochemistry. 1997;36:347–355. doi: 10.1021/bi960601w. [DOI] [PubMed] [Google Scholar]
- 32.Wu Y, Perisic O, Williams RL, Katan M, Roberts MF. Phosphoinositide-specific phospholipase C delta1 activity toward micellar substrates, inositol 1,2-cyclic phosphate, and other water-soluble substrates: a sequential mechanism and allosteric activation. Biochemistry. 1997;36:11223–11233. doi: 10.1021/bi971039s. [DOI] [PubMed] [Google Scholar]
- 33.Zhou C, Garigapati V, Roberts MF. Short-chain phosphatidylinositol conformation and its relevance to phosphatidylinositol-specific phospholipase C. Biochemistry. 1997;36:15925–15931. doi: 10.1021/bi9716175. [DOI] [PubMed] [Google Scholar]
- 34.Feng J, Roberts M, Drin G, Scarlata S. Dissection of the steps of phospholipase Cb2 activity that are enhanced by Gbg subunits. Biochem. 2005;44 doi: 10.1021/bi0482607. in press. [DOI] [PubMed] [Google Scholar]
- 35.Feng J, Roberts MF, Drin G, Scarlata S. Dissection of the steps of phospholipase C beta 2 activity that are enhanced by G beta gamma subunits. Biochemistry. 2005;44:2577–2584. doi: 10.1021/bi0482607. [DOI] [PubMed] [Google Scholar]
- 36.Romoser V, Ball R, Smrcka AV. Phospholipase C b2 Association with Phospholipid Interfaces Assessed by Fluorescence Resonance Energy Transfer. G Protein bg subunit-mediated translocation is not required for enzyme activation. Journal of Biological Chemistry. 1996;271:25071–25078. doi: 10.1074/jbc.271.41.25071. [DOI] [PubMed] [Google Scholar]
- 37.Alberts B, Bray D, Lewis J, Raff M, Roberts K, Watson J. Molecular Biology of the Cell. Garland; New York: 1994. [Google Scholar]
- 38.Guo Y, Golebiewska U, D'Amico S, Scarlata S. The Small G Protein Rac1 Activates Phospholipase Cδ1 through Phospholipase Cβ2. Journal of Biological Chemistry. 2010;285:24999–25008. doi: 10.1074/jbc.M110.132654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang W, Neer EJ. Reassembly of Phospholipase C-β2 from Separated Domains. Journal of Biological Chemistry. 2001;276:2503–2508. doi: 10.1074/jbc.M003562200. [DOI] [PubMed] [Google Scholar]
- 40.Essen LO, Perisic O, Cheung R, Katan M, Williams RL. Crystal structure of a mammalian phosphoinositide-specific phospholipase C delta. Nature. 1996;380:595–602. doi: 10.1038/380595a0. [DOI] [PubMed] [Google Scholar]
- 41.Drin G, Douguet D, Scarlata S. The pleckstrin homology domain of phospholipase Cbeta transmits enzymatic activation through modulation of the membrane-domain orientation. Biochemistry. 2006;45:5712–5724. doi: 10.1021/bi052317n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Runnels LW. Regulation of phospholipase C beta isozymes by heterotrimeric GTP binding proteins. State University of NY; Stony Brook: 1997. [Google Scholar]
- 43.Philip F, Scarlata S. Influence of membrane components in the binding of G proteins to membrane surfaces. Biochem. 2004;43:11691–11700. doi: 10.1021/bi049381+. [DOI] [PubMed] [Google Scholar]
- 44.Park D, Jhon DY, Lee CW, Lee KH, Rhee SG. Activation of phospholipase C isozymes by G protein beta gamma subunits. J Biol Chem. 1993;268:4573–4576. [PubMed] [Google Scholar]
- 45.Morris AJ, Scarlata S. Regulation of effectors by G-protein alpha- and beta gamma-subunits. Recent insights from studies of the phospholipase c-beta isoenzymes. Biochem Pharmacol. 1997;54:429–435. doi: 10.1016/s0006-2952(97)00032-4. [DOI] [PubMed] [Google Scholar]
- 46.Lyon AM, Tesmer VM, Dhamsania VD, Thal DM, Gutierrez J, Chowdhury S, Suddala KC, Northup JK, Tesmer JJG. An autoinhibitory helix in the C-terminal region of phospholipase C-β mediates Gαq activation. Nat Struct Mol Biol. 2011 doi: 10.1038/nsmb.2095. advance online publication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kuang Y, Wu Y, Smrcka A, Jiang H, Wu D. Identification of a phospholipase Cb2 region that interacts with Gbg. Proc Natl Acad Sci USA. 1996;93:2964–2968. doi: 10.1073/pnas.93.7.2964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sankaran B, Osterhout J, Wu D, Smrcka AV. Identification of a structural element in phospholipase C beta2 that interacts with G protein betagamma subunits. Journal of Biological Chemistry. 1998;273:7148–7154. doi: 10.1074/jbc.273.12.7148. [DOI] [PubMed] [Google Scholar]
