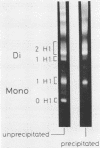
Abstract
Exonuclease III was used to uniformly trim DNA ends of micrococcal nuclease-prepared chromatin fragments down to the first major impediment encountered by the enzyme, which arises from the interaction of H1 with the nucleosome. This trimming, when performed on nucleosome dimers, allowed one to quantitatively determine the center-to-center distance of nucleosomes. This distance, of mean 198 base pairs, was found to essentially vary between about 180 and 215 base pairs, with extremes of 165 and 230 base pairs. Trimming of trimers further revealed that the overall arrangement of nucleosome center-to-center distances along the chromatin fiber is that expected on a statistical basis.
Full text
PDF


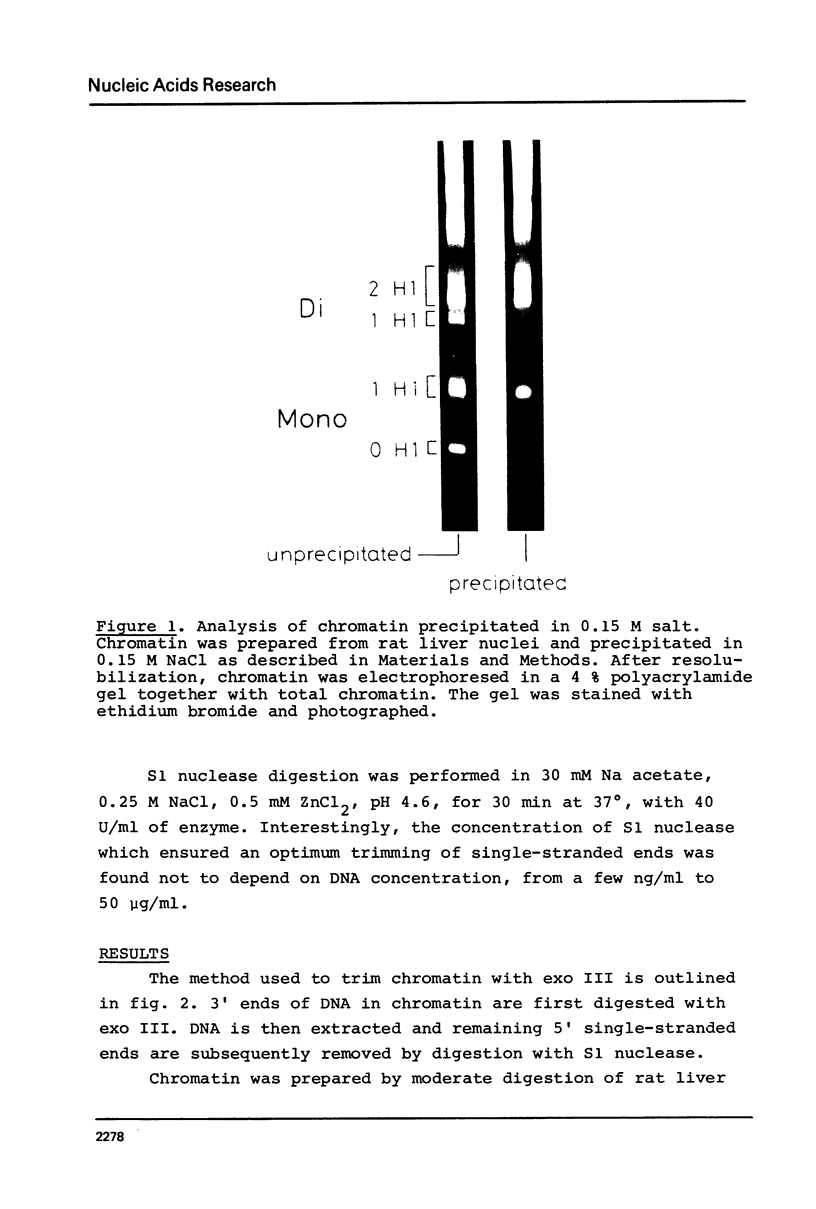
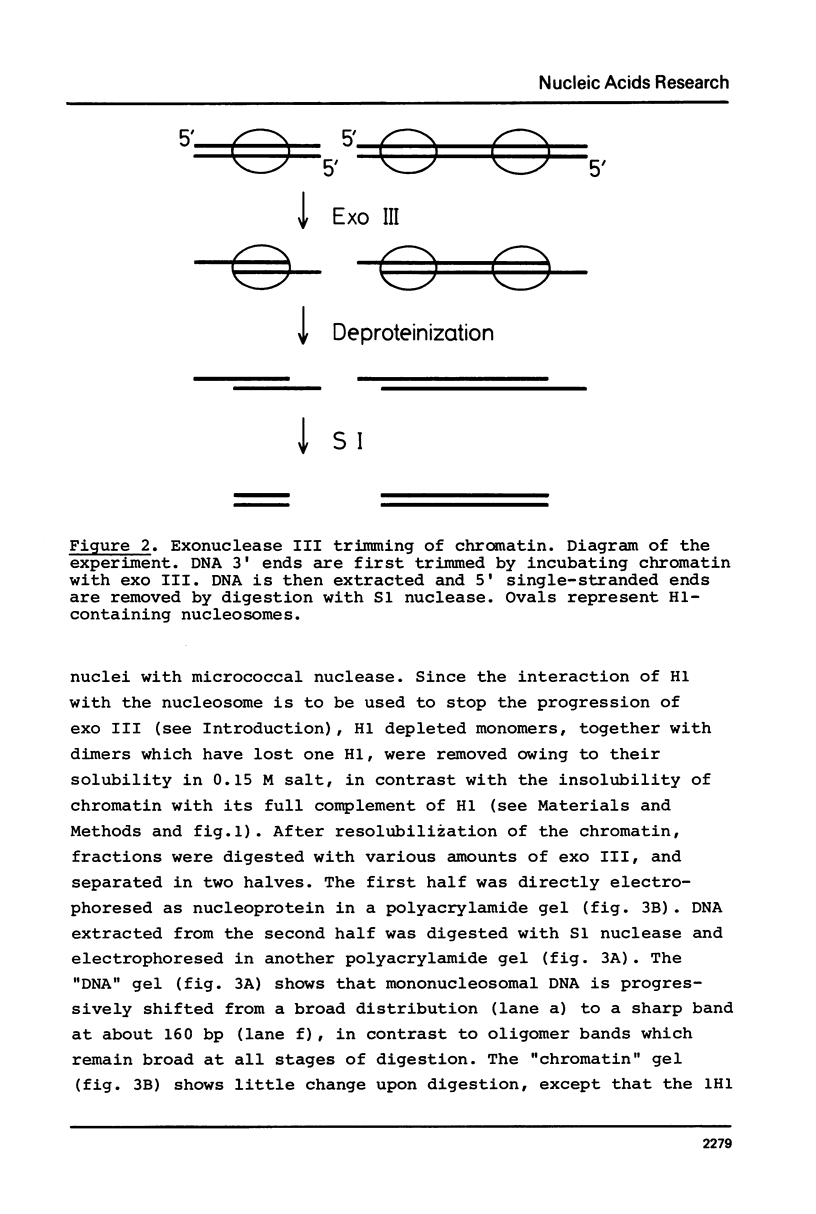
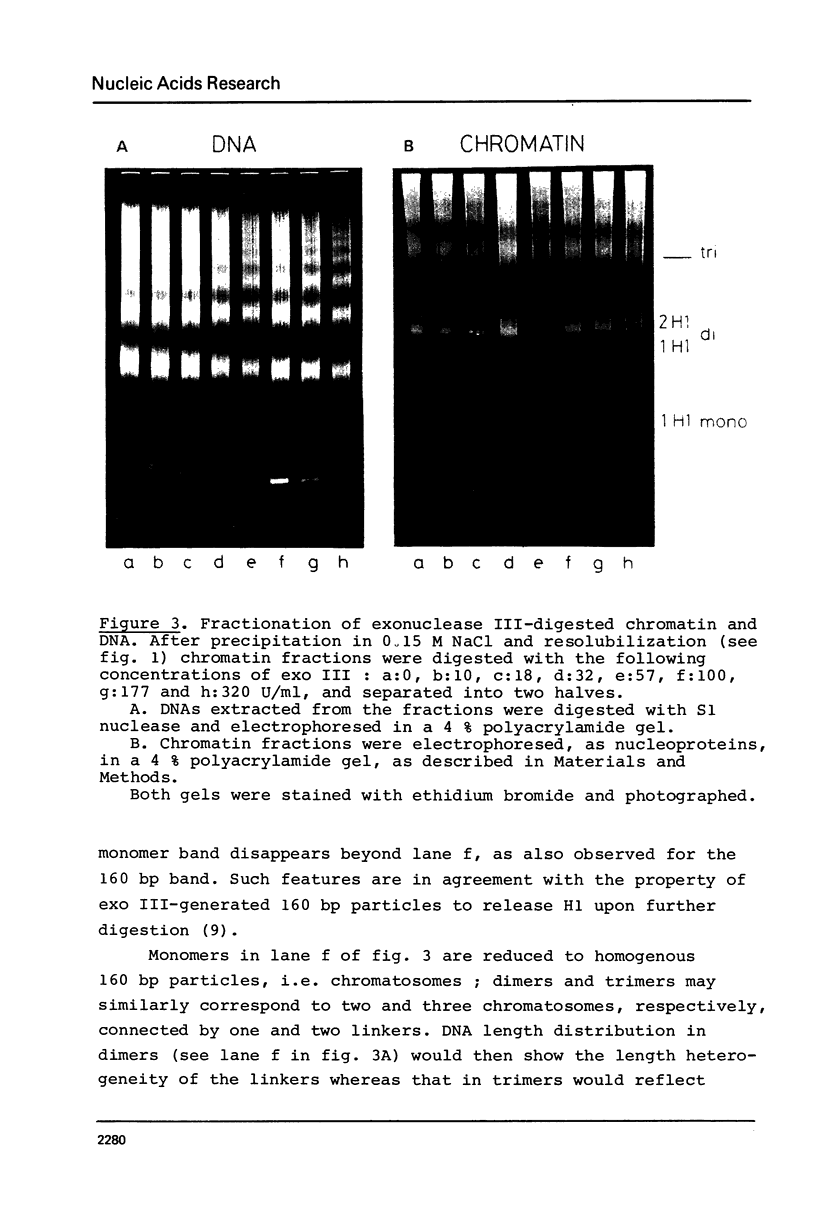






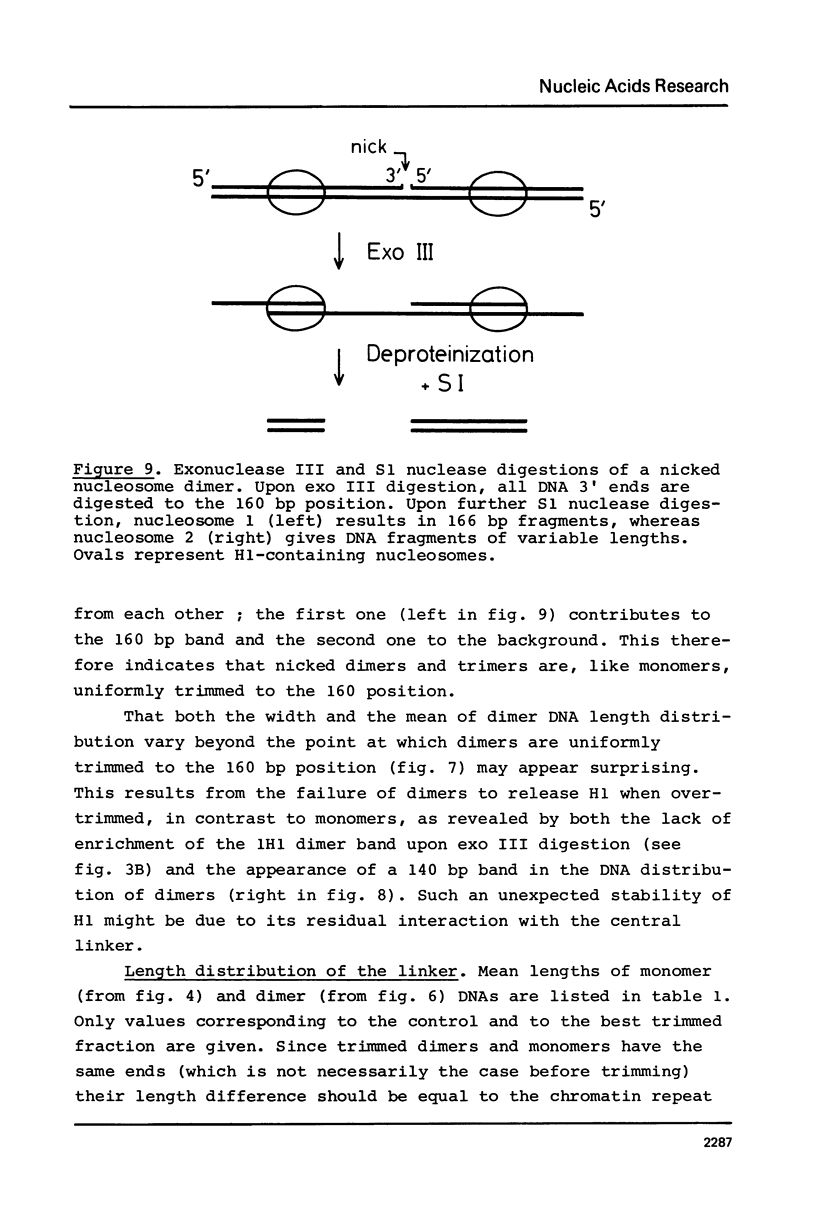






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Blobel G., Potter V. R. Nuclei from rat liver: isolation method that combines purity with high yield. Science. 1966 Dec 30;154(3757):1662–1665. doi: 10.1126/science.154.3757.1662. [DOI] [PubMed] [Google Scholar]
- Compton J. L., Bellard M., Chambon P. Biochemical evidence of variability in the DNA repeat length in the chromatin of higher eukaryotes. Proc Natl Acad Sci U S A. 1976 Dec;73(12):4382–4386. doi: 10.1073/pnas.73.12.4382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewish D. R., Burgoyne L. A. Chromatin sub-structure. The digestion of chromatin DNA at regularly spaced sites by a nuclear deoxyribonuclease. Biochem Biophys Res Commun. 1973 May 15;52(2):504–510. doi: 10.1016/0006-291x(73)90740-7. [DOI] [PubMed] [Google Scholar]
- Igo-Kemenes T., Omori A., Zachau H. G. Non-random arrangement of nucleosomes in satellite I containing chromatin of rat liver. Nucleic Acids Res. 1980 Nov 25;8(22):5377–5390. doi: 10.1093/nar/8.22.5377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keichline L. D., Wassarman P. M. Developmental study of the structure of sea urchin embryo and sperm chromatin using micrococcal nuclease. Biochim Biophys Acta. 1977 Mar 2;475(1):139–151. doi: 10.1016/0005-2787(77)90348-3. [DOI] [PubMed] [Google Scholar]
- Keichline L. D., Wassarman P. M. Structure of chromatin in sea urchin embryos, sperm, and adult somatic cells. Biochemistry. 1979 Jan 9;18(1):214–219. doi: 10.1021/bi00568a033. [DOI] [PubMed] [Google Scholar]
- Kornberg R. D. Structure of chromatin. Annu Rev Biochem. 1977;46:931–954. doi: 10.1146/annurev.bi.46.070177.004435. [DOI] [PubMed] [Google Scholar]
- Kornberg R. The location of nucleosomes in chromatin: specific or statistical. Nature. 1981 Aug 13;292(5824):579–580. doi: 10.1038/292579a0. [DOI] [PubMed] [Google Scholar]
- Loening U. E. The fractionation of high-molecular-weight ribonucleic acid by polyacrylamide-gel electrophoresis. Biochem J. 1967 Jan;102(1):251–257. doi: 10.1042/bj1020251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohr D., Corden J., Tatchell K., Kovacic R. T., Van Holde K. E. Comparative subunit structure of HeLa, yeast, and chicken erythrocyte chromatin. Proc Natl Acad Sci U S A. 1977 Jan;74(1):79–83. doi: 10.1073/pnas.74.1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniatis T., Jeffrey A., van deSande H. Chain length determination of small double- and single-stranded DNA molecules by polyacrylamide gel electrophoresis. Biochemistry. 1975 Aug 26;14(17):3787–3794. doi: 10.1021/bi00688a010. [DOI] [PubMed] [Google Scholar]
- Martin D. Z., Todd R. D., Lang D., Pei P. N., Garrard W. T. Heterogeneity in nucleosome spacing. J Biol Chem. 1977 Nov 25;252(22):8269–8277. [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. A new method for sequencing DNA. Proc Natl Acad Sci U S A. 1977 Feb;74(2):560–564. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Noll M., Kornberg R. D. Action of micrococcal nuclease on chromatin and the location of histone H1. J Mol Biol. 1977 Jan 25;109(3):393–404. doi: 10.1016/s0022-2836(77)80019-3. [DOI] [PubMed] [Google Scholar]
- Ozaki H. Developmental studies of sea urchin chromatin. Chromatin isolated from spermatozoa of the sea urchin Strongylocentrotus purpuratus. Dev Biol. 1971 Oct;26(2):209–219. doi: 10.1016/0012-1606(71)90123-0. [DOI] [PubMed] [Google Scholar]
- Palau J., Ruiz-Carrillo A., Subirana J. A. Histones from sperm of the sea urchin Arbacia lixula. Eur J Biochem. 1969 Jan;7(2):209–213. doi: 10.1111/j.1432-1033.1969.tb19593.x. [DOI] [PubMed] [Google Scholar]
- Prunell A., Kornberg R. D. Relation of nucleosomes to DNA sequences. Cold Spring Harb Symp Quant Biol. 1978;42(Pt 1):103–108. doi: 10.1101/sqb.1978.042.01.011. [DOI] [PubMed] [Google Scholar]
- Prunell A., Kornberg R. D. Relation of nucleosomes to nucleotide sequences in the rat. Philos Trans R Soc Lond B Biol Sci. 1978 May 11;283(997):269–273. doi: 10.1098/rstb.1978.0023. [DOI] [PubMed] [Google Scholar]
- Prunell A. Random arrangement of nucleosome on DNA in chromatin. FEBS Lett. 1979 Nov 15;107(2):285–287. doi: 10.1016/0014-5793(79)80391-9. [DOI] [PubMed] [Google Scholar]
- Riley D., Weintraub H. Nucleosomal DNA is digested to repeats of 10 bases by exonuclease III. Cell. 1978 Feb;13(2):281–293. doi: 10.1016/0092-8674(78)90197-6. [DOI] [PubMed] [Google Scholar]
- Simpson R. T. Structure of the chromatosome, a chromatin particle containing 160 base pairs of DNA and all the histones. Biochemistry. 1978 Dec 12;17(25):5524–5531. doi: 10.1021/bi00618a030. [DOI] [PubMed] [Google Scholar]
- Spadafora C., Bellard M., Compton J. L., Chambon P. The DNA repeat lengths in chromatins from sea urchin sperm and gastrule cells are markedly different. FEBS Lett. 1976 Oct 15;69(1):281–285. doi: 10.1016/0014-5793(76)80704-1. [DOI] [PubMed] [Google Scholar]
- Spadafora C., Oudet P., Chambon P. Rearrangement of chromatin structure induced by increasing ionic strength and temperature. Eur J Biochem. 1979 Oct;100(1):225–235. doi: 10.1111/j.1432-1033.1979.tb02053.x. [DOI] [PubMed] [Google Scholar]
- Staynov D. Z., Pinder J. C., Gratzer W. B. Molecular weight determination of nucleic acids by gel electrophoresis in non-aqueous solution. Nat New Biol. 1972 Jan 26;235(56):108–110. doi: 10.1038/newbio235108a0. [DOI] [PubMed] [Google Scholar]
- Sutcliffe J. G. pBR322 restriction map derived from the DNA sequence: accurate DNA size markers up to 4361 nucleotide pairs long. Nucleic Acids Res. 1978 Aug;5(8):2721–2728. doi: 10.1093/nar/5.8.2721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoma F., Koller T., Klug A. Involvement of histone H1 in the organization of the nucleosome and of the salt-dependent superstructures of chromatin. J Cell Biol. 1979 Nov;83(2 Pt 1):403–427. doi: 10.1083/jcb.83.2.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varshavsky A. J., Bakayev V. V., Georgiev G. P. Heterogeneity of chromatin subunits in vitro and location of histone H1. Nucleic Acids Res. 1976 Feb;3(2):477–492. doi: 10.1093/nar/3.2.477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zachau H. G., Igo-Kemenes T. Face to phase with nucleosomes. Cell. 1981 Jun;24(3):597–598. doi: 10.1016/0092-8674(81)90084-2. [DOI] [PubMed] [Google Scholar]