INTRODUCTION
N-terminalomics identifies proteins by selectively enriching for and sequencing their N-terminal peptides by mass spectrometry (MS) in a high-throughput manner. Several N-terminalomics procedures have been developed to identify protein cleavage sites in a variety of cell signaling events, such as apoptosis and N-terminal methionine excision. In this protocol, a newly developed N-terminalomics approach, N-CLAP (N-terminalomics by Chemical Labeling of the α-Amine of Proteins), is described. This procedure utilizes Edman chemistry to modify all the amines in proteins, followed by the generation of a new unmodified amine at the N-terminus after the removal of the first amino acid by peptide bond cleavage. A commercially available N-hydroxysuccinimide reagent is used to label the α-amine at the protein N-terminus with a cleavable biotin affinity tag, which facilitates the downstream purification of the N-terminal peptides. Peptides are eluted by cleaving the biotin affinity tag using reducing agent, and identified by tandem mass spectrometry (MS/MS). N-CLAP can be used for the identification of signaling peptides for mature proteins as well as for global profiling of cleavage events that occur during cell signaling, such as apoptosis.
RELATED INFORMATION
Proteolytic processing of proteins is a major regulatory mechanism in signal transduction (Ehrmann and Clausen, 2004). Unfortunately, in most cases, the specific proteins targeted by a protease are unknown and the cleavage events that occur during a signaling event are difficult to identify. Profiling of protein N-termini in a complex protein sample can potentially identify cleavages occurred during cell signaling. Several techniques to enrich for N-terminal peptides have been developed in proteomics study. Some of them use “negative selection” in which non-N-terminal peptides are removed and only the N-terminal peptides are retained in the tryptic digest (Gevaert et al., 2003; McDonald and Beynon, 2006; McDonald et al., 2005; Staes et al., 2008). However, the non-N-terminal peptides are much more abundant than the N-terminal peptides, adversely affecting the efficiency of these approaches. Instead, “positive selection” directly purifies N-terminal peptides from a mixture of peptides by selective labeling of the N-termini of proteins (Mahrus et al., 2008; Timmer et al., 2007). The challenge of positive selection is to selectively label the α-amine but not the abundant ε-amine on the lysine residues.
The procedure described here is a newly developed proteomic strategy, N-terminalomics by chemical labeling of the α-amine of proteins (N-CLAP), which selectively labels and profiles protein N-termini in a high-throughput manner (Fig. 1). This procedure utilizes Edman chemistry (Edman, 1956) to block all the amines and to subsequently generate a new free amine at the N-terminus under acidic conditions (Fig. 2). Thus, the sole α-amine in a protein can be labeled by a cleavable biotin N-hydroxysulfosuccinimide reagent. This labeling facilitates the capture of the N-terminal peptides from proteins in complex biological samples using biotin affinity chromatography. N-CLAP can be used to characterize the diversity of N-terminal processing and protein processing that occurs during cell signaling, such as apoptosis (Xu et al., 2009).
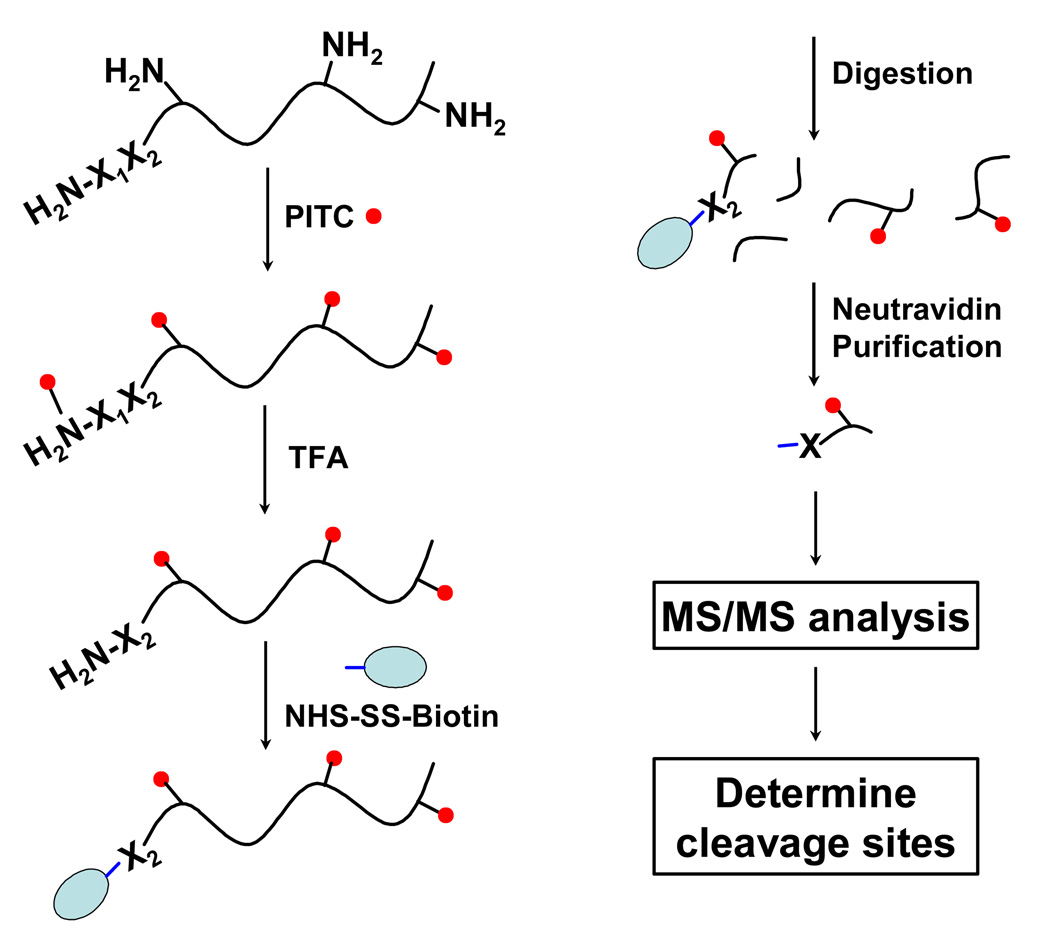
Figure 1.
A flow chart for global profiling of N-termini by N-CLAP. Amines in proteins are blocked by PITC under basic conditions and a free amine is generated by the TFA cleavage of the first amino acid modified by PITC. The new free amine is labeled by a cleavable biotin reagent and proteins are digested with trypsin for affinity purification on Neutravidin agarose beads. Peptides are eluted from the beads by the cleavage of a linker in the biotin tag, and then are identified by MS/MS. Internal cleavage sites that occur during cell signaling events can be determined.
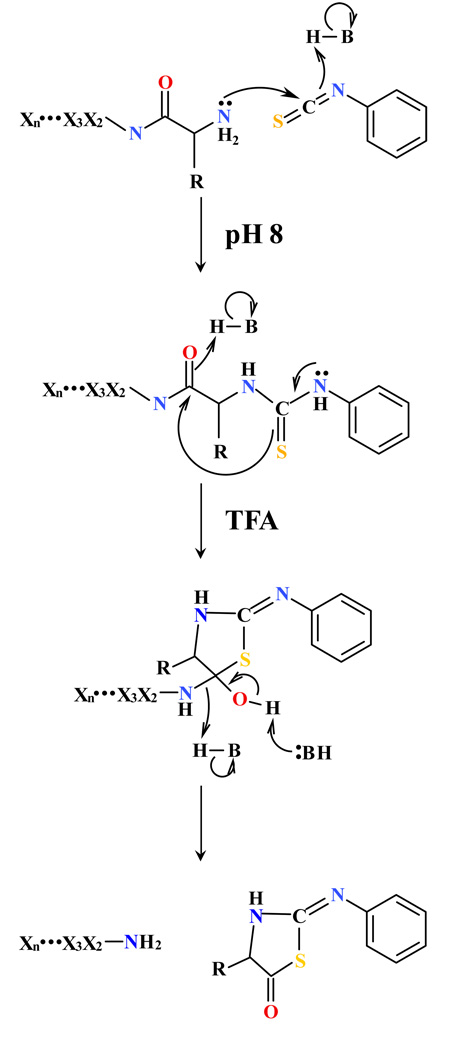
Figure 2.
Scheme of the chemical reactions in Edman chemistry. Amines at protein N-terminus and lysine residues are blocked by PITC. TFA treatment releases the first amino acid with a PITC modification and generates a new unmodified N-terminal amine whereas the modification on the ε-amines is retained. The amino acid sequence in the peptide from the second amino acid are denoted as X2X3…Xn. H-B indicates an acid, with H reflecting the proton bound to B, the base.
A typical experiment from this procedure can result in many N-CLAP peptides that originate from the N-terminus of proteins and from the N-terminus of the C-terminal portion of the protease-cleaved proteins (Xu et al., 2009). By incorporating with stable isotope labeling by amino acids in cell culture (SILAC) (Ong et al., 2002), N-CLAP can potentially quantify the degree of protease processing events under different cellular conditions.
A method similar to SDS-PAGE and in-gel digestion used in this procedure can be found in other protocols (Link and Labaer, 2009; Shevchenko et al., 2006).
MATERIALS
Reagents
Acetonitrile
Ammonium bicarbonate
Anti-biotin antibody or streptavidin-HRP
Benzamidine
Benzene <!>
Bradford protein assay
Dimethyl sulfoxide (DMSO)
Ether <!>
Ethylenediamine tetraacetic acid (EDTA)
Formic acid <!>
Gel washing solution (50 % acetonitrile/25 mM ammonium bicarbonate in distilled water, freshly prepared)
Hydroxylamine hydrochloride
4-(2-Hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES)
Liquid chromatography (LC)-MS solvent A (0.1 % formic acid in LC-MS grade H2O)
LC-MS solvent B (90 % acetonitrile/0.1 % formic acid in LC-MS grade H2O)
LDS sample buffer (4X, Invitrogen)
Lysis buffer (150 mM NaCl, 50 mM HEPES, 2 mM EDTA, 1 % triton X-100, pH 7.4)
Methanol
Neutravidin agarose beads (Pierce)
Nitrogen gas
NuPage 4–12 % Tris-Glycine mini-gel (Invitrogen)
Protein digestion solution (15 µg/mL trypsin solution in 25 mM ammonium bicarbonate and 1 mM CaCl2 in distilled water; aliquot and store at −20 °C; minimize freeze/thaw cycles up to three times)
Peptide extraction solvent (5 % formic acid/50 % acetonitrile in distilled water)
Phenyl isothiocyanate (PITC, sequencing grade) <!>
Phosphate buffered saline (PBS)
Protease inhibitor cocktail (Roche)
Pyridine <!>
SDS-PAGE reagents
Sodium chloride
Sodium hydroxide (5 M in distilled water) <!>
Sulfosuccinimidyl 2-(biotinamido) ethyl-1, 3′ dithiopropionate (EZ-Link Sulfo-NHS-SS-biotin) (Pierce)
Triethylamine <!>
Trifluoroacetic acid (TFA, anhydrous) <!>
Tris(2-carboxyethyl)phosphine hydrochloride (TCEP)
Triton X-100
Trypsin, sequencing grade-modified (Promega)
Urea
Caution: These chemicals are volatile, corrosive, or harmful when inhaled or contacted with skin. Wear gloves and use these chemicals in a chemical hood.
Equipment
Centrifuge (Benchtop centrifuge and Ultracentrifuge)
Centrifuge tubes, thick wall polycarbonate (Beckman)
Eppendorf centrifuge tubes, 1.6-mL
Conical tubes, 50 mL (polypropylene)
Mass spectrometer (LC-MS/MS with online nanoflow reversed-phase liquid chromatography)
Peptide identification software (available commercially from MS instrument vendors or online programs: such as X!Tandem, OMSSA, and Mascot)
pH meter or pH strips
Pipette tips
SDS-PAGE apparatus
0.2 µm Spin column (Pall Science)
Thermomixer (Eppendorf)
Vortex
METHOD
Preparation of Cell Lysate (1 h)
-
1.
Lyse cells in 1 mL lysis buffer (150 mM NaCl, 50 mM HEPES, 2 mM EDTA, 1 % triton X-100, pH 7.4) by gentle sonication in the presence of protease inhibitor cocktail.
Depending on the cell types, the amount of cells used in the experiment may vary. Use appropriate amount of cells which produce about 2 mg proteins.
-
2.
Centrifuge cell lysate at 100,000 g for 30 min in an ultracentrifuge using thick wall polycarbonate centrifuge tubes.
-
3.
Collect supernatant and measure protein concentration using Bradford protein assay (Olson and Markwell, 2007). The recommended protein concentration is about 2 mg/mL.
PAUSE POINT: Store sample at − 20 °C.
PITC modification of amines (3 ~ 4 h)
-
4.
Prepare twenty 1.6 mL Eppendorf centrifuge tubes in a chemical hood.
-
5.
Add 50 µL of cell lysate and 1 mL of a mixed solvent consisting of methanol, pyridine, triethylamine and distilled water in a ratio of 7:1:1:1 to each tube. Mix sample thoroughly by vortex.
-
6.
Add 200 µL PITC into each tube and slowly blow nitrogen gas over each tube for 5 s to remove oxygen. Caution: Adjust nitrogen gas flow to a low rate to avoid sample loss. Carry out this step in a chemical hood.
-
7.
Incubate the tubes at 55 °C in a thermomixer for 1 h with constant shaking (~800 RPM).
-
8.
Add an additional 200 µL PITC to each tube and continue the reaction for 1 h at 55 °C in a thermomixer for 1 h with constant shaking (~800 RPM). Alternatively, two 50 mL conical tubes can be used for the above reaction if a suitable heat block is available to fit the 50 mL conical tubes in the thermomixer.
-
9.
Transfer the reaction solution to two 50 mL polypropylene conical tubes.
-
10.
Add 40 mL of benzene to each 50 mL conical tube in a chemical hood, vortex for 30 s, and centrifuge at 3000 g for 2 min at 25 °C. Remove the top organic phase and discard it to a waste solvent bottle in the chemical hood. Repeat this step twice. Caution: Benzene is a toxic and flammable liquid. Carry out this step and handle benzene in a chemical hood all the time. Dispose waste benzene properly.
-
11.
Transfer the protein phase to a 1.6 mL centrifuge tube and evaporate residual benzene by slowly blowing air in the chemical hood for 15 min or until the residual organic layer on the top disappears. Caution: Make sure the air flow is sufficiently low to avoid sample loss.
-
12.
Dry the sample in a SpeedVac to remove residual water.
PAUSE POINT: Store sample at 4 °C overnight.
Note: Carry out steps 4–11 in a chemical hood. Cap, label, and appropriately discard the waste solvent bottle in accordance with institutional regulations after each use.
Cleavage of PITC modified N-terminal amino acids (1 ~ 2 h)
-
13.
Add 500 µL anhydrous TFA to the centrifuge tube from step 12 and slowly flush the tube for 5 s with nitrogen gas in a chemical hood to remove oxygen.
-
14.
Vortex the centrifuge tube briefly in a chemical hood at 25 °C to completely dissolve the protein pellet. Caution: Wear gloves and carry out this step in a chemical hood. A cap lock can be applied to the tube to ensure that the cap does not open during vortexing or subsequent heating steps.
-
15.
Carry out the TFA cleavage reaction at 45 °C for 15 min with constant shaking in a thermomixer (800 RPM) in a chemical hood.
-
16.
Reduce the volume of the TFA solution to between 50 – 100 µL by gently blowing nitrogen gas over the sample in a chemical hood for 15 min. Caution: Control nitrogen gas to a low flow rate and avoid sample loss.
-
17.
Carry out protein precipitation and pellet washing. Add 1 mL of ice-cold anhydrous ether to the sample tube in a chemical hood. Vortex for 10 s and centrifuge for 30 s at 2000 g at 4 °C in a chemical hood. Use pipette tip to remove ether and dispose to the waste solvent bottle in the chemical hood. Use 1 mL of ice-cold anhydrous ether to wash the pellet twice using the same procedure for protein precipitation to remove residual TFA.
PAUSE POINT: Store sample at 4 °C overnight.
Note: Perform steps 13–17 in a chemical hood.
Affinity labeling of N-terminus with biotin-SS-NHS (6 h ~ 1 d)
-
18.
Dissolve the sample in 500 µL of 0.2 M HEPES buffer (pH 8.5) in the presence of 2 % SDS and 150 mM NaCl and keep 5 µL for Western blotting analysis. Use pH meter or pH strips to measure the pH of the sample. If the pH of the sample is acidic due to the presence of trace amounts of residual TFA in the pellet, use 1 M NaOH to adjust pH to 8.5. Incubate the sample at 25 °C in a thermomixer with constant shaking (1000 RPM).
Note: This step may take several hours. Alternatively, sonicate sample in a water bath for 30 min. Make sure there are no free amines (e.g. Tris-HCl buffer) and do not add any reducing agent in the buffer.
-
19.
Add EZ-Link Sulfo-NHS-SS-biotin (200 mM in DMSO, freshly made) to the sample. The final concentration of EZ-Link Sulfo-NHS-SS-biotin is 5 mM.
-
20.
Incubate the sample at 37 °C for 4 h.
PAUSE POINT: The reaction can also be carried out at 4 °C overnight.
-
21.
Prepare fresh 1 M hydroxylamine solution and adjust the pH to 8.5 with 5 M NaOH. Treat the sample from step 20 with 1 M hydroxylamine (final concentration is 100 mM) and incubate at 25 °C for 30 min. Keep 5 µL for Western blotting analysis.
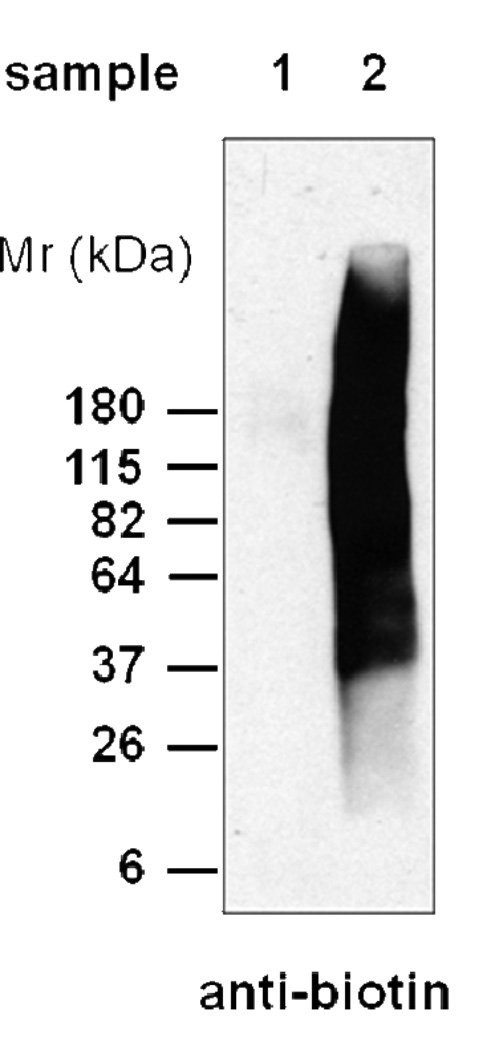
Note: Make sure pH does not exceed 8.5 for the 1 M hydroxylamine solution. Use anti-biotin Western blotting (no reducing reagent) to confirm the labeling of N-terminus by EZ-Link Sulfo-NHS-SS-biotin (samples from steps 18 and 21). A representative anti-biotin Western blotting for a complex cell lysate obtained in steps 18 and 21 is shown in Fig. 3.
Figure 3.
A typical anti-biotin Western blotting for complex protein mixtures obtained before and after reacting with biotin-SS-NHS in the N-CLAP approach. Samples 1 and 2 are from step 18 and step 21, respectively.
SDS-PAGE and in-gel trypsin digestion (1 d)
-
22.
Add the appropriate amount of 4 × LDS sample buffer (without reducing agent) to adjust the protein sample to 1 ×, and incubate at 37 °C for 10 min.
Note: Make sure no reduce agent, such as dithiothreitol and β-mercaptoethanol, is added in the LDS sample buffer.
-
23.
Load the sample on to a NuPage 4–12 % Tris-Glycine mini-gel (Use multiple lanes if necessary). After electrophoresis, wash the gel twice with distilled water for 10 min each with constant shaking in a plate shaker.
Note: The gel can be stained with Coomassie Brilliant blue for visualization during the gel excision step. The gel pieces can be destained by incubating with gel washing solvent with constant shaking in a thermomixer (1000 RPM) and by changing gel washing solvent a few times.
-
24.
Divide the gel strip into five fractions to reduce the complexity of proteins in each fraction. Cut each fraction into 1 mm × 1 mm pieces.
-
25.
Wash gel pieces twice with gel washing solvent in a thermomixer with constant shaking (1000 RPM, 25 °C) for 20 min each time.
-
26.
Dehydrate gel pieces with 100 % acetonitrile for 5 min in a thermomixer with constant shaking (1000 RPM, 25 °C). Dry gel pieces completely in a SpeedVac for 5 min at 25 °C.
-
27.
Add protein digestion solution to gel pieces until the gel becomes fully swollen and covered by protein digestion solution.
-
28.
Perform digestion at 37 °C overnight in a thermomixer with constant shaking (600 RPM).
PAUSE POINT: Continue digestion at 37 °C overnight.
-
29.
Transfer digested solution to a new Eppendorf tube.
-
30.
Add 1 mL of peptide extraction solvent to the gel pieces. Incubate the samples in a thermomixer with constant shaking (1000 RPM, 25 °C) for 30 min. Sonicate the samples in a water bath sonicator for 20 min. Transfer the peptide solution to the tube from step 29. Repeat this extraction one more time. Finally, use 100 % acetonitrile to incubate the gel pieces in the thermomixer with constant shaking (1000 RPM) for 5 min. Combine all the peptide solution to the centrifuge tube from step 29.
-
31.
Dry the sample in a SpeedVac at 25 °C for 4 ~ 6 h. Alternatively, lyophilize sample overnight.
Sample preparation for mass spectrometry analysis (6 h)
-
32.
Dissolve peptide pellet from step 31 in 800 µL of re-suspension buffer (100 mM HEPES, 1 M NaCl, pH 7.4) in the presence of 250 mM benzamidine and incubate the sample at 25 °C for 30 min with constant shaking.
-
33.
Centrifuge in a benchtop centrifuge at 21,000 g for 15 min at 25 °C to remove any undissolved material.
-
34.
Add 30 µL of 50 % Neutravidin agarose beads to each sample and incubate at 4 °C for 2 h.
-
35.
Wash Neutravidin beads (in an end-over-end rotator) sequentially with 1 mL of each of the following buffers for 10 min each: 5 M NaCl in PBS (pH 7.4); 3 M NaCl in PBS; 1 M NaCl in PBS; 2 M urea in PBS; PBS; 20 % methanol; distilled water.
-
36.
Incubate beads with 100 µL of 10 mM TCEP (pH 7.0) in a thermomixer with constant shaking (1000 RPM) at 25 °C for 2 h.
-
37.
Spin down the beads in a benchtop centrifuge at 2000 g for 30 s and collect the supernatant.
-
38.
Wash the beads once with distilled H2O and combine the liquid with the supernatant obtained from step 37.
-
39.
Filter the sample through a 0.2 µm spin column by centrifuging in a benchtop centrifuge at 14,000 g for 2 min.
-
40.
Dry the sample in SpeedVac and re-suspend it in 20 µL of 5 % acetonitrile/0.1 % TFA aqueous solvent.
Optional: Remove salts and other impurities using C18 reverse-phase desalting tips, such as StageTips (Proxeon) (Rappsilber et al., 2007).
PAUSE POINT: Store sample at − 20 °C overnight.
Mass spectrometry analysis and identification of N-terminal cleavages (2 h ~ 1 d)
-
41.
Carry out LC-MS/MS analysis in an Agilent Q-TOF MS or other MS instrument with similar mass accuracy (below 20 p.p.m. mass accuracy for precursor ions) with following parameter setting. The Agilent Q-TOF LC-MS/MS system used in this experiment consists of an 1100 Series HPLC, HPLC-Chip Cube MS interface, and 6520 series Q-TOF mass spectrometer. The HPLC-Chip contains a 40 nL enrichment column and a 43 mm×75 Pm analytical column packed with Zorbax 300SB-C18 (5 µm particles). Eight microliters of each sample are injected onto the MS instrument and peptides are loaded onto the enrichment column with 97 % LC-MS solvent A and 3 % LC-MS solvent B with a flow rate of 4 µL/min. Peptides are eluted with a gradient from 3 % to 45 % LC-MS solvent B in 45 min, followed by a steep gradient to 90 % solvent B for 5 min and by an equilibration step (10 min) with 3 % solvent B at a flow rate of 0.3 µL/min. Mass spectra are acquired in the positive-ion mode with automated data-dependent MS/MS on the five most intense ions from precursor MS scans and every selected precursor peak is analyzed twice within 2 min. Each sample is analyzed twice and the peptides identified in the first run are excluded for MS/MS fragmentation in the second run in order to increase the identification of low abundant peptides.
-
42.
The database search is performed using search programs from instrument vendors, such as Sequest and Spectrum Mill, or using commercial or open source search programs, such as Mascot, OMSSA and X!Tandem. The modifications that are used during the search are: PITC modification for lysine residues (C7H6NS; the monoisotopic mass difference caused by this modification is 135.01 Da), cleavable biotin modification (C3H5OS; the monoisotopic mass difference caused by this modification is 88.00 Da) for the N-terminus, and a variable modification for methionine oxidation (the mass difference is 15.99 Da). Note: A hydrogen atom at the ε-amine of the lysine residue and at the N-terminus is subtracted during the chemical reaction with PITC and cleavable biotin reagent, which should be reflected on the modification used for the database search. Because all the lysines in the proteins are modified by PITC, trypsin cleaves only after arginine residues. Therefore, set Arg-C as the digestion enzyme and N-terminal cleavage as nonspecific during the database search. If this option is not available, use trypsin as the enzyme, N-terminal cleavage as nonspecific and allow additional missed cleavage sites, such as up to four missed cleavage sites. Set methionine oxidation as a variable modification due to potential oxidation of this residue.
-
43.
Manually validate the MS/MS spectra and determine the N-terminal amino acid. Make sure all the major peaks match the fragment ions. Confirm the fragmentation of identified peptides fits the common rules, such as preferred cleavages at the proline residues. Verify PITC modification on lysine and -COCH2CH2SH modification at N-terminus. In addition, all the peptides have a C-terminal arginine. The only exceptions are the peptides derived from the C-terminus of proteins, which is a very rare case. If manual validation is not practical for all the identified peptides due to the large number of hits, a target-decoy search strategy (Elias and Gygi, 2007) can be used to evaluate the false positive rate of the identification. In this strategy, a protein database, such as Swiss-Prot protein database, with concatenated reverse or random decoy database with same entries and same protein lengths, is used during the database search. The hits from the decoy database are regarded as false identification. The relative number of hits obtained from the decoy database and from the normal protein database can be used to determine the cutoff scores for positive identifications. The false positive rate is normally set below 5 % or below 1 % of the total identification.
-
44.
Reconstitute the first amino acid in the original peptides by using a protein database, such as Swiss-Prot database (Boeckmann et al., 2003), NCBI database (Pruitt et al., 2007), and IPI protein database (Kersey et al., 2004), or the peptide information obtained from database search because the first amino acid in the protein is removed during the N-CLAP procedure. Determine the protein cleavage sites which occur during cell signaling.
TROUBLESHOOTING
Problem: Aqueous phase and organic phase do not separate well.
[Step 10]
Solution:
Make sure to add enough benzene during the extraction step.
Mix the solution thoroughly and centrifuge the sample briefly (3,000 g for 2 min).
Problem: Sample is not completely dissolved.
[Step 18]
Solution:
Make sure pH is about 8.5. The pH may decrease due to the presence of trace amounts of TFA.
Make sure 2 % SDS is present in the re-suspension buffer.
Sonicate sample in a water bath sonicator for 30 min.
Incubate sample for a longer period of time or overnight.
Increase the incubation temperature to 37 °C.
Increase the volume of re-suspension buffer to 1 mL.
Problem: No signal upon anti-biotin Western blotting.
[Step 21]
Solution:
Make sure no reducing agent is present during the biotin labeling, SDS-PAGE and Western blotting analysis.
Maintain pH at ~ 8.5. Avoid significant pH deviation from 8.5 in steps 18–20.
Make sure no amines, such as Tris-HCl, are introduced in steps 18–20.
Make sure the pH of 1 M hydroxylamine solution is about 8.5 in step 21.
The cleavage biotin NHS reagent may be hydrolyzed after long-time storage. Use new cleavage biotin NHS reagent.
Problem: No signal is detected in MS analysis.
[Step 41]
Solution:
Trypsin digestion is not working well. Make sure the pH of the digestion solution is about 8.0. Prepare fresh ammonium bicarbonate to dissolve trypsin for digestion. Use sequencing grade modified trypsin from Promega. Increase trypsin concentration. Incubate a longer time, such as 24 h, during trypsin digestion. Make sure that 1 mM CaCl2 is included in the protein digestion solution.
Peptides are not efficiently extracted. Make sure that fresh formic acid is included in the extraction solvent. Incubate for a longer time in a thermomixer with constant shaking (1000 RPM) during peptide extraction step. Sonicate samples for 20 min in a water bath sonicator after each incubation. Carry out extraction step three times before using 100 % acetonitrile for final extraction.
Peptides do not bind to Neutravidin beads. Maintain pH ~ 7.4. Make sure no reducing agents and biotin-containing reagents are present in steps 32 – 35.
Peptide is not eluted from Neutravidin beads. Make sure the following condition is maintained during the peptide elution step: maintain pH at ~ 7.0; prepare fresh TCEP solution for peptide elution; slightly increase incubation temperature to 37 °C and increase incubate time to 4 h.
MS instrument is not in working condition. Use a standard, such as bovine serum albumin (BSA) digest (New England BioLabs), to diagnose problems associated with the instrument.
DISCUSSION
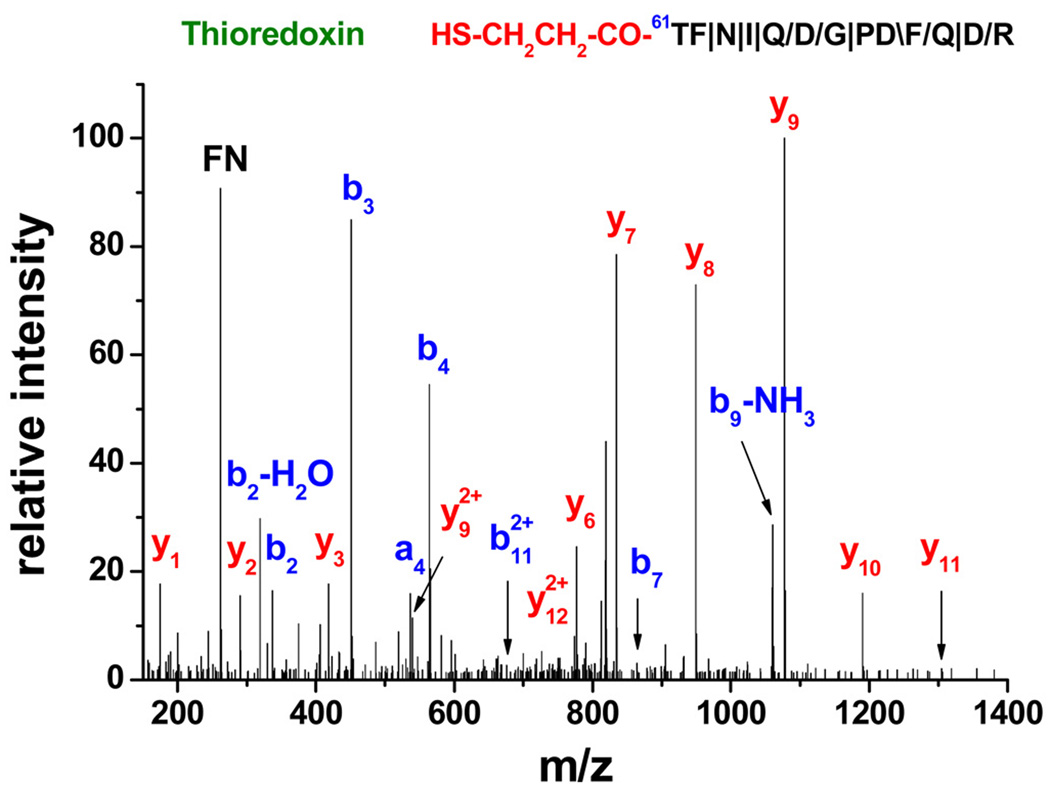
Profiling of N-termini by N-CLAP described in this protocol utilizes simple chemical reactions developed by Edman (Edman, 1956) to block amines and generate a new unmodified α-amine in each protein (Fig. 2). A biotin affinity tag is then conjugated to the N-terminus of proteins and the N-terminal peptides are enriched using biotin affinity chromatography after the digestion of a protein mixture. All the chemicals used in this protocol are commercially available and this approach can be carried out in any laboratory with biochemistry expertise. The whole process does not require advanced training in MS data analysis. Since N-CLAP only retains one peptide for each protein, it significantly simplifies a peptide mixture for downstream MS analysis, which can increase the proteome coverage in a complex protein mixture. Because all the lysines are modified by PITC, trypsin only cleaves proteins after arginine. Therefore, the C-terminus of all N-CLAP peptides is arginine, which provides a signature y1-ion of 175 m/z. A representative annotated MS/MS spectrum is shown (Fig. 4). The only exceptions are peptides derived from the C-terminus of proteins. These peptides end with the native C-terminal residue. This feature can be used to confirm the specificity of peptides identified by the N-CLAP approach (Xu et al., 2009). Typically in a Q-TOF MS instrument, N-CLAP can profile several hundreds of N-terminal peptides, processed N-terminal peptides, and neo-N-terminal peptides, which are derived from internal cleavages in signaling events, in a complex cell lysate. Overall, ~ 80 % of identified peptides are N-terminal peptides and the rest of ~ 20 % are non-N-terminal peptides that are retained on avidin beads and co-eluted with the N-terminal peptides. In addition, N-CLAP has been demonstrated to identify protein cleavage events in signaling pathways (Xu et al., 2009).
Figure 4.
A representative annotated MS/MS spectrum obtained from N-CLAP profiling of N-termini. The spectrum is acquired using an Agilent 6520 series Q-TOF mass spectrometry. The sequence map of the peptide, the N-terminal modification, the position of the first amino acid in the coding sequence, and the protein name are indicated in the spectrum. The sequence indicated in red, HS-CH2CH2CO-, is the portion of the biotin tag that remains on the peptide after TCEP cleavage. This N-CLAP peptide starts from the 61st amino acid from the coding sequence, which indicates that the first 59 amino acids are removed in the nascent chain of mitochondrial thioredoxin since the N-CLAP removes one additional N-terminal amino acid. The result is consistent with the annotation in Swiss-Prot database (Accession #: Q99757). The symbols, \, / and |, represent b-ions, y-ions, and both b-ions and y-ions, respectively.
It has been reported that some N-terminal amines are blocked, e.g. acetylated (Meinnel et al., 2005), in vivo. These blocked proteins would not be detected since they cannot be labeled on their N-termini. This problem may be overcome by using enzymes or chemical approaches to remove N-terminal blocking groups (Hirano and Kamp, 2003). However, certain N-terminal modifications, such as methylation, are expected to be detectable, because they are compatible with Edman degradation (Chen et al., 1977). The second limitation is that protein digested by trypsin may generate N-terminal peptides which are not suitable (either too long or too short) for MS identification, which is a common problem for proteomics studies. In this case, different enzymes, such as Glu-C, chymotrypsin, and the combination of multiple enzymes, can be used during the sample digestion step in order to increase the proteome coverage. In addition, some protein homologues have the same amino acid sequence at their N-termini. In such case, additional biochemical verification is required to further confirm the exact protein(s) cleaved in signaling pathways.
ACKNOWLEDGMENTS
This work was supported by NIH RR19355 and RR22615 (MS instrumentation), training grant T32CA062948 from the National Cancer Institute (GX), NIAID (AI068639) and the Dorothy Rodbell Sarcoma Foundation (SRJ).
Footnotes
COMPETING FINANCIAL INTERESTS STATEMENT
The authors declare that they have no competing financial interests.
REFERENCES
- Boeckmann B, Bairoch A, Apweiler R, Blatter MC, Estreicher A, Gasteiger E, Martin MJ, Michoud K, O'Donovan C, Phan I, et al. The SWISS-PROT protein knowledgebase and its supplement TrEMBL in 2003. Nucleic Acids Res. 2003;31:365–370. doi: 10.1093/nar/gkg095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R, Brosius J, Wittmann-Liebold B. Occurrence of methylated amino acids as N-termini of proteins from Escherichia coli ribosomes. J Mol Biol. 1977;111:173–181. doi: 10.1016/s0022-2836(77)80121-6. [DOI] [PubMed] [Google Scholar]
- Edman P. Mechanism of the Phenyl Isothiocyanate Degradation of Peptides. Nature. 1956;177:667–668. [Google Scholar]
- Ehrmann M, Clausen T. Proteolysis as a regulatory mechanism. Annu Rev Genet. 2004;38:709–724. doi: 10.1146/annurev.genet.38.072902.093416. [DOI] [PubMed] [Google Scholar]
- Elias JE, Gygi SP. Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat Methods. 2007;4:207–214. doi: 10.1038/nmeth1019. [DOI] [PubMed] [Google Scholar]
- Gevaert K, Goethals M, Martens L, Van Damme J, Staes A, Thomas GR, Vandekerckhove J. Exploring proteomes and analyzing protein processing by mass spectrometric identification of sorted N-terminal peptides. Nat Biotechnol. 2003;21:566–569. doi: 10.1038/nbt810. [DOI] [PubMed] [Google Scholar]
- Hirano H, Kamp RM. Deblocking of N-terminally modified proteins. Methods Mol Biol. 2003;211:355–363. doi: 10.1385/1-59259-342-9:355. [DOI] [PubMed] [Google Scholar]
- Kersey PJ, Duarte J, Williams A, Karavidopoulou Y, Birney E, Apweiler R. The International Protein Index: an integrated database for proteomics experiments. Proteomics. 2004;4:1985–1988. doi: 10.1002/pmic.200300721. [DOI] [PubMed] [Google Scholar]
- Link AJ, Labaer J. In-gel trypsin digest of gel-fractionated proteins. Cold Spring Harb Protoc. 2009;2009 doi: 10.1101/pdb.prot5110. pdb prot5110. [DOI] [PubMed] [Google Scholar]
- Mahrus S, Trinidad JC, Barkan DT, Sali A, Burlingame AL, Wells JA. Global sequencing of proteolytic cleavage sites in apoptosis by specific labeling of protein N termini. Cell. 2008;134:866–876. doi: 10.1016/j.cell.2008.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald L, Beynon RJ. Positional proteomics: preparation of amino-terminal peptides as a strategy for proteome simplification and characterization. Nat Protoc. 2006;1:1790–1798. doi: 10.1038/nprot.2006.317. [DOI] [PubMed] [Google Scholar]
- McDonald L, Robertson DH, Hurst JL, Beynon RJ. Positional proteomics: selective recovery and analysis of N-terminal proteolytic peptides. Nat Methods. 2005;2:955–957. doi: 10.1038/nmeth811. [DOI] [PubMed] [Google Scholar]
- Meinnel T, Peynot P, Giglione C. Processed N-termini of mature proteins in higher eukaryotes and their major contribution to dynamic proteomics. Biochimie. 2005;87:701–712. doi: 10.1016/j.biochi.2005.03.011. [DOI] [PubMed] [Google Scholar]
- Olson BJ, Markwell J. Assays for determination of protein concentration. Curr Protoc Protein Sci. 2007;Chapter 3 doi: 10.1002/0471140864.ps0304s48. Unit 3 4. [DOI] [PubMed] [Google Scholar]
- Ong SE, Blagoev B, Kratchmarova I, Kristensen DB, Steen H, Pandey A, Mann M. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol Cell Proteomics. 2002;1:376–386. doi: 10.1074/mcp.m200025-mcp200. [DOI] [PubMed] [Google Scholar]
- Pruitt KD, Tatusova T, Maglott DR. NCBI reference sequences (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2007;35:D61–D65. doi: 10.1093/nar/gkl842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rappsilber J, Mann M, Ishihama Y. Protocol for micro-purification, enrichment, pre-fractionation and storage of peptides for proteomics using StageTips. Nat Protoc. 2007;2:1896–1906. doi: 10.1038/nprot.2007.261. [DOI] [PubMed] [Google Scholar]
- Shevchenko A, Tomas H, Havlis J, Olsen JV, Mann M. In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat Protoc. 2006;1:2856–2860. doi: 10.1038/nprot.2006.468. [DOI] [PubMed] [Google Scholar]
- Staes A, Van Damme P, Helsens K, Demol H, Vandekerckhove J, Gevaert K. Improved recovery of proteome-informative, protein N-terminal peptides by combined fractional diagonal chromatography (COFRADIC) Proteomics. 2008;8:1362–1370. doi: 10.1002/pmic.200700950. [DOI] [PubMed] [Google Scholar]
- Timmer JC, Enoksson M, Wildfang E, Zhu W, Igarashi Y, Denault JB, Ma Y, Dummitt B, Chang YH, Mast AE, et al. Profiling constitutive proteolytic events in vivo. Biochem J. 2007;407:41–48. doi: 10.1042/BJ20070775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu G, Shin SB, Jaffrey SR. Global profiling of protease cleavage sites by chemoselective labeling of protein N-termini. Proc Natl Acad Sci U S A. 2009;106:19310–19315. doi: 10.1073/pnas.0908958106. [DOI] [PMC free article] [PubMed] [Google Scholar]