- 49.Barr AJ, Ali H, Haribabu B, Snyderman R, Smrcka AV. Identification of a region at the N-terminus of phospholipase C-beta 3 that interacts with G protein beta gamma subunits. Biochemistry. 2000;39:1800–1806. doi: 10.1021/bi992021f. [DOI] [PubMed] [Google Scholar]
- 50.Yoshikawa DM, Bresciano K, Hatwar M, Smrcka AV. Characterization of a phospholipase C beta 2-binding site near the amino-terminal coiled-coil of G protein beta gamma subunits. J Biol Chem. 2001;276:11246–11251. doi: 10.1074/jbc.M006073200. [DOI] [PubMed] [Google Scholar]
- 51.Bonacci TM, Ghosh M, Malik S, Smrcka AV. Regulatory interactions between the amino terminus of G-protein betagamma subunits and the catalytic domain of phospholipase Cbeta2. J Biol Chem. 2005;280:10174–10181. doi: 10.1074/jbc.M412514200. [DOI] [PubMed] [Google Scholar]
- 52.Guo Y, Philip F, Scarlata S. The Pleckstrin homology domains of phospholipases C-beta and -delta confer activation through a common site. J Biol Chem. 2003;278:29995–30004. doi: 10.1074/jbc.M301438200. [DOI] [PubMed] [Google Scholar]
- 53.Wang T, Dowal L, El-Maghrabi MR, Rebecchi M, Scarlata S. The pleckstrin homology domain of phospholipase C-beta(2) links the binding of Gbetagamma to activation of the catalytic core. J Biol Chem. 2000;275:7466–7469. doi: 10.1074/jbc.275.11.7466. [DOI] [PubMed] [Google Scholar]
- 54.Lomasney JW, Cheng HF, Wang LP, Kuan Y, Liu S, Fesik SW, King K. Phosphatidylinositol 4,5-bisphosphate binding to the pleckstrin homology domain of phospholipase C-delta1 enhances enzyme activity. Journal of Biological Chemistry. 1996;271:25316–25326. doi: 10.1074/jbc.271.41.25316. [DOI] [PubMed] [Google Scholar]
- 55.Illenberger D, Schwald F, Pimmer D, Binder W, Maier G, Dietrich A, Gierschik P. Stimulation of phospholipase C-beta2 by the Rho GTPases Cdc42Hs and Rac1. Embo J. 1998;17:6241–6249. doi: 10.1093/emboj/17.21.6241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Illenberger D, Stephan I, Gierschik P, Schwald F. Stimulation of phospholipase C-beta 2 by Rho GTPases. Methods Enzymol. 2000;325:167–177. doi: 10.1016/s0076-6879(00)25441-4. [DOI] [PubMed] [Google Scholar]
- 57.Harden TK, Sondek J. Regulation of phospholipase C isoenzymes by Ras superfamily GTPases. Annu Rev Pharmacol Toxicol. 2006;26:355–379. doi: 10.1146/annurev.pharmtox.46.120604.141223. [DOI] [PubMed] [Google Scholar]
- 58.Simoes AP, Schnabel P, Pipkom R, Camps M, Gierschik P. A peptide corresponding to a potential polyphosphoinositide binding site of phospholipase C-[beta]2 enhances its catalytic activity. FEBS Lett. 1993;331:248–251. doi: 10.1016/0014-5793(93)80346-v. [DOI] [PubMed] [Google Scholar]
- 59.Piiper A, Stryjekkaminska D, Illenberger D, Klengel R, Schmidt JM, Gierschik P, Zeuzem S. Synthetic Peptides Containing a Bxbxxxb(B) Motif Activate Phospholipase C-Beta-1. Biochemical Journal. 1997;326:669–674. doi: 10.1042/bj3260669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Buck E, Li J, Chen Y, Weng G, Scarlata S, Iyengar R. Resolution of a signal transfer region from a general binding domain in G-b for stimulation of phospholipase C-b2. Science. 1999;283:1332–1335. doi: 10.1126/science.283.5406.1332. [DOI] [PubMed] [Google Scholar]
- 61.Buck E, Schatz P, Scarlata S, Iyengar R. Role of dynamic interactions in effective signal transfer for Gbeta stimulation of phospholipase C-beta 2. J Biol Chem. 2002;277:49707–49715. doi: 10.1074/jbc.M205553200. [DOI] [PubMed] [Google Scholar]
- 62.Litosch I. Regulation of Phospholipase Cb-1 Activity by Phosphatidic Acid. Biochem. 2000;39:7736–7743. doi: 10.1021/bi000022y. [DOI] [PubMed] [Google Scholar]
- 63.Wu D, Jiang H, Katz A, Simon MI. Identification of critical regions on phospholipase C-b1 required for action by G-proteins. Journal of Biological Chemistry. 1993;268:3704–3709. [PubMed] [Google Scholar]
- 64.Park D, Jhon DY, Kriz R, Knopf J, Rhee SG. Cloning, sequencing, expression, and Gq-independent activation of phospholipase C-beta 2. J Biol Chem. 1992;267:16048–16055. [PubMed] [Google Scholar